Abstract
Oxysterols are a class of endogenous signaling molecules that can activate the Hedgehog pathway, which plays critical roles in development, regeneration and cancer. However, it has been unclear how oxysterols influence Hedgehog signaling, including whether their effects are mediated through a protein target or indirectly through effects on membrane properties. To answer this question, we synthesized the enantiomer and an epimer of the most potent oxysterol, 20(S)-hydroxycholesterol. Using these molecules, we show that the effects of oxysterols on Hedgehog signaling are exquisitely stereoselective, consistent with their function through a specific protein target. We present several lines of evidence that this protein target is the 7-pass transmembrane protein Smoothened, a major drug target in oncology. Our work suggests that these enigmatic sterols, which have multiple effects on cell physiology, may act as ligands for signaling receptors and provides a generally applicable framework for probing their mechanism of action.
Embryonic development is directed by a core set of signaling pathways such as the Hedgehog (Hh), Wnt and Notch pathways. As their roles in both development and adult life have been unraveled, these pathways have become important therapeutic targets in oncology and regenerative medicine. The Hh pathway, the focus of our work, has been implicated in human birth defects as well as in a variety of familial and sporadic cancers1. In fact, a recent survey of clinicaltrials.gov revealed that over twenty clinical trials are testing small-molecule inhibitors of the Hh pathway against tumors of the lung, skin, brain, pancreas, and prostate2.
Despite extensive work in vertebrate and invertebrate systems, the biochemical mechanisms regulating many steps in signaling remain incompletely understood. The initiating step in Hh signaling is controlled by a poorly characterized interaction between two multi-pass transmembrane proteins, Patched1 (Ptc1), a tumor suppressor protein, and Smoothened (Smo), an oncoprotein. In the absence of Hh ligands, Ptc1, the receptor, inhibits the activity of Smo3. Upon binding of a Hh ligand, Sonic, Indian or Desert Hedgehog (Shh, Ihh, Dhh), Ptc1 is inactivated, unleashing Smo and allowing the Gli transcription factors to initiate target gene transcription4,5. Despite the fact that the interaction between Ptc1 and Smo is the most commonly damaged step in Hh-related diseases6, the biochemical basis of this step remains unknown. The plant alkaloid cyclopamine, the foundational small-molecule inhibitor of Hh signaling, directly binds Smo7,8. In addition, a large number of cell-based screens against the Hh pathway have identified small molecules that directly bind Smo, either as agonists such as SAG9 and purmorphamine10 or antagonists such as the SANTs9 and GDC-044911. All of these small molecules are assumed to interact with the same site on Smo since they compete with cyclopamine for binding to Smo9–11. The presence of this seemingly easily “druggable” site has incited a search for an endogenous small molecule ligand that may regulate the activity of Smo.
Oxysterols, naturally occurring molecules derived from enzymatic and non-enzymatic oxidation of cholesterol, have emerged as useful probes of this critical step in signaling. In cultured fibroblasts, mesenchymal stem cells and medulloblastoma cells, specific oxysterols can activate Hh signaling, trigger the transcription of target genes, and drive the differentiation of osteogenic cells in culture and in animals12–14. Oxysterols can induce the dramatic accumulation of Smo in the primary cilium15, a key change in sub-cellular localization that allows Smo to activate downstream signaling16. The elimination of Smo abrogates the effects of oxysterols on Hh signaling, suggesting that they function at the level of Smo, Ptc1 or an undiscovered intermediate step13. However, unlike most synthetic Hh effectors, oxysterols do not compete with cyclopamine for binding to Smo, a property used to argue that Smo is not a direct target for oxysterols13. The open question remains, however, how do oxysterols activate Hh signaling?
In addition to their role in the Hh pathway, oxysterols are a class of fascinating and understudied endogenous small molecules that likely constitute an important class of signaling molecules. They have been implicated in vesicle and lipid trafficking17, cholesterol homeostasis signaling18, the activation of nuclear receptors19,20, and leukocyte chemotaxis21,22. However, they have many other effects, such as induction of apoptosis23 and regulation of macrophage function24 that remain to be understood at a molecular level. While their cellular concentrations are orders of magnitude below that of cholesterol, their increased hydrophilicity endows them with the ability to rapidly move between membrane compartments25. Thus, we embarked on an effort to understand how these enigmatic oxysterols influence the Hh signaling pathway. The study of molecules like oxysterols is complicated by their ability to influence cellular processes in two distinct ways that are often difficult to disentangle. First, oxysterols can bind directly to proteins and affect their activity, as exemplified by their interactions with the liver X receptor (LXR)19,20 and the INSIG protein18. Alternatively, because of their lipophilicity, oxysterols can incorporate into host membranes and alter their physical properties, thereby indirectly influencing membrane proteins such as Ptc1 or Smo26,27.
