Abstract
A significant need exists for improved biomarkers for differential diagnosis, prognosis and monitoring of therapeutic interventions for mucopolysaccharidoses (MPS), inherited metabolic disorders that involve lysosomal storage of glycosaminoglycans. Here, we report a simple reliable method based on the detection of abundant non-reducing ends of the glycosaminoglycans that accumulate in cells, blood, and urine of MPS patients. In this method, glycosaminoglycans were enzymatically depolymerized releasing unique mono-, di-, or trisaccharides from the non-reducing ends of the chains. The composition of the released mono- and oligosaccharides depends on the nature of the lysosomal enzyme deficiency, and therefore they serve as diagnostic biomarkers. Analysis by liquid chromatography/mass spectrometry allowed qualitative and quantitative assessment of the biomarkers in biological samples. We provide a simple conceptual scheme for diagnosing MPS in uncharacterized samples and a method to monitor efficacy of enzyme replacement therapy or other forms of treatment.
Keywords: Lysosomal storage disorders, mucopolysaccharidoses, glycosaminoglycans, mass spectrometry, Sensi-Pro assay
Introduction
Mucopolysaccharidoses (MPS) are recessive inherited disorders characterized by a failure to degrade glycosaminoglycans (GAGs), which leads to lysosomal storage. MPS are chronic and progressive diseases with a large number of clinical manifestations, including skeletal and joint abnormalities, development of course facial features, hepatomegaly and splenomegaly, cardiovascular and respiratory dysfunction, hearing and vision loss, and neurological dysfunction1. To date eleven different MPS sub-classes have been identified, each affecting a specific lysosomal hydrolase. Six MPS subclasses affect sulfatases (MPS II, MPS IIIA, MPS IIID, MPS IVA, and MPS VI), five subclasses affect glycosidases (MPS I, MPS IIIB, MPS IVB, MPS VII, and MPS IX) and one subclass affects an acetyltransferase (MPS IIIC). Variation occurs in the severity of symptoms dependent on the specific MPS subclass, the extent of enzyme inactivation, and genetic background of the individual.
A number of therapies, including enzyme replacement therapy (ERT) and hematopoietic stem cell transplantation, positively affect disease outcome2,3. However, the optimization of these therapies has proved challenging due to the lack of effective biomarkers. A promising approach is the measurement of GAG accumulation, but two underlying considerations have historically challenged the utility of GAG accumulation as a biomarker. First, GAGs are present in variable amounts in healthy subjects, creating a high background4. Second, the GAGs that accumulate in MPS patients are dramatically heterogeneous with respect to length, sulfation patterns, and other structural variations5–9. As a result, the identification and detection of specific analytes that are present in abundance in MPS subjects, but not in healthy normal subjects, remains problematic.
Lysosomal degradation of GAGs occurs in an ordered manner from the non-reducing end of the chains10. Thus, the absence of any one enzyme in the pathway results in the accumulation of characteristic non-reducing terminal carbohydrate structures. Although this fact has been appreciated for many years, no method has been reported that allows selective quantification of these terminal structures that is broadly applicable to all MPS disorders. In this report, we describe a novel approach for liberating sets of disease-specific biomarkers derived from the non-reducing ends of the GAGs that accumulate in MPS. These carbohydrate biomarkers were readily distinguished from internal segments of the chain by liquid chromatography/mass spectrometry and in several examples they were quantitated. In principle, the biomarkers provide a tool for diagnosis and detection of MPS amenable to various types of samples including tissues, blood, and urine. The method should also provide an effective technique for monitoring the efficacy of enzyme replacement and other therapies.
Material and Methods
Cell Lines and Culture Conditions
Human foreskin fibroblasts (HFF) were obtained from the American Type Culture Collection (CRL-1634, Manassas, VA). Human dermal fibroblasts derived from patient biopsies were purchased from Coriell Institute (Camden, New Jersey): normal clinically healthy individuals, GM00408, GM00409, GM00200 (clinically unaffected sibling of a patient with metachromatic leukodystrophy), GM05659, GM08398, and GM15871 (clinically unaffected sibling of an unclassified Ehlers-Danlos patient); heterozygous MPS I carriers, GM01392, GM01393 and GM0003; homozygous MPS I individuals, GM06214, GM11495, GM00034, GM01391 and GM01256; MPS II patients, GM01896, GM03181, GM00615 and GM00298; MPS IIIA patients, GM00879, GM00643, GM00934, GM06110, and GM00629; MPS IIIB, GM01426; MPS IIIC, GM05157; MPS IIID, GM05093; MPS VI, GM00519; and MPS VII, GM00121. All cells were grown in DMEM containing 50 Units/ml penicillin, 50 µg/ml streptomycin, 2 mM glutamine and 10% fetal bovine serum. Cells were seeded on 15 cm diameter tissue culture dishes, grown to confluence and maintained in culture for up to 8 weeks to ensure sufficient amounts of GAG for analysis8,11. Sulfamidase activity in cell extracts was measured with 4-methylumbelliferyl-α-d-N-sulfoglucosaminide according the vendors instructions substituting Tris-acetate buffer (pH 6.5) (Moscerdam, The Netherlands).
MPS IIIA mice (Sgsh−/−) were obtained from Jackson Laboratory (B6.Cg-Sgsh) and were housed in Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved vivaria in the School of Medicine, University of California San Diego, following standards and procedures approved by the local Institutional Animal Care and Use Committee for the ethical use of animals in experiments. Canine samples were provided by Dr. Patricia Dickson (University of California, Harbor). Samples from MPS IIIB mice were provided by Elizabeth Neufeld (University of California, Los Angeles). Human urine samples without personal identifying information were obtained by Zacharon Pharmaceuticals, Inc. from donors from an MPS patient advocacy group with informed consent.
Purification and Enzymatic Depolymerization of GAGs
Cell monolayers were washed with phosphate buffered saline and detached by treatment with GIBCO trypsin/EDTA (Invitrogen, Carlsbad, CA). After centrifugation, the supernatant was removed and the cells were lysed in 0.5 ml of 0.1 N sodium hydroxide. The amount of protein was determined by bicinchoninic acid (BCA assay, Thermo Scientific, San Jose, CA). GAGs were isolated by anion exchange chromatography and digested with heparin lyase (IBEX) or chondroitinase ABC (Seikagaku) as described12.
For determining susceptibility of NRE structures to iduronidase, 1 µg of heparan sulfate was treated with 300 µU α-L-iduronidase (Genzyme) for 2 h at 37 °C in reaction buffer (0.1 mg/ml BSA, 50 mM sodium formate pH 3.0, 10 mM NaCl). For determining susceptibility of NRE structures to iduronate-2-sulfatase, purified heparan sulfate was treated with 0.5 µU iduronate 2-sulfatase (R and D Systems) in sulfatase reaction buffer (0.1 M sodium acetate, pH 5.5, 0.1 mg/ml of pH-inactivated BSA) at 37 °C for 16 h. For determining susceptibility of NRE structures to sulfamidase, purified heparan sulfate was treated with 300 µU N-sulfoglucosamine sulfohydrolase (sulfamidase, R and D Systems) in sulfatase reaction buffer at 37 °C for 16 h.
Non-Reducing End Analysis
Enzymatically depolymerized GAG preparations were differentially mass labeled by reductive amination with [12C6]aniline as previously described13. Briefly, HS and CS disaccharides (1–10 pmoles) were dried down in a centrifugal evaporator and reacted with [12C6]aniline or [13C6]aniline (15 µl, 165 µmol) and 15 µl of 1 M NaCNBH3 (Sigma-Aldrich) freshly prepared in dimethylsulfoxide:acetic acid (7:3, v/v) was added to each sample. Reactions were carried out at 65 °C for 4 h or alternatively at 37 °C for 16 h and then dried in a centrifugal evaporator. Unsubstituted amines were reacted with propionic anhydride (Sigma-Aldrich). Dried samples were reconstituted in 20 µl of 50% methanol and 3 µl of propionic anhydride (Sigma-Aldrich, 23.3 µmol) was added. Reactions were carried out at room temperature for 2 h. Acylated disaccharides were subsequently aniline-tagged as described above. Each sample was mixed with commercially available standard unsaturated disaccharides (Seikagaku), standard N-sulfoglucosamine, glucosamine-6-sulfate, N-acetylgalactosamine-4-sulfate and N-acetylgalactosamine-6-sulfate (Sigma-Aldrich), and/or β-d-idopyranosyluronate)-(1→4)-(2-N-acetyl-2-deoxy-α/β-d-glucopyranoside (I0S0) that was synthesized (Compound 7, Supplementary Methods). All standards were tagged with [13C6]aniline (Sigma/Aldrich). Samples were then analyzed by liquid chromatography-mass spectrometry using an LTQ Orbitrap Discovery electrospray ionization mass spectrometer (Thermo Scientific) equipped with quaternary high-performance liquid chromatography pump (Finnigan Surveyor MS pump) and a reverse-phase capillary column as previously described13.
Results
Structural analysis of GAG non-reducing ends
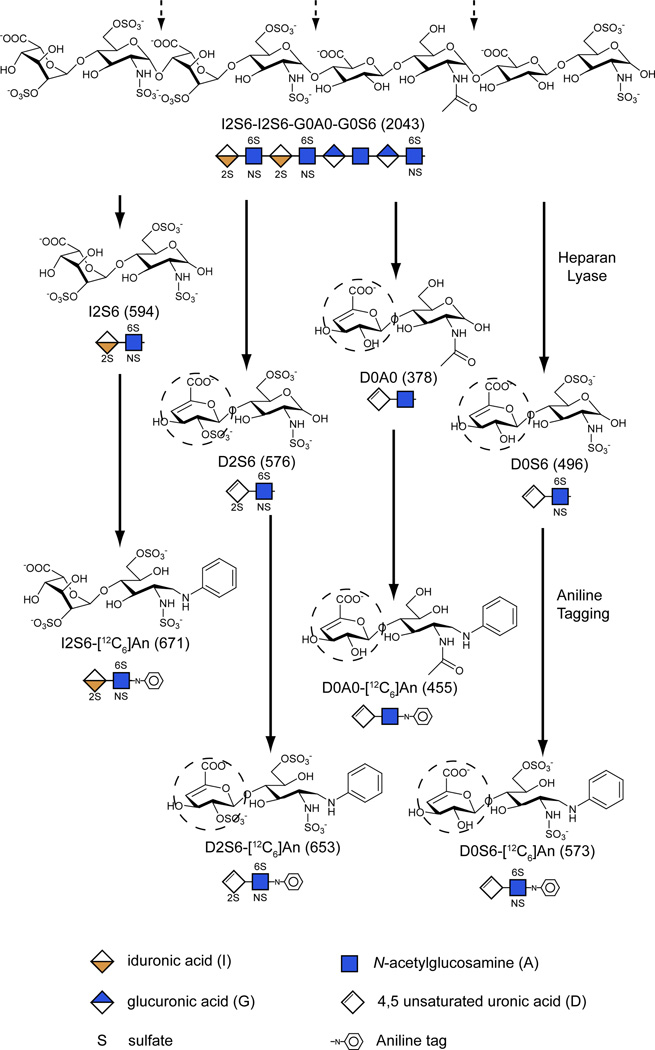
We previously described the application of glycan reductive isotope labeling/liquid chromatography tandem mass spectrometry (GRIL-LC/MS) for disaccharide profiling of GAGs13. The method involves depolymerization of GAG chains with endolytic enzymes and reductive amination with isotopically tagged aniline14. Because many of these enzymes act by elimination (bacterial heparin lyases and chondroitinases), they generate disaccharides containing a Δ4,5-unsaturated uronic acid and a hexosamine containing a variable number of sulfate groups12. Figure 1 provides a pictorial representation of the cleavage reaction and several disaccharide products derived from a fragment of heparan sulfate. For simplicity, the disaccharides are represented by symbols and a simple four digit Disaccharide Structural Code (DSC) as explained in the figure legend15. Enzyme digestion also results in the liberation of non-reducing end (NRE) carbohydrates (the sugars present on left end of the chains as drawn in the figure). In the idealized example shown in Fig. 1, the NRE consists of 2-sulfo-iduronic (I2) linked to N-sulfoglucosamine-6-sulfate (S6), which is designated I2S6. Thus, cleavage of the chain would yield one equivalent of I2S6 relative to three unsaturated disaccharides (D2S6, D0A0, D0S6). Note that I2S6 differs from the comparably sulfated internal disaccharide (D2S6) by 18 atomic mass units (amu) due to the saturated uronic acid in the NRE disaccharide versus the unsaturated uronic acid in the disaccharide derived from internal portions of the chain (dashed circles, Fig. 1). This difference in mass therefore distinguishes the terminal disaccharide at the NRE of the glycan from internal disaccharide subunits and acts as a mass tag uniquely identifying the NRE. In theory, NRE structures can be monosaccharides, disaccharides or trisaccharides depending on the composition of the terminal structure and the specificity of the heparin or chondroitin lyase used for depolymerization (12 and see below).
Figure 1. Scheme for determining non-reducing ends and internal disaccharides.
(A) Eliminative depolymerization of a heparan sulfate oligosaccharide with heparan lyase results in the release of internal disaccharide residues (dashed arrows) that contain an unsaturated uronic acid moiety (dotted circles). Because of its terminal location, the non-reducing end liberated from the left end of the chain as drawn lacks the Δ4,5-double bond and is 18 amu larger than a corresponding internal disaccharide. Reductive amination with aniline ([12C6]An) facilitates separation of the various disaccharides by LC/MS, yielding the m/z values for the molecular ions indicated within the parentheses. The glycan structures are graphically represented by geometric symbols, which are defined in the lower part of the figure43. To simplify the representation of constituent oligosaccharides from glycosaminoglycans, we use a Disaccharide Structure Code (DSC)15. In DSC, a uronic acid is designated as U, G, I, or D for an unspecified hexuronic acid, d-glucuronic acid, l-iduronic acid, or Δ4,5-unsaturated uronic acid, respectively. The hexosamines are designated in upper case for glucosamine and lower case for galactosamine, and the N-substituent is either H, A, S or R for hydrogen, acetate, sulfate, or some other substituent, respectively. The presence and location of ester linked sulfate groups are depicted by the number of the carbon atom on which the sulfate group is located or by 0 if absent. For example, I2S6 refers to a disaccharide composed of 2-sulfoiduronic acid-N-sulfoglucosamine-6-sulfate, whereas D2S6 refers to same disaccharide, but bearing a Δ4,5-double bond in the uronic acid.
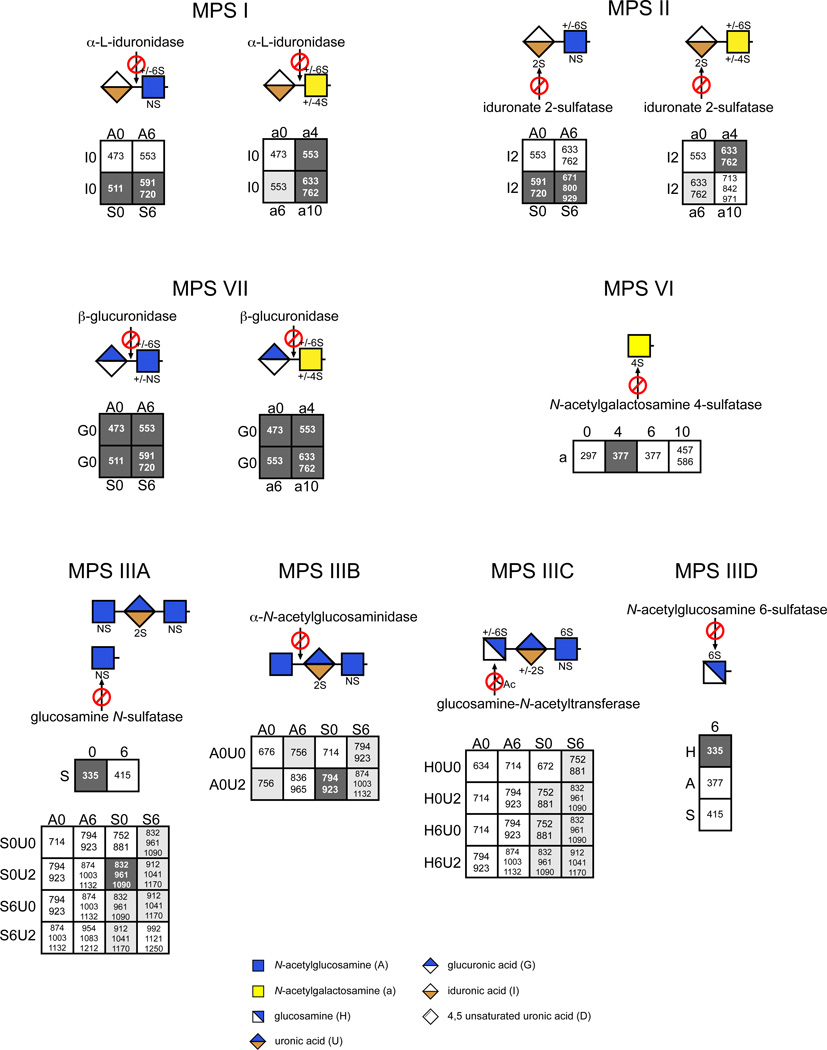
In this adaptation of GRIL-LC/MS, which is called the Sensi-Pro assay, the various liberated mono-, di-, and trisaccharides were derivatized with aniline, which provides the advantage of improved resolution by reverse phase chromatography and the ability to quantitate recovery of the various components with available standards13. Known amounts of standards tagged with [13C6]aniline were admixed with [12C6]aniline-tagged digestion products. Thus, monitoring the recovery of the isotopically tagged samples and standards by mass spectrometry provides a way to quantitate all of the digestion products. We rationalized all possible candidate structures assuming the enzymes liberate a terminal disaccharide if the chain ends in a uronic acid, or a monosaccharide residue (hexosamine) and/or trisaccharide (hexosamine-uronic acid-hexosamine) if the chains end in a hexosamine. The various possible structures and their calculated m/z values are provided in Fig. 2. Because MPS disorders result from a deficiency of glycosidases or sulfatases that act strictly at the NRE of the glycans, each lysosomal disorder should in theory give rise to a unique set of NRE structures, which then define candidate biomarkers for each subclass of MPS.
Figure 2. MPS Non-reducing end carbohydrates.
The defective enzyme for each MPS subclass is displayed along with the liberated NRE carbohydrates characteristic of MPS I, II, IIIA, IIIB, IIIC, IIID, VI and VII using geometric symbols. The matrices show all NRE carbohydrates that are theoretically possible for each MPS subclass using the DSC. The boxes with a black background and whiteface font depict structures that were detected and whose identities were confirmed by their co-chromatography and identical mass spectra as standards, as well as their sensitivity to exoglycosidases or propionic acid anhydride. Suspected structures shown in boxes with a gray background are implied from the liquid chromatography/mass spectra data, i.e. their size and content of sulfate and acetate groups are consistent with the proposed structures. The structures in boxes with a white background are theoretically possible, but have not been observed. The m/z values for both the free molecular ions and adduction ions formed with the ion pairing reagent dibutylamine (DBA) are listed. The single letter designations for the variously modified sugars are described in Fig. 1. The glycan structures are graphically represented by geometric symbols, which are defined in the lower part of the figure43.
NRE Analysis of MPS I, II, VI and VII cells
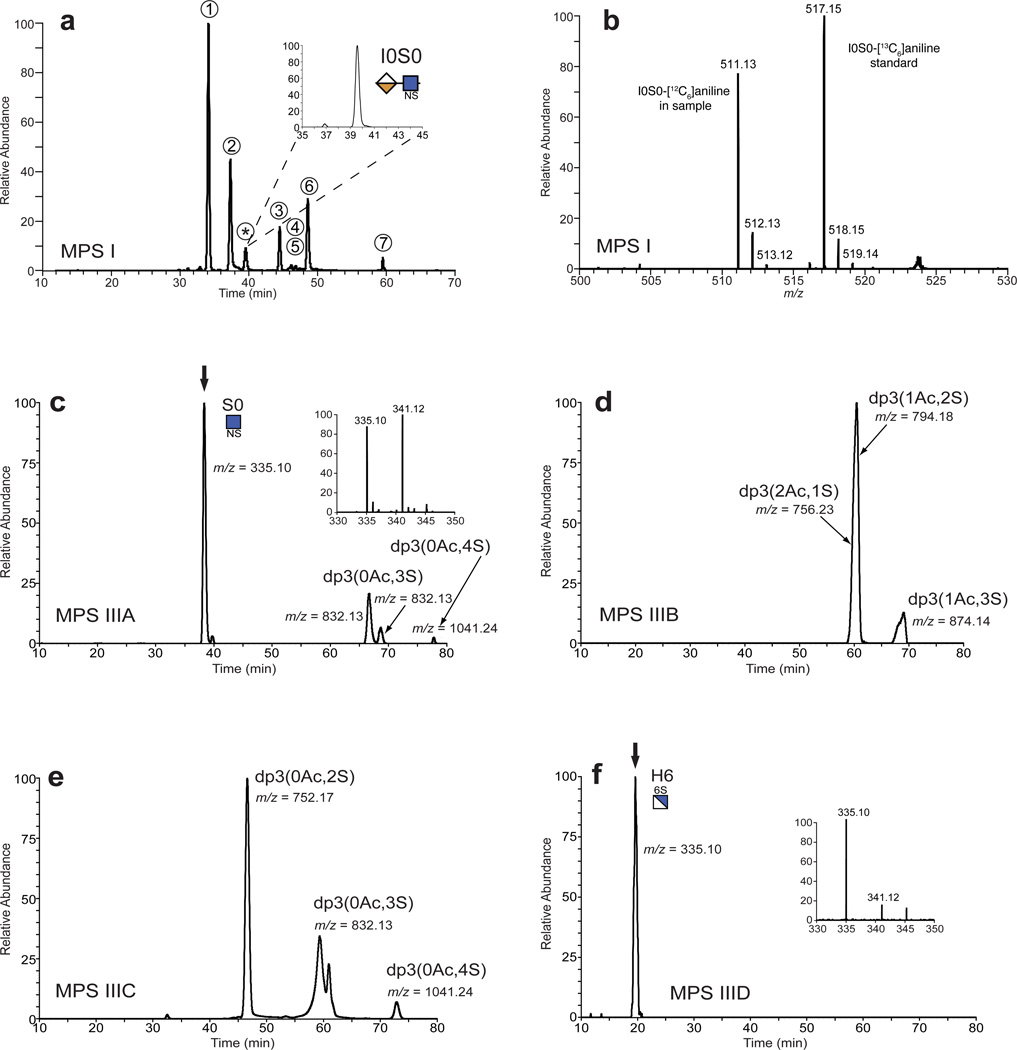
To demonstrate the potential utility of this approach, we grew up dermal fibroblasts from human MPS I patients (α-iduronidase [IDUA] deficiency) and from normal human donors. Cells were expanded and kept in culture up to 8 weeks to allow for significant lysosomal accumulation8,11. GAGs remaining in the cell layer were extracted and subjected to enzymatic depolymerization followed by reductive amination with [12C6]aniline. Samples were mixed with 10 pmoles of each [13C6]aniline unsaturated disaccharide standard and [13C6]aniline-tagged I0S0 that was synthesized (Supplementary Methods). In our initial studies we searched for all possible candidate structures (Fig. 2) and obtained the extracted ion chromatogram shown in Fig. 3a. Peaks 1–7 comigrated with known unsaturated disaccharides and had the expected m/z values. The MPS I sample had an additional peak marked by an asterisk that was not present in the normal fibroblast sample. This peak had the same elution position as the aniline-tagged I0S0 standard (expanded inset in Fig. 3a). The mass spectrum for this peak gave an m/z = 511.1 and isotopic cluster consistent with the proposed structure I0S0, and a corresponding m/z = 517.1 and isotopic cluster expected for the [13C6]aniline tagged standard (Fig. 3b). Further verification was carried out by collision-induced dissociation (CID), which demonstrated structural identity with the I0S0 standard (Supplementary Fig. 1). Digestion of heparan sulfate from cells derived from MPS I patients also yielded a disaccharide of m/z = 591.1 (Supplementary Fig. 2a), consistent with the structure I0S6. This material comigrated with the internal disaccharide D2S0 and thus was contained within peak 6 in the chromatogram shown in Fig. 3a. However, it was easily discriminated from D2S0 by the mass detector given the 18 amu difference. Pretreatment of an MPS I sample with α-l-iduronidase led to the loss of the native NRE structures confirming their identity (Supplementary Figure 3). Fibroblasts from three different MPS I patients exhibited NRE species identified as I0S0 and I0S6. These entities were not observed in samples prepared from normal human fibroblasts.
Figure 3. Analysis of non-reducing ends found in MPS I and Sanfilippo heparan sulfate.
(a) Depolymerized heparan sulfate from MPS I fibroblasts (GM01391) was tagged with [12C6]aniline and mixed with standard [13C6]aniline-labeled unsaturated disaccharides and I0S0. The sample was analyzed by LC/MS and the extracted ion current for all known NRE and internal disaccharides was recorded: peak 1, D0A0; peak 2, D0S0; peak 3, D0A6; peak 4, D0S6; peak 5, D2A0; peak 6, D2S0; and peak 7, D2S6. The asterisk marks the [12C6]aniline tagged NRE, which comigrated with [13C6]aniline-labeled I0S0 standard (inset). (b) Mass spectrum for the I0S0 peak shown in panel A. GAGs purified from (c) MPS IIIA (GM00643), (d) MPS IIIB (GM01426), (e) MPS IIIC (GM05157) and (f) MPS IIID (GM17495) fibroblasts were subjected to NRE analysis. For simplicity, only the extracted ion current for m/z values corresponding to monosaccharide and trisaccharide (dp3) NREs are shown for each sample. When the NRE structure was identified by comparison with commercially available standards, the name is indicated in DSC and glycan symbols. The dp3(0Ac,4S) NRE residues in the MPS IIIA and the MPS IIIC samples were detected as adduction ions with the ion pairing reagent ([M−2H + DBA]−1); hence their m/z values were increased by 129 amu (see Fig. 2). The insets in panels c and f show the mass spectra for the monosaccharide biomarkers S0 and H6, respectively, and the corresponding [13C6]aniline tagged standards (arrows in panels c and f).
MPS II (idurono-2-sulfatase [IDS] deficiency) and MPS VII (β-d-glucuronidase [GLCA] deficiency) also affect heparan sulfate degradation due to defects in processing the non-reducing terminal uronic acid (desulfation of iduronate-2-sulfate and removal of glucuronic acid, respectively). Saturated NRE disaccharides were detected after digestion of GAGs derived from fibroblasts of MPS II and MPS VII patients (Fig. 2 and Supplementary Figs. 2b and 2c). The mass spectra for the MPS II NREs and their elution positions were consistent with the expected disaccharide biomarkers I2S0 and I2S6. Analysis of MPS VII heparan sulfate was more complex, yielding four disaccharides tentatively identified as G0A0, G0A6, G0S0 and G0S6. Treatment of the MPS II samples with recombinant IDS converted the NRE to those found in MPS I (I0S0 and I0S6, respectively; Supplementary Fig. 4).
MPS I, MPS II, and MPS VII also affect the degradation of chondroitin sulfate and dermatan sulfate. Analysis of these GAGs using chondroitinase ABC yielded a set of NRE disaccharides diagnostic for each disorder (Fig. 2). MPS I yielded the monosulfated NRE disaccharides I0a4 and I0a6 in addition to the disulfated disaccharide I0a10 (Supplementary Fig. 2d). MPS II yielded I2a4 and I2a6 (Supplementary Fig. 2e) and MPS VII yielded G0a0, G0a4, G0a6 and G0a10 (Supplementary Fig. 2f). Mass standards are not yet available for these entities.
Analysis of chondroitin sulfate and dermatan sulfate from MPS VI (N-acetylgalactosamine 4-sulfatase [G4S] deficiency) demonstrated accumulation of N-acetylgalactosamine-4-sulfate (a4, Fig. 2), which co-migrated with an aniline tagged a4 standard (Supplementary Fig. 5). Note that a4 resolves partially by liquid chromatographically from 6-sulfo-N-acetylgalactosamine (a6) (lower panel). However, the biological sample yielded predominantly a4, consistent with the deficiency in the N-acetylgalactosamine 4-sulfatase in these cells. No trisaccharides species were detected.
NRE Analysis of MPS III cells
Using the same approach, we analyzed the Sanfilippo family of MPS disorders: MPS IIIA (sulfamidase [SGSH] deficiency), MPS IIIB (α-N-acetylglucosaminidase [NAGLU] deficiency), MPS IIIC (N-acetyltransferase [HGSNAT] deficiency) and MPS IIID (glucosamine-6-sulfatase [GNS] deficiency). These disorders only affect lysosomal degradation of heparan sulfate and have in common deficiencies in the enzymes that process the NRE glucosamine residue. Because heparin lyases cleave linkages between a glucosamine unit and a uronic acid, we predicted that Sensi-Pro analysis of Sanfilippo heparan sulfate should yield diagnostic monosaccharides (glucosamine derivatives) or trisaccharides (glucosamine - uronic acid - glucosamine derivatives) from the NRE as opposed to the disaccharides observed in MPS I, II and VII.
Analysis of MPS IIIA samples showed the typical unsaturated disaccharides generated from internal segments of the chains and a unique peak at 38.5 minutes not present in control or other MPS samples. This material had the characteristic mass spectrum expected for [12C6]aniline-tagged N-sulfoglucosamine (S0, m/z = 335.1) and comigrated with an authentic [13C6]aniline-tagged standard (m/z = 341.1; Fig. 3c, inset). Treatment of two different MPS IIIA samples with sulfamidase prior to heparinase depolymerization destroyed the S0 biomarker, consistent with its proposed identity (Supplementary Fig. 6). Digestion of MPS IIIA heparan sulfate also yielded trisaccharides that varied in the number of acetate and sulfate groups (Fig. 3c, dp3). The most prominent species dp3(0Ac,3S) was analyzed by CID and gave a fragmentation pattern consistent with S0U2S0 (Supplementary Fig. 7a). Although the uronic acid could be glucuronic acid, iduronic acid predominates in segments of the chain containing repeating N-sulfoglucosamine units 16.
Analysis of MPS IIIB samples yielded three NRE trisaccharides, with m/z values consistent with the presence of 1–2 acetate groups and 1–3 sulfates (Fig. 3d). Since MPS IIIB is characterized by the lack of α-N-acetylglucosaminidase, the terminal sugar should be N-acetylglucosamine, which was confirmed by CID analysis of the predominant trisaccharide (m/z = 794) identified as A0U2S0 (Supplementary Fig. 7b). Similarly, the NREs from MPS IIIC were predicted to contain a free unsubstituted amine due to the deficiency of glucosamine N-acetyltransferase. Four trisaccharides were detected, all lacking acetate groups (Fig. 3e). CID analysis of dp3(0Ac,3S) and derivatization with propionyl anhydride suggested structures consistent with H6U0S6 or H0U2S6 (Supplementary Fig. 7c). We also noted free N-acetylglucosamine and unsubstituted glucosamine in digested MPS IIIB and MPS IIIC samples, respectively, but the utility of these monosaccharide markers was diminished because of environmental contamination by N-acetylglucosamine and other contaminants in the low mass range.
MPS IIID cells lack the 6-sulfatase that can remove the 6-O-sulfate group from terminal N-acetylglucosamine units. NRE analysis of MPS IIID heparan sulfate detected a single monosaccharide species with m/z value of 335 corresponding to N-unsubstituted GlcNH26S (H6) (Fig. 3f). While H6 is isobaric with S0 found in MPS IIIA, its retention time was significantly less due to the presence of the unsubstituted amine and consequently these markers were easily discriminated. Furthermore, H6 in MPS IIID co-eluted with [13C6]aniline-labeled standard H6 verifying its identity (Fig. 3f, inset). Surprisingly, no N-acetylglucosamine-6-sulfate was detected nor were any trisaccharide NRE species bearing a non-reducing terminal 6-O-sulfated N-acetylglucosamine unit (Fig. 3f).
Use of NRE Biomarkers
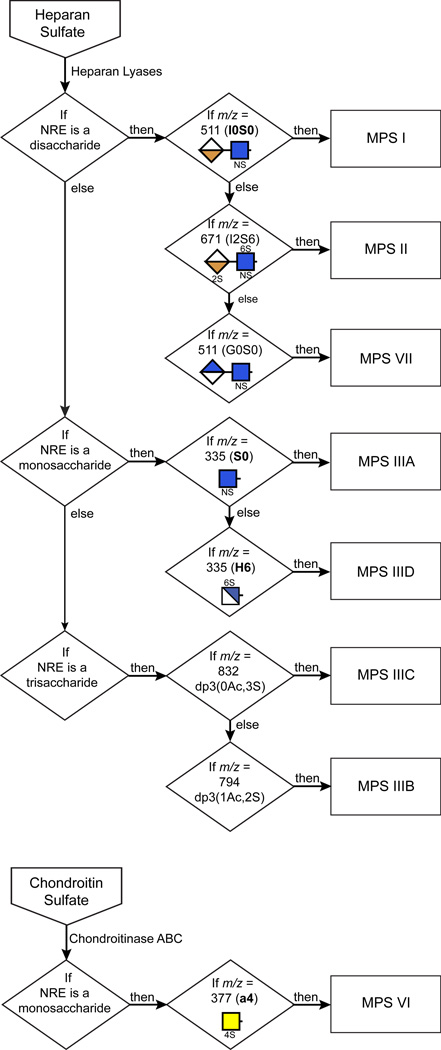
Although most MPS GAG samples yielded multiple NRE carbohydrates (Fig. 2), it was possible to select single unique NREs as biomarkers for each MPS disorder and then combine them into a decision tree based on size of NRE structures (mono-, di- and trisaccharides), degree of sulfation, and retention time during liquid chromatography (Fig. 4). The diagnostic decision tree becomes even more robust by inclusion of other carbohydrate biomarkers (Fig. 2), but the specific NREs indicated in Fig. 4 are sufficient to diagnose the eight MPS disorders. To determine the potential utility of these markers for diagnosis, we screened nine different human urine samples from normal control subjects and patients suffering from various Sanfilippo disorders as well as two canine urine samples (one normal and one with MPS I) and liver, brain and kidney GAGs from MPS IIIB mice. Using the scheme outlined in Fig. 4 we correctly diagnosed all of the samples (Table 1). Analysis of multiple MPS IIIA cell lines showed striking accumulation of the S0 biomarker, which corresponded well with the level of heparan sulfate storage (Supplementary Table 1). Normal fibroblasts yielded minute amounts of S0. In general, samples from normal cells, tissues and urine exhibited less than 1% of the amount of NRE biomarkers observed in samples from affected patients or animals.
Figure 4. Systematic diagnostic screening of GAG samples for various MPS disorders.
Shown is a flow chart for MPS discovery based on the detection of diagnostic non-reducing end glycans present in GAG samples extracted from patient or animal model sources. The detection criteria are based on NRE size (monosaccharide, disaccharide and trisaccharide), m/z value, and structural features (number of acetates (Ac) or sulfates (S)). For a complete unknown, portions of the sample are analyzed in parallel for heparan sulfate and chondroitin/dermatan sulfate NREs.
Table 1. Analysis of GAG samples purified from mouse tissues and human and canine urine.
GAG was extracted as described in the Methods and analyzed for MPS diagnostic biomarkers using the scheme shown in Figure 4.
| Sample | Sample Identity | NRE Biomarkers Detected |
Sensi-Pro Analysis |
|---|---|---|---|
| Liver, Mouse-1 | Unaffected, MPS IIIB Carrier (Het) | Trace | Normal |
| Liver, Mouse-2 | MPS IIIB | A0U2S0 | MPSIIIB |
| Brain, Mouse-1 | Unaffected, MPS IIIB Carrier (Het) | Trace | Normal |
| Brain, Mouse-2 | MPS IIIB | A0U2S0 | MPSIIIB |
| Kidney, Mouse-1 | MPS IIIB | A0U2S0 | MPSIIIB |
| Urine, Human-1 | Normal | Trace | Normal |
| Urine, Human-2 | Normal | Trace | Normal |
| Urine, Human-3 | Normal | Trace | Normal |
| Urine, Human-4 | Normal | Trace | Normal |
| Urine, Human-5 | MPS IIIC | dp3(0Ac,3S04) | MPSIIIC |
| Urine, Human-6 | MPS IIIC | dp3(0Ac,3S04) | MPSIIIC |
| Urine, Human-7 | MPS IIIC | dp3(0Ac,3S04) | MPSIIIC |
| Urine, Human-8 | MPS IIIA | S0, S0U2S0 | MPSIIIA |
| Urine, Human-9 | MPS IIIB | A0U2S0 | MPSIIIB |
| Urine, Canine-1 | Unaffected, MPS I Carrier (Het) | Trace | Normal |
| Urine, Canine-2 | MPS I | I0S0 | MPS I |
Diagnostic markers: MPS I, I0S0; MPS II, I2S6; MPS IIIA, S0 and S0U2S0; MPS IIIB, A0U2S0; MPS IIIC dp3(0Ac,3S04), a trisaccharide containing no acetate groups and three sulfate groups; MPS IIID, H6; MPS VI, a4; MPS VII, G0S0.
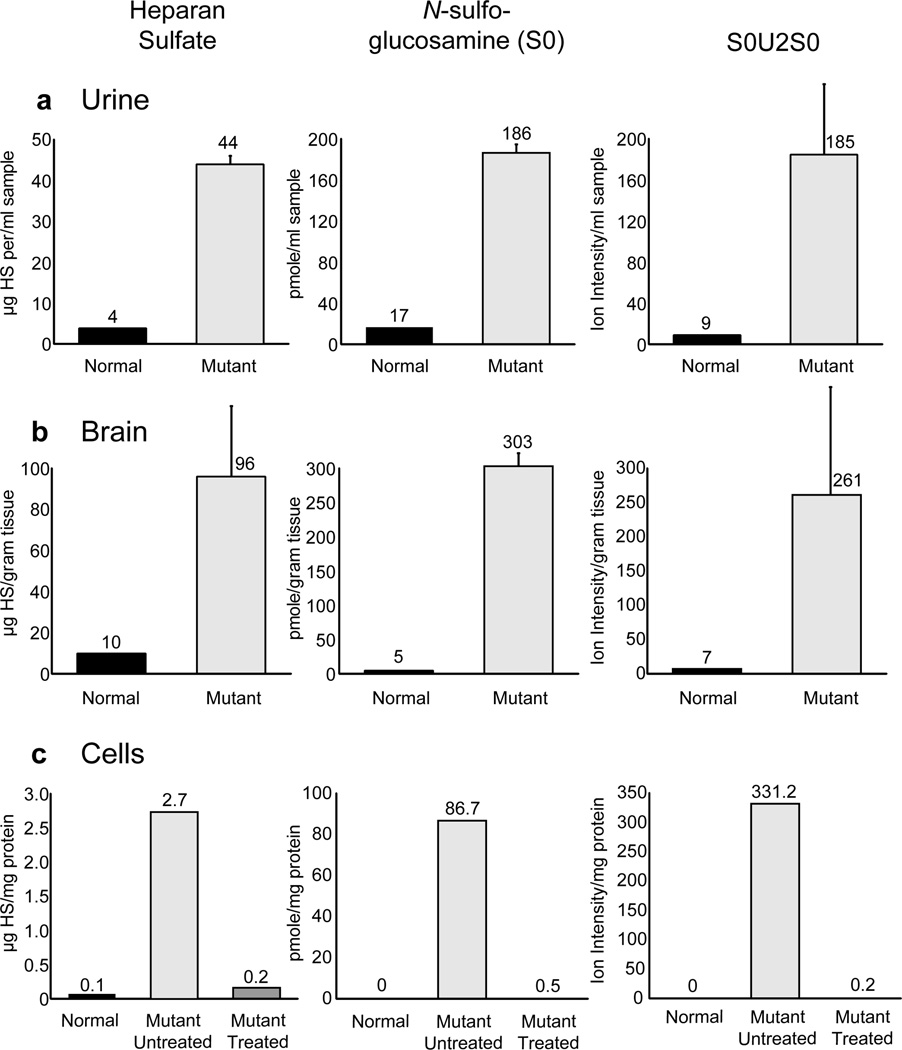
The detection of lysosomal storage based on GAG content in the brain and urine has been challenging due to various methods used for detection and quantitation, in particular indirect techniques based on dye binding or displacement. To determine the utility of Sensi-Pro analysis, we analyzed urine and brain samples from MPS IIIA (Sgsh−/−) and wildtype mice and MPS IIIA human fibroblasts. Using this method, we noted that total heparan sulfate and the biomarker S0 showed a 12-fold accumulation in MPS IIIA urine samples compared to the wildtype (Fig. 5a). The trisaccharide biomarker S0U2S0 was readily detectable in the Sgsh−/− urine, but virtually undetectable in wildtype urine. In brain samples the heparan sulfate level was elevated 12-fold, whereas the biomarker S0 increased 60-fold compared to the wildtype (Fig 5b). The trisaccharide marker was essentially present only in the Sgsh−/− sample.
Figure 5. Comparison of total heparan sulfate to N-sulfoglucosamine (S0) in MPS IIIA samples.
(a) Heparan sulfate from normal (black bars) and MPS IIIA (light grey bars) urine was analyzed. The individual disaccharides and NRE N-sulfoglucosamine (S0) was quantitated relative to standards. Since trisaccharide standards are not available, the values of the extracted ion current for S0U2S0 are shown. (b) Analysis of normal (black bars) and MPS IIIA (light grey bars) brain heparan sulfate as in panel A. (c) MPS IIIA cells underwent enzyme replacement by incubation with 0.06 mU/ml of sulfamidase for 48 hours prior to GAG extraction and subsequent analysis. The amount of heparan sulfate (black bars), the monosaccharide biomarker S0 (light grey bars), and the trisaccharide biomarker S0U2S0 (dark grey bars) were measured and compared to samples from cells without enzyme supplementation. The bars represent the average ± standard deviation, n=3.
Based on these encouraging findings, we tested whether the NRE structures afford a more precise and sensitive assay to monitor therapeutic enzyme replacement. Cultures of MPS IIIA human fibroblasts were supplemented with recombinant sulfamidase for 48 hours prior to GAG extraction and analysis. Enzyme replacement led to a significant drop in heparan sulfate and both the biomarkers, S0 and S0U2S0 (Fig. 5c). Thus, the NRE biomarkers, in particular the trisaccharides, have the potential for being used to monitor therapeutic efficacy of intervention strategies.
Discussion
In this paper we define a set of biomarkers specific for eight MPS disorders based on the structures of the NRE of the chains that accumulate. Monitoring of the various biomarkers potentially provides multiple improvements over existing methods for diagnosis and monitoring of therapeutic interventions: (i) The biomarkers are mutually exclusive and thus specific for each MPS subtype, allowing a deductive process for diagnosis (Fig. 4); (ii) All of the NRE biomarkers can be assessed simultaneously in a single sample, while simultaneously providing quantitative evaluation of total GAG storage and biomarker prevalence; (iii) The biomarkers are present in high abundance in cells, tissues and fluids from affected individuals, dogs and mice compared to control subjects and wildtype animals; (iv) The method is extremely sensitive, limited only by parameters dictated by the configuration of the mass spectrometer. Thus, biomarker analysis of other tissues and fluids, such as blood, cerebrospinal fluid, saliva, and tears should be consistently possible. As a result, the method could prove useful for differential diagnosis, disease prognosis, measurement of response to treatment, and optimization of therapies.
Other diagnostic approaches based on glycan detection and analysis have been described previously6,7,17–22. Some of these methods depend on detection of naturally occurring monosaccharides and oligosaccharides that accumulate in blood and urine8,23–25 or they involve the use of exoglycosidases to alter the structure of the non-reducing end of GAGs to confirm their origin from aberrant degradation26,27. Some methods focus on quantitation of internal unsaturated disaccharides, which may be best suited for measuring total GAG content28,29. Analyses that concentrate solely on gross composition obscure the sensitivity and specificity afforded by analysis of the NRE. The method described here has the advantage of simultaneous detection of multiple NRE biomarkers for each MPS disorder in a single analytical run and provides the benefits of orthogonal approaches for determining structure (liquid chromatography and tandem mass spectrometry with mass standards).
An interesting feature of NRE analysis is its high signal in MPS GAG samples compared to samples from unaffected animals or patients. Lysosomal degradation of GAGs normally occurs efficiently, converting the chains into free uronic acids, N-acetylhexosamines and inorganic sulfate. Because catabolism proceeds sequentially, a deficiency in any one of the enzymes stalls the process, resulting in accumulation of undegraded fragments with defined ends. Thus, diagnostic NREs accumulate relative to the internal disaccharides. This cumulative effect is enhanced further by the action of endogenous endoglycosidases, such as heparanase or endo-β-hexosaminidases that cleave heparan sulfate and chondroitin/dermatan sulfate chains, respectively, at selective sites 30,31. These endolytic cleavage reactions amplify the number of NREs at least several-fold depending on the frequency of the cleavage sites and the size of the chains. However, they do not affect the total amount of GAG present in the sample, thus making the NRE a more sensitive and specific biomarker than merely measuring the extent of GAG accumulation.
Another interesting outcome of this work is the finding that the GAGs accumulating in MPS IIID cells contain NRE glucosamine-6-sulfate (H6), as opposed to a mixture of H6 and N-acetylglucosamine-6-sulfate (A6). Since free glucosamine units are relatively rare in heparan sulfate13,32, this observation suggests that the 6-sulfatase (GNS) acts preferentially after the sulfamidase, but prior to N-acetylation of the glucosamine unit. N-acetylglucosamine-6-sulfate or trisaccharides containing terminal N-acetylglucosamine-6-sulfate were not detected in the two MPS IIID samples analyzed, which may reflect the low abundance of N-acetylglucosamine-6-sulfate compared N-sulfoglucosamine-6-sulfate in typical heparan sulfate preparations. The preference of heparanase to cleave the chains within heavily sulfated domains may further accentuate the accumulation of H6 30. The original studies of MPS IIID demonstrated loss of 6-sulfatase activity using oligosaccharides terminating in N-acetylglucosamine-6-sulfate33, which later became the basis for a fluorometric assay using 4-methylumbelliferyl-α-N-acetylglucosamine-6-sulfate34. To our knowledge detailed studies of the enzyme using glucosamine-6-sulfate have not been reported. The detection of H6 as the primary NRE in MPS IIID suggests that lysosomal degradation most likely proceeds in the order: sulfamidase (IIIA) → 6-sulfatase (IIID) → N-acetyltransferase (IIIC) → N-acetylglucosaminidase (IIIB).
Diagnosis of MPS without prior family history of disease typically relies on clinical observation by astute physicians, and subsequent measurement of urinary or plasma GAGs1. Unfortunately, the onset of clinical symptoms often occurs after severe, sometimes irreversible developmental damage has taken place, making early detection critical for successful therapeutic intervention. Thus, great interest exists in developing early diagnostic methods applicable to blood or urine. To date, most of these diagnostic approaches have focused on the detection of residual enzyme activity or quantity in dried blood spots using mass spectrometry and fluorometric assays35–41. However, diagnosis is more difficult in individuals with milder forms of the disease. Variation in the content and composition of GAGs in blood and urine in the general population according to ethnicity, activity and health condition also makes analysis of GAGs in samples unreliable4,42. More field experience will be needed to determine if NRE biomarkers or combinations of GAG analysis and enzyme assays will provide a more reliable way to diagnose individuals with intermediate-severity MPS disease. One clear-cut advantage of NRE biomarkers is their utility for monitoring the efficacy of therapeutic treatments based on enzyme replacement therapy (Fig. 5). Evaluating disease severity and therapy will depend on the availability of patient samples, additional quantitative disaccharide and trisaccharide standards (standards are available currently for MPS I, MPS IIIA, MPS IIID and MPS VI), and further development of the Sensi-Pro assay for clinical use.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health grants R01 GM077471 to J.D.E. and P41 RR005351 to G.J.B. for synthesis of disaccharide standards, a Kirschstein National Research Service Award (NRSA) DK085905 to W.C.L. and grants from the National MPS Society (to J.D.E. and B.E.C.).
Footnotes
Author Contributions. The project was conceived by B.E.C., J.R.Brown, and J.D.E. Execution and interpretation of non-reducing end analysis was carried out by R.L. and W.C.L. Carbohydrate standards were synthesized by K.A. and G.B. The manuscript was written by R.L., W.C.L., J.R.Beitel., B.E.C. and J.D.E.
Competing Financial Interest. Brett E. Crawford, Jillian R. Brown and James R. Beitel are employees of and hold equity positions in Zacharon Pharmaceuticals, Inc. Jeffrey D. Esko is a founder of and has an equity position in Zacharon Pharmaceuticals. Both he and Roger Lawrence consult with Zacharon Pharmaceuticals.
References Cited
- 1.Neufeld EF, Muenzer J. The Mucopolysaccharidoses. In: Scriver CR, et al., editors. Metabolic and Molecular Basis of Inherited Disease. Vol. 3. San Francisco: MacGraw-Hill; 2001. pp. 3421–3452. [Google Scholar]
- 2.Clarke LA. The mucopolysaccharidoses: a success of molecular medicine. Expert Rev Mol Med. 2008;10:e1. doi: 10.1017/S1462399408000550. [DOI] [PubMed] [Google Scholar]
- 3.Beck M. Therapy for lysosomal storage disorders. IUBMB Life. 2010;62:33–40. doi: 10.1002/iub.284. [DOI] [PubMed] [Google Scholar]
- 4.Wei W, Ninonuevo MR, Sharma A, Danan-Leon LM, Leary JA. A comprehensive compositional analysis of heparin/heparan sulfate-derived disaccharides from human serum. Anal Chem. 2011;83:3703–3708. doi: 10.1021/ac2001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomatsu S, et al. Dermatan sulfate and heparan sulfate as a biomarker for mucopolysaccharidosis I. J Inherit Metab Dis. 2010;33:141–150. doi: 10.1007/s10545-009-9036-3. [DOI] [PubMed] [Google Scholar]
- 6.Tomatsu S, et al. Heparan sulfate levels in mucopolysaccharidoses and mucolipidoses. J Inherit Metab Dis. 2005;28:743–757. doi: 10.1007/s10545-005-0069-y. [DOI] [PubMed] [Google Scholar]
- 7.Fuller M, Chau A, Nowak RC, Hopwood JJ, Meikle PJ. A defect in exodegradative pathways provides insight into endodegradation of heparan and dermatan sulfates. Glycobiology. 2006;16:318–325. doi: 10.1093/glycob/cwj072. [DOI] [PubMed] [Google Scholar]
- 8.Fuller M, Meikle PJ, Hopwood JJ. Glycosaminoglycan degradation fragments in mucopolysaccharidosis I. Glycobiology. 2004;14:443–450. doi: 10.1093/glycob/cwh049. [DOI] [PubMed] [Google Scholar]
- 9.Holley RJ, et al. Mucopolysaccharidosis type I: Unique structure of accumulated heparan sulfate and increased N-sulfotransferase activity in mice lacking alpha-L-iduronidase. J Biol Chem. 2011 doi: 10.1074/jbc.M111.287474. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeze HH. Genetic Disorders of Glycan Degradation. In: Varki A, et al., editors. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press; 2009. pp. 567–583. [PubMed] [Google Scholar]
- 11.Lamanna WC, Lawrence R, Sarrazin S, Esko JD. Secondary storage of dermatan sulfate in Sanfilippo disease. J Biol Chem. 2011;286:6955–6962. doi: 10.1074/jbc.M110.192062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linhardt RJ. Analysis of glycosaminoglycans with polysaccharide lyases. Curr Protoc Mol Biol. 2001;Chapter 17(Unit17):13B. doi: 10.1002/0471142727.mb1713bs48. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence R, et al. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]
- 16.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J.Clin.Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjoberg I, Fransson LA, Matalon R, Dorfman A. Hunter's syndrome: a deficiency of L-idurono-sulfate sulfatase. Biochem Biophys Res Commun. 1973;54:1125–1132. doi: 10.1016/0006-291x(73)90809-7. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien JF, Cantz M, Spranger J. Maroteaux-Lamy disease (mucopolysaccharidosis VI), subtype A: deficiency of a N-acetylgalactosamine-4-sulfatase. Biochem Biophys Res Commun. 1974;60:1170–1177. doi: 10.1016/0006-291x(74)90435-5. [DOI] [PubMed] [Google Scholar]
- 19.Kimura A, Hayashi S, Tsurumi K. Chemical structure of urinary dermatan sulfate excreted by a patient with the Hunter syndrome. Tohoku J Exp Med. 1980;131:241–247. doi: 10.1620/tjem.131.241. [DOI] [PubMed] [Google Scholar]
- 20.Kimura A, Hayashi S, Koseki M, Tsurumi K. Characteristics of urinary glycosaminoglycans excreted by a patient with the Hurler-Scheie compound syndrome. Tohoku J Exp Med. 1982;136:61–66. doi: 10.1620/tjem.136.61. [DOI] [PubMed] [Google Scholar]
- 21.Kimura A, Hayashi S, Koseki M, Kochi H, Tsurumi K. Fractionation and characterization of urinary heparan sulfate excreted by patients with Sanfilippo syndrome. Tohoku J Exp Med. 1984;144:227–236. doi: 10.1620/tjem.144.227. [DOI] [PubMed] [Google Scholar]
- 22.Toma L, Dietrich CP, Nader HB. Differences in the nonreducing ends of heparan sulfates excreted by patients with mucopolysaccharidoses revealed by bacterial heparitinases: A new tool for structural studies and differential diagnosis of Sanfilippo's and Hunter's syndromes. Lab.Invest. 1996;75:771–781. [PubMed] [Google Scholar]
- 23.Ramsay SL, Meikle PJ, Hopwood JJ. Determination of monosaccharides and disaccharides in mucopolysaccharidoses patients by electrospray ionisation mass spectrometry. Mol Genet Metab. 2003;78:193–204. doi: 10.1016/s1096-7192(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 24.King B, Savas P, Fuller M, Hopwood J, Hemsley K. Validation of a heparan sulfate-derived disaccharide as a marker of accumulation in murine mucopolysaccharidosis type IIIA. Mol Genet Metab. 2006;87:107–112. doi: 10.1016/j.ymgme.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 25.Mason KE, Meikle PJ, Hopwood JJ, Fuller M. Characterization of sulfated oligosaccharides in mucopolysaccharidosis type IIIA by electrospray ionization mass spectrometry. Anal Chem. 2006;78:4534–4542. doi: 10.1021/ac052083d. [DOI] [PubMed] [Google Scholar]
- 26.Byers S, Rozaklis T, Brumfield LK, Ranieri E, Hopwood JJ. Glycosaminoglycan accumulation and excretion in the mucopolysaccharidoses: characterization and basis of a diagnostic test for MPS. Mol Genet Metab. 1998;65:282–290. doi: 10.1006/mgme.1998.2761. [DOI] [PubMed] [Google Scholar]
- 27.Bhaumik M, et al. A mouse model for mucopolysaccharidosis type III A (Sanfilippo syndrome) Glycobiology. 1999;9:1389–1396. doi: 10.1093/glycob/9.12.1389. [DOI] [PubMed] [Google Scholar]
- 28.Oguma T, Tomatsu S, Montano AM, Okazaki O. Analytical method for the determination of disaccharides derived from keratan, heparan, and dermatan sulfates in human serum and plasma by high-performance liquid chromatography/turbo ionspray ionization tandem mass spectrometry. Anal Biochem. 2007;368:79–86. doi: 10.1016/j.ab.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen TC, Rozek T, Hopwood JJ, Fuller M. Determination of urinary oligosaccharides by high-performance liquid chromatography/electrospray ionization-tandem mass spectrometry: Application to Hunter syndrome. Anal Biochem. 2010;402:113–120. doi: 10.1016/j.ab.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285:14504–14513. doi: 10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneiwa T, Mizumoto S, Sugahara K, Yamada S. Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology. 2010;20:300–309. doi: 10.1093/glycob/cwp174. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. J Biol Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kresse H, Paschke E, von Figura K, Gilberg W, Fuchs W. Sanfilippo disease type D: deficiency of N-acetylglucosamine-6-sulfate sulfatase required for heparan sulfate degradation. Proc Natl Acad Sci U S A. 1980;77:6822–6826. doi: 10.1073/pnas.77.11.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He W, Voznyi Ya V, Boer AM, Kleijer WJ, van Diggelen OP. A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease type D (MPS IIID) J Inherit Metab Dis. 1993;16:935–941. doi: 10.1007/BF00711508. [DOI] [PubMed] [Google Scholar]
- 35.Dean CJ, Bockmann MR, Hopwood JJ, Brooks DA, Meikle PJ. Detection of mucopolysaccharidosis type II by measurement of iduronate-2-sulfatase in dried blood spots and plasma samples. Clin Chem. 2006;52:643–649. doi: 10.1373/clinchem.2005.061838. [DOI] [PubMed] [Google Scholar]
- 36.Blanchard S, Sadilek M, Scott CR, Turecek F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: application to screening newborns for mucopolysaccharidosis I. Clin Chem. 2008;54:2067–2070. doi: 10.1373/clinchem.2008.115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe BJ, et al. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: application to screening newborns for mucopolysaccharidosis II (Hunter Syndrome) Anal Chem. 2010;83:1152–1156. doi: 10.1021/ac102777s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duffey TA, et al. A tandem mass spectrometry triplex assay for the detection of Fabry, Pompe, and mucopolysaccharidosis-I (Hurler) Clin Chem. 2010;56:1854–1861. doi: 10.1373/clinchem.2010.152009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camelier MV, et al. Practical and reliable enzyme test for the detection of Mucopolysaccharidosis IVA (Morquio Syndrome type A) in dried blood samples. Clin Chim Acta. 2011;412:1805–1808. doi: 10.1016/j.cca.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Fuller M, et al. Screening patients referred to a metabolic clinic for lysosomal storage disorders. J Med Genet. 2011;48:422–425. doi: 10.1136/jmg.2010.088096. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H, Fernhoff P, Vogt RF. Newborn bloodspot screening for lysosomal storage disorders. J Pediatr. 2011;159:7–13. doi: 10.1016/j.jpeds.2011.02.026. e1. [DOI] [PubMed] [Google Scholar]
- 42.Komosinska-Vassev KB, Winsz-Szczotka K, Kuznik-Trocha K, Olczyk P, Olczyk K. Age-related changes of plasma glycosaminoglycans. Clin Chem Lab Med. 2008;46:219–224. doi: 10.1515/CCLM.2008.048. [DOI] [PubMed] [Google Scholar]
- 43.Varki A, et al. Symbol nomenclature for glycan representation. Proteomics. 2009;9:5398–5399. doi: 10.1002/pmic.200900708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.