Abstract
Two distinct classes of methicillin-resistant Staphylococcus aureus (MRSA) are spreading in hospitals (as hospital-acquired MRSA, HA-MRSA) and in the community (as community-acquired MRSA, CA-MRSA). Multilocus sequence type (ST) 239 MRSA, one of the most worldwide-disseminated lineages, has been noted as a representative HA-MRSA. Here, we isolated ST239 MRSA (spa type 3 [t037] and staphylococcal cassette chromosome mec [SCCmec] type III.1.1.1) and its novel variant with ST239/spa351 (t030)/SCCmecIII.1.1.4 (SCCmecIIIR) not only from hospitals but also from patients with urethritis in the community in Russia. The Russian variant (strain 16K) possessed a hybrid genome consisting of CC8 and CC30, similar to the ST239/spa3/SCCmecIII.1.1.1 HA-MRSA (TW20) genome, but with marked diversity. The 16K′ CC30 section had SCCmecIIIR carrying the dcs-carrying unit (which corresponded to the SCCmecIVc J3 joining region of ST30 CA-MRSA), lacked SCCmercury, and possessed a novel mobile element structure (MES16K) carrying the ccrC-carrying unit (with the recombinase gene ccrC1 allele 3) and drug resistance tranposons. The Russian variant included strains with a high ability to transfer its multiple drug resistance by conjugation; e.g., for strain 16K, the transfer frequency of a chloramphenicol resistance plasmid (p16K-1 with 2.9 kb in size) reached 1.4×10−2, followed by Tn554 conjugative transfer at 3.6×l0−4. The Russian variant, which has been increasing recently, included divergent strains with different plasmid patterns and pulsed field gel electrophoresis profiles. The data demonstrate the alternative nature of ST239 MRSA as CA-MRSA and also as a drug resistance disseminator, and its micro but dynamic evolution in Russia.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) was isolated in the early 1960s and has continued to be a life-threatening multiple drug-resistant bacterium in hospitals [1], [2]. MRSA is generated from methicillin-susceptible S. aureus (MSSA) by the acquisition of staphylococcal cassette chromosome mec (SCCmec) at the 3′ end of orfX (SCCmec insertion site, att) [3]. It is considered that this has occurred only a limited number of times, resulting in the current epidemics in hospital settings [3]–[5].
Of these clones, ST239 MRSA is one of the most worldwide-disseminated lineages [4], [6], and is evolutionarily of interest, because it is the first example of a bacterial hybrid, consisting of two distinct MRSA belonging to clonal complex (CC) 30 (founder, ST30) and CC8 (founder, ST8) [7], [8]. The ST239 MRSA lineage exhibits marked geographic variations [6] in terms of the protein A gene (spa) type, SCCmec type III structures, and pulsed-field gel electrophoresis (PFGE) patterns. Many terms have been used to accommodate this variation [4], e.g., Brazilian clone [9], Portuguese clone [10], [11], Hungarian clone [12]–[14], Viennese clone [15], and British EMRSA-1, -4, and -11 clones [16], [17].
Of those, the Brazilian clone (first ST239 MRSA) spread among hospitals in Brazil in 1992 [18], and then caused intercontinental spread to hospitals in Portugal, possibly linked to Brazil-to-Portugal migration of human populations since 1992–1993 [11], [19]. The Hungarian clone emerged in hospitals in Hungary in 1993, became predominant in hospitals until 1998, and then almost disappeared in 2003–2004 from Hungary [12]–[14]; however, there is no single reference addressing the molecular typing or characteristics of MRSA strains (Hungarian clone) isolated in Hungary. SCCmec types were SCCmecIIIA for the Brazilian clone (e.g., strain HU25) while SCCmecIII for the Hungarian clone (e.g., strain HU106) [19].
Moreover, the ST239 TW clone (strain TW20) was noted as intensive care unit (ICU)-associated MRSA in London between 2002 and 2004 [20], and the complete genome of TW20 was described [8]. Harris et al. [6] have recently described the comparative genomics of globally-collected ST239 strains using Illumina genome analysis and the TW20 genome as a reference, demonstrating the global geographic structure within ST239 MRSA, based on genome-wide single nucleotide polymorohisms (SNPs). The ST239 lineage consisted of more than five MRSA clades reflecting the continental origin, such as Asia, North America, South America, Europe (with marked divergence), and Australia, albeit with some intercontinental transmission cases. According to the study of Harris et al. [6], TW20 clustered within the Thai clade (most probably suggesting transmission from southeast Asia to London), the Brazilian clone (e.g., strain HU25) clustered within the South America clade, and Hungarian isolates (during 1993–1996) exhibited divergent European phylogeny in SNPs.
In the community, another class of MRSA, called community-acquired MRSA (CA-MRSA), emerged during the period from 1997 to 1999 [2], [21], [22]. CA-MRSA includes clones belonging to (e.g.) ST8 (USA300), ST30, and ST80 [2], [22], [23]. CA-MRSA generally exhibits SCCmecIV or V, a narrow range of drug resistance, and mecA-mediated low-level resistance to β-lactam agents (oxacillin and imipenem) [1], [2], [22]–[24].
The term, hospital-acquired MRSA (HA-MRSA), which includes MRSA isolated in the nosocomial environment, has been used since the description of CA-MRSA. In this study, we isolated MRSA, including ST239 MRSA, from patients in hospitals and also from patients with nongonococcal urethritis in the community in Russia, and examined the molecular characteristics of these MRSA strains. We also investigated the comparative genomics of an ST239 variant (strain 16K) with unique features in terms of evolution and drug resistance using the TW20 genome as a reference.
Materials and Methods
Patients and bacterial strains
Thirteen outpatients were enrolled in this study, all of whom were male outpatients (age range, 19–40 years; mean age, 29 years) of two hospitals in Vladivostok, Russia, through the period from 2006 to 2008. All outpatients had discharge (pus) from the urethra, discomfort inside the urethra, and discomfort while passing urine. Examination of swabs (discharge) from the urethra revealed the presence of polymorphonuclear neutrophils and MRSA in all cases; no other pathogens, such as Neisseria gonorrhea, Chlamydia, Ureaplasma, Mycoplasma, Candia, Trichomonas and STD-associated viruses, were detected; therefore, outpatients were diagnosed with nongonococcal urethritis due to MRSA. No nosocomial or familial transmissions were observed. The 13 MRSA (including strain 16K) were epidemiologically diagnosed as CA-MRSA; in this study, CA-MRSA was defined as MRSA isolated from outpatients who had no history of hospitalization within at least the past year and presented with no other established risk factors for MRSA infections, such as surgery, residence in a long-term care facility, dialysis, or indwelling percutaneous medical devices and catheters; and HA-MRSA was defined as MRSA isolated from inpatients 48 h after hospitalization [2], [22].
In this study, 18 HA-MRSA strains, isolated from inpatients (age, 34–71 years; mean age, 51.8 years) in four hospitals in Vladivostok between 2004 and 2008, were also examined; they were isolated from surgical wound infections, pneumonia, and blood stream infection (including sepsis). Five HA-MRSA strains were isolated from inpatients (age, 3–64 years; mean age, 26.6 years) in two hospitals in Vladivostok in 2011 from surgical wound infections and burn infections. This study was complied with the ethics review board Niigata University School of Medicine, Niigata, Japan. Written informed consent was obtained from patients.
The epidemic HA-MRSA-type strains included ANS46 (reference strain isolated in 1982 in Australia; ST239/SCCmecIII), HU25 (Brazilian clone; ST239/SCCmecIIIA), BK2464 (New York/Japan clone; ST5/SCCmecII), BM18 (Pediatric clone; ST5/SCCmecIVa), HDE288 (Pediatric clone; ST5/SCCmecVI), HAR22 (EMRSA-15 clone; ST22/SCCmecIV), HAR24 (EMRSA-16 clone; ST36/SCCmecII), HAR38 (Berlin clone; ST45/SCCmecIVa), HPV107 (Iberian clone; ST247/SCCmecIA), and COL (Archaic clone; ST250/SCCmecI); they were kindly provided by H. de Lencastre. CA-MRSA strain NN1 (ST30/SCCmecIVc), which was positive for the collagen adhesin gene (cna), was isolated from a child with bullous impetigo in the community [25], [26].
Genotyping and virulence gene analysis
MRSA typing was performed as described previously [27]. The spa type was analyzed by PCR using reference strains, and determined using public spa type databases, eGenomics (http://tools.egenomics.com/) or Ridom SpaServer (http://spaserver.ridom.de/). Typing of agr was carried out by PCR with previously reported primers [28], [29] or by sequencing the variable region [30]. SCCmec types (I to V) were analyzed by PCR using reference strains [31], [32]. The subtypes of SCCmecIII were described expressing the differences in the J1, J2, and J3 joining regions, according to the guidelines described in 2009 [33]. For virulence gene analysis, the target genes included 46 genes; 3 leukocidin genes (lukPVSF, lukE-lukD, and lukM), 5 hemolysin genes (hla, hlb, hlg, hlg-v, and hld), 18 staphylococcal enterotoxin (SE) genes (tst, sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, sep, seq, and ser), 1 putative staphylococcal enterotoxin gene (seu), 3 exfoliative toxin genes (eta, etb, and etd), an exotoxin-like gene cluster (set), the epidermal cell differentiation inhibitor gene (edin), and 14 adhesin genes (icaA, icaD, eno, fib, fnbA, fnbB, ebpS, clfA, clfB, sdrC, sdrD, sdrE, cna, and bbp) [27].
PFGE analysis
Bacterial DNA was digested with SmaI and the digested DNA was applied to PFGE (1.2% agarose), as described previously [34]. A lambda ladder (Bio-Rad Laboratories, Tokyo, Japan) was used as the molecular size standard.
Plasmid analysis
Plasmid DNA of MRSA strains was prepared using a Plasmid Midi Kit (QIAGEN Sciences, Tokyo). Plasmid DNA was introduced into MRSA by electroporation using a Gene Pulser II electroporator (Bio-Rad).
Conjugative transfer
Donor strains were mated with S. aureus RN2677 (recipient strain, which is restriction-negative and resistant to rifampicin and novobiocin) on membrane filters on tryptic soy agar (Difco, Sparks, MD, USA) (filter mating), as previously described [25]; RN2677 was used as a recipient because it carried no plasmids and had non-transmissible drug resistance (recipient) markers. In some experiments, donor and recipient cultures were mixed at 1∶2, centrifuged, and spotted on tryptic soy agar without filters (non-filter mating). Alternatively, donor and recipient cultures were mixed in tryptic soy broth (Difco) (liquid mating). The resistance genes of donor strains and transconjugants were examined by PCR as previously described [27].
Susceptibility testing
Bacterial susceptibility testing was carried out according to previous procedures [35]. Breakpoints for drug resistance were those described by the CLSI [35].
Genome analysis
The MRSA 16K genome was analyzed by pyrosequencing using a genome sequencer FLX system with the assembler software GS De Novo Assembler version 2.0 (Roche Diagnostics, Branford, CT, USA); pyrosequencing analysis yields contigs of larger sizes than Illumina genome analysis. In this study, 274,241 reads yielded 111 Mb raw sequences, corresponding to approximately 37-fold of the genome size; GenBank accession numbers for the 16K genome (107 contigs with ≥20 bp in size) are BABZ01000001-BABZ01000107. Constructed contigs were mapped on the 3,043,210-bp complete TW20 genome [GenBank accession number FN433596; 6] using MUMmer software (http://mummer.sourceforge.net/). The gene or open reading frame (orf) was searched for using the software in silico MolecularCloning (version 4.2) (In Silico Biology, Yokohama, Japan).
Phylogenetic and homology analysis
Phylogenetic tree analysis was performed using TreeViewX software (version 0.5.0) (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The bootstrap value (1000) represents the accuracy of a branch in the phylogenetic tree (analysis was repeated 1,000 times). Homology analysis was performed using the software BLAST (http://blast.ddbj.nig.ac.jp/top-e.html) and FASTA (http://fasta.ddbj.nig.ac.jp/top-j.html).
Results
Characteristics of MRSA from outpatients with urethritis in the community
Data are summarized in Table 1 and Figure 1. Thirteen MRSA strains were classified into four groups (A1 to A4) (Table 1). Group A1 (n = 6) exhibited ST239/spa3/SCCmecIII.1.1.1 and shared the same genotype with ST239 reference strains, TW20 and ANS46, but was distinct from HU25. Group A1 strains shared the same PFGE pattern (Figure 1).
Table 1. Characteristics of MRSA isolated from out patients with nongonococcal urethritis and from inpatients in Russia compared with ST239 MRSA reference strains.
| MRSA from urethritis(CA-MRSA)2006–2008 | MRSA from inpatients (HA-MRSA)2004–2008 | MRSA from inpatients (HA-MRSA) 2011 | ST239 MRSA reference strains | |||||||||
| Type, virulence gene or drug resistance | Group A1 | Group A2 | Group A3 | Group A4 | Group B1 | Group B2 | Group B3 | Group C1 | Group C2 | TW20a UK 2003 | ANS46 Australia 1982 | HU25 Brazil 1993 |
| Type | (n = 6) | (n = 4) | (n = 2) | (n = 1) | (n = 11) | (n = 6) | (n = 1) | (n = 1) | (n = 4) | |||
| CC | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| ST | 239 | 239 | 8 | 72 | 239 | 239 | 8 | 239 | 239 | 239 | 239 | 239 |
| spa | 3 (t037) | 351 (t030) | 826 (tUK) | 451 (t324) | 3 (t037) | 351 (t030) | 826 (tUK) | 3 (t037) | 351 (t030) | 3 (t037) | 3 (t037) | 390 (t138) |
| agr | 1 | 1 (3/4), 1′ (1/4) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| SCCmec type | III.1.1.1 | III.1.1.4 (IIIR) | IVc | IVc | III.1.1.1 | III.1.1.4 (IIIR) | IVc | III.1.1.1 | III.1.1.4 (IIIR) | III.1.1.1 | III.1.1.1 | III.1.1.2 (IIIA) |
| Coagulase type | IV | IV | III | V | IV | IV | III | IV | IV | IV | IV | IV |
| Virulence gene | ||||||||||||
| Leukocidin | ||||||||||||
| lukE-lukD | + | + | + | + | + | + | + | + | + | + | + | + |
| Hemolysin | ||||||||||||
| hla, hlb (split), hlg, hlg-v | + | + | + | + | + | + | + | + | + | + | + | + |
| hld | + | + | + | + | + | + | + | + | + (1/4) | + | + | + |
| Enterotoxin | ||||||||||||
| egc (seg, sei, sem, sen, seo) | − | − | − | + | − | − | − | − | − | − | − | − |
| sea | + | + | + | − | + | + | + | + | + | + | + | − |
| SaPI1 (sek, seq) | + | + | − | − | + | + | − | + | + | + | + | − |
| Adhesin | ||||||||||||
| c12ag b | + | + | + (1/2) | + | + | + | + | + | + | + | + | + |
| cna | + | + | − | − | + | + | − | + | + | + | + | + |
| Susceptibilityc | ||||||||||||
| Oxa (MIC, µg/ml) | ≥256 | ≥256 | 32 | 32 | ≥256 | ≥256 | 64 | ≥256 | ≥256 | ND | 128 | ≥256 |
| Ipm (MIC, µg/ml) | 32–64 | 64 | 0.06 | 0.5 | 32–64 | 64–128 | 1 | 32 | 64–128 | ND | 16 | 64 |
| Resistance to non β-lactam | Gen, Kan, Str, Spt, Tet, Ery, Cli, Lvx, Sul, Tmp | Gen, Kan, Str, Spt, Tet, Ery, Cli, Lvx, Sul, Chl, Rif | Gen, Kan, Ery (1/2), Cli (1/2), Chl | Kan | Gen, Kan, Str, Spt, Tet, Ery, Cli, Lvx, Sul (10/11), Tmp, Chl (1/11) | Gen, Kan, Str, Spt, Tet, Ery, Cli, Lvx(R, 4/6; I 2/6), Sul, Chl, Rif | Gen, Kan, Ery, Cli, Chl, Rif | Gen, Kan, Str, Spt, Tet, Ery, Cli, Lvx, Sul, Tmp | Gen, Kan, Str, Spt, Tet, Ery, Cli (1/4), Lvx, Sul, Chl, Rif | Gen, Kan, Str, Tet, Ery, Cli, Lvx, Tmp | Kan, Str, Spt, Tet, Ery, Cli, Sul, Tmp, Chl | Gen, Kan, Str, Tet, Ery, Cli, Lvx, Sul, Tmp |
| Plasmidsd (kb) | 38* (5/6), 2.9 (4/6) | 32 (1/4), 4.4 (1/4), 2.9** (d), 2.4 | 34, 2.9**, 2.6 (1/2) | 3.6, 2.7 | 41** (1/11),38 (7/11), 2.9 (10/11), 2.6 (1/11) | 41** (5/6), 30** (1/6), 2.9 (d), 2.4 | - | 38, 2.9 | 41** (1/4) 38*(3/4), 2.9 (3/4)** (d), 2.4 | 29.6*, 3.0 | 4.4* | - |
MRSA groups A2, B2, and C2 represent the Russian variant of ST239 MRSA.
The data of TW20 are from GenBank accession numbers FN433596, FN433597, and FN433598. A 29.6-kb plasmid (pTW20_1) is a heavy metal resistance plasmid [8].
c12ag, core 12 adhesin genes shared by all (or most) strains: icaA, icaD (for biofilm formation); eno (for laminin-adhesin); fnbA, fnbB (for fibronectin-adhesin); ebpS (for elastin-adhesin); clfA, clfB, fib, sdrC, sdrD, sdrE (for fibrinogen).
Oxa, oxacillin; Imp, imipenem; Gen, gentamicin; Kan, kanamycin; Str, streptomycin; Spt, spectinomycin; Ery, erythromycin; Cli, clindamycin; Tet, tetracycline; Lvx, levofloxacin; Sul, sulfamethoxazole; Tmp, trimethoprim; Chl, chloramphenicol; Rif, rifampicin. MICs of rifampicin: 256 µg/ml for groups A2, B2, and C2; 8 µg/ml for group B3. ND, not determined.
Resistance plasmids were examined by conjugation and electroporation.
*, plasmid encoding for cadmium resistance;
**, plasmid encoding for chloramphenicol resistance.
In group A2, strain 49K carried 32-kb and 4.4-kb plasmids in addition to 2.9-kb and 2.4-kb plasmids. (−), no plasmid.
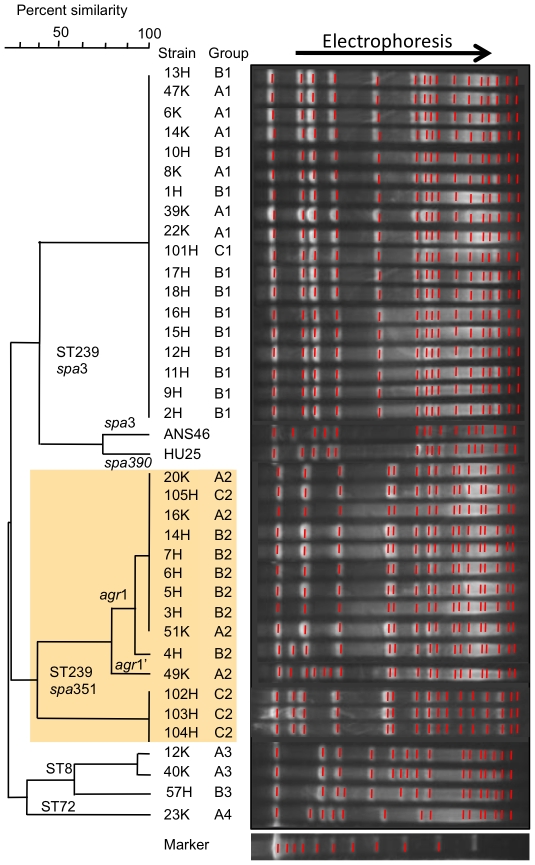
Figure 1. PFGE patterns of MRSA isolated from outpatients with nongonococcal urethritis and from inpatients in Russia compared with ST239 reference strains.
MRSA groups (A1–A4, B1–B3, and C1 and C2) are described in Table 1. The Russian variant of ST239 MRSA (groups A2, B2, and C2) is marked with shading (yellow). Strains 6K to 51K (n = 13) are isolated from outpatients during 2006–2008: (group A1) 6K, 8K, 14K, 22K, 39K, 47K; (group A2) 16K, 20K, 49K, 51K; (group A3) 12K and 40K; (group A4) 23K. Strains 1H to 18H (n = 18) are isolates from inpatients during 2004–2008: (group B1) 1H, 2H, 8H, 9H, 11H, 12H, 13H, 15H, 16H, 17H, 18H; (group B2) 3H, 4H, 5H, 6H, 7H, 14H; (group B3) 10H. Strains 101H to 105H (n = 5) are isolates from inpatients in 2011: (group C1) 101H; (group C2) 102H, 103H, 104H, 105H.
Group A2 (n = 4) was a variant of the ST239 lineage. These strains (including 16K) exhibited spa351/SCCmecIII.1.1.4 (named SCCmecIIIR), were resistant to chloramphenicol and rifampicin and susceptible to trimethoprim, and possessed a 2.9-kb chloramphenicol-resistance plasmid (Table 1). Group A2 strains shared the same PFGE pattern, except for one strain (49K) (Figure 1); 49K exhibited a divergent agr subtype (named agr1'; see Fig. S1 for detailed analysis) and plasmid profile (Table 1).
Group A3 (n = 2; ST8/SCCmecIVc) and group A4 (n = 1; ST72/SCCmecIVc) exhibited a narrow range of drug resistance and low oxacillin and imipenem resistance levels (Table 1), similarly to CA-MRSA [24].
The ST239 lineage strains, but not ST8 or ST72 strains, were positive for the collagen adhesin gene (cna). Of the epidemic-type HA-MRSA clones, EMRSA-15 (ST22), EMRSA-16 (ST36), and Berlin (ST45) were also cna-positive.
Characteristics of MRSA from inpatients
Data are summarized in Table 1 and Figure 1. Of 18 MRSA strains epidemiologically diagnosed as HA-MRSA during 2006–2008, 11 strains belonged to group B1 (ST239/spa3/SCCmecIII.1.1.1) and corresponded to group A1; one strain (strain 2H) possessed a 41-kb chloramphenicol-resistance plasmid. Six strains belonged to group B2 (ST239/spa351/SCCmecIIIR) and corresponded to group A2, although most B2 strains possessed a 41-kb chloramphenicol-resistance plasmid. The remaining strain belonged to group B3 (ST8/SCCmecIVc) and resembled group A3.
Therefore, around 2006, the ST239/spa3/SCCmecIII.1.1.1 type was the most prevalent and the variant (ST239/spa351/SCCmecIIIR) type was the second most prevalent, not only in the community but also in hospitals.
Of the five MRSA strains epidemiologically diagnosed as HA-MRSA in 2011, one strain (group C1) corresponded to group A1 or B1. Four strains (group C2) exhibited the same genotype as group A2 or B2, indicating the increase of the ST239 variant type in 2011; however, of four C2 strains, three strains exhibited slightly distinct PFGE profiles and plasmid patterns (containing 2.9-kb chloramphenicol resistance and 38-kb cadmium resistance plasmids) (Table 1 and Figure 1).
Comparative genomics of the ST239 Russian variant (strain 16K)
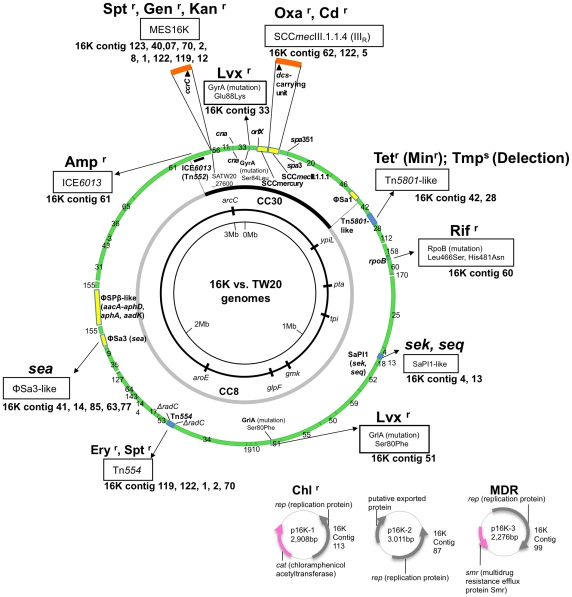
The 16K genome sequence was compared with the TW20 genome (Figure 2). The 16K genome was estimated to be at least 2.8 Mb in size, showing approximately 99.9% homology with the TW20 region (in green), excluding four large non- or less homologous regions (283.6-Kb, in yellow). The CC30 and CC8 hybrid regions on the genome were assigned according to Holden et al. [8].
Figure 2. Genome information for ST239 MRSA strain 16K in comparison with ST239 MRSA strain TW20.
The genome information includes drug resistance genetic traits (related structures and mutations), virulence genes, superantigen-associated phages, and plasmids. The 16K genome contigs, obtained by pyrosequencing, were mapped on the 3,043,210-bp TW20 genome (GenBank accession number FN433596; shown as a circle). The information on the 16K and TW20 genomes is presented outside and inside the genome circle, respectively. Colored regions in the 16K genome map: green, highly homologous to TW20; yellow, non- or less homologous to TW20; blue, slightly divergent from TW20; brown, insertion. Drug resistance (r): Oxa, oxacillin; Tet, tetracycline; Min, minocycline; Rif, rifampicin; Lvx, levofloxacin; Chl, chloramphenicol; Ery, erythromycin; Spt, spectinomycin; Gen, gentamicin; Kan, kanamycin; Amp, ampicillin; Ery, erythromycin. MDR, multiple drug resistance. The three plasmids of strain 16K are shown at the right bottom of the figure. The CC30 and CC8 genome sections are from Holden et al. [8].
In the CC30 section, 16K possessed SCCmecIIIR instead of SCCmecIII.1.1.1. Moreover, 16K lacked SCCmercury, but possessed a novel mobile element structure (MES16K), which carried the recombinase gene (ccrC)-carrying unit [36], similarly to SCCmercury.
There were several phage deletions in16K: ΦSa1 and ΦSPβ-like were absent; and a part of ΦSa3 was lacking (Figure S2A). Moreover, marked divergence was observed with SaPI1 (Figure S2B).
In 16K, the blaz gene (encoding ampicillin resistance) was located in ICE6013 (Figure 2), and the tetM gene (encoding tetracycline and minocycline resistance) was in a smaller Tn5801-like structure, which lacked dfrG (encoding trimethoprim resistance) (Figure S2C). Rifampicin resistance (MIC, 256 µg/ml) was due to Leu 466Ser and His481Asn mutations in rpoB, and levofloxacin resistance (MIC, 4 µg/ml) was due to mutations Ser80Phe in grlA and Glu88Lys in gyrA (Figure 2).
Strain 16K possessed three plasmids, 2.9-kb chloramphenicol resistance plasmid (p16K-1), 3.0-Kb putative exported protein plasmid (p16K-2), which corresponded to 3.0-Kb pTW20_2 of TW20, and 2.3-Kb multidrug resistance efflux protein plasmid (p16K-3), which showed similarity to 2.4-Kb pPSK108 of S. epidermidis.
SCCmecIIIR structure of strain 16K
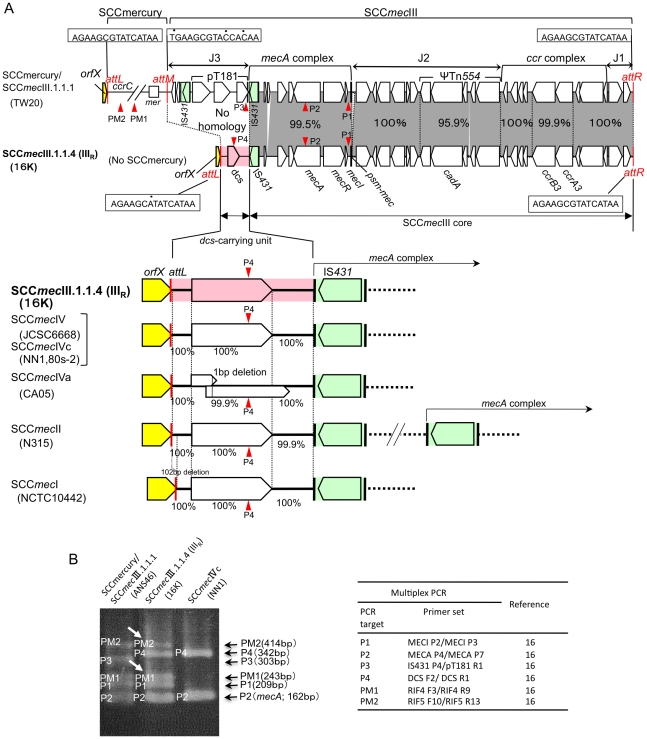
The complete sequence of 16K SCCmecIIIR was determined (Figure 3A). It was 31,198 bp in size, and consisted of 15-bp attL (adjacent to orfX), 2,208-bp dcs-carrying unit (J3 region), 28,960-bp SCCmecIII core region (carrying mecA complex and ccr complex), and 15-bp attR. The J3 region of 16K showed no homology with the 6,266-bp J3 region of TW20. The dcs-carrying unit (J3) was shared by 16K SCCmecIIIR, SCCmecI, SCCmecII, and SCCmecIV (Figure 3A); of those, the sequences of 16K SCCmecIIIR and SCCmecIVc (of ST30 CA-MRSA strains NN1 and 80s-2) were identical, suggesting that the dcs-carrying unit (J3) of 16K SCCmecIIIR was acquired by recombination. The mosaic SCCmecIIIR structure was identified by multiplex PCR (Figure 3B), although PCR (PM1 and PM2) detected MES16K in strain 16K (which lacked SCCmercury).
Figure 3. The SCCmecIIIR structure of ST239 MRSA strain 16K.
GenBank accession number for SCCmecIIIR is AB539727. The data for strain TW20 are from GenBank accession number FN433596. Homologous regions are shaded. In A, when the structure of SCCmecIIIR was compared with SCCmercury/SCCmecIII.1.1.1 of TW20 (upper part of figure), the SCCmecIII core was shared by the two, but the left side regions (flanked by att and IS431) of SCCmecIIIR and SCCmecIII.1.1.1 showed no homology. Moreover, Strain 16K lacked SCCmercury. The dcs-carrying unit (marked in pink) was shared by SCCmecIIIR, SCCmecIV, SCCmecII, and SCCmecI (lower part of figure). The data for strains JCSC6668, NN1, 80s-2, CA05, N315, and NCTC10442 were from GenBank accession numbers AB425823, AB245470, AB245471, AB063172, NC_002745, and AB033763, respectively. In B, the mosaic SCCmecIIIR structure was specifically detected by multiplex PCR. Multiplex PCR detects targets: P1 and P2 in the SCCmecIII core, P3 in the left side region (attM-pT181) of SCCmecIII.1.1.1, P4 in the left side region (dcs-carrying unit) of SCCmecIIIR, and PM1 (in an orf) and PM2 in SCCmercury or MES16K (PCR targets are shown in this figure [in A] and Fig. 4 for SCCmercury, and in Fig. 4 for MES16K); strain 16K was negative for the mer operon (lacked SCCmercury). ST239/SCCmecIII.1.1.1 MRSA strain ANS46 gave five PCR bands; three bands (P1, P2, and P3) for SCCmecIII.1.1.1 and two bands (PM1 and PM2) for SCCmercury. In contrast, the Russian variant (strain 16K) gave different combinations of five PCR bands; three bands (P1, P2, and P4) for SCCmecIIIR and two bands (PM1 and PM2, marked with arrows) for MES16K. PCR band P4 was shared by SCCmecIIIR and SCCmecIVc (and SCCmecIVa, SCCmecII, and SCCmecI). The combination of P1, P2, P4, PM1, and PM2 was unique to the Russian variant (strain 16K).
MES16K structure of strain 16K
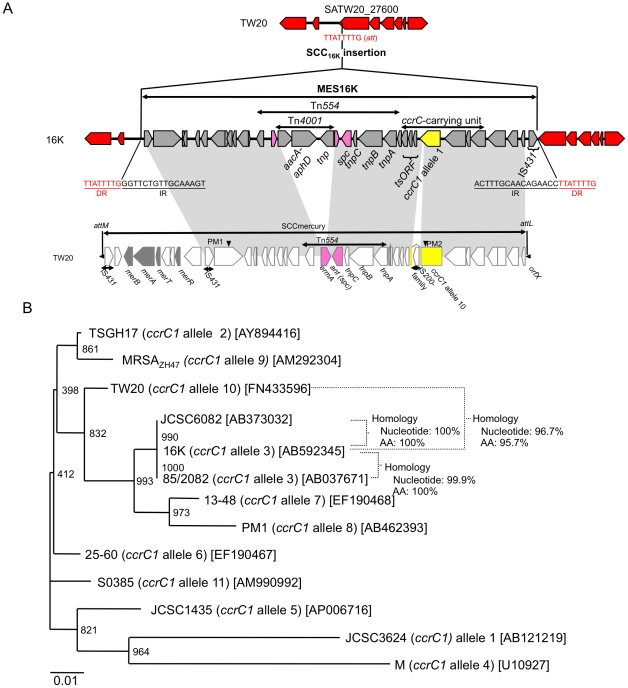
The complete sequence of MES16K was determined (Figure 4A). It was 30,818 bp in size and flanked by 16-bp inverted repeats (IRs). MES16K was inserted into the orf on the 16K genome, corresponding to SATW20_27600 of TW20; the att (target) sequence was 8-bp long, and was duplicated at both ends of MES16K. The 8-bp att and 16-bp IR sequences showed no homology to the 15-bp att sequences for SCCmecIII or SCCmercury. MES16K shared a similar ccrC-carrying unit with SCCmercury, albeit with ccrC1 allele 3 for 16K and ccrC1 novel allele 10 for TW20 (Figure 4B) and also with IS insertion in ccrC1 in TW20 (but not in 16K). Tn554 was present in both MES16K and SCCmercury, but in MES16K, Tn4001 was inserted into the ermA gene of Tn554.
Figure 4. MES16K structure and phylogenetic tree analysis for the ccrC genes.
The SCCmercury data for strain TW20 are from GenBank accession number FN433596. The GenBank accession number of MES16K is AB666466. Homologous regions are shaded. In A, for strain TW20, the ccrC-carrying unit was located within SCCmercury; the ccrC gene (marked in yellow) was split by the insertion of IS (IS200 family). For strain 16K, the ccrC-carrying unit was located within a mobile element structure MES16K, which was inserted into the orf (corresponding to SATW20_27600 of TW20) on the 16K genome, as shown in Fig. 2; the ccrC gene (marked in yellow) was intact. The attachment sequence (att) of MES16K showed no homology to that (att) of the orfX, SCCmecIII.1.1.1 or SCCmercury. Arrowheads PM1 and PM2 represent PCR targets for detection of SCCmercury of ST239/SCCmecIII.1.1.1 MRSA and MES16K of the Russian variant (strain 16K) by multiplex PCR (Fig. 3). In B, the ccrC gene sequence of strain 16K and that of strain TW20 (expected intact ccrC gene sequence, obtained by excluding the inserted IS200-family sequence) were analyzed for phylogenetic diversity, as described previously [36]. The ccrC gene of strain TW20 was a novel ccrC1 allele (named ccrC1 allele 10) and that of strain 16K was assigned as ccrC1 allele 3, unambiguously demonstrating evolutionary diversity between them; moreover, the ccrC1 gene of ST398 MRSA strain S0385 was also a novel allele (named ccrC1 allele 11).
Conjugative transfer of drug resistance
The ST239/spa351/SCCmecIIIR variant (especially group A2) transferred its drug resistance to S. aureus RN2677 by bacterial mating on agar plates, in the order of chloramphenicol, erythromycin, gentamicin, cadmium, and tetracycline resistance (Table 2).
Table 2. Conjugative transfer of drug resistance from Russian MRSA strains to S. aureus RN2677.
| Donor MRSA strainsa | Strains examined (n) | Transfer frequencyb: drug-resistant recipients/donor | |||||
| (recipient, RN2677) | |||||||
| Chloramphenicol resistance | Erythromycin resistance | Gentamicin resistance | Tetracycline resistance | Oxacillin resistance | Cadmium resistance | ||
| Russian isolates | |||||||
| Group A1 | 6 | –c | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 |
| (ST239/spa3) | |||||||
| Group A2 | 4 | 1.4×10−2 – | 3.6×10−4 – | 1.0×10−5 – | 2.8×10−8 – | <1.0×10−9 | 2.1×10−6 – |
| (ST239/spa351) | 5.3×10−4 d | 6.7×10−5 | 6.9×10−6 | 5.6×10−9 | 1.8×10−7 | ||
| ( cat /p2.9) | ( ermA ) | ( aacA-aphD ) | ( tetM ) | ( cadA ) | |||
| Group A3 | 2 | 9.5×10−5 – | 1.8×10−5 | <1.0×10−9 | –c | <1.0×10−9 | <1.0×10−9 |
| (ST8/spa826) | 3.0×10−6 | ( ermA ) | |||||
| ( cat /p2.9) | |||||||
| Group A4 | 1 | –c | –c | –c | –c | <1.0×10−9 | <1.0×10−9 |
| (ST72/spa3451) | |||||||
| Group B1 | |||||||
| (ST239/spa3) | 10 | –c | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 |
| (ST239/spa3) | 1e | 2.3×10−3 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 |
| (−/p41) | |||||||
| Group B2 | 6 | 1.1×10−2 – | 4.2×10−7 – | 3.7×10−8 – | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 |
| (ST239/spa351) | 9.7×10−3 | 1.6×10−8 | 1.6×10−8 | ||||
| (−/p41 or p30) | ( ermA ) | ( aacA-aphD ) | |||||
| Group C1 | 1 | –c | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 |
| (ST239/spa3) | |||||||
| Group C2 | |||||||
| (ST239/spa351) | 1f | 6.5×10−3 | 5.5×10−4 | 5.7×10−5 | <1.0×10−9 | <1.0×10−9 | 3.9×10−7 |
| (−/p41) | ( ermA ) | ||||||
| (ST239/spa351) | 3g | 3.3×10−6 – | 5.2×10−7 – | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | 6.4×10−7 – |
| 1.2×10−7 | 2.0×10−7 | 1.5×10−7 | |||||
| ( cat /p2.9) | ( ermA ) | ( cadA,cadD/p38 ) | |||||
| ST239 reference strains | |||||||
| ANS46 | <1.0×10−9 | 9.5×10−8 | –c | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | |
| ( ermA ) | |||||||
| HU25 | –c | 6.0×10−6 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | <1.0×10−9 | |
| ( ermA ) | |||||||
Groups of MRSA strains from Russia are from Table 1.
Drug resistance genes acquired by transconjugants are shown in parentheses (cat, chloramphenicol resistance; ermA, erythromycin and clindamycin resistance; aacA-aphD, gentamicin and kanamycin resistance; tetM, tetracycline and minocycline resistance; cadA, cadmium resistance; −, no previously-described sequences). Plasmid size was described after the resistance gene in parentheses.
Donor MRSA strains were susceptible to each drug (resulting in no transconjugants).
Resistance transfer frequencies (average data in two or three experiments) for four group2 isolates were 1.4×10−2, 6.5×10−3, 1.0×10−3, and 5.3×10−4.
Strain 2H; 105H; and 102H, 103H, and 104H, respectively.
Plasmids were detected only in chloramphenicol-resistant transconjugants (Tables 1 and 2). For group A2 MRSA (isolated from the community during 2006–2008), the size of chloramphenicol-resistance plasmids was 2.9-kb. The sequence of the chloramphenicol-resistance plasmid (GenBank accession number, AB539725), donated from strain 16K (group A2), was consistent with p16K-1 in Figure 2. In contrast, for MRSA strains from hospitals during 2004–2008 (one B1 strain [strain 2H] and B2 strains), the plasmid size was 41-kb or 30-kb. And, for MRSA from hospitals in 2011, the plasmid size in one strain (strain 105H) was 41-kb, while the plasmid size in three strains (strains 102H, 103H, and 104H) was 2.9-kb, indicating periodical changes of chloramphenicol-resistance plasmids.
No transfer or low-level transfer was observed for ST239/spa3/SCCmecIII.1.1.1 strains (groups A1, B1, and C1), except for strain 2H (group B1) which donated a 41-kb chlorampenicol resistance plasmid (Table 2). Moreover, group A1 strain 6K carrying p16K-1, which was constructed by introduction of p16K-1 DNA into strain 6K by electroporation, gave no transconjugants, indicating that p16K-1 transfer occurred in a host (strain 16K)-dependent manner.
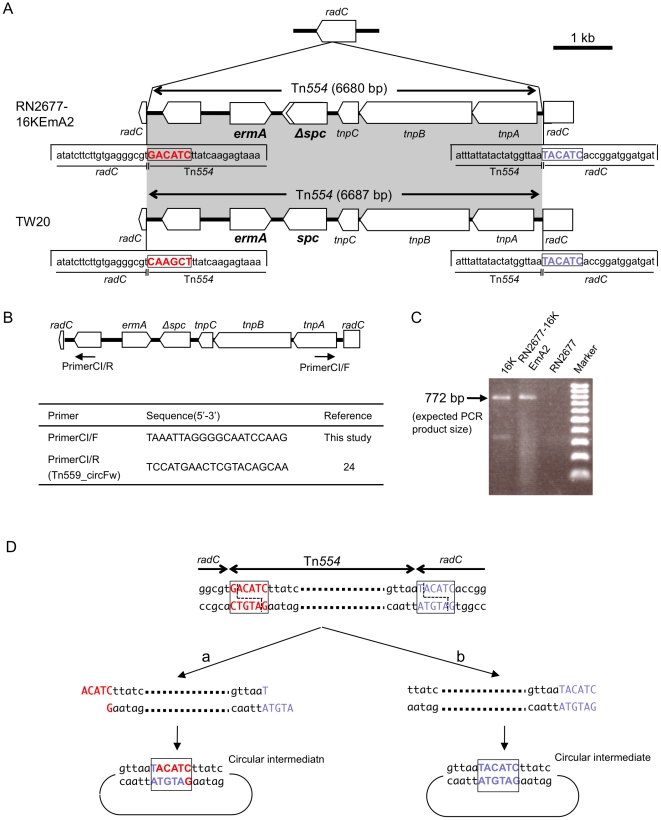
In mating between 16K and RN2677, erythromycin-resistant transconjugants (including RN2677-16KEmA2) possessed Tn554 (GenBank accession number, AB539726) inserted into the radC gene (Figures 2 and 5A). There were no repeats at either terminus of Tn554 and no repeats of the “target” sequence, unlike other transposons (as reported previously [37]–[39]). The hexanucleotide sequence at the left end of Tn554 in RN2677-16KEmA2 (5′-GACATC) was distinct from that of Tn554 within the TW20 radC gene (5′-CAAGCT) (Figure 5A; sequence in red), reflecting the former target site sequence [37]–[39] in each case. The free circular intermediate of Tn554 was present for both donor strain 16K and transconjugant RN2677-16KEmA2 (Figure 5B–D). For Tn554 in the transconjugant, there was a deletion in spc (Figure 5A), although it still conferred spectinomycin resistance.
Figure 5. The structure of Tn554 and its circular intermediate in erythromycin-resistant transconjugants (RN2677-16KEmA2).
Transconjugant RN2677-16KEmA2 was obtained by mating between strain 16K (donor) and S. aureus RN2677 (recipient). The data for strain TW20 are from GenBank accession number FN433596. In A, Tn554 was integrated into the radC gene in RN2677, at exactly the same site as the Tn554 integration site in radC for strain TW20; the radC right-side hexanucleotide sequence, immediately adjacent to Tn554, is marked in blue. The Tn554 left-side hexanucleotide sequence (marked in red) was distinct between the two Tn554 sequences of strains RN2677-16KEmA2 and TW20, reflecting previous target site sequences for each Tn554 (note that the orientation of the Tn554 structure in the figure is opposite, when compared with previously described Tn554 structures [37]–[39]). The spc gene on Tn554 in a transconjugant (RN2677-16KEmA2) had a single base substitution, resulting in a truncated gene product (lacking 27-aa at the 3′-end); the mutated spc gene still conferred spectinomycin resistance. In B, since Tn554 transposes through a circular intermediate [38]–[39], PCR primers to detect a Tn554 circular intermediate were designed for Tn554 originated in strain 16K. In C, the PCR primer set (CI/F and CI/R, shown in B) exactly detected a Tn554 circular intermediate in strains 16K and RN2677-16KEmA2 (PCR product size, 772 bp) in PCR assay; there were no such bands for strain RN2677. In D, when the sequence of the PCR product, obtained in C, was determined, the Tn554 circular intermediate in RN2677-16KEmA2 possessed the radC hexanucleotide (5′-TACATC; marked in blue), but not the Tn554 hexanucleotide sequence (5′-GACATC; marked in red).
Conjugative transfer of erythromycin resistance was also observed for group A3, group B2, group C2, and ST239 reference strains ANS46 and HU25 (Table 2); these transconjugants also carried spectinomycin resistance gene (spc) and were positive for a Tn554 circular intermediate by PCR (data not shown).
Discussion
The ST239 MRSA lineage is a representative multiple drug-resistant HA-MRSA circulating worldwide [2], [4]–[6], [8]; for example, it includes TW20, which emerged in an ICU in London [20]. Harris et al. [6] recently demonstrated, based on genome-wide SNPs, that the ST239 lineage consisted of more than five MRSA clades, such as Asia, North America, South America, Europe, and Australia. In this study, we isolated two divergent ST239 MRSA groups with spa3/SCCmecIII.1.1.1 and spa351/SCCmecIIIR from Russia; the latter has tended to increase recently. According to Harris et al. [6], spa3 (t037) represents the ancestral (most prevalent) ST239 spa type, and the spa3 type is distributed worldwide, including North and South America, Australia, Europe, and Asia. In contrast, the spa351 (t030) type has been found only in areas of Europe. Based on this, together with the deletion patterns of genomic islands and mobile genetic elements by Harris et al. [6], the Russian ST239/spa351/SCCmecIIIR variant may belong to the European clade, suggesting possible transmission from Europe to Far Eastern Russia (although SCCmecIII subtypes or sequences have not been described by Harris et al. [6]).
The two ST239 MRSA groups in Russia were isolated not only from patients in hospitals but also from patients with nongonococcal urethritis in the community; therefore, MRSA may be an agent of sexually transmitted disease in Russia, which would cause great concern about the potential to spread in the community. When the Russian ST239 MRSA strains (spa3/SCCmecIII.1.1.1 or spa351/SCCmecIIIR) from hospitals and the community were compared, there was a marked divergence in plasmid patterns; e.g., a 2.9-kb chloramphenicol-resistance plasmid in group A2 vs. a 41-kb chloramphenicol-resistance plasmid in group B2. Moreover, the ST239/spa351/SCCmecIIIR variant isolated recently (in 2011) showed a slightly divergent PFGE pattern, suggesting micro but dynamic evolution in Russia.
The CC30 genome section of the ST239 hybrid genome (16K) carried two mobile elements, SCCmecIIIR and MES16K, and three potential virulence genes, spa [40], [41], cna [26], [42], and psm-mec (encoding a peptide cytolysin [43]). SCCmecIIIR was a mosaic SCCmec, consisting of the SCCmecIII core and dcs-carrying unit (as a J3 region). An identical dcs-carrying unit was found in SCCmecIVc of ST30 CA-MRSA (strains NN1 and 80s-2 in Japan [25], [44] and also strain RS08 in Russia [45]), indicating that the dcs-carrying unit of the Russian variant could originate in ST30 CA-MRSA. The dcs-carrying unit corresponds to a region with the ccrC-carrying unit in SCCmecVII [36] (now renamed SCCmecV [33]). This region flanked by orfX(attL) and IS431 seems to be a hot spot for recombination.
In strain 16K, the ccrC-carrying unit, found in SCCmecV and SCCmercury [36], was located in MES16K, which carried a composite transposon. MES16K is markedly distinguished from SCCmer or SCCmercury, even in terms of the attachment sequence (att). The ccrC gene is highly variable; for example, it is ccrC1 allele 3 for strain 16K, ccrC1 allele 8 for ST59 CA-MRSA strain PM1 [36], and ccrC1 allele 10 (novel allele type) for ST239 HA-MRSA strain TW20.
One of the most striking features of the Russian variant is its behavior as a multiple drug resistance disseminator. To our knowledge, this is the first demonstration of multiple drug resistance transfer in the ST239 MRSA lineage. For mating, close contact of donor and recipient cells on agar plates is needed, but the use of filters is not essential. There is a possibility that during close-contact mating, plasmids in donor (16K) cells are preferably transferred into recipient bacterial cells, as in the case of p16K-1. For Tn554, which transposes through the circular intermediate [38], [39], the circular intermediate may be transferred, followed by subsequent integration into the recipient genome, thus exhibiting lower transfer frequency than a chloramphenicol plasmid (p16K-1).
In conclusion, the ST239/spa3/SCCmecIII.1.1.1 MRSA and its variant (ST239/spa351/SCCmecIIIR) have emerged not only in hospitals but also in the community, associated with nongonococcal urethritis, in Russia. The Russian variant showed high similarity to TW20 in terms of the bulk of the hybrid genome, but was markedly divergent from TW20, such as spa351 vs. spa3, novel mosaic SCCmecIIIR vs. SCCmecIII.1.1.1, MES16K vs. SCCmercury, and the presence vs. absence of small chloramphenicol-resistance plasmids. The Russian variant behaved as a drug resistance disseminator, mediating chloramphenicol-resistance plasmid transfer and conjugative Tn554 transposition. Micro but dynamic evolution of ST239 MRSA is proceeding in Russia.
Supporting Information
The agr subtyping for the Russian variant strain 49K. In A, PCR primers for agr typing are shown as an arrow. Strains: agr1, agr1 type strain; agr2, agr2 type strain; 49K, the Russian variant strain 49K. Strain 49K produced a product only for primer sets (Pan and agr1), and not for (Pan and agr2), assigning strain 49K as agr1. However, in the other PCR assay, strain 49K produced a product both for primer sets (agr1 forward and agr1 reverse) and for agr2 forward and agr2 reverse, resulting in no precise assignment. In B, the agr subtypes were examined by phylogenetic tree analysis, based on the nucleotide sequences of the agr variable region, as described previously [30]. GenBank accession number for the variable region sequence of strain 49K is AB592345. Based on the data, strain 49K was assigned as agr1′.
(TIF)
Deletions in drug resistance or virulence genetic traits in 16K, in comparison with TW20. The data of strain TW20 are from GenBank accession number FN433596. Homologous regions are shaded. In A, contigs 41, 14, 85, and 63 (and probably 77) of strain 16K constituted the bulk of the ΦSa3 phage sequence, showing the presence of the sea gene (encoding superantigen SEA, a virulence factor associated with severe invasive infections) in strain 16K; no right side sequence of ΦSa3 was present. In B, contigs 4 and 13 of strain 16K constituted the left and right side regions of the SaPI1 superantigen-associated pathogenicity island, showing the presence of the superantigen genes sek and seq in strain 16K. In C, contigs of 42 and 28 of strain 16K constituted the Tn5801-like sequence (including 20-bp direct repeat at both ends), but with deletion of the dfrG gene sequence (encoding trimethoprim resistance) in contig 28. Strain 16 was also negative for dfrG by PCR assay.
(TIF)
Acknowledgments
We thank Wei-Chun Hung and Akihito Nishiyama for technical assistance and H. de Lencastre for epidemic HA-MRSA-type strains.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported, in part, by a grant from Heiwa Nakajima Foundation, Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.rundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto T, Nishiyama A, Takano T, Yabe S, Higuchi W, et al. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J Infect Chemother. 2010;16:225–254. doi: 10.1007/s10156-010-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noto MJ, Kreiswirth BN, Monk AB, Archer GL. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J Bacteriol. 2008;190:1276–1283. doi: 10.1128/JB.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires de Sousa M, Conceição T, Simas C, de Lencastre H. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J Clin Microbiol. 2005;43:5150–5157. doi: 10.1128/JCM.43.10.5150-5157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DA, Enright MC. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol. 2004;186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holden MT, Lindsay JA, Corton C, Quail MA, Cockfield JD, et al. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol. 2010;192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teixeira LA, Resende CA, Ormonde LR, Rosenbaum R, Figueiredo AM, et al. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanches IS, Aires de Sousa M, Cleto L, de Campos MB, de Lencastre H. Tracing the origin of an outbreak of methicillin-resistant Staphylococcus aureus infections in a Portuguese hospital by molecular fingerprinting methods. Microb Drug Resist. 1996;2:319–329. doi: 10.1089/mdr.1996.2.319. [DOI] [PubMed] [Google Scholar]
- 11.Aires de Sousa M, Sanches IS, Ferro ML, Vaz MJ, Saraiva Z, et al. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lencastre H, Severina EP, Milch H, Thege MK, Tomasz A. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin Microbiol Infect. 1997;3:289–296. doi: 10.1111/j.1469-0691.1997.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 13.Aires de Sousa M, de Lencastre H. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol. 2004;40:101–111. doi: 10.1016/S0928-8244(03)00370-5. [DOI] [PubMed] [Google Scholar]
- 14.Conceição T, Aires-de-Sousa M, Füzi M, Tóth Á, Pászti J, et al. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin Microbiol Infect. 2007;13:971–979. doi: 10.1111/j.1469-0691.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- 15.Witte W. Antibiotic resistance in gram-positive bacteria: epidemiological aspects. J Antimicrob Chemother. 1999;44(Suppl A):1–9. doi: 10.1093/jac/44.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 16.Marples RR, Cooke EM. Workshop on methicillin-resistant Staphylococcus aureus held at the headquarters of the Public Health Laboratory Service on 8 January 1985. J Hosp Infect. 1985;6:342–348. [PubMed] [Google Scholar]
- 17.Kerr S, Kerr GE, Mackintosh CA, Marples RR. A survey of methicillin-resistant Staphylococcus aureus affecting patients in England and Wales. J Hosp Infect. 1990;16:35–48. doi: 10.1016/0195-6701(90)90047-r. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira D, Santos-Sanches I, Mato R, Tamayo M, Ribeiro G, et al. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the ‘Iberian’ and the ‘Brazilian’ clones of MRSA. Clin Microbiol Infect. 1998;4:373–384. doi: 10.1111/j.1469-0691.1998.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 20.Edgeworth JD, Yadegarfar G, Pathak S, Batra R, Cockfield JD, et al. An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin Infect Dis. 2007;44:493–501. doi: 10.1086/511034. [DOI] [PubMed] [Google Scholar]
- 21.From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus - Minnesota and North Dakota, 1997-1999. MMWR Morb Mortal Wkly Rep. 1999;48:707–710. [PubMed] [Google Scholar]
- 22.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 23.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takano T, Higuchi W, Yamamoto T. Superior in vitro activity of carbapenems over anti-methicillin-resistant Staphylococcus aureus (MRSA) and some related antimicrobial agents for community-acquired MRSA but not for hospital-acquired MRSA. J Infect Chemother. 2009;15:54–57. doi: 10.1007/s10156-008-0665-5. [DOI] [PubMed] [Google Scholar]
- 25.Takizawa Y, Taneike I, Nakagawa S, Oishi T, Nitahara Y, et al. A Panton-Valentine leucocidin (PVL)-positive community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J Clin Microbiol. 2005;43:3356–3363. doi: 10.1128/JCM.43.7.3356-3363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi D, Ishii S, Sato T, Yamazaki H, Matsunaga M, et al. Staphylococcal scalded skin syndrome in an extremely low-birth-weight neonate: molecular characterization and rapid detection by multiplex and real-time PCR of methicillin-resistant Staphylococcus aureus. Pediatr Int. 2010;53:211–217. doi: 10.1111/j.1442-200X.2010.03246.x. [DOI] [PubMed] [Google Scholar]
- 27.Takano T, Higuchi W, Otsuka T, Baranovich T, Enany S, et al. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob Agents Chemother. 2008;52:837–845. doi: 10.1128/AAC.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40:4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strommenger B, Cuny C, Werner G, Witte W. Obvious lack of association between dynamics of epidemic methicillin-resistant Staphylococcus aureus in central Europe and agr specificity groups. Eur J Clin Microbiol Infect Dis. 2004;23:15–19. doi: 10.1007/s10096-003-1046-8. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi W, Hung WC, Takano T, Iwao Y, Ozaki K, et al. Molecular characteristics of the Taiwanese multiple drug-resistant ST59 clone of Panton-Valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus from pediatric cellulitis. J Infect Chemother. 2010;16:144–149. doi: 10.1007/s10156-010-0029-9. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto T, Dohmae S, Saito K, Otsuka T, Takano T, et al. Molecular characteristics and in vitro susceptibility to antimicrobial agents, including the des-fluoro(6) quinolone DX-619, of Panton-Valentine leucocidin-positive methicillin-resistant Staphylococcus aureus isolates from the community and hospitals. Antimicrob Agents Chemother. 2006;50:4077–4086. doi: 10.1128/AAC.00847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing: 18th informational supplement M100-S18, Wayne, PA, USA: Clinical and Laboratory Standards Institute.
- 36.Higuchi W, Takano T, Teng LJ, Yamamoto T. Structure and specific detection of staphylococcal cassette chromosome mec type VII. Biochem Biophys Res Commun. 2008;377:752–756. doi: 10.1016/j.bbrc.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Murphy E, Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984;307:292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- 38.Haroche J, Allignet J, El Solh N. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin a and related compounds including dalfopristin. Antimicrob Agents Chemother. 2002;46:2337–2343. doi: 10.1128/AAC.46.8.2337-2343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadlec K, Schwarz S. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother. 2010;54:3475–3477. doi: 10.1128/AAC.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gómez MI, O'Seaghdha M, Magargee M, Foster TJ, Prince AS. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem. 2006;281:20190–20196. doi: 10.1074/jbc.M601956200. [DOI] [PubMed] [Google Scholar]
- 41.Kahl BC, Peters G. Microbiology. Mayhem in the lung. Science. 2007;315:1082–1083. doi: 10.1126/science.1139628. [DOI] [PubMed] [Google Scholar]
- 42.de Bentzmann S, Tristan A, Etienne J, Brousse N, Vandenesch F, et al. Staphylococcus aureus isolates associated with necrotizing pneumonia bind to basement membrane type I and IV collagens and laminin. J Infect Dis. 2004;190:1506–1515. doi: 10.1086/424521. [DOI] [PubMed] [Google Scholar]
- 43.Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5:e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taneike I, Otsuka T, Dohmae S, Saito K, Ozaki K, et al. Molecular nature of methicillin-resistant Staphylococcus aureus derived from explosive nosocomial outbreaks of the 1980s in Japan. FEBS Lett. 2006;580:2323–2334. doi: 10.1016/j.febslet.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 45.Baranovich T, Potapov V, Yamamoto T. The first isolation of Panton-Valentine leukocidin (PVL) positive community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) in Russia. Euro Surveill. 2007;12:E070315 4. doi: 10.2807/esw.12.11.03157-en. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The agr subtyping for the Russian variant strain 49K. In A, PCR primers for agr typing are shown as an arrow. Strains: agr1, agr1 type strain; agr2, agr2 type strain; 49K, the Russian variant strain 49K. Strain 49K produced a product only for primer sets (Pan and agr1), and not for (Pan and agr2), assigning strain 49K as agr1. However, in the other PCR assay, strain 49K produced a product both for primer sets (agr1 forward and agr1 reverse) and for agr2 forward and agr2 reverse, resulting in no precise assignment. In B, the agr subtypes were examined by phylogenetic tree analysis, based on the nucleotide sequences of the agr variable region, as described previously [30]. GenBank accession number for the variable region sequence of strain 49K is AB592345. Based on the data, strain 49K was assigned as agr1′.
(TIF)
Deletions in drug resistance or virulence genetic traits in 16K, in comparison with TW20. The data of strain TW20 are from GenBank accession number FN433596. Homologous regions are shaded. In A, contigs 41, 14, 85, and 63 (and probably 77) of strain 16K constituted the bulk of the ΦSa3 phage sequence, showing the presence of the sea gene (encoding superantigen SEA, a virulence factor associated with severe invasive infections) in strain 16K; no right side sequence of ΦSa3 was present. In B, contigs 4 and 13 of strain 16K constituted the left and right side regions of the SaPI1 superantigen-associated pathogenicity island, showing the presence of the superantigen genes sek and seq in strain 16K. In C, contigs of 42 and 28 of strain 16K constituted the Tn5801-like sequence (including 20-bp direct repeat at both ends), but with deletion of the dfrG gene sequence (encoding trimethoprim resistance) in contig 28. Strain 16 was also negative for dfrG by PCR assay.
(TIF)