Using a series of structure-activity studies that exploit the fundamental properties of regio- and stereochemical isomerism, we show that oxysterols influence the Hh pathway through a protein target. Through extensive pharmacological analysis and the development of a novel click-chemistry compatible oxysterol analogue, we then provide compelling evidence that oxysterols activate Smo, a major drug target and oncoprotein, through an allosteric mechanism mediated through a distinct site from the canonical cyclopamine binding site.
Results
Cholesterol, the precursor to oxysterols, is an amphiphile composed of a fused tetracyclic ring system, which bears a single hydrophilic moiety at one end (3-hydroxyl), and an iso-octyl chain (IOC) at the opposite end (Fig. 1a). We focused our analysis on the class of dihydroxylated oxysterols that carry, in addition to the 3-hydroxyl in cholesterol, a second hydroxyl group on the IOC (Fig. 1a). This decision was based on the prior findings that IOC-hydroxylated oxysterols are the strongest inducers of Hh signaling12,13.
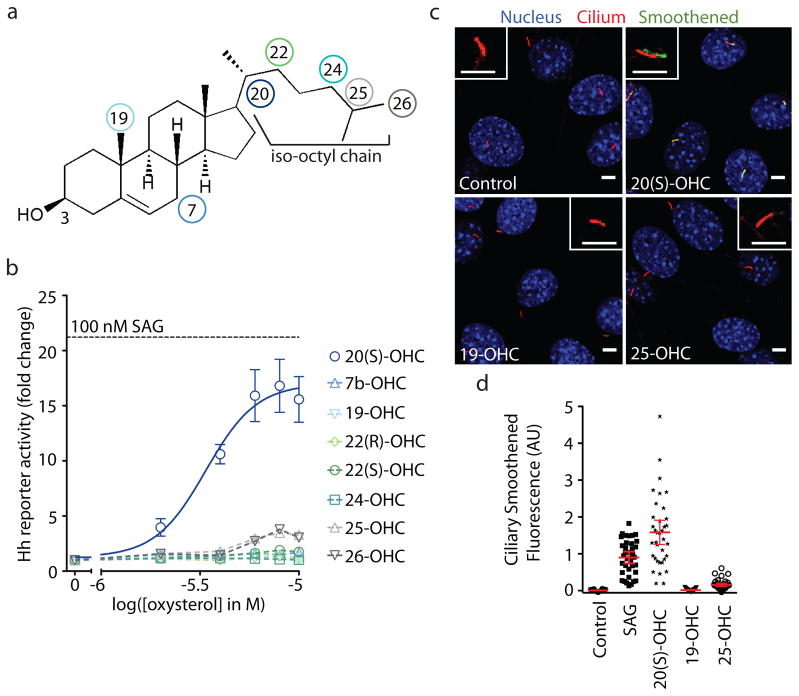
Figure 1. Activation of Hh signaling by oxysterols is regioselective.
(a) Structure of cholesterol, which carries a hydroxyl group at the 3 position, with colored circles marking the positions of the second hydroxyl group on the oxysterols tested. (b) The mean (± SEM) Hh luciferase reporter activity from triplicate wells was measured at various concentration of the indicated oxysterols and plotted as the fold-change compared to activity measured from control cells treated with solvent (ethanol). The black dotted line shows the reporter activity produced by a saturating concentration (100 nM) of the agonist SAG. A non-linear curve fit to the 20(S)-OHC response (blue line) yields an EC50 of ~3 μM. (c) Confocal images of ciliated NIH 3T3 cells treated with solvent (control) or the indicated oxysterol (10 μM). The cilia marker acetylated tubulin (red) and Smoothened (green) were both detected by immunofluorescence; nuclei (blue) were stained with DAPI. The inset is a zoomed image of a single cilium where the green channel is shifted relative to the red channel. Scale bars: 5 μm. (d) Each dot represents Smo fluorescence at a single cilium. Red bars depict the mean and 95% confidence interval (n~35).
Hh activation by oxysterols is regioselective
We first explored the importance of regiochemistry in the ability of IOC-hydroxylated oxysterols to activate Hh signaling by testing a set of commercially available oxysterols (Fig. 1b), each possessing a hydroxyl group at a different position on the IOC. All sterols were tested at a range of doses in NIH 3T3 cells, a Shh-responsive mouse embryonic fibroblast cell line, using the activity of a Hh-responsive firefly luciferase reporter gene as a metric for signaling28. Out of the eight oxysterols tested, 20(S)-hydroxycholesterol (20(S)-OHC), was both the most potent (EC50, or concentration at which 50% activation is achieved, is ~3 μM) and the most effective inducer of Hh reporter gene transcription (Fig. 1b). In comparison, two oxysterols bearing hydroxyls on the tetracyclic ring system (7β-OHC and 19-OHC) were ineffective12,13. As a negative control, we confirmed that 20(S)-OHC did not induce transcription of a Wnt pathway reporter gene (Supplementary Results, Supplementary Fig. 1). Activation was remarkably regioselective, as even related structural isomers such as 22(R)- or 22(S)-OHC did not significantly induce reporter gene transcription. This level of regioselectivity is stricter than that noted previously for the binding of oxysterols to other protein targets, such as LXR19 or the INSIG18 and NPC1 transmembrane proteins29.
