Abstract
Background
In addition to clinical characteristics, DNA aneuploidy has been identified as a prognostic factor in epithelial malignancies in general and in endometrial cancers in particular. We mapped ploidy-associated chromosomal aberrations and identified corresponding gene and protein expression changes in endometrial cancers of different prognostic subgroups.
Methods
DNA image cytometry classified 25 endometrioid cancers to be either diploid (n = 16) or aneuploid (n = 9), and all uterine papillary serous cancers (UPSC) to be aneuploid (n = 8). All samples were subjected to comparative genomic hybridization and gene expression profiling. Identified genes were subjected to Ingenuity pathway analysis (IPA) and were correlated to protein expression changes.
Results
Comparative genomic hybridization revealed ploidy-associated specific, recurrent genomic imbalances. Gene expression analysis identified 54 genes between diploid and aneuploid endometrioid carcinomas, 39 genes between aneuploid endometrioid cancer and UPSC, and 76 genes between diploid endometrioid and aneuploid UPSC to be differentially expressed. Protein profiling identified AKR7A2 and ANXA2 to show translational alterations consistent with the transcriptional changes. The majority of differentially expressed genes and proteins belonged to identical molecular functions, foremost Cancer, Cell Death, and Cellular Assembly and Organization.
Conclusions
We conclude that the grade of genomic instability rather than the histopathological subtype correlates with specific gene and protein expression changes. The identified genes and proteins might be useful as molecular targets for improved diagnostic and therapeutic intervention and merit prospective validation.
Keywords: aneuploidy, endometrial carcinoma, genomic instability, comparative genomic hybridization, expression arrays, pathway analysis, UPSC
Background
Endometrial cancer is the most common malignancy of the female genital tract in the Western world and the fourth common cancer in women [1]. In general it is considered to have a favorable prognosis since it usually becomes symptomatic at an early tumor stage. Thus, about 70% of the affected women are detected at tumor stage I. At this stage, the mean survival of five years has been estimated to be 87%. However, one histopathological subtype, uterine papillary serous cancer (UPSC), presents with an aggressive clinical course characterized by early metastasis, reduced survival rates and inferior prognosis compared to endometrioid carcinomas [2]. Next to histopathology, tumor stage and tumor grade are known to be the most influencing prognostic factors [3].
In breast, prostate and colorectal cancer, also DNA aneuploidy has been reported to be an independent prognostic marker [4-6]. In endometrial cancer, patients with diploid cell populations have a more favorable 5-year survival rate of 94% as opposed to those with aneuploid malignancies (83%) [7]. Aneuploidy can be assessed at the chromosomal level by comparative genomic hybridization (CGH) [8]. Interestingly, CGH results have shown a conserved pattern of chromosomal gains and losses that is distinct and characteristic for different epithelial malignancies [9]. In carcinomas of the vagina the most frequent aberration detected is a gain of 3q [10], while in endometrial carcinomas, copy number gains were mapped to chromosome arms 1q, 3q, 8q, and 10q [11-13]. The predominance of these tumor entity specific chromosomal alterations leads to increased expression of resident genes that seems to be independent of tissue and/or cell type [14] and gives an irreversible disturbance of transcriptional regulation in aneuploid cells [15].
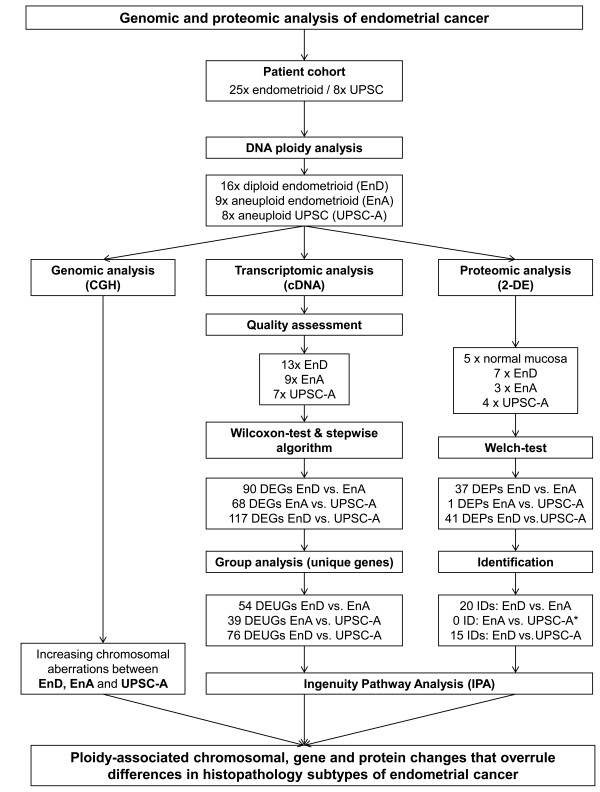
Against this background we now evaluated whether genomic instability correlates with chromosomal alterations and impacts on gene and protein expression changes in endometrial carcinomas. We utilized well-characterized surgical specimens of endometrial cancer representing different histopathological subtypes which are associated with a distinct prognosis (Figure 1).
Figure 1.
Technical workflow of the study design. * No protein was identified in the EnA vs. UPSC-A comparison due to extremely weak abundance of the protein spot in the polyacrylamide gel.
Results
Here we describe a comprehensive evaluation of aneuploidy-associated alterations of the genome, transcriptome, and proteome in different histopathological subtypes of endometrial cancer. We were particularly interested in identifying chromosomal alterations that underlay aneuploidy and how these might impact on transcriptional and translational changes and thereby influence patients' prognosis.
Genomic instability
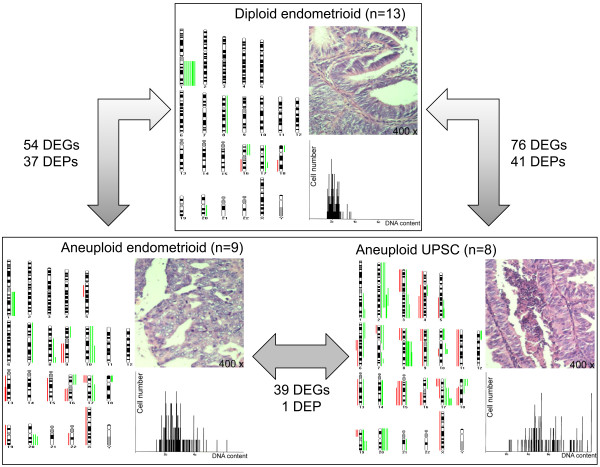
Of the cancerous samples, 16 of the 25 endometrioid carcinomas showed diploid cell distribution pattern (EnD) and nine presented with aneuploid cell populations (EnA), while all eight UPSC tumors were classified as aneuploid (UPSC-A). Representative histograms for each group are provided in Figure 2. The mean value of the DNA stem line increased from 2.23c in the EnD group to 2.98c in the EnA and 3.06c in the UPSC-A group (p < 0.004).
Figure 2.
Examples of ploidy types and number of differential expressed genes (DEGs) and proteins (DEPs). DNA histograms show DNA content on the x-axis and the total number of cell on the y-axis.
The "stemline scatter index" (SSI) measures the clonal heterogeneity of the constituent tumor cells and is calculated as the sum of (a) the percentage of cells with DNA content values in the S-phase region (S-phase), (b) the percentage of cells with DNA content values exceeding twice the modal value plus 1c (G2 exceeding rate), and (c) the coefficient of variation (CV) of the respective tumor stemline [16]. In our study, all but one diploid and all aneuploid carcinomas showed SSI values above the threshold of overall genomic instability (of 8.8) found for breast carcinomas [16]. The mean SSI values of 26.9 (EnD), 45.4 (EnA), and 53.5 (UPSC-A) now found indicate increasing genomic instability for aneuploid and UPSC subtypes (p < 0.004). The increasing degree of genomic instability in the EnA and UPSC-A groups compared to the EnD group was also reflected in an increase in chromosomal copy number changes as measured by CGH. A detailed summary and comparison of chromosomal aberrations found is presented in Figure 2. Chromosomal imbalances in the EnD tumors were mostly restricted to gains of chromosome 1q (33%) and 16p (11%). In contrast, EnA tumors showed diverse changes, including a gain of 10q, 20q (both 33%), and 1q, 8q, 10p, 16p and 17q (all 22%). These changes were accompanied by frequent losses of 9q, 16q, 17p, 19p, 19q and 22q (all 22%) (Additional file 1). In the UPSC-A malignancies, chromosomal aberrations affected also chromosomal regions that were not changed in the endometrioid tumors. UPSCs presented in 62% a gain of 2q, 8q, 17q, and 20p and in 50% a gain of 20q. The most frequent loss was observed for 15q (50%).
The increasing number of chromosomal aberrations between the EnD and EnA carcinomas and between the EnA and UPSC-A malignancies was furthermore reflected by the average number of copy number alterations (ANCA), calculated as the sum of all detected chromosomal aberrations and divided by the number of cases analyzed. The ANCA value increased from 0.041 (EnD) to 0.145 (EnA) and up to 0.429 (UPSC-A) (p < 0.001). The same trend was found for the average number of regional amplifications (ANRA, calculated as the sum of all amplifications and divided by the number of cases analyzed). The ANRA amounted to 0.003 in the EnD group, increased to 0.014 in the EnA group and to 0.065 in the UPSC-A group (p < 0.002).
Gene expression profiling
We applied the Wilcoxon test with permutation test and the stepwise algorithm [17] to identify differentially expressed genes for pair-wise comparisons of the three groups. Considering only those genes that were commonly detected by both approaches, we found that 54 genes were differentially expressed between EnD and EnA samples. A total of 39 differentially expressed genes (DEGs) defined expression differences between the aneuploid malignancies of endometrioid or UPSC histology (EnA and UPSC-A). However, the vast majority-76 genes-was differentially expressed between the groups distinguished by different histology and ploidy status, namely EnD and UPSC-A (Table 1). All genes were unique for pair-wise group comparison. The gene lists describing differences between all groups are listed in Additional file 2.
Table 1.
Overview of significantly expressed genes
| Wicoxon test (p < 0.05) | ||
|---|---|---|
| EnD versus EnA | EnD versus UPSC-A | EnA versus UPSC-A |
| 478 | 576 | 276 |
| Stepwise Analysis (Up/Down) | ||
| EnD versus EnA | EnD versus UPSC-A | EnA versus UPSC-A |
| 140/118 | 195/179 | 165/154 |
| Genes present in both analyses | ||
| EnD versus EnA | EnD versus UPSC-A | EnA versus UPSC-A |
| 90 | 117 | 68 |
| Genes unique for each group comparison | ||
| EnD versus EnA | EnD versus UPSC-A | EnA versus UPSC-A |
| 54 | 76 | 39 |
EnD, endometrioid diploid
EnA, endometrioid aneuploid
UPSC-A, UPSC aneuploid
When mapping the differentially expressed genes to their chromosomal location, we found that the deregulation of 114 out of 275 (41.45%) genes could be attributed to chromosomal copy number changes.
Correlation of Gene and Protein expression changes
Mapping differentially expressed proteins (DEPs) previously detected by two-dimensional gel electrophoresis [18] to the chromosomal location of their corresponding genes showed that in the EnD versus EnA comparison 11 of 20 proteins (55%) and in the EnD versus UPSC-A comparison 7 of 15 (47%) mapped to positions that were affected by copy number changes (Additional file 3).
For 5 of the 35 identified proteins, the corresponding cDNA was included on our microarray platform. Two of the corresponding genes, AKR7A2 and ANXA2, showed a similar trend in transcriptional expression as observed for the translational changes. However, both genes did not reach our significance levels of the gene expression analysis: AKR7A2 was down-regulated in EnD versus UPSC-A, while ANXA2 showed an up-regulation in EnA versus EnD and in UPSC-A versus EnD.
Functional annotation of DEGs and DEPs that discern the EnD, EnA, and UPSC-A tumors using Ingenuity Pathway Analysis
Using Ingenuity Pathway Analysis (IPA), differentially expressed genes and proteins that discerned EnD, EnA, and UPSC-A were functionally annotated. An overview of all networks found by transcriptomic and proteomic profiling is provided in Table 2.
Table 2.
IPA analysis overview
| Comparison | Analysis | Top networks | Score | Top Diseases and Disorders | p-Value | # of Molecules | Top Molecular and Cellular Functions | p-Value | # of Molecules |
|---|---|---|---|---|---|---|---|---|---|
| EnD vs. EnA | Transcriptomics | Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism | 48 | Cancer | < 0.0161 | 23 | Lipid Metabolism | < 0.0184 | 6 |
| Lipid Metabolism, Small Molecule Biochemistry, Genetic Disorder | 30 | Hematological Disease | < 0.0132 | 12 | Small Molecule Biochemistry | < 0.0184 | 10 | ||
| Gene Expression, Nutritional Disease, Cellular Development | 22 | Gastrointestinal Disease | < 0.0105 | 9 | Vitamin and Mineral Metabolism | < 0.0175 | 12 | ||
| Proteomics & | Cellular Assembly and Organization, Nucleic Acid Metabolism, Small Molecule Biochemistry | 25 | Neurological Disease | < 0.0106 | 7 | Cellular Growth and Proliferation | < 0.0382 | 7 | |
| Genetic Disorder | < 0.0486 | 8 | Cell Morphology | < 0.0297 | 4 | ||||
| Cancer | < 0.0394 | 7 | Cellular Assembly and Organization | < 0.0346 | 6 | ||||
| EnD vs. UPSC-A | Transcriptomics | Organism Injury and Abnormalities, Cardiac Necrosis/Cell Death, Cell Death* | 44 | Cancer | < 0.0196 | 31 | Cellular Growth and Proliferation | < 0.0212 | 28 |
| Organ Morphology, Reproductive System Development and Function, Skeletal and Muscular Disorders # | 28 | Genetic Disorder | < 0.0234 | 47 | Cell-To-Cell Signaling and Interaction | < 0.0234 | 11 | ||
| Cellular Development, Cellular Growth and Proliferation, Cancer | 20 | Reproductive System Disease | < 0.0196 | 12 | Cell Death | < 0.0234 | 26 | ||
| Cardiovascular Disease, Hematological Disease, Skeletal and Muscular Disorders | 19 | ||||||||
| Endocrine System Development and Function, Small Molecule Biochemistry, Gene Expression | 15 | ||||||||
| Proteomics & | Lipid Metabolism, Small Molecule Biochemistry, Cell Morphology *, # | 25 | Cancer | < 0.0428 | 7 | Amino Acid Metabolism | < 0.0150 | 2 | |
| Gastrointestinal Disease | < 0.0214 | 4 | Cell Morphology | < 0.010 | 1 | ||||
| Inflammatory Disease | < 0.0479 | 2 | Cellular Assembly and Organization | < 0.0125 | 2 | ||||
| EnA vs. UPSC-A | Transcriptomics | Cardiovascular System Development and Function, Cell Cycle, Lipid Metabolism | 28 | Cardiovascular Disease | < 0.0430 | 12 | Gene Expression | < 0.0245 | 7 |
| Cell Death, Cellular Movement, Hematological System Development and Function | 28 | Development Disorder | < 0.0301 | 5 | Cell Death | < 0.0338 | 15 | ||
| Cellular Assembly and Organization, Cellular Function and Maintenance, Cell Signaling | 22 | Connective Tissue Disorder | < 0.0487 | 9 | Cellular Movement | < 0.0485 | 8 | ||
& Refers to Gemoll et al. [18]
* Overlapping of marked networks via platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) dimer (PDGFBB)
# Overlapping of marked networks via beta-Actin (ACTB)
EnD, endometrioid diploid
EnA, endometrioid aneuploid
UPSC-A, UPSC aneuploid
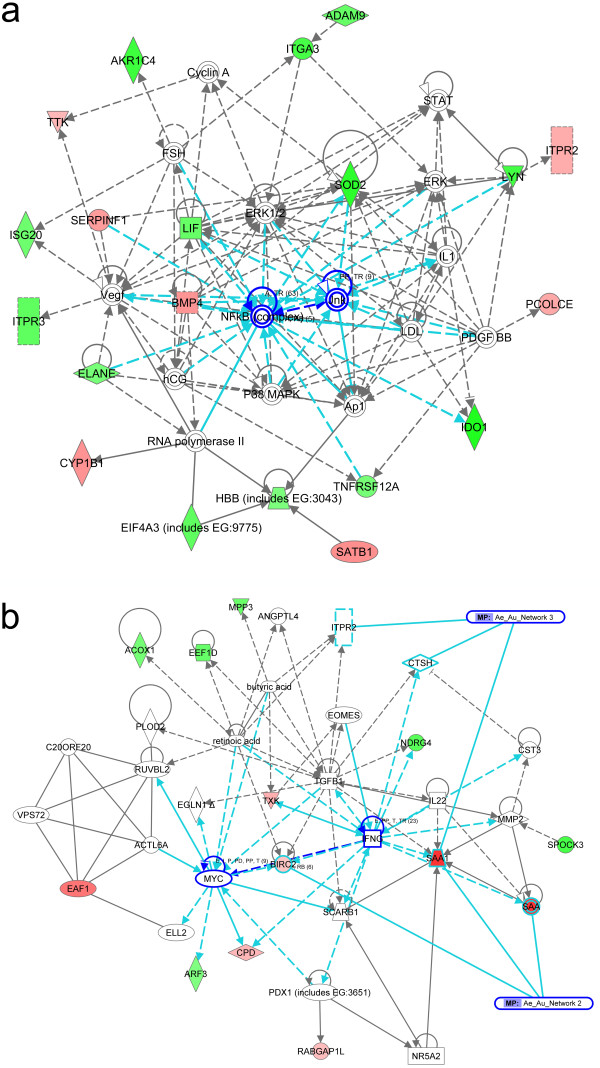
For the comparison of EnD versus EnA, 45 (83%) of the 54 DEGs were recognized in the IPA database and resulted in three networks. The highest ranked network with a score of 48 comprised 20 of the DEGs. These genes interacted in a network (Figure 3a) associated with Lipid Metabolism, Small Molecule Biochemistry, and Vitamin and Mineral Metabolism. NFkB, Jnk and ERK1/2 were central nodes of this network and associated with diseases and functions regarding Cancer, Hematological Disease, and Gastrointestional Disease (p < 0.00001 to p < 0.0161). The second highest network (score of 30) comprised 14 of the DEGs and was associated with Lipid Metabolism, Small Molecule Biochemistry, and Genetic Disorder. The third network (score of 22) comprised 11 of the DEGs and was associated with Gene Expression, Nutritional Disease and Cellular Development.
Figure 3.
IPA-based pathway analysis of differential expressed genes. (a) Red and green designations indicate over- and underexpressed genes in the respective groups. Blue arrows and circulations indicate central nodes of the networks. (b) Red and green designations indicate over- and underexpressed genes in the respective groups. Blue arrows and circulations indicate central nodes of the networks.
The comparison between the aneuploid carcinomas (EnA versus UPSC-A) allowed 33 (84.6%) of all 39 DEGs for IPA analysis. Three overlapping networks reached the level of significance, with a score of 28 to 22. The top network (score of 28) was associated with Cardiovascular System Development and Function, Cell Cycle, Lipid Metabolism and particularly with the canonical pathways Gene Expression (p < 0.0245) and Cell Death (p < 0.0338). This network interacts via BIRC2, SAA, and SAA1 with the second highest network (score of 28), associated with Cell Death, Cellular Movement, and Haematological System Development and Function, and via CTSH, ITPR2, and SAA1 with the third highest network (score of 22), associated with Cellular Assembly and Organization, Cellular Function and Maintenance, and Cell Signalling) (Figure 3b). Interestingly, SAA1 connected as well network 2 and network 3 with each other. IFNG, TGFB, MYC, and NFkB act as central nodes in these networks.
In comparisons of EnD versus UPSC-A, a total of 67 (88.2%) of the 76 DEGs were part of the IPA database. Here, we could define five overlapping networks with the first one reaching a score of 44 including 20 of the DEGs being associated with pathways of Organismal Injury and Abnormalities, Cardiac Necrosis/Cell Death, and Cell Death. Network 2 obtained a score of 28, comprised 14 DEGs and revealed Organ Morphology, Reproductive System Development and Function, and Skeletal and Muscular Disorders pathways. The third network reached a score of 20, comprised 11 of the DEGs and was associated with Cellular Development, Cellular Growth and Proliferation, and Cancer. The fourth highest ranked network reached a score of 19, comprised 11 of the DEGs and was associated with Cardiovascular Disease, Hematological Disease, and Skeletal and Muscular Disorders. The fifth network consists of 9 DEGs and reached a score of 15. Associated network functions were Endocrine System Development and Function, Small Molecule Biochemistry, and Gene Expression. All networks were associated with Cancer (p < 0.0196), Genetic Disorder (p < 0.0234), Cellular Growth and Proliferation (p < 0.0212), and Cell-To-Cell Signalling and Interaction (p < 0.0234). Three remaining networks obtained one focus gene only and failed the level of significance with a score < 5. Involved genes of all networks are presented in Additional file 4.
A network comparison analysis between all significant networks mentioned above and their corresponding protein profiling networks showed three interacting networks connected via ACTB and PDGFBB (Table 2), thus representing a close relation of transcriptomics and proteomics data.
Discussion
We mapped ploidy-associated chromosomal aberrations and identified corresponding gene and protein expression changes in endometrial cancers of different prognostic subgroups, including diploid (n = 16) and aneuploid (n = 9) endometrioid, and aneuploid uterine papillary serous cancer (n = 8) malignancies. CGH revealed ploidy-associated specific, recurrent genomic imbalances comprising gains of chromosome arms 1q, 3q, and 8q, as well as losses of 4q and 15q. Since these alterations dominate recurrent pattern of chromosomal imbalances characterizing endometrial malignancies, the genes located on such aberrant chromosome loci might play a key role in initiation and/or progression of endometrial malignancies.
The identified genes with altered expression changes belong to several functional groups. These genes are involved in functions related to fundamental biological processes known to be affected in cancer. For instance, ATF3, DNMT3B, LMo2, and TCF12 affect DNA binding, transcriptional activation and proliferation. Interestingly, a higher proliferation rate of aneuploid as compared to diploid endometrial carcinomas has been observed [19].
Another group includes genes that code for enzymatic proteins like proteases, reductases, and transferases. Many of these genes, such as aflatoxin B 1 aldehyde reductase member 2 (AKR7A2), v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (LYN), or cytochrome b reductase 1 (CYP1B1) have not been reported in prior studies of endometrial cancer and thus provide novel potential targets for diagnosis and treatment. AKR7A2 (located at 1p36.13) is a Golgi-associated AKR7 family member and is expressed in a broad range of tissues [20]. AKR7A2 is involved in the detoxification of aldehydes and ketones in the phase I metabolism. AKR7A2 protein levels are elevated in the cerebral cortex of patients with Alzheimer disease [21]. Our results of the expression analysis indeed proved the proteomic data that demonstrate an AKR7A2 overexpression in EnD compared to UPSC-A [18]. LYN (located at 8q13) is among the highly ranked signature genes overexpressed in EnD versus EnA carcinomas and documents a strong correlation between DNA and RNA analysis. LYN is a member of the Src-family kinases, a family of non-receptor tyrosine kinases. LYN was of particular interest because as a kinase, it is "druggable" and might provide a therapeutic opportunity for targeting endometrial cancer. Choi et al. identified LYN as a possible new therapeutic target with particular relevance to clinically aggressive basal-like breast cancer [22].
A further group consists of genes and proteins involved in mechanisms of transport and protein binding. Annexins are characterized by their capacity to bind to phospholipids in the presence of calcium ions and their susceptibility to phosphorylation and dephosphorylation. Belonging to subfamily A of annexins, human annexins are further classified as annexin A1-A11 and A13. One isoform, ANXA2 (Annexin A2; located at 15q22.2) was found significantly upregulated as detected in our proteomic approach and proved the same trend with respect to gene expression [18]. ANXA2 is located in the cytoplasm as a monomer (heavy chain of 36 kDa, or p36) or in a complex with a member of the S100A10 [23]. There is evidence of a relationship between proteases and extracellular matrix (ECM) proteins through ANXA2 whereby ANXA2 may facilitate the reorganization of the ECM in physiological and pathological processes such as tumor invasion [24]. Therefore, the over-expression of ANXA2 in colorectal and gastric carcinomas alludes to a correlation with invasiveness and poor prognosis and is in line with our analysis [25,26]. Further, we found Wnt-7a (wingless-type MMTV integration site family, member 7A; located at 3p25) to be more expressed in EnA samples compared to EnD cancers. Wnt genes are associated with cellular responses such as proliferation, cell fate determination or specification, apoptosis and oncogenesis. The Wnt-7a gene is detected at high levels in the female reproductive tract [27]. Observations suggest that disruption of normal Wnt-7a expression by diethylstilbestrol and other estrogenic compounds leads to altered uterine cytoarchitecture and might be a mechanism ultimately causing neoplasia in the reproductive tract [28].
In the comparison of EnD and UPSC-A cases we found that the progesterone receptor (PGR; located at 11q22-q23) gene was significantly lower expressed in UPSC-A cases. PGR showed the highest ratio of differential expression overall. Furthermore, lower expression in UPSC-A was associated with a loss of the whole chromosome 11 in 25% of the UPSC-A cases whereas in diploid cases no alteration could be detected. PGR encodes a member of the steroid receptor superfamily. The encoded protein mediates the physiological effects of progesterone, which plays a central role in reproductive events associated with the establishment and maintenance of pregnancy. Excessive estrogen stimulation, unbalanced by progesterone, might play a central role in the development of endometrial cancer across all ethnic populations [29,30]. Progesterone ameliorates estrogen-induced proliferation by interacting with its receptor [31,32], primarily through two functionally distinct PGR isoforms. It could be shown that hormone therapy of PGR-positive patients increases their response rate substantially from 8-17% to 37-89% [33]. Our data exhibit that particularly patients with diploid carcinomas could benefit from hormone therapy and that ploidy status assessment could assist in patient stratification.
The comparison between EnA and UPSC-A revealed one network including the differentially expressed genes BIRC2 (baculoviral IAP repeat-containing 2; located at 11q22), BIRC3 (baculoviral IAP repeat-containing 3; located at 11q22), MAP3K5 (mitogen-activated protein kinase kinase kinase 5; located at 6q22.33), and SAA1 (serum amyloid A1, located at 11p15.1). All four genes are associated with different cancer types in general but a connection to endometrial cancer has not been described so far. Interestingly, all four genes are lower expressed in the UPSC-A cancers. These findings are in accordance with a loss of the according gene loci on chromosome 6 and 11 in up to 25% of the UPSC-A cases whereas in the EnA cases no alteration could be detected. BIRC2 and BIRC3 are members of a family of proteins that inhibit apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2, probably by interfering with activation of ICE-like proteases. These encoded proteins inhibit apoptosis induced by serum deprivation and menadione, a potent inducer of free radicals. Cheng et al. showed that BIRC2 overexpression might play a critical role for mammary carcinogenesis associated with p53 mutations [34]. It is of interest that overexpression of BIRC2 has been recently proposed to be associated with luminal subtype B of breast cancer [35]. MAP3K5 (also known as apoptosis signal-regulating kinase 1; ASK1) has been widely accepted as one of the key components regulating reactive oxygen species (ROS) [36]. There is evidence suggesting that oxidative stress contributes to the pathogenesis of prostate cancer [37]. SAA1, a high-density lipoprotein (HDL)-associated apolipoprotein, is an acute-phase protein which is elevated in response to trauma, inflammation, and neoplasia [38]. SAA1 has several functions such as tumor cell invasion and metastasis by induction of cell adhesion and migration through induction of enzymes degrading the ECM and inhibition of cell attachment to ECM proteins by SAA derived peptides [39-41]. Cumulatively, these properties might place SAA as an ECM-associated adhesion protein, with a potential role in tumor pathogenesis.
Conclusions
We detected that chromosomal copy number alterations do impact on gene expression changes. In addition, differentially expressed genes and proteins interacted within overlapping networks (Table 2). This was proved further by two differentially expressed proteins, AKR7A2 and ANXA2, showing similar gene expression alterations. Overall, we have identified different and specific expressions patterns between 16 diploid endometrioid-, 9 aneuploid endometrioid-cancers, and 8 aneuploid UPSCs with CGH, RNA microarray and two-dimensional gel electrophoresis. We have revealed a number of aberrantly regulated genes and proteins that are potential biomarkers for an improved diagnosis and prognostication in endometrial cancers.
Methods
Patient Samples
Fresh tumor material was collected from women who underwent hysterectomy for endometrial cancer at the Department of Obstetrics and Gynecology at the Karolinska University Hospital, Stockholm, Sweden, during 1997 and 2003. Clinical material was collected from surgically removed tumors adhering to the approval of the local ethical review board. Carcinomas were diagnosed on H&E-stained tissue sections and graded according to the FIGO classification [42]. Patients treated with neoadjuvant therapy were excluded. A total of 25 endometrioid carcinomas and 8 UPSCs were randomly selected for ploidy assessment as well as genomic, and transcriptomic analysis. Proteomic analysis of according samples was reported earlier [18]. Data on ploidy, histopathologic subtype, stage, age, observation time, and survival status are provided in Table 3. After surgery, clinical tissues were first used for touch preparation slides for ploidy assessment and then snap frozen until further processing. Snap frozen specimens were divided into one part for protein expression and one part for DNA and RNA extraction. In addition, paraffin-embedded specimens of the same tumors were used for histopathology and immunohistochemistry.
Table 3.
Clinical data and ploidy assessment
| Case | Histo-pathology | Ploidy | SSI | Age | FIGO 1988 | FIGO 2010 | Grade | Metastasis | CGH | ANCA | ANRA | Observation time | Died at month |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D01 | endometrioid | diploid | 35, 0 | 83 | 1c | 1b | 1 | no | 3191 | 0.043 | 0 | 74 | |
| D02 | endometrioid | diploid | 31, 7 | 59 | 1b | 1a | 1 | no | 3192 | 0.087 | 0 | 42 | 42 |
| D03 | endometrioid | diploid | 45, 3 | 60 | 1c | 1b | 1 | no | 3193 | 0.130 | 0 | 94 | |
| D04 | endometrioid | diploid | 16, 1 | 52 | 1b | 1a | 1 | no | 3194 | 0 | 0 | 92 | |
| D05 | endometrioid | diploid | 6, 8 | 87 | 1c | 1b | 2 | no | 3195 | 0.043 | 0.043 | 15 | 15 |
| D06* | endometrioid | diploid | 13, 2 | 67 | 1b | 1a | 2 | no | 3196 | 0.043 | 0 | 109 | |
| D07* | endometrioid | diploid | 17, 6 | 78 | 3a | 3a | 2 | yes | 3197 | 0.174 | 0 | 4 | 4 |
| D08* | endometrioid | diploid | 13, 7 | 78 | 1b | 1a | 1 | no | 3198 | 0.043 | 0 | 104 | |
| D09 | endometrioid | diploid | 17, 3 | 59 | 1b | 1a | 1 | no | 3193 | 0 | 0 | 94 | |
| D10 | endometrioid | diploid | 12, 4 | 55 | 1b | 1a | 1 | no | 3200 | 0 | 0 | 94 | |
| D11 | endometrioid | diploid | 14, 5 | 81 | 1b | 1a | 1 | no | 3201 | 0 | 0 | 86 | |
| D12 | endometrioid | diploid | 46, 5 | 72 | 1b | 1a | 1 | no | 3202 | 0 | 0 | 75 | |
| D13 | endometrioid | diploid | 35, 6 | 63 | 1c | 1b | 2 | no | 3203 | 0 | 0 | 26 | 26 |
| D14 | endometrioid | diploid | 56, 4 | 51 | 1b | 1a | 1 | no | 3204 | 0 | 0 | 88 | |
| D15 | endometrioid | diploid | 31, 0 | 53 | 1b | 1a | 2 | no | 3205 | 0 | 0 | 55 | |
| D16 | endometrioid | diploid | 38, 4 | 66 | 1b | 1a | 3 | no | 3206 | 0.087 | 0 | 53 | |
| Ae1 | endometrioid | aneuploid | 68, 0 | 82 | 1c | 1b | 1 | no | 3207 | 0.043 | 0 | 146 | |
| Ae2 | endometrioid | aneuploid | 20, 4 | 80 | 1b | 1a | 1 | no | 3208 | 0.304 | 0 | 71 | |
| Ae3 | endometrioid | aneuploid | 35, 2 | 69 | 1b | 1a | 1 | no | 3203 | 0.043 | 0 | 107 | |
| Ae4 | endometrioid | aneuploid | 26, 9 | 46 | 1b | 1a | 2 | no | 3210 | 0.565 | 0.087 | 81 | |
| Ae5 | endometrioid | aneuploid | 35, 5 | 55 | 2a | 2 | 3 | yes | 3211 | 0.087 | 0 | 73 | |
| Ae6 | endometrioid | aneuploid | 30, 7 | 66 | 1b | 1a | 2 | no | 3212 | 0 | 0 | 63 | |
| Ae7 | endometrioid | aneuploid | 71, 0 | 79 | 1b | 1a | 2 | no | 3213 | 0.130 | 0.043 | 55 | |
| Ae8 | endometrioid | aneuploid | 59, 6 | 60 | 1b | 1a | 3 | no | 3214 | 0.043 | 0 | 51 | |
| Ae9 | endometrioid | aneuploid | 61, 7 | 72 | 1c | 1b | 3 | yes | 5521 | 0.087 | 0 | 42 | |
| Au1 | UPSC | aneuploid | 70, 9 | 77 | 3a | 3a | 3 | yes | 3215 | 0.565 | 0.174 | 36 | 36 |
| Au2 | UPSC | aneuploid | 42, 0 | 78 | 4b | 4b | 0 | yes | 3216 | 0.609 | 0 | 51 | |
| Au3 | UPSC | aneuploid | 28, 5 | 66 | 1c | 1b | 3 | no | 3217 | 0.652 | 0.043 | 19 | 19 |
| Au4 | UPSC | aneuploid | 59, 7 | 80 | 3a | 3a | 3 | yes | 3218 | 0.565 | 0 | 27 | 27 |
| Au5 | UPSC | aneuploid | 48, 6 | 90 | 3 | 3a | 3 | yes | 3219 | 0.130 | 0 | 9 | 9 |
| Au6 | UPSC | aneuploid | 27, 5 | 88 | 3b | 3b | 0 | yes | 3220 | 0.478 | 0.13 | 17 | 17 |
| Au7* | UPSC | aneuploid | 59, 2 | 80 | 1a | 1a | 0 | no | 3359 | 0.348 | 0.13 | 7 | 7 |
| Au8 | UPSC | aneuploid | 91, 3 | 83 | 3a | 3a | 0 | yes | 5522 | 0.087 | 0.043 | 10 | 10 |
* Arrays did not pass visual quality criteria and were thus excluded from further analysis.
Staging was assessed according to FIGO Committee on Gynecologic Oncology. Ploidy status was defined by image cytometry according to the Auer classification. Observation time is given in months. For CGH, the case number of the database is given http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi. SSI, Stem line scatter index.
Image Cytometry
Image cytometry was performed on Feulgen-stained paraffin-embedded histopathological slides. The staining procedure, internal standardization, and tumor cell selection were based on methods described previously [4]. All DNA-values were expressed in relation to the corresponding staining controls which were given the value 2c, denoting the normal diploid DNA-content. The tumors were classified as belonging to three groups: (i) diploid cases with a distinct peak in the normal 2c region and no cells exceeding 5c, (ii) aneuploid cases with a main peak different from 2c and a stemline scatter index (SSI) below or equal 8.8, and (iii) aneuploid samples with a varying numbers of cells (> 5%) exceeding 5c (SSI above 8.8). This novel classification system adheres to the parameters established by Kronenwett and colleagues, who defined the stemline scatter index (SSI) as a measurement of clonal heterogeneity in the tumor cell population [16].
The degree of genomic instability status in the three groups (EnD, EnA, UPSC-A) was compared with metric parameters (stemline, SSI, ANCA, ANRA) using ANOVA test. The threshold of significance was set to p < 0.05.
Comparative Genomic Hybridization (CGH)
DNA was extracted from fresh frozen tissue using TRIzol. CGH was performed as described in detail http://www.riedlab.nci.nih.gov. Fluorescence intensity ratio plots were generated using Leica CW4000 Karyo V1.0 software (Leica Imaging Systems, Cambridge, UK). Interpretation of changes at 1pter, 16, 19, and 22 required careful examination because these loci are prone to artifacts due to the high proportion of repetitive sequences. CGH profiles of individual cases as well as the summary display of all cases can be found at http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
Microarray analysis
Total RNA was extracted using TRIzol (Invitrogen) followed by Qiagen RNeasy column purification (Qiagen, Valencia, CA). All samples were hybridized against the universal human reference RNA (Stratagene, La Jolla, CA, USA) using a slightly modified protocol from Hedge and colleagues [43]. Extraction and hybridization protocols used can be viewed in detail at http://www.riedlab.nci.nih.gov.
In brief, 20 μg of total RNA was reverse transcribed using random primers and converted into cDNA using reverse transcriptase. After incorporation of aminoallyl-conjugated nucleotides, the RNA was indirectly labelled with Cy3 (tumor RNA) and Cy5 (reference RNA, Amersham, Piscataway, NJ). Each sample was hybridized against the reference RNA in a humid chamber (Arraylt™ Hybridization Cassette, TeleChem Intl., Sunnyvale, CA, USA) for 16 hours at 42°C, washed, and scanned by the Axon GenePix 4000B Scanner (Axon Instruments, Union City, CA, USA). In order to account for potential amplification bias, total RNA was hybridized following the same protocol for 11 samples (20 μg each). We used customized arrays obtained from the National Cancer Institute's microarray core facility. Arrays were used from one print batch and composed of 9, 128 cDNAs denatured and immobilized on a poly-L-lysine-coated glass surface. The gene annotation file (GAL file) used (Hs-UniGEM2-v2px-32Bx18Cx18R.gal) can be found at the facility's website http://nciarray.nci.nih.gov. GenePix software version 4.0.1.17 was used to apply the GAL file through an interactive gridding process. All images of the scanned microarray slides were meticulously inspected for artifacts. Empty spots and aberrant spots and slide regions were flagged for exclusion from analyses [44].
Microarray quality assessment and data analysis (two-group class comparison)
After discarding arrays that did not pass our visual quality filtering, a total of 13 EnD, 9 EnA, and 7 UPSC-A malignancies could be processed for further analysis. All values that did not meet the quality control criteria were treated as missing values as described in supplemental materials (Additional file 5). Intensity ratios were calculated using the background corrected median intensities that were normalized with the locally weighted scatter plot smoother (LOWESS) algorithm for each print-tip group. The fraction of data points used in the local regression (f) was 0.2 and other parameters were adjusted as suggested by Cleveland [45]. The value of f was determined using self versus self experiment. All within-slide normalized ratios were log-transformed (natural base). A total of 4, 995 genes were identified that did not show any missing values across all samples. Out of those 4, 995 genes, differentially expressed genes were identified with pair-wise analysis. To identify differentially expressed genes we used Wilcoxon rank-sum test with a permutation test (p < 0.05) and a step-wise gene selection procedure [17,46]. Genes that were identified by both approaches were selected for further analysis. Further details of applied algorithms can be found in Additional file 6.
Biological pathway analysis
We used IPA software (v8.7, Ingenuity, Mountain View, CA) to assess the involvement of significantly differentially expressed genes and proteins in known pathways and networks. IPA determined groups of genes that together constitute networks. Such networks indicate how the genes and/or proteins of interest may influence each other above and beyond canonical pathways that are described in the Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg). The IPA generated networks are listed in a certain order, with the top networks having a lower likelihood that the generation of the networks was serendipitous.
List of abbreviations
ANCA: Average number of copy number alterations; ANRA: Average number of regional amplifications; CGH: Comparative genomic hybridization; DEG: Differentially expressed gene; DEP: Differentially expressed protein; ECM: Extracellular matrix; EnA: Aneuploid endometrioid; EnCa: Endometrial cancer; FIGO: Fédération Internationale de Gynécologie et d'Obstétrique; IPA: Ingenuity pathways analysis; KEGG: Kyoto encyclopedia of genes and genomes; PCA: Principal component analysis; SSI: Stem line scatter index; UPSC: Uterine papillary serous cancer; UPSC-A: Aneuploid uterine papillary serous cancer.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JKH designed the study, performed genomic and transcriptomic experiments, and wrote the manuscript. NB performed genomic and transcriptomic experiments, analyzed results, and drafted the manuscript. TG performed result analysis and proteomics experiments, and wrote the manuscript. SH evaluated statistical data. CL drafted the manuscript and collected clinical data. DW contributed to genomics and transcripomic experiments. JD performed ploidy experiments. HPB designed the study and drafted the manuscript. BN contributed to the sample collection and ploidy experiments. UJR designed the study and drafted the manuscript. HJ helped to design the study, was involved in mass spectrometric experiments and contributed substantially to the manuscript. GA helped to design the study and drafted the manuscript. TR designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
CGH data details. Detailed list of all chromosomal imbalances of all endometrial tumors.
Unique DEG list. List of differentially expressed genes that were unique for all pair-wise group comparisons.
Differential expressed protein lists. List of all significant proteins of all pair-wise group comparisons.
Involved genes in pathway analysis. Presentation of all genes that are involved in identified pathways.
Quality assessment criteria. Quality control criteria for microarray values that had to be fulfilled for further analysis.
Microarray data analysis (two-group class comparison). Details of applied algorithms for microarray data analysis.
Contributor Information
Jens K Habermann, Email: Jens.Habermann@gmail.com.
Nana K Bündgen, Email: nanabuendgen@googlemail.com.
Timo Gemoll, Email: timo.gemoll@googlemail.com.
Sampsa Hautaniemi, Email: sampsa.hautaniemi@helsinki.fi.
Caroline Lundgren, Email: caroline.lundgren@karolinska.se.
Danny Wangsa, Email: wangsad@mail.nih.gov.
Jana Doering, Email: jana.doering@gmx.de.
Hans-Peter Bruch, Email: Bruch@uni-luebeck.de.
Britta Nordstroem, Email: nordstrom.britta@telia.com.
Uwe J Roblick, Email: drdr.ujroblick@me.com.
Hans Jörnvall, Email: hans.jornvall@ki.se.
Gert Auer, Email: gert.auer@karolinska.se.
Thomas Ried, Email: riedt@mail.nih.gov.
Acknowledgements
We thank Inga Maurin and Ulla Aspenblad for assistance with immunohistochemical analysis and Buddy Chen, Tom Ellerman and Joseph Cheng for IT support. Grants from the Boehringer Ingelheim Fonds, the Werner and Clara Kreitz Foundation, the North German Tumorbank of Colorectal Cancer (DKH #108446), and the Ad Infinitum Foundation are gratefully acknowledged. This study was performed in connection with the Surgical Center for Translational Oncology-Lübeck (SCTO-L). This study was supported in part by the intramural research program of the National Institutes of Health, National Cancer Institute.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Zaino RJ, Kurman RJ, Diana KL, Morrow CP. The utility of the revised International Federation of Gynecology and Obstetrics histologic grading of endometrial adenocarcinoma using a defined nuclear grading system. A Gynecologic Oncology Group study. Cancer. 1995;75:81–86. doi: 10.1002/1097-0142(19950101)75:1<81::AID-CNCR2820750114>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Creasman WT, Odicino F, Maisonneuve P, Beller U, Benedet JL, Heintz AP, Ngan HY, Sideri M, Pecorelli S. Carcinoma of the corpus uteri. J Epidemiol Biostat. 2001;6:47–86. [PubMed] [Google Scholar]
- Auer G, Eriksson E, Azavedo E, Caspersson T, Wallgren A. Prognostic significance of nuclear DNA content in mammary adenocarcinomas in humans. Cancer Res. 1984;44:394–396. [PubMed] [Google Scholar]
- Gerling M, Meyer KF, Fuchs K, Igl BW, Fritzsche B, Ziegler A, Bader F, Kujath P, Schimmelpenning H, Bruch HP, High Frequency of Aneuploidy Defines Ulcerative Colitis-Associated Carcinomas: A Comparative Prognostic Study to Sporadic Colorectal Carcinomas. Ann Surg. 2010. [DOI] [PubMed]
- Lexander H, Palmberg C, Hellman U, Auer G, Hellstrom M, Franzen B, Jornvall H, Egevad L. Correlation of protein expression, Gleason score and DNA ploidy in prostate cancer. Proteomics. 2006;6:4370–4380. doi: 10.1002/pmic.200600148. [DOI] [PubMed] [Google Scholar]
- Lundgren C, Auer G, Frankendal B, Moberger B, Nilsson B, Nordstrom B. Nuclear DNA content, proliferative activity, and p53 expression related to clinical and histopathologic features in endometrial carcinoma. Int J Gynecol Cancer. 2002;12:110–118. doi: 10.1046/j.1525-1438.2002.01079.x. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science (New York, NY. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(SICI)1098-2264(199907)25:3<195::AID-GCC1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Habermann JK, Hellman K, Freitag S, Heselmeyer-Haddad K, Hellstrom AC, Shah K, Auer G, Ried T. A recurrent gain of chromosome arm 3q in primary squamous carcinoma of the vagina. Cancer Genet Cytogenet. 2004;148:7–13. doi: 10.1016/S0165-4608(03)00245-0. [DOI] [PubMed] [Google Scholar]
- Muresu R, Sini MC, Cossu A, Tore S, Baldinu P, Manca A, Pisano M, Loddo C, Dessole S, Pintus A. et al. Chromosomal abnormalities and microsatellite instability in sporadic endometrial cancer. Eur J Cancer. 2002;38:1802–1809. doi: 10.1016/S0959-8049(02)00152-1. [DOI] [PubMed] [Google Scholar]
- Muslumanoglu HM, Oner U, Ozalp S, Acikalin MF, Yalcin OT, Ozdemir M, Artan S. Genetic imbalances in endometrial hyperplasia and endometrioid carcinoma detected by comparative genomic hybridization. Eur J Obstet Gynecol Reprod Biol. 2005;120:107–114. doi: 10.1016/j.ejogrb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Pere H, Tapper J, Wahlstrom T, Knuutila S, Butzow R. Distinct chromosomal imbalances in uterine serous and endometrioid carcinomas. Cancer Res. 1998;58:892–895. [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP. et al. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, Alaiya A, Sennerstam R, Zetterberg A, Auer G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–909. doi: 10.1158/0008-5472.CAN-03-2451. [DOI] [PubMed] [Google Scholar]
- Xiong M, Li W, Zhao J, Jin L, Boerwinkle E. Feature (gene) selection in gene expression-based tumor classification. Mol Genet Metab. 2001;73:239–247. doi: 10.1006/mgme.2001.3193. [DOI] [PubMed] [Google Scholar]
- Gemoll T, Habermann JK, Lahmann J, Szymczak S, Lundgren C, Bundgen NK, Jungbluth T, Nordstrom B, Becker S, Lomnytska MI, Protein profiling of genomic instability in endometrial cancer. Cellular and molecular life sciences: CMLS. 2011. in press . [DOI] [PMC free article] [PubMed]
- Nordstrom B, Strang P, Bergstrom R, Nilsson S, Tribukait B. A comparison of proliferation markers and their prognostic value for women with endometrial carcinoma. Ki-67, proliferating cell nuclear antigen, and flow cytometric S-phase fraction. Cancer. 1996;78:1942–1951. doi: 10.1002/(SICI)1097-0142(19961101)78:9<1942::AID-CNCR15>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- O'Connor T, Ireland LS, Harrison DJ, Hayes JD. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem J. 1999;343(Pt 2):487–504. [PMC free article] [PubMed] [Google Scholar]
- Picklo MJ, Olson SJ, Hayes JD, Markesbery WR, Montine TJ. Elevation of AKR7A2 (succinic semialdehyde reductase) in neurodegenerative disease. Brain Res. 2001;916:229–238. doi: 10.1016/S0006-8993(01)02897-9. [DOI] [PubMed] [Google Scholar]
- Choi YL, Bocanegra M, Kwon MJ, Shin YK, Nam SJ, Yang JH, Kao J, Godwin AK, Pollack JR. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res. 2010;70:2296–2306. doi: 10.1158/0008-5472.CAN-09-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo Tapia JP, Pena Alonso E, Garcia-Pedrero JM, Florentino Fresno M, Suarez Nieto C, Owen Morgan R, Fernandez MP. [Annexin A2 expression in head and neck squamous cell carcinoma] Acta Otorrinolaringol Esp. 2007;58:257–262. doi: 10.1016/S0001-6519(07)74923-3. [DOI] [PubMed] [Google Scholar]
- Mai J, Waisman DM, Sloane BF. Cell surface complex of cathepsin B/annexin II tetramer in malignant progression. Biochim Biophys Acta. 2000;1477:215–230. doi: 10.1016/S0167-4838(99)00274-5. [DOI] [PubMed] [Google Scholar]
- Emoto K, Sawada H, Yamada Y, Fujimoto H, Takahama Y, Ueno M, Takayama T, Uchida H, Kamada K, Naito A. et al. Annexin II overexpression is correlated with poor prognosis in human gastric carcinoma. Anticancer Res. 2001;21:1339–1345. [PubMed] [Google Scholar]
- Emoto K, Yamada Y, Sawada H, Fujimoto H, Ueno M, Takayama T, Kamada K, Naito A, Hirao S, Nakajima Y. Annexin II overexpression correlates with stromal tenascin-C overexpression: a prognostic marker in colorectal carcinoma. Cancer. 2001;92:1419–1426. doi: 10.1002/1097-0142(20010915)92:6<1419::AID-CNCR1465>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Miller C, Sassoon DA. Wnt-7a maintains appropriate uterine patterning during the development of the mouse female reproductive tract. Development. 1998;125:3201–3211. doi: 10.1242/dev.125.16.3201. [DOI] [PubMed] [Google Scholar]
- Kitajewski J, Sassoon D. The emergence of molecular gynecology: homeobox and Wnt genes in the female reproductive tract. Bioessays. 2000;22:902–910. doi: 10.1002/1521-1878(200010)22:10<902::AID-BIES5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Siiteri PK. Steroid hormones and endometrial cancer. Cancer Res. 1978;38:4360–4366. [PubMed] [Google Scholar]
- Key TJ, Pike MC. The dose-effect relationship between 'unopposed' oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. Br J Cancer. 1988;57:205–212. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen SC, Hanekamp EE, Hanifi-Moghaddam P, Sijbers AM, van Gool AJ, Burger CW, Blok LJ, Huikeshoven FJ. Growth regulation and transcriptional activities of estrogen and progesterone in human endometrial cancer cells. Int J Gynecol Cancer. 2006;16:110–120. doi: 10.1111/j.1525-1438.2006.00279.x. [DOI] [PubMed] [Google Scholar]
- Smid-Koopman E, Kuhne LC, Hanekamp EE, Gielen SC, De Ruiter PE, Grootegoed JA, Helmerhorst TJ, Burger CW, Brinkmann AO, Huikeshoven FJ, Blok LJ. Progesterone-induced inhibition of growth and differential regulation of gene expression in PRA- and/or PRB-expressing endometrial cancer cell lines. J Soc Gynecol Investig. 2005;12:285–292. doi: 10.1016/j.jsgi.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Decruze SB, Green JA. Hormone therapy in advanced and recurrent endometrial cancer: a systematic review. Int J Gynecol Cancer. 2007;17:964–978. doi: 10.1111/j.1525-1438.2007.00897.x. [DOI] [PubMed] [Google Scholar]
- Cheng L, Zhou Z, Flesken-Nikitin A, Toshkov IA, Wang W, Camps J, Ried T, Nikitin AY. Rb inactivation accelerates neoplastic growth and substitutes for recurrent amplification of cIAP1, cIAP2 and Yap1 in sporadic mammary carcinoma associated with p53 deficiency. Oncogene. 2010. [DOI] [PMC free article] [PubMed]
- Frasor J, Weaver A, Pradhan M, Dai Y, Miller LD, Lin CY, Stanculescu A. Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer. Cancer Res. 2009;69:8918–8925. doi: 10.1158/0008-5472.CAN-09-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa A, Ichijo H. Stress-responsive protein kinases in redox-regulated apoptosis signaling. Antioxid Redox Signal. 2005;7:472–481. doi: 10.1089/ars.2005.7.472. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Parr RL, Costello LC, Franklin RB, Thayer RE. Altered metabolism and mitochondrial genome in prostate cancer. J Clin Pathol. 2006;59:10–16. doi: 10.1136/jcp.2005.027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7:64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- Badolato R, Wang JM, Murphy WJ, Lloyd AR, Michiel DF, Bausserman LL, Kelvin DJ, Oppenheim JJ. Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med. 1994;180:203–209. doi: 10.1084/jem.180.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkoviz R, Preciado-Patt L, Lider O, Fridkin M, Dastych J, Metcalfe DD, Mekori YA. Extracellular matrix-anchored serum amyloid A preferentially induces mast cell adhesion. Am J Physiol. 1997;273:C179–187. doi: 10.1152/ajpcell.1997.273.1.C179. [DOI] [PubMed] [Google Scholar]
- Migita K, Kawabe Y, Tominaga M, Origuchi T, Aoyagi T, Eguchi K. Serum amyloid A protein induces production of matrix metalloproteinases by human synovial fibroblasts. Lab Invest. 1998;78:535–539. [PubMed] [Google Scholar]
- Shepherd JH. Revised FIGO staging for gynaecological cancer. Br J Obstet Gynaecol. 1989;96:889–892. doi: 10.1111/j.1471-0528.1989.tb03341.x. [DOI] [PubMed] [Google Scholar]
- Hedge P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Earle-Hughes J, Snesrud E, Lee NH, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–554. doi: 10.2144/00293bi01. 556, 558-560. [DOI] [PubMed] [Google Scholar]
- Korn EL, Habermann J, Upender MB, Ried T, McShane LM. Objective method of comparing DNA microarray image analysis systems. Biotechniques. 2004;36:960–967. doi: 10.2144/04366BI01. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Robust Locally Weighted Fitting and Smoothing Scatterplots. Journal of the American Statistical Association. 1979;74:829–836. doi: 10.2307/2286407. [DOI] [Google Scholar]
- Kauraniemi P, Hautaniemi S, Autio R, Astola J, Monni O, Elkahloun A, Kallioniemi A. Effects of Herceptin treatment on global gene expression patterns in HER2-amplified and nonamplified breast cancer cell lines. Oncogene. 2004;23:1010–1013. doi: 10.1038/sj.onc.1207200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CGH data details. Detailed list of all chromosomal imbalances of all endometrial tumors.
Unique DEG list. List of differentially expressed genes that were unique for all pair-wise group comparisons.
Differential expressed protein lists. List of all significant proteins of all pair-wise group comparisons.
Involved genes in pathway analysis. Presentation of all genes that are involved in identified pathways.
Quality assessment criteria. Quality control criteria for microarray values that had to be fulfilled for further analysis.
Microarray data analysis (two-group class comparison). Details of applied algorithms for microarray data analysis.