Abstract
Autophagy is a highly conserved homeostatic pathway by which cells transport damaged proteins and organelles to lysosomes for degradation. Dysregulation of autophagy contributes to the pathogenesis of clinically important disorders in a variety of organ systems but, until recently, little was known about its relationship to diseases of the lung. However, there is now growing evidence at the basic research level that autophagy is linked to the pathogenesis of important pulmonary disorders such as chronic obstructive pulmonary disease, cystic fibrosis, and tuberculosis. In this review, we provide an introduction to the field of autophagy research geared to clinical and research pulmonologists. We focus on the best-studied autophagic mechanism, macroautophagy, and summarize studies that link the regulation of this pathway to pulmonary disease. Last, we offer our perspective on how a better understanding of macroautophagy might be used for designing novel therapies for pulmonary disorders.
Keywords: autophagy, macroautophagy, lung, disease, chronic obstructive pulmonary disease
At the cellular level, homeostasis is maintained by a series of deeply conserved pathways of which “autophagy” is one central pillar. Autophagy refers to a collection of catabolic pathways that transport components of the cytoplasm to lysosomes for degradation (1). In addition to proteins, autophagy can target both carbohydrates (2, 3) and lipids (4) for digestion as well as entire organelles, such as mitochondria and peroxisomes (5–7). The products of digestion, such as free amino acids, are recycled back to the cytoplasm for use in various biosynthetic pathways (8). Autophagy contributes to cellular homeostasis via three basic mechanisms. First, autophagy provides an alternative source of metabolic fuel (9). Second, autophagy removes damaged cellular components, such as dysfunctional mitochondria and aggregated proteins that would otherwise be toxic to the cell (7, 10, 11). Finally, autophagy is entwined at the signal transduction level with the apoptotic pathway and impacts the decision of a cell to undergo programmed cell death (12, 13). To study autophagy is to study an aspect of the core operating system that enables eukaryotic cells to function.
Which begs the question: what does autophagy have to do with patients suffering from pulmonary disease? Interestingly, the answer may be “quite a lot.” The purpose of this review is twofold: to provide a basic introduction to the rapidly expanding field of autophagy research with an emphasis on the most studied autophagic pathway (macroautophagy), and also to review recent studies that link autophagic regulation to pulmonary diseases.
How Does Autophagy Work?
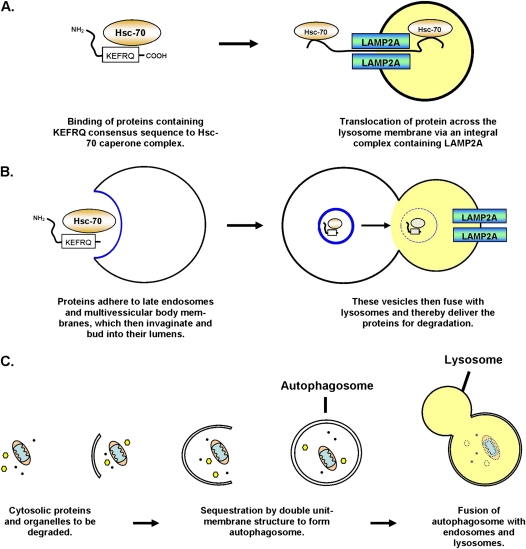
At present there are three known mechanisms by which autophagy can occur (Figure 1). The first mechanism is called chaperone-mediated autophagy (CMA) and was originally described in lung fibroblasts (Figure 1A) (14). This mechanism involves the direct translocation of proteins across the lysosomal membrane via a complex that includes Hsc70 and the lysosome transmembrane protein Lamp2A (15, 16). CMA is difficult to monitor in vivo and at this point we do not have any information about whether this pathway is affected in pulmonary disease. However, CMA plays an important role in the pathogenesis of neurodegeneration and aging (17, 18), and is still an evolving field of biomedical research. The second autophagic pathway is called microautophagy and involves the direct invagination of cytosolic material into late endosomes or into multivesicular bodies, which subsequently either degrade the material on site or deliver the material to lysosomes for degradation (19). Until more recently nothing was known about microautophagy at the molecular level, and the existence of this process was suspected in mammalian tissues only on the basis of electron micrographs (EMs). However, it was shown that microautophagy also employs Hsc70 but, unlike CMA, it targets proteins to late endosomal membranes through electrostatic interactions between this chaperone and the lipid phosphatidylserine, rather than by binding to Lamp2A (Figure 1B) (20). Microautophagy is similarly difficult to measure in vivo and so little is understood about its physiological significance at this point, although this may change as the molecular mechanism is better defined.
Figure 1.
Schematic depiction of the three known autophagy pathways. (A) Chaperone mediated autophagy based on Dice (120). (B) Microautophagy based on Sahu and colleagues (20). (C) Macroautophagy based on Mizushima (121).
The final pathway, macroautophagy (21), receives the most attention in the literature and its contribution to human disease is the best explored of the three pathways. In fact, macroautophagy is so much better studied that it is often referred to in many papers as simply “autophagy,” even though it is only one of the pathways involved in lysosome-dependent degradation. This is in large part because macroautophagy can be visualized at both the light microscopic level (using fluorescent fusion proteins), and at the EM level making it relatively easy to detect (22). In macroautophagy, a vesicular membrane is constructed around a volume of cytoplasm that is intended for degradation (Figure 1C). This novel structure, called an autophagosome, is distinct from other vesicles on electron micrographs because it contains a double-unit limiting membrane (21). Autophagosomes then deliver their cargo for degradation by fusing with late endosomes and lysosomes, gradually losing their distinctive membrane structure (23). The products of digestion, such as free amino acids, are recycled back to the cytoplasm via lysosomal permeases (8). Of the three known autophagic pathways, macroautophagy is notable for its ability to process large intracellular structures such as organelles (5, 6), invasive bacteria in the cytoplasm (24, 25), and large protein aggregates (10). Macroautophagy is sensitive to nutrient availability (19, 26), and is altered in a variety of nonpulmonary diseases, such as neurodegeneration (27), myopathy (28, 29), and cancer (30). All of the current literature about the role of autophagy in pulmonary disease focuses on macroautophagy, and therefore we focus on this mechanism for the remainder of this review.
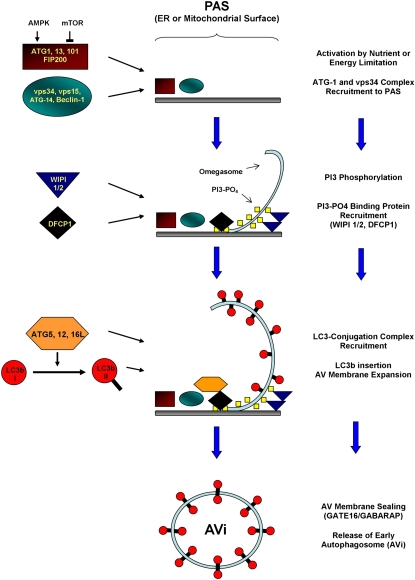
The last decade has seen a dramatic increase in our understanding of the molecular mechanisms underlying macroautophagy (Figure 2). The current paradigm is that macroautophagy is mediated by a highly intricate mechanism that requires the contribution of at least 20–30 core proteins that are physically grouped into several distinct multiprotein complexes, each with a distinct functional specialization (31). The most upstream complex described so far is composed of the proteins ATG1 (autophagy-related-1), FIP200 (FAK [focal adhesion kinase] family interacting protein of 200 kD), ATG101, and ATG13 and possesses serine/threonine kinase activity (32, 33). This complex is regulated by mTOR (mammalian target of rapamycin) and AMPK (AMP-activated protein kinase), which sense nutrient and intracellular ATP availability and, through phosphorylation of ATG1, adjust the rate of macroautophagy to the metabolic needs of the cell (34). The primary function of the ATG1-containing complex is to control the activity and localization of another structure called the vps34 multiprotein complex, although the mechanism by which this is accomplished is unknown. The vps34 complex has phosphatidylinositol-3-kinase (PI3K) activity and the production of PI3 moieties is required for autophagosome formation (35, 36). One important constituent of the vps34 complex is the protein ATG6/Beclin-1, which is an adaptor protein that is crucial for the participation of the vps34 complex in autophagosome formation (37, 38). In yeast, the vps34 complex localizes to a distinct perinuclear structure called the phagophore assembly site (39), whereas in mammalian cells this complex localizes to many sites distributed throughout the cell (40). The precise location of these sites is controversial but there are microscopy data suggesting the vps34 complex can nucleate autophagosomes on the surface of both the endoplasmic reticulum (ER) (40) and mitochondria (41), depending on the cell type and experimental conditions. The PI3-phosphate moieties laid down by vps34 recruit binding proteins that are critical for autophagosome maturation and may also intrinsically produce local changes in the membrane physical properties that make it easier to mold (42–44). The result is a cuplike structure that has been termed the “omegasome” (40, 45, 46). The final group of proteins is an ubiquitin-like ligase system composed of the proteins ATG3, ATG7, and ATG10, and a trimeric complex composed of ATG5, ATG12, and ATG16L that localizes to the omegasome (47, 48). The function of this system is to conjugate a protein called ATG8/LC3b to the lipid phosphatidylethanolamine (PE), which is abundant in autophagosome membranes (49). The PE-conjugated form of LC3b (called LC3b-II) is inserted into both sides of the autophagosome membrane (50), and is required for autophagosome membrane elongation (51, 52). LC3b is actually just one of at least five homologous proteins in mammalian cells (53, 54), all of which are targeted to the autophagosome by PE conjugation and cooperate in autophagosome maturation and sealing (52). Luminal-facing LC3b-II is degraded by lysosomal hydrolases (55), while protein on the cytosolic-facing side of the autophagosome is recovered via delipidation (Figure 3) (56). The cytoplasmic, nonlipidated form of LC3b, called LC3b-I, can then be recruited for the construction of new autophagosomes (50, 56). LC3b also plays an important diagnostic role in macroautophagy research: because it is targeted uniquely to autophagosome membranes it serves as a convenient marker for this structure (50).
Figure 2.
Proposed molecular mechanism for autophagosome formation, based on Itakura and Mizushima (122), Hayashi-Nisino and colleagues (45, 46), and Weidberg and colleagues (52). AMPK = AMP-activated protein kinase; ATG = autophagy-related protein; AV = autophagic vacuole; DFCP1 = double FYVE-containing protein 1; ER = endoplasmic reticulum; FIP200 = FAK [focal adhesion kinase] family interacting protein of 200 kD; GABARAP = γ-aminobutyric acid type A receptor–associated protein; GATE16 = Golgi-associated ATPase enhancer of 16 kD; mTOR = mammalian target of rapamycin; PAS = phagophore assembly site; PI3 = 3-phosphatidylinositol; WIPI = WD repeat domain, phosphoinositide-interacting protein.
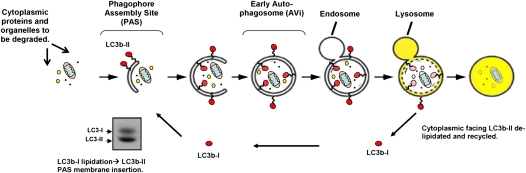
Figure 3.
Schematic of the macroautophagy pathway. Red circles depict LC3b; orange ovals depict mitochondria; yellow hexagons and black dots depict ribosomes and cytosolic proteins, respectively. A representative Western blot of LC3b-I and LC3b-II is displayed at lower left.
Although the above-described molecular mechanism proposed for macroautophagy is now impressively detailed, much of this information comes from genetically deleting individual pathway members and then observing the results. As such we know a great deal about the hierarchal relationships between macroautophagy proteins, but we have little insight into how the functional groups cooperate on a biochemical level. It is not even certain that macroautophagy represents a single unified process, which is the current paradigm, or a collection of processes that happen to produce similar-looking vesicles under EM, but only partially overlap on the molecular level. Indeed, autophagosomes have been observed in mammalian cells even when critical proteins in the “canonical pathway” described above have been genetically knocked out (57). That said, it is clear from genetic deletion of macroautophagy-related (ATG) proteins in mice that protein turnover via this process has physiological consequences that are relevant to human disease, including pulmonary disease.
How Is Macroautophagy Measured?
In the current literature a diverse and somewhat confusing variety of approaches are used to examine the macroautophagy pathway (22). They can be grouped into three basic approaches: microscopy (see Table E1 in the online supplement), static biochemical measurements (such as Western blot analysis; Table E2), and dynamic biochemical measurements (Table E3).
Microscopy was the first technique used to study macroautophagy and the discovery of autophagosomes on EM images is what gave rise to the field in the first place (58). At the ultrastructural level, early-stage autophagosomes (AVis) can be distinguished by their double-unit membrane structure and by the consistency of its luminal material, which resembles that of the adjacent cytoplasm (59). However, EM image analysis is less sensitive for picking up late-stage autophagosomes (AVds), as they tend to have lost their distinguishing physical characteristics. Fluorescence-based methods get around this problem by tagging specific proteins that are found on both early- and late-stage autophagosomes. The most commonly used reporter is green fluorescent protein–tagged microtubule-associated protein-1 light chain-3 (GFP–LC3) (22). On induction of macroautophagy by nutrient starvation, GFP–LC3 changes its cellular localization from a diffuse cytosolic pattern to a punctate pattern as it is recruited to newly formed autophagosomes (22). This phenomenon can be observed both in mammalian tissue culture cells and in vivo, using GFP–LC3–overexpressing transgenic mice (60). Fluorescence-based reporter systems can be used in fixed cells or observed dynamically by video microscopy (61).
The most widely used assay to examine macroautophagy is Western blot analysis of LC3b protein (62). During the autophagosome maturation process LC3b is conjugated to the lipid phosphatidylethanolamine (PE) and then inserted into the inner and outer leaflets of the autophagosome membrane. Despite its higher molecular weight the PE-conjugated form of LC3b (called LC3b-II) migrates more rapidly in sodium dodecyl sulfate–polyacrylamide gels than the unconjugated version (LC3b-I), and can be easily distinguished on Western blot (Figure 3). Levels of LC3b-II on Western blot were shown to correlate with autophagosome abundance and so this method provides a convenient alternative to microscopy and can also be applied to tissue biopsies (50, 63). Other macroautophagy markers that are amenable to Western blot analysis include p62 (64), which is an adaptor protein that targets ubiquitinated proteins to the autophagosome by interacting with LC3b-II (65). Under starvation conditions, the steady state level of p62 tends to fall as a result of increased consumption of this protein within autophagosomes (64), and so levels of p62 are generally assumed to inversely correlate with macroautophagic activity (or flux).
A common feature of the assays mentioned so far is that they are primarily static estimates of the levels of autophagosomes or macroautophagy proteins at a given point in time. However, some authors have begun to question the assumption that such static measurements reliably equate with increased protein turnover (66, 67), citing examples where increased amounts of autophagosomes and LC3b-II proved instead to represent a decrease in the fusion of autophagosomes with lysosomes and hence a lower rate of flux (68, 69). As such, there has been an increased emphasis on dynamic “activity assays” that directly measure macroautophagic flux.
At present the most popular method for measuring macroautophagic flux is the “LC3b turnover assay,” in which cells are cultured in the presence or absence of an inhibitor of lysosome proteases for a fixed length of time (67). Because LC3b-II is inserted on the luminal surface of autophagosomes as well as the exterior, a portion of LC3b-II is degraded when autophagosomes fuse with lysosomes (Figure 3). By comparing the difference in the amount of LC3b-II on Western blot in the presence versus the absence of lysosome inhibitors, macroautophagic flux can be measured as the amount of LC3b-II degraded per unit time (67). Other groups have produced turnover assays that are based on specific substrates of macroautophagy besides LC3b-II (64, 70–72).
In summary, there are a wide variety of ways to examine individual components of the macroautophagy pathway as well as the proteolytic flux that is a product of this pathway. What is missing from the field is any kind of consensus about what represents the minimal acceptable set of measurements on which conclusions should be based. For basic researchers interested in pulmonary disease the problem is even more complex because the lung is a heterogeneous organ composed of up to 40 different cell types (73). As a result, changes in the macroautophagy system that occur in a subset of cell types can be obscured when analyzing whole lung homogenates. Even when a stimulus globally alters the macroautophagy machinery in the lung the physiological significance of that regulation might vary by cell type (see the section on chronic obstructive pulmonary disease [COPD] below). Our recommendation is to carefully define the cell type being studied (e.g., respiratory epithelia, fibroblasts), and to include at least one macroautophagic flux measurement in addition to static measurements of autophagosomes or LC3b-II. Given the improvements in methodology and the pioneering studies described below, the stage is now set to rapidly explore the significance of macroautophagy in pulmonary disease.
Why Study Macroautophagy In The Lung?
As mentioned previously, macroautophagy is just one catabolic pathway among several that can degrade proteins in lysosomes. For the pulmonologist it is fair to ask whether there is any evidence that would justify focusing on macroautophagy as opposed to any other catabolic pathway. Inoue and colleagues (74) described the first lung-specific macroautophagy knockout mouse, in which atg7 is specifically deleted from Clara cells. They found that atg7 deletion produced severe abnormalities in airway epithelial cells that included cellular swelling, a loss of rough ER, loss of cilia, and abnormal-looking mitochondria (74). The cellular swelling was physiologically significant because it produced increased airway resistance after challenging the mice with methacholine (74). This effect was related to exaggerated changes in airway caliber in the atg7-deleted lungs rather than a true asthma phenotype, as there was no evidence of the inflammation or airway remodeling characteristic of that disease. Nevertheless, these results suggest that macroautophagy is critical for maintaining the internal organization of small-airway epithelia, which has important implications for human airway diseases such as asthma. Whether macroautophagy is also important for the basal functioning of other parenchymal cells, such as alveolar epithelium, will hopefully be addressed with new tissue-specific knockout mice currently in development. In summary, there is now evidence to support a crucial role specifically for macroautophagy in pulmonary cellular physiology. Moreover, macroautophagy has also been linked to the pathogenesis of several important adult pulmonary disorders such as COPD, cystic fibrosis, and tuberculosis, as described below.
Chronic Obstructive Pulmonary Disease
COPD is among the most prevalent adult disorders worldwide (75), and at present, is also the pulmonary disorder with the most intensively studied link to macroautophagy. It is important to note first that, at the histological level, COPD is an umbrella term meant to capture a range of pathologies that result in irreversible obstructive ventilatory deficits in patients. At one histological extreme is a chronic bronchitis picture characterized by small-airway remodeling, chronic inflammatory cell infiltration, goblet cell hyperplasia, and frequent bacterial colonization (76). At the other extreme is the emphysematous phenotype characterized by alveolar membrane destruction and air space enlargement without inflammatory infiltrates (76). It is presumed that most patients fall on a continuum between these stereotypical presentations. Much of the macroautophagy literature has primarily been connected to emphysema as this phenotype can be easily reproduced in mice (77).
The first indication that macroautophagy was regulated in patients with COPD came from Western blot analysis of lung biopsy specimens, which showed elevated levels of LC3b-II protein in patients with COPD compared with non-COPD control patients (78). The amount of LC3b-II induction correlated positively with clinical severity as measured by GOLD (Global Initiative for Chronic Obstructive Pulmonary Disease) score (78). To further investigate this, our laboratory turned to smoke exposure models in tissue culture cells and in mice, given that cigarette smoke represents the major risk factor for COPD development. Our group as well as others found that exposure of lung epithelial cell lines and fibroblasts to cigarette smoke extract (CSE) induced the accumulation of autophagosomes on electron micrographs and enhanced levels of LC3b-II protein, similar to what was seen in lung biopsies from patients with COPD (79, 80). The appearance of autophagosomes occurred quickly, on the order of a few hours, and preceded cell death, which is seen with prolonged exposure to CSE (79). Importantly, genetic depletion of two macroautophagy pathway members, Beclin-1 and LC3b, reduced the rate of cell death in CSE-exposed cells (78, 79), and mice deficient in LC3b were resistant to emphysematous changes caused by chronic cigarette smoke exposure (81). The overall interpretation was that the macroautophagy pathway facilitated lung epithelial and fibroblast cell death in response to cigarette smoke, which then contributed to the development of emphysema. Given that cigarette smoke exposure stimulated the formation of the death-inducing signaling complex, a complex containing the protein Fas receptor, we concluded that macroautophagy must mediate emphysematous change through programmed cell death (79).
While our group focused on the role of macroautophagy in lung epithelial cells during COPD, others have focused on alveolar macrophages. Macrophages derived from patients with COPD often have impaired antimicrobial activities (82, 83), but at the same time they are prone to hypersecrete proinflammatory cytokines when presented with a stimulant such as bacterial endotoxin (84). The combination of these two functional alterations in macrophages may help to make patients with COPD vulnerable both to airway bacterial colonization and chronic bronchitis (85). Monick and colleagues (86) examined the role of macroautophagy in alveolar macrophages from actively smoking patients (with a greater than 10 pack-year smoking history), or macrophages of nonsmokers who were exposed to CSE. Importantly, their study employed direct measurements of macroautophagic flux and so could directly examine the effect of smoking on protein turnover via this pathway. They found that macroautophagic flux was strongly inhibited in the alveolar macrophages of patients with COPD, and this effect could be reproduced in the macrophages of nonsmokers by exposing them to CSE (86). The level of inhibition was at a downstream step in the pathway: autophagosomes could form but then were not degraded as fast as normal, leading to an overall accumulation of autophagosomes. The inhibition of macroautophagy by smoke exposure coincided with alterations in the energy metabolism of alveolar macrophages: including increased numbers of depolarized, nonfunctional mitochondria, decreased ATP levels, and a greater dependence on anaerobic respiration for ATP production (86). An identical metabolic phenotype can be produced, in the absence of smoke exposure, by inhibiting macroautophagy through genetic or chemical means (87–89). Taken together, the results of Monick and colleagues (86) suggest that a loss of macroautophagic flux is at least partly responsible for the metabolic disturbances seen in the alveolar macrophages of smokers and patients with COPD. Moreover, because mitochondrial damage has been implicated in immune dysfunction in septic patients (90), their results may help to explain the increased susceptibility of patients with COPD to infection.
Macroautophagy inhibition also may help to explain why macrophages from patients with COPD secrete higher amounts of IL-1β compared with healthy individuals. Multiple groups have found that inhibiting macroautophagy leads to excessive secretion of multiple cytokines when cells are challenged with microbe-derived compounds such as endotoxin (91–93). We and others investigated the mechanism by which macroautophagy inhibition leads to increased cytokine production by focusing on caspase-1 activation, which is the rate-limiting step in the secretion of IL-1β and IL-18 in macrophages (88, 89). We found that, in macroautophagy-deficient macrophages, the mitochondrial population was dysfunctional because of a failure in organelle quality control. Mitochondria from macroautophagy-deficient cells produce excessive amounts of reactive oxygen species and are more prone to undergo membrane permeabilization when stressed by endotoxin exposure (89). As a result internal components of mitochondria, such as mitochondrial DNA, leak into the cytoplasm and lead to caspase-1 activation, followed by excessive IL-1β and IL-18 secretion (89). Our overall conclusion was that macroautophagy exerts a moderating effect on at least some aspects of the innate inflammatory response by maintaining mitochondrial health. Given that smoking inhibits macroautophagy and impairs mitochondrial homeostasis, it is tempting to speculate that the inhibition of macroautophagy contributes to the excessive secretion of proinflammatory cytokines observed in some patients with COPD.
Taken together, there are now multiple studies to suggest that macroautophagy plays a significant and complex role in COPD pathogenesis. In lung epithelial cells, macroautophagy may contribute to apoptosis and thus promote the emphysematous phenotype, whereas in alveolar macrophages macroautophagy inhibition by cigarette smoke may contribute to airway inflammation and bacterial infection. From a clinical perspective, the contributions of macroautophagy to COPD development have potential therapeutic and diagnostic implications. From a therapeutic standpoint, the possibility that macroautophagy may play different physiological roles depending on the cell type may mean that simply providing a chemical stimulator of macroautophagy to patients with COPD could have unpredictable consequences, improving some symptoms of this disease while making others worse. From a diagnostic standpoint, the fact that macroautophagy marker proteins such as LC3b are increased before the onset of apoptosis suggests that they might prove useful as early biomarkers of COPD progression. Future research will focus how we can rationally apply what we have learned about macroautophagy in COPD pathogenesis to ameliorate the clinical consequences of this important disease.
Cystic Fibrosis
It now well enshrined in textbooks that the lung pathology caused by cystic fibrosis (CF) is primarily a consequence of impaired mucociliary clearance caused by mutations in the cftr chloride channel, leading to chronic bacterial colonization, bronchiectasis, and obstructive physiology (94). On the basis of this paradigm, the arena where the crucial steps in CF pathogenesis occur is in the extracellular space—within the lumen of small airways. As such, the idea that CF is also a disease of defective intracellular housekeeping within airway epithelial cells may come as a surprise. However, this is the conclusion of a series of cell biology articles that propose that impaired macroautophagy is a factor in epithelial cell dysfunction in CF.
The basis of these articles derives from research showing that inactivating cftr mutations lead to exaggerated proinflammatory cytokine secretion in airway epithelial cells when challenged with Pseudomonas aeruginosa (95). To investigate why cystic fibrosis transmembrane conductance regulator (CFTR)–deficient cells are intrinsically prone to airway inflammation, Maiuri and colleagues (96) focused on the most common cftr mutant, Δ508, which folds improperly and is therefore retained in the ER. Expression of Δ508 mutant protein led to ER stress and elevated reactive oxygen species production followed by activation of the enzyme tissue transglutaminase (TG2), an enzyme that can act as a “natural fixative” by forming covalent bridges between lysine and glutamine residues (96). TG2 activation in this context led to the production of protein aggregates that sequestered the antiinflammatory regulatory proteins peroxisome proliferator-activated receptor (PPAR)-γ and IκBα (96, 97). It was proposed that the loss of these proteins explained the proinflammatory phenotype seen in epithelial cells expressing mutant CFTR.
Intracellular protein aggregates are typically cleared by macroautophagy, and the failure to do so is critical for the pathogenesis of important CNS diseases such as Huntington's disease (98). Therefore Luciani and colleagues (99) examined why protein aggregates induced by the Δ508CFTR mutant were not disposed of by macroautophagy. They found that that expressing Δ508CFTR in epithelial cells inhibited macroautophagic flux (99). The level of inhibition was at the level of autophagosome formation and was caused by depletion of the protein Beclin-1. Beclin-1 is a multifunctional adaptor protein that is a critical component of the vps34 complex that is responsible for autophagosome membrane initiation (100). Luciani and colleagues found that Beclin-1 was a substrate for TG2 and, in the context of Δ508CFTR expression, was cross-linked by TG2 and sequestered in protein aggregates similar to PPAR-γ (99). This led to the sequestration of the entire vps34 complex and inhibition of macroautophagy (99). The loss of macroautophagy was significant because this pathway appears to be responsible for degrading CFTR protein that becomes aggregated within the ER (101). As such macroautophagy inhibition leads to a vicious cycle wherein Δ508CFTR protein accumulates unchecked and leads to continual TG2 activity, protein aggregation in the cytoplasm, and excessive inflammation. Importantly, restoring macroautophagy by overexpressing Beclin-1 led to protein aggregate clearance in the cytoplasm and marked reduction in airway inflammation in a mouse model of CF (99). Presumably, restoring macroautophagy somehow led to improved folding of the Δ508 mutant in the ER, because Beclin-1 overexpression allowed some of this mutant protein, which is otherwise functional, to reach the cell surface.
Taken together, there is rapidly emerging evidence at the basic research level that impaired macroautophagy may be important to airway disease in patients with CF. Translating this knowledge into novel CF therapies is complicated because of the nature of macroautophagy blockade in this disease: simply providing a chemical stimulant of the process, such as rapamycin, is unlikely to be effective. However, a gene therapy approach in which macroautophagy genes are overexpressed in airway epithelia might be feasible and perhaps might enhance the effectiveness of prior attempts at gene-targeted therapy for CF.
Mycobacterium tuberculosis
Mycobacterium tuberculosis (MTb) remains one of the most prevalent infectious agents worldwide and represents a highly significant disease for pulmonologists. Despite the high disease burden imposed by MTb it is important to note that the majority of immunocompetent patients successfully resolve their infection. Studies suggest that macroautophagy contributes to the eradication of this pathogen.
The initial stages of MTb infection occur within alveolar macrophages that encounter inhaled droplets containing the organism. These previously unstimulated macrophages succeed in internalizing MTb bacteria by phagocytosis. However, the phagocytic vesicles containing MTb fail to fuse with lysosomes (102), because of MTb virulence factors that alter the phospholipid structure of the phagosome membrane (103–105). As such the vesicles become protected niches that support unchecked MTb replication, which leads to the eventual lysis of the infected macrophages, often within mediastinal lymph nodes. In immunocompetent hosts this ultimately leads to an inflammatory response directed by helper T cell type 1–polarized CD4+ T cells, which contains or eliminates the MTb infection (106). Crucial to the success of the cellular immune response to MTb is the elaboration of IFN-γ, which activates macrophages and enables them to kill MTb bacteria or else sequester them in granulomas (107). Macrophages stimulated by IFN-γ acquire several antimicrobial activities not observed in naive cells, but most notably they are able to overcome the block in fusion between MTb-containing phagocytic vesicles and lysosomes (108). As a result IFN-γ–activated macrophages are able to kill the MTb bacteria they ingest, which contributes to clinical resolution.
Interestingly, the ability of IFN-γ to stimulate the clearance of MTb organisms within macrophages requires an intact macroautophagy pathway. IFN-γ was shown to stimulate macroautophagy and inhibition of this process by chemical or genetic means leads to elevated bacterial MTb titers within macrophages (108, 109). Conversely, activation of macroautophagy by means other than IFN-γ exposure (such as nutrient starvation or rapamycin), similarly leads to bacterial clearance (108, 110). Interestingly, the bulk macroautophagy pathway alone is insufficient to promote MTb clearance. Rather, autophagosomes export specific cytosolic proteins to the MTb-containing vesicle, which is mediated by the adaptor protein p62 (109). Among the proteins that are delivered are ubiquitin and a ribosomal protein (rps30) that, when cleaved by lysosomal hydrolases, are degraded to cationic peptides that are potent MTb antibiotics (109, 111). This novel mechanism of reprocessing cytosolic proteins to generate endogenous antimicrobial peptides may apply to other infections besides MTb.
Furthermore there are genetic data to suggest that macroautophagy contributes to MTb immunity in humans. IFN-γ appears to activate macroautophagy by up-regulating a 47-kD GTPase called IRGM-1 (108, 112–114). IRGM-1 is required for macroautophagy stimulated by both IFN-γ and starvation and appears to work by manipulating the life cycle and redox state of mitochondria (114). Interestingly, polymorphisms in the IRGM-1 gene were linked to increased susceptibility to MTb infection, at least in some cohorts (115, 116). Other polymorphisms in the same gene were also linked to Crohn's disease, another disorder in which macroautophagy is thought to influence pathogenesis (117).
How can this knowledge be exploited to improve MTb therapeutics? One possibility is to employ drugs that stimulate macroautophagy in persistently infected individuals, with the goal of reducing latent infection or the development of drug resistance. Although some of these drugs have undesirable immunosuppressive effects (such as rapamycin), there may be other candidates with better side effect profiles (such as metformin [118] or carbamazepine [119]). Alternatively, because the role of macroautophagy is to bring antimicrobial peptides into contact with MTb, administering these protein fragments exogenously may have therapeutic value as it would bypass the block in phagosome maturation. It will be interesting to see if our growing knowledge of macroautophagy influences new approaches to MTb therapies.
Summary
In summary, autophagy plays an important role in many diseases relevant to pulmonologists. Although progress has been made in elucidating the role of macroautophagy in some pulmonary diseases, research into other autophagic pathways (CMA and microautophagy) is still very much in its infancy. As with any other core cellular processes, turning basic science knowledge about autophagy into therapies is difficult because of the interdependent nature of biochemical pathways. For the practicing pulmonologist, autophagy-based approaches to therapy are not yet on the horizon, but there is now sufficient evidence to speculate that exploiting this pathway will lead to novel therapies for important pulmonary disorders. For the pulmonary scientific community, research into the role of autophagy in pulmonary diseases is just beginning and will yield new insights and perhaps even challenge paradigms that shape our approach to pulmonary disease.
Supplementary Material
Acknowledgments
The authors thank Joshua Englert, Hilaire Lam, Stephan Ryter, and Robyn Haspel for critical review of this manuscript.
Footnotes
Supported by NIH T32 HL007633 and R03 HL097005.
Author Contributions: J.A.H. and A.M.K.C. cowrote this review article.
This article has an online supplement, which is available from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201106-0966CI on August 11, 2011
Author Disclosure: No author has a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Forms available with the text of this article at www.atsjournals.org
References
- 1.Kelekar A. Autophagy. Ann N Y Acad Sci 2005;1066:259–271 [DOI] [PubMed] [Google Scholar]
- 2.Kondomerkos DJ, Kalamidas SA, Kotoulas OB. An electron microscopic and biochemical study of the effects of glucagon on glycogen autophagy in the liver and heart of newborn rats. Microsc Res Tech 2004;63:87–93 [DOI] [PubMed] [Google Scholar]
- 3.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract 2006;202:631–638 [DOI] [PubMed] [Google Scholar]
- 4.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009;458:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem 2006;281:4035–4041 [DOI] [PubMed] [Google Scholar]
- 6.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA 2008;105:20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA 2010;107:832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Klionsky DJ. Permeases recycle amino acids resulting from autophagy. Autophagy 2007;3:149–150 [DOI] [PubMed] [Google Scholar]
- 9.Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 2011;7:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamark T, Kirkin V, Dikic I, Johansen T. Nbr1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009;8:1986–1990 [DOI] [PubMed] [Google Scholar]
- 11.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 2005;12:1613–1621 [DOI] [PubMed] [Google Scholar]
- 12.Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 2008;4:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006;8:1124–1132 [DOI] [PubMed] [Google Scholar]
- 14.Dice JF. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem 1982;257:14624–14627 [PubMed] [Google Scholar]
- 15.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 2000;113:4441–4450 [DOI] [PubMed] [Google Scholar]
- 16.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci 2001;114:2491–2499 [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 2008;14:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis 2011;43:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA 1983;80:2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011;20:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008;4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ericsson JL. Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp Cell Res 1969;55:95–106 [DOI] [PubMed] [Google Scholar]
- 24.Fujita N, Yoshimori T. Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 2011;23:492–497 [DOI] [PubMed] [Google Scholar]
- 25.Ogawa M, Nakagawa I, Yoshikawa Y, Hain T, Chakraborty T, Sasakawa C. Streptococcus-, Shigella-, and Listeria-induced autophagy. Methods Enzymol 2009;452:363–381 [DOI] [PubMed] [Google Scholar]
- 26.Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 1977;270:174–176 [DOI] [PubMed] [Google Scholar]
- 27.Metcalf DJ, Garcia-Arencibia M, Hochfeld WE, Rubinsztein DC. Autophagy and misfolded proteins in neurodegeneration. Exp Neurol (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masiero E, Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 2010;6:307–309 [DOI] [PubMed] [Google Scholar]
- 29.Raben N, Schreiner C, Baum R, Takikita S, Xu S, Xie T, Myerowitz R, Komatsu M, Van der Meulen JH, Nagaraju K, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder—murine Pompe disease. Autophagy 2010;6:1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol 2009;3:366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008;132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 2009;20:1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009;20:1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007;12:209–218 [DOI] [PubMed] [Google Scholar]
- 36.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997;243:240–246 [DOI] [PubMed] [Google Scholar]
- 37.Funderburk SF, Wang QJ, Yue Z. The Beclin 1–VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 2010;20:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin–VPS34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci 2006;119:259–270 [DOI] [PubMed] [Google Scholar]
- 39.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 2008;19:2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008;182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010;141:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy (In press) [DOI] [PubMed] [Google Scholar]
- 43.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA 2011;108:7769–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14l. J Cell Biol 2010;190:511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009;11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 46.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy 2010;6:301–303 [DOI] [PubMed] [Google Scholar]
- 47.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008;19:2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem 2004;279:40584–40592 [DOI] [PubMed] [Google Scholar]
- 49.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004;36:2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4b mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 2008;19:4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010;29:1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004;117:2805–2812 [DOI] [PubMed] [Google Scholar]
- 54.Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, Perez V, Jouvenot M, Boyer-Guittaut M. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy (In press) [DOI] [PubMed] [Google Scholar]
- 55.Ueno T, Takahashi K. A cathepsin l-specific inhibitor preferentially inhibits degradation of autophagosomal LC3 and GABARAP in HeLa and Huh-7 cells. Autophagy 2009;5:878–879 [DOI] [PubMed] [Google Scholar]
- 56.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor–associated protein–phospholipid conjugates. J Biol Chem 2004;279:36268–36276 [DOI] [PubMed] [Google Scholar]
- 57.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009;461:654–658 [DOI] [PubMed] [Google Scholar]
- 58.Eskelinen EL, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy 2011;7:935–956 [DOI] [PubMed] [Google Scholar]
- 59.Swanlund JM, Kregel KC, Oberley TD. Investigating autophagy: quantitative morphometric analysis using electron microscopy. Autophagy 2010;6:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol 2009;452:13–23 [DOI] [PubMed] [Google Scholar]
- 61.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 2005;1:23–36 [DOI] [PubMed] [Google Scholar]
- 62.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007;3:542–545 [DOI] [PubMed] [Google Scholar]
- 63.Jiang ZF, Shao LJ, Wang WM, Yan XB, Liu RY. Decreased expression of Beclin-1 and LC3 in human lung cancer. Mol Biol Rep (In press) [DOI] [PubMed] [Google Scholar]
- 64.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009;452:181–197 [DOI] [PubMed] [Google Scholar]
- 65.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131–24145 [DOI] [PubMed] [Google Scholar]
- 66.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer.” Autophagy 2009;5:585–589 [DOI] [PubMed] [Google Scholar]
- 67.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet 2008;17:3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide–sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell 2010;21:1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ju JS, Miller SE, Jackson E, Cadwell K, Piwnica-Worms D, Weihl CC. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy 2009;5:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dennis PB, Mercer CA. The GST-BHMT assay and related assays for autophagy. Methods Enzymol 2009;452:97–118 [DOI] [PubMed] [Google Scholar]
- 72.Seglen PO, Overbye A, Saetre F. Sequestration assays for mammalian autophagy. Methods Enzymol 2009;452:63–83 [DOI] [PubMed] [Google Scholar]
- 73.Sorokin SP. The cells of the lungs. : Nettesheim P, Hanna MG, Jr, Deatherage JW, Jr, Morphology of experimental respiratory carcinogenesis, proceedings of a Biology Division, Oak Ridge National Laboratory conference. Oak Ridge, TN: U.S. Atomic Energy Commission, Division of Technical Information; 1970. pp 3–43 Available from National Technical Information Service (Library of Congress Catalog Card Number 73609398) [Google Scholar]
- 74.Inoue D, Kubo H, Taguchi K, Suzuki T, Komatsu M, Motohashi H, Yamamoto M. Inducible disruption of autophagy in the lung causes airway hyper-responsiveness. Biochem Biophys Res Commun 2011;405:13–18 [DOI] [PubMed] [Google Scholar]
- 75.Halbert RJ, Natoli JL, Gano A, Badamgarav E, Buist AS, Mannino DM. Global burden of COPD: systematic review and meta-analysis. Eur Respir J 2006;28:523–532 [DOI] [PubMed] [Google Scholar]
- 76.Szilasi M, Dolinay T, Nemes Z, Strausz J. Pathology of chronic obstructive pulmonary disease. Pathol Oncol Res 2006;12:52–60 [DOI] [PubMed] [Google Scholar]
- 77.Wright JL, Cosio M, Churg A. Animal models of chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol 2008;295:L1–L15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, et al. EGR-1 regulates autophagy in cigarette smoke–induced chronic obstructive pulmonary disease. PLoS ONE 2008;3:e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim HP, Wang X, Chen ZH, Lee SJ, Huang MH, Wang Y, Ryter SW, Choi AM. Autophagic proteins regulate cigarette smoke–induced apoptosis: protective role of heme oxygenase-1. Autophagy 2008;4:887–895 [DOI] [PubMed] [Google Scholar]
- 80.Hwang JW, Chung S, Sundar IK, Yao H, Arunachalam G, McBurney MW, Rahman I. Cigarette smoke–induced autophagy is regulated by SIRT1-PARP-1–dependent mechanism: implication in pathogenesis of COPD. Arch Biochem Biophys 2010;500:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, Ifedigbo E, Parameswaran H, Ryter SW, Choi AM. Autophagy protein microtubule-associated protein 1 light chain-3b (LC3B) activates extrinsic apoptosis during cigarette smoke–induced emphysema. Proc Natl Acad Sci USA 2010;107:18880–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hodge S, Hodge G, Ahern J, Jersmann H, Holmes M, Reynolds PN. Smoking alters alveolar macrophage recognition and phagocytic ability: implications in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2007;37:748–755 [DOI] [PubMed] [Google Scholar]
- 83.Doyle I, Ratcliffe M, Walding A, Vanden Bon E, Dymond M, Tomlinson W, Tilley D, Shelton P, Dougall I. Differential gene expression analysis in human monocyte–derived macrophages: impact of cigarette smoke on host defence. Mol Immunol 2010;47:1058–1065 [DOI] [PubMed] [Google Scholar]
- 84.Xu J, Xu F, Lin Y. Cigarette smoke synergizes lipopolysaccharide-induced interleukin-1β and tumor necrosis factor-α secretion from macrophages via substance P–mediated nuclear factor-κB activation. Am J Respir Cell Mol Biol 2011;44:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baginski TK, Dabbagh K, Satjawatcharaphong C, Swinney DC. Cigarette smoke synergistically enhances respiratory mucin induction by proinflammatory stimuli. Am J Respir Cell Mol Biol 2006;35:165–174 [DOI] [PubMed] [Google Scholar]
- 86.Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A, Hansdottir S, Hunninghake GW. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol 2010;185:5425–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, et al. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011;469:221–225 [DOI] [PubMed] [Google Scholar]
- 89.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011;12:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belikova I, Lukaszewicz AC, Faivre V, Damoisel C, Singer M, Payen D. Oxygen consumption of human peripheral blood mononuclear cells in severe human sepsis. Crit Care Med 2007;35:2702–2708 [DOI] [PubMed] [Google Scholar]
- 91.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16l1 enhances endotoxin-induced IL-1β production. Nature 2008;456:264–268 [DOI] [PubMed] [Google Scholar]
- 92.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species–dependent amplification of RLR signaling. Proc Natl Acad Sci USA 2009;106:2770–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, et al. Autophagy controls IL-1β secretion by targeting pro–IL-1β for degradation. J Biol Chem 2011;286:9587–9597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pilewski JM, Frizzell RA. How do cystic fibrosis transmembrane conductance regulator mutations produce lung disease? Curr Opin Pulm Med 1995;1:435–443 [DOI] [PubMed] [Google Scholar]
- 95.Heeckeren A, Walenga R, Konstan MW, Bonfield T, Davis PB, Ferkol T. Excessive inflammatory response of cystic fibrosis mice to bronchopulmonary infection with Pseudomonas aeruginosa. J Clin Invest 1997;100:2810–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maiuri L, Luciani A, Giardino I, Raia V, Villella VR, D'Apolito M, Pettoello-Mantovani M, Guido S, Ciacci C, Cimmino M, et al. Tissue transglutaminase activation modulates inflammation in cystic fibrosis via PPARγ down-regulation. J Immunol 2008;180:7697–7705 [DOI] [PubMed] [Google Scholar]
- 97.Luciani A, Villella VR, Vasaturo A, Giardino I, Raia V, Pettoello-Mantovani M, D'Apolito M, Guido S, Leal T, Quaratino S, et al. Sumoylation of tissue transglutaminase as link between oxidative stress and inflammation. J Immunol 2009;183:2775–2784 [DOI] [PubMed] [Google Scholar]
- 98.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 2002;11:1107–1117 [DOI] [PubMed] [Google Scholar]
- 99.Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, Gavina M, Pulze L, Giardino I, Pettoello-Mantovani M, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol 2010;12:863–875 [DOI] [PubMed] [Google Scholar]
- 100.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for VPS34 binding, autophagy and tumor suppressor function. Autophagy 2005;1:46–52 [DOI] [PubMed] [Google Scholar]
- 101.Fu L, Sztul E. ER-associated complexes (ERACs) containing aggregated cystic fibrosis transmembrane conductance regulator (CFTR) are degraded by autophagy. Eur J Cell Biol 2009;88:215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 1971;134:713–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shui W, Petzold CJ, Redding A, Liu J, Pitcher A, Sheu L, Hsieh TY, Keasling JD, Bertozzi CR. Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J Proteome Res 2011;10:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang PB, Azad AK, Torrelles JB, Kaufman TM, Beharka A, Tibesar E, DesJardin LE, Schlesinger LS. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan–mediated phagosome biogenesis. J Exp Med 2005;202:987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hmama Z, Sendide K, Talal A, Garcia R, Dobos K, Reiner NE. Quantitative analysis of phagolysosome fusion in intact cells: inhibition by mycobacterial lipoarabinomannan and rescue by an 1α,25-dihydroxyvitamin D3-phosphoinositide 3-kinase pathway. J Cell Sci 2004;117:2131–2140 [DOI] [PubMed] [Google Scholar]
- 106.Harris J, Master SS, De Haro SA, Delgado M, Roberts EA, Hope JC, Keane J, Deretic V. Th1–Th2 polarisation and autophagy in the control of intracellular mycobacteria by macrophages. Vet Immunol Immunopathol 2009;128:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hansch HC, Smith DA, Mielke ME, Hahn H, Bancroft GJ, Ehlers S. Mechanisms of granuloma formation in murine Mycobacterium avium infection: the contribution of CD4+ T cells. Int Immunol 1996;8:1299–1310 [DOI] [PubMed] [Google Scholar]
- 108.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004;119:753–766 [DOI] [PubMed] [Google Scholar]
- 109.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HW, Kyei GB, Johansen T, Vergne I, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010;32:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Floto RA, Sarkar S, Perlstein EO, Kampmann B, Schreiber SL, Rubinsztein DC. Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy, reduce toxicity in Huntington's disease models and enhance killing of mycobacteria by macrophages. Autophagy 2007;3:620–622 [DOI] [PubMed] [Google Scholar]
- 111.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA 2007;104:6031–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-γ–inducible LRG-47. Science 2003;302:654–659 [DOI] [PubMed] [Google Scholar]
- 113.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006;313:1438–1441 [DOI] [PubMed] [Google Scholar]
- 114.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 2010;12:1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Che N, Li S, Gao T, Zhang Z, Han Y, Zhang X, Sun Y, Liu Y, Sun Z, Zhang J, et al. Identification of a novel IRGM promoter single nucleotide polymorphism associated with tuberculosis. Clin Chim Acta 2010;411:1645–1649 [DOI] [PubMed] [Google Scholar]
- 116.King KY, Lew JD, Ha NP, Lin JS, Ma X, Graviss EA, Goodell MA. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One 2011;6:e16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. A synonymous variant in IRGM alters a binding site for mir-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 2011;43:242–245 [DOI] [PubMed] [Google Scholar]
- 118.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 2007;67:6745–6752 [DOI] [PubMed] [Google Scholar]
- 119.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin z and reduces hepatic fibrosis. Science 2010;329:229–232 [DOI] [PubMed] [Google Scholar]
- 120.Dice JF. Chaperone-mediated autophagy. Autophagy 2007;3:295–299 [DOI] [PubMed] [Google Scholar]
- 121.Mizushima N. Autophagy: process and function. Genes Dev 2007;21:2861–2873 [DOI] [PubMed] [Google Scholar]
- 122.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010;6:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.