We assayed this same set of oxysterols for the ability to drive the accumulation of Smo in the primary cilium, a hallmark of Hh pathway activation15,16. A representative subset of this group is shown (Fig. 1c,d). This cilia localization assay, which measures the ciliary levels of Smo using quantitative fluorescence microscopy, is an important complement to the Hh reporter activity assay described above because it monitors an early, non-transcriptional step in signaling. Consistent with the results from the reporter assay, 20(S)-OHC induced Smo protein accumulation in the primary cilium to a much greater extent than the other oxysterols tested (Fig. 1c,d). These results also confirmed that 20(S)-OHC most likely works by influencing the sub-cellular localization of Smo by acting on the pathway at the level of Ptc1, Smo or an intermediate step.
Hh activation by oxysterols is stereoselective
The degree of regioselectivity observed for Hh pathway activation suggests that oxysterols act on a protein target, since ligand binding sites of proteins have specific structural requirements. However, regioisomers of IOC-hydroxylated oxysterols have dramatically different effects on the physical properties of membranes, such as membrane thickness and area, and the distributions of ordered and disordered domains30. Thus, regioselectivity cannot be interpreted as definitive evidence for a protein-oxysterol interaction.
Using 20(S)-OHC as a substrate for all subsequent studies, we next analyzed the stereochemical requirements for the ability of oxysterols to activate Hh signaling. 20(S)-OHC has eight chiral centers (Fig. 2a), potentially yielding 256 diastereomers. Since sterol synthesis in cells proceeds under the strict steric control of enzymes, only one stereo-isomer is found in nature, hereafter called natural or nat-20(S)-OHC (1; Fig. 2a). Given that modification at the C-20 position, a chiral center, was a key determinant of activity (Fig. 1b), we synthesized and characterized an epimer of nat-20(S)-OHC, nat-20(R)-OHC (2; Fig. 2a; Supplementary Methods, Supplementary Scheme 1). nat-20(S)-OHC and nat-20(R)-OHC are diastereomers that have opposite stereochemical orientation only at the C-20 position but share the same configuration at the remaining 7 stereocenters. Strikingly, a simple switch from the 20(S) to the 20(R) stereochemistry was sufficient to completely abrogate its ability to activate Hh signaling, as measured by either the activation of a luciferase reporter gene (Fig. 2b) or the induction of an endogenous target gene, Gli1 (Fig. 2c). These results point to the critical importance of stereochemistry at the C-20 position.
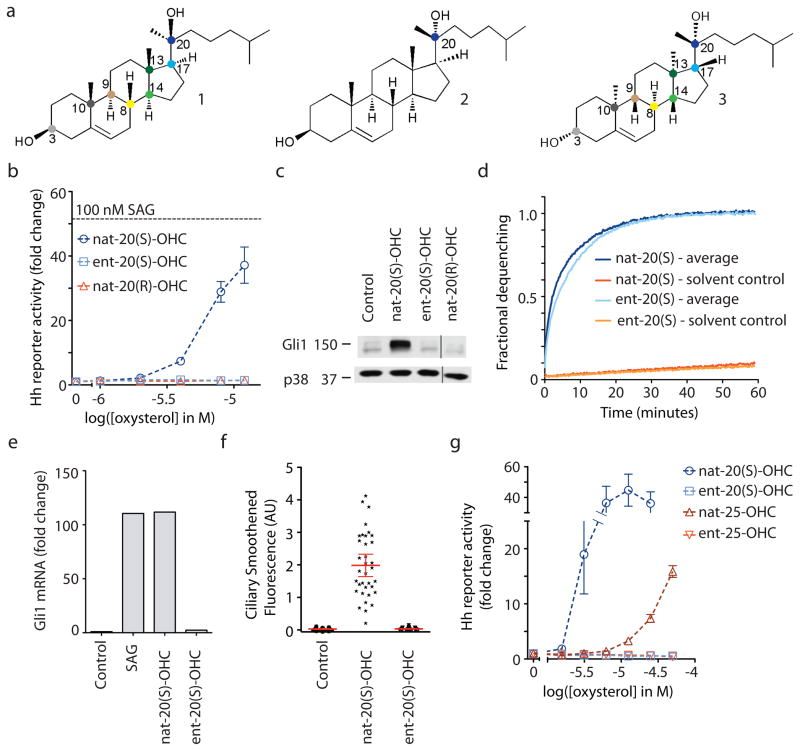
Figure 2. Activation of Hh signaling by nat-20(S)-OHC is stereoselective.
(a) Structures of nat-20(S)-OHC (1), nat-20(R)-OHC (2) and ent-20(S)-OHC (3). Colored dots denote chiral carbon centers. ent-20(S)-OHC is the mirror image of nat-20(S)-OHC, such that the stereochemistry at each of the eight chiral centers is reversed. nat-20(R)-OHC is a diastereomer of nat-20(S)-OHC, with only the stereochemistry at the C-20 position reversed. (b) Mean (± SEM) Gli reporter activity, expressed as fold-change relative to a control treated with ethanol alone in NIH 3T3 cells treated with oxysterols. Results with this Gli reporter construct were confirmed by measuring increases in endogenous Gli1 protein by immunoblotting (c, Supplementary Fig. 10) and Gli1 RNA (e) by q-RT-PCR after treatment with oxysterols (10 μM). (d) nat- and ent-20(S)-OHC produce identical levels of fluorescence dequenching when added to carboxyfluorescein-loaded unilamellar vesicles. Controls were run in parallel with each oxysterol. (f) Quantification of ciliary Smo fluorescence in NIH 3T3 cells treated with solvent control or 10 μM of the indicated oxysterols (n~35). Each dot represents one cilium and the red bar represents the mean bracketed by the 95% confidence interval. (g) Hh reporter activation was measured at various concentrations of the indicated isomers of 20(S)-OHC and 25-OHC.
While epimers such as nat-20(S)-OHC and nat-20(R)-OHC are very similar in structure, they also have different physicochemical properties that could potentially affect both an interaction with a protein and with a membrane domain31. In contrast, enantiomers are molecules that are non-superimposable mirror images of each other, possessing identical physicochemical properties, such as thermodynamic and spectroscopic properties, and identical interactions with achiral substrates. Most protein binding sites are exquisitely chiral and thus enantioselective for their cognate ligands. Though membrane lipids are also chiral, a large body of literature has shown that the interactions of sterol enantiomers with membrane lipids are indistinguishable (i.e. non-enantioselective), perhaps because the mobility of lipids in membranes precludes a structurally defined chiral binding site for a sterol31–33. Thus, enantioselectivity can be used as a simple but powerful criterion to distinguish between the effects of sterols on proteins or on membranes.
To this end, we synthesized the enantiomer of nat-20(S)-OHC, ent-20(S)-OHC (3; Fig. 2a; Supplementary Scheme 2). To assess the effects of these enantiomers on membranes, we employed a fluorescence dequenching assay using unilamellar vesicles composed of a bilayer of dioleoylphosphatidic acid (DOPC). This assay has been previously used to show that the oxysterols nat-25-OHC and ent-25-OHC induce similar degrees of membrane expansion in DOPC vesicles34. nat-20(S)-OHC and ent-20(S)-OHC produced essentially identical levels of fluorescence dequenching when added to carboxyfluorescein-loaded DOPC liposomes, suggesting that both members of this enantiomeric pair were equally capable of expanding a lipid bilayer (Fig. 2d and Supplementary Fig. 2).
In contrast to their identical behavior when added to liposomes, nat- and ent-20(S)-OHC had drastically different effects on Hh signaling, assayed both by the transcription of target genes (Fig. 2b,c,e) and by the accumulation of Smo in primary cilia (Fig. 2f, Supplementary Fig. 3). ent-20(S)-OHC was completely inactive in all assays, suggesting exquisite enantioselectivity in the ability of nat-20(S)-OHC to influence Hh signaling. We confirmed that neither nat- nor ent- 20(S)-OHC affected cell viability at the concentrations used in these experiments (Supplementary Fig. 4).
While this is the first reported study of ent-20(S)-OHC, a prior study rigorously demonstrated that nat-25-OHC and ent-25-OHC have identical effects on model membranes using a battery of assays with phospholipid bilayers and monolayers34. Since nat-25-OHC produced a small degree of Hh pathway activation at higher concentrations (Fig. 1b), we tested whether the activation of Hh signaling by 25-OHC is enantioselective. Consistent with the 20(S)-OHC results, only nat-25-OHC was able to activate Hh reporter genes; ent-25-OHC was completely inactive (Fig. 2g).
Taken together, the regioselectivity, enantioselectivity, and diastereoselectivity displayed at the C-20 position provided a compelling case for the existence of a protein receptor for nat-20(S)-OHC that plays an important role in Hh signaling.
Cyclopamine non-competitively inhibits nat-20(S)-OHC
What protein target mediates the effects of nat-20(S)-OHC on Hh signaling? Previous studies have clearly established that oxysterols act at the level of Ptc1, Smo, or an intermediate step between Ptc1 and Smo13 (Fig. 3a). nat-20(S)-OHC seems to depend on the Ptc1-Smo module for its function because it does not influence target gene transcription in cells lacking Ptc1, leading to a high level of constitutive signaling13 (Supplementary Fig. 5). However, the study of nat-20(S)-OHC interacting proteins has been difficult due to the lack of radiolabeled or otherwise modified analogs.
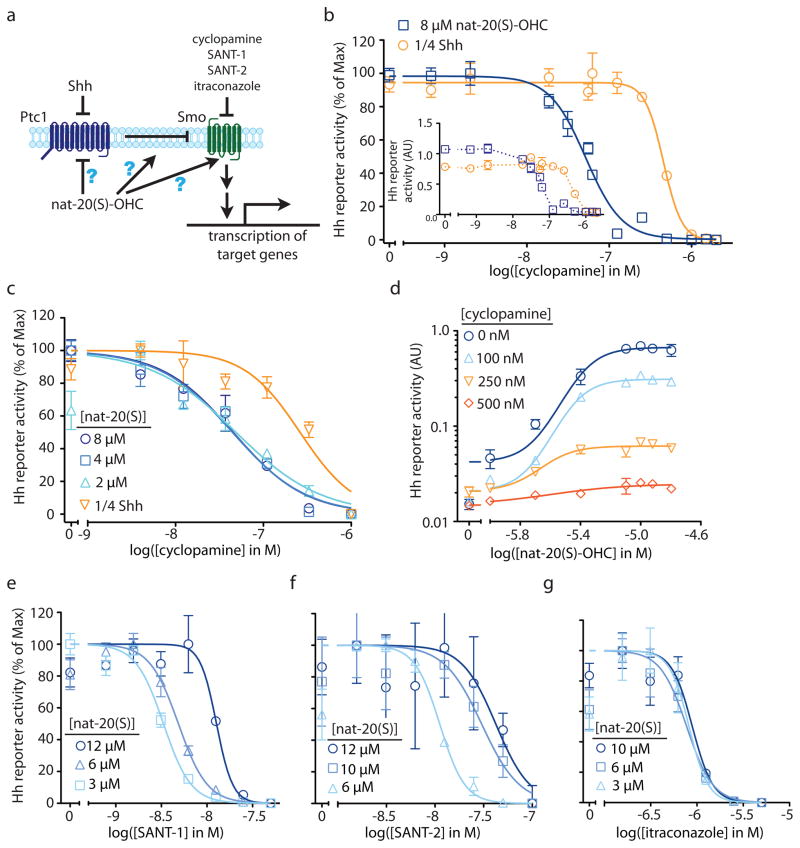
Figure 3. Distinct pharmacological interactions of Smo inhibitors with nat-20(S)-OHC.
(a) nat-20(S)-OHC could activate the Hh signaling by directly inhibiting Ptc1, directly activating Smo or by modulating an intermediate step. (b) Cyclopamine inhibition curves in cells treated with nat-20(S)-OHC (8 μM) or Shh-containing conditioned media (Shh-CM) used at a ¼ dilution (1/4 Shh). The main graph shows Gli luciferase reporter activity, normalized such that the reporter activities with no cyclopamine and maximal cyclopamine are set to 100% and 0% respectively. Inset shows the ratio of Gli to Renilla luciferase activity without normalization (AU: arbitrary units). (c) The IC50 of cyclopamine is not affected by the dose of nat-20(S)-OHC used to activate Hh signaling. (d) Cyclopamine reduces the maximum level of Hh pathway activity driven by saturating concentrations of nat-20(S)-OHC. The IC50 of SANT-1 (e) and SANT-2 (f) increases with increasing doses of nat-20(S)-OHC, but the IC50 of itraconazole is unaffected (g).
Despite these limitations, the availability of many Hh pathway agonists and antagonists with defined targets motivated us to analyze the pharmacological interactions between these molecules and nat-20(S)-OHC. We measured the IC50 (concentration at which 50% inhibition is achieved) of cyclopamine, a direct Smo inhibitor, under conditions where Hh target genes were activated to similar levels by the application of either the endogenous activator Shh, or nat-20(S)-OHC. Cyclopamine was chosen for the initial analysis because the majority of Smo agonists and antagonists have been shown to compete with cyclopamine for binding to Smo-containing membranes, thus defining the cyclopamine-binding site as an important regulatory site on the Smo35. We reasoned that if nat-20(S)-OHC functions by inactivating Ptc1 in a manner analogous to Shh, or by influencing a step between Ptc1 and Smo, the IC50 of cyclopamine should be the same in the presence of equal activation by either agonist because the cyclopamine target, Smo, is downstream of these steps. Contrary to this prediction, the IC50 of cyclopamine was 9-fold lower when the pathway was activated with nat-20(S)-OHC (IC50: 50 nM) compared to when it was activated with Shh (IC50: 460 nM) (Fig. 3b). Importantly, we used doses of Shh and nat-20(S)-OHC that produced near equivalent levels of target gene transcription, ensuring that that this difference was not simply because nat-20(S)-OHC activated the pathway to a lesser extent (Fig. 3b). Our findings suggest that the mechanism of activation of Hh signaling by nat-20(S)-OHC is different from that of Shh. Smo is significantly more sensitive to inhibition by cyclopamine when cells are activated by nat-20(S)-OHC compared to when they are activated by Shh, suggesting that Smo is in distinctly different conformations under these two conditions. These data make Ptc1 an unlikely candidate for the nat-20(S)-OHC receptor, motivating us to question the prevailing model and ask if nat-20(S)-OHC binds to Smo after all?
We further characterized the antagonistic interaction between nat-20(S)-OHC and cyclopamine by determining the IC50 of cyclopamine at different doses of nat-20(S)-OHC. In the case of competitive inhibition, increasing doses of nat-20(S)-OHC should produce a progressively higher IC50 for cyclopamine36. This is exactly the interaction observed between cyclopamine and the small molecule SAG, which activates Smo by competing for the cyclopamine binding site9 (Supplementary Fig. 6). In contrast, we found that the IC50 of cyclopamine does not significantly change with increasing doses of nat-20(S)-OHC (Fig. 3c), indicating that this is not a competitive interaction and disfavoring the idea that nat-20(S)-OHC and cyclopamine bind to the same site on Smo. A prior study reached a similar conclusion regarding the lack of a competitive interaction between nat-20(S)-OHC and cyclopamine, both at the level of binding to Smo and at the level of Hh pathway activation, leading the authors to suggest that Smo is not a target of oxysterols13.
However, an alternative possibility is that cyclopamine and nat-20(S)-OHC bind to Smo at distinct sites and that cyclopamine is a non-competitive, “insurmountable” inhibitor of nat-20(S)-OHC-induced Hh signaling36. Formally, this would imply an allosteric interaction between the two molecules since cyclopamine binding at one site would influence either the binding or the efficacy of nat-20(S)-OHC at a different site. The kinetic signature of non-competitive antagonism is a reduction in the maximum achievable level of activation by the agonist36. Indeed, consistent with non-competitive inhibition, cyclopamine reduced the maximum extent of activation produced by saturating doses of nat-20(S)-OHC (Fig. 3d). The data presented to this point are consistent with the model that nat-20(S)-OHC binds and activates Smo, albeit through a site different from the canonical cyclopamine binding site.
nat-20(S)-OHC interactions with other Smo antagonists
SANT-1 and SANT-2 are two direct Smo antagonists that compete with cyclopamine for binding to Smo9,35. Surprisingly, unlike cyclopamine, both antagonists show a competitive interaction with nat-20(S)-OHC (Fig. 3e,f). Increasing doses of nat-20(S)-OHC led to a progressive rightward shift in the SANT-1 and SANT-2 inhibition curves and a ~3–4 fold increase in their IC50 values (Fig. 3e,f). Since the SANT molecules inhibit cyclopamine binding9 and nat-20(S)-OHC has no effect on cyclopamine binding13, it is unlikely that the SANTs occupy the same binding site as nat-20(S)-OHC; instead, the interaction between them is likely mediated by an allosteric mechanism.
Amongst the Smo antagonists described to date, the anti-fungal itraconazole is the only one that does not compete with cyclopamine for Smo binding37. Itraconazole was inferred to be a direct Smo antagonist based on its ability to synergize with cyclopamine to inhibit Hh signaling. Given that nat-20(S)-OHC also does not inhibit the cyclopamine-Smo interaction13 (Fig. 3c), we considered the possibility that itraconazole and nat-20(S)-OHC bind to the same site on Smo, a mechanism which predicts a competitive interaction between these molecules. However, itraconazole and nat-20(S)-OHC likely bind to different sites because increasing doses of nat-20(S)-OHC did not cause a rightward shift in the itraconazole inhibition curve or alter its IC50 (Fig. 3g).
Taken together, the interaction of nat-20(S)-OHC with a panel of direct Smo antagonists demonstrates the property of “probe specificity”-- nat-20(S)-OHC impacts the IC50s of SANT-1 and SANT-2 but has no effect on the IC50s of cyclopamine and itraconazole. Probe specificity is considered a hallmark of allosteric ligands because they can stabilize an ensemble of conformations of a protein that can have a large impact on one receptor probe but little impact on others36. In summary, the distinct pharmacological interactions between nat-20(S)-OHC and various Smo ligands provided additional evidence for its direct interaction with Smo.
Smo agonists synergize with nat-20(S)-OHC
The model that nat-20(S)-OHC and cyclopamine bind to distinct sites on Smo predicts that nat-20(S)-OHC should show allosteric interactions with Smo activators, such as SAG or purmorphamine, that compete with cyclopamine9,10. On the other hand, if nat-20(S)-OHC activated signaling in a completely different way, without a direct interaction with Smo, it would be unlikely to show an allosteric interaction with these agonists. We measured the dose-response curve for activation by nat-20(S)-OHC in the absence or presence of low concentrations of SAG (Fig. 4a). Notably, the concentration of SAG used here (0.3 nM) is 10-fold lower than its EC509 and results in less than 10% of the maximum degree of Hh pathway activation. In the presence of this miniscule concentration of SAG, the EC50 of nat-20(S)-OHC sharply declined over 10-fold to from 3.2 μ to 0.24 μM (Fig. 4a), providing compelling evidence for a synergistic interaction between these agonists. In the reciprocal experiment, low concentrations of nat-20(S)-OHC also significantly decreased the EC50 of SAG from 2 nM to 0.4 nM (Fig. 4b). We confirmed the synergy between SAG and nat-20(S)-OHC by measuring endogenous Gli1 protein levels (Fig. 4c) and by measuring the accumulation of Smo in primary cilia (Fig. 4d, Supplementary Fig. 7).
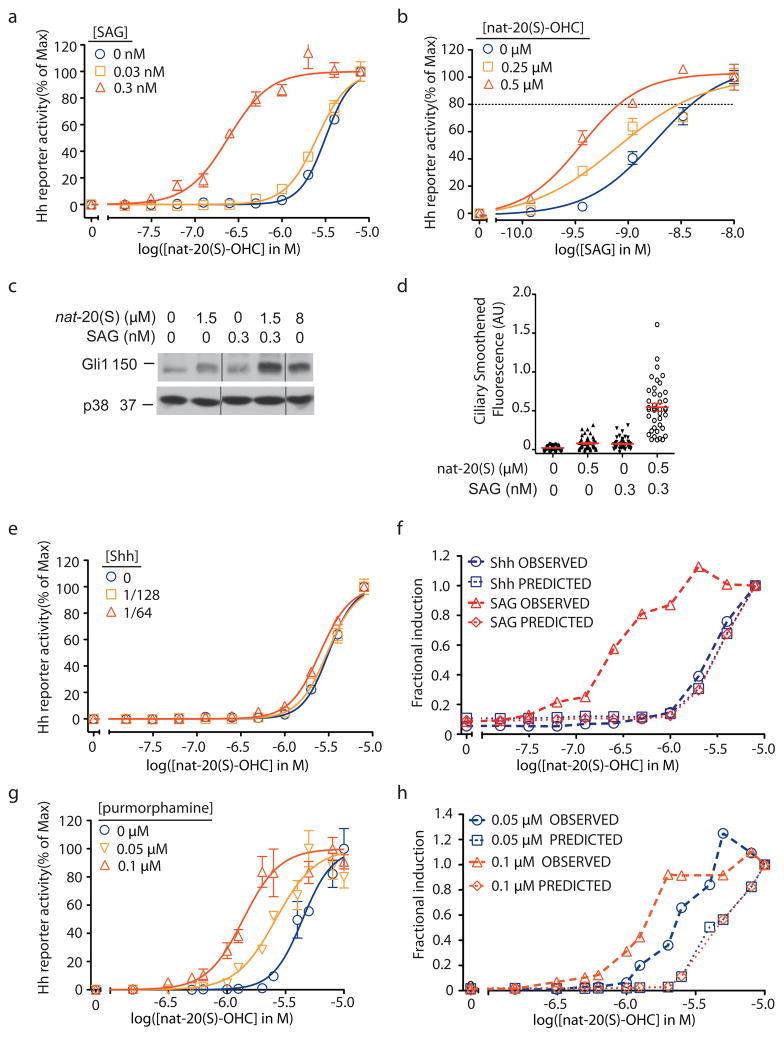
Figure 4. Synergistic activation of Hh signaling by nat-20(S)-OHC and SAG.
(a) Dose response curves of nat-20(S)-OHC in the presence of low concentrations of SAG. The EC50 of nat-20(S)-OHC shifts from 3 μM without SAG to 0.24 μM with 0.3 nM SAG. (b) The reciprocal experiment shows that the EC50 of SAG drops from 2 nM with no nat-20(S)-OHC to 0.4 nM with 0.5 μM nat-20(S)-OHC. The dotted line denotes reporter activity produced by 8μM nat-20(S)-OHC. A combination of low concentrations of SAG and nat-20(S)-OHC can fully activate Hh signaling as measured by the induction of endogenous Gli1 protein levels (c, Supplementary Fig. 10) and the accumulation of Smo in cilia (d). The low concentrations of each agonist used have minimal effects when added individually to cells (a–d). (f) Low doses of Shh-CM (1/64 and 1/128 dilutions) do change the EC50 of nat-20(S)-OHC. (f) The observed Hh reporter activity from (a) and (e) and the predicted reporter activity based on a purely additive Bliss model is plotted at various doses of nat-20(S)-OHC in combination with either low Shh or low SAG. (g) Low doses of purmorphamine can reduce the EC50 of nat-20(S)-OHC. (h) Bliss independence analysis of the data in (g) suggests synergy between nat-20(S)-OHC and purmorphamine. Error bars in all graphs represent SEM.
The simplest explanation for the synergy seen between SAG and nat-20(S)-OHC is that both agonists bind to Smo at different sites and display a positive allosteric interaction. In comparison, the combination of nat-20(S)-OHC with Shh, which does not bind to Smo but indirectly leads to its activation by inactivating Ptc1, did not show evidence of synergy (Fig. 4e). The EC50 of nat-20(S)-OHC did not change drastically when combined with low concentrations of Shh (EC50 - Shh: 3.2 μM, EC50 + Shh: 2.5 μM). We used Bliss independence analysis38 to compare the interaction between nat-20(S)-OHC and SAG with the interaction between nat-20(S)-OHC and Shh (Fig. 4f). The Bliss score uses the activities of each agonist to calculate the predicted response if the agonists behaved in a purely additive manner with respect to each other, as might be expected if the two molecules influenced different proteins. Our observed responses when nat-20(S)-OHC and SAG were combined were much stronger than this Bliss prediction (Fig. 4f), consistent with synergy. In contrast, the Bliss prediction is virtually superimposable with the observed response when Shh and nat-20(S)-OHC were combined (Fig. 4f).
Based on dose-response data and Bliss analysis, nat-20(S)-OHC also displayed a synergistic interaction with purmorphamine, a direct Smo agonist that is ~100-fold less potent than SAG (Fig. 4g,h)10. Trace doses of purmorphamine (~10-fold below its EC50) decreased the EC50 of nat-20(S)-OHC by ~3-fold (Fig. 4g). Thus, two different direct Smo agonists, varying in potency over two orders of magnitude, displayed a positive allosteric interaction with nat-20(S)-OHC.
Smo binds to nat-20(S)-OHC immobilized on beads
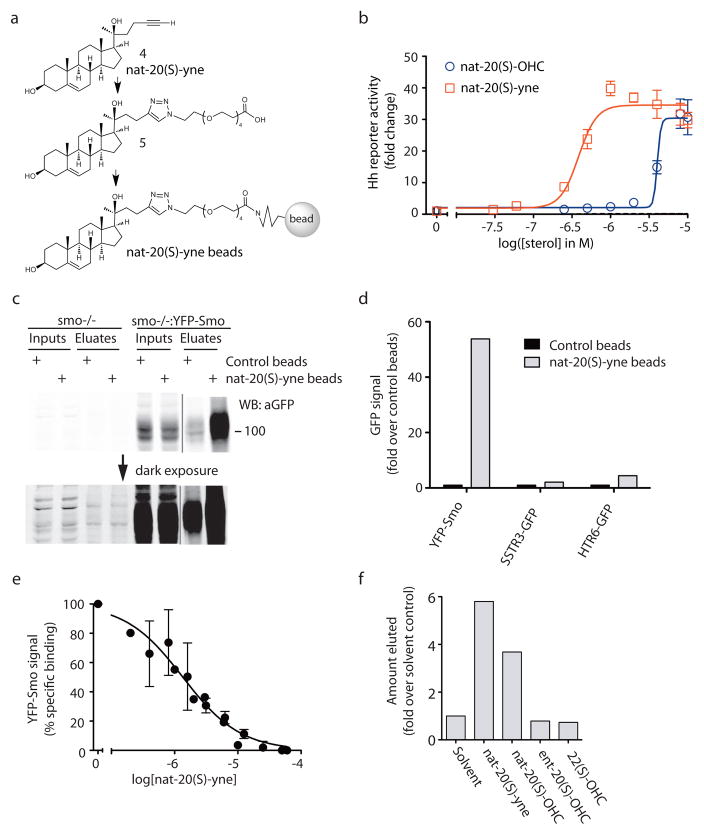
Our pharmacological analyses suggested that Smo was the receptor for nat-20(S)-OHC. To test this prediction, we synthesized an analog of nat-20(S)-OHC with a terminal alkyne group (4; nat-20(S)-yne) compatible with click chemistry techniques for coupling the molecule to a matrix (Fig. 5a; Supplementary Scheme 3). Remarkably, this analog was 8-fold more potent than nat-20(S)-OHC itself (Fig. 5b) when tested in the Hh reporter assay, making it an ideal moleculre for ligand affinity chromatography. nat-20(S)-yne induced Smo accumulation in cilia, confirming that its mechanism of action was the same as nat-20(S)-OHC (Supplementary Fig. 8a). Using click chemistry, nat-20(S)-yne was covalently coupled to magnetic beads through an intermediate molecule (5) that contained a polyethylene glycol (PEG) linker interposed to minimize steric inhibition of receptor binding (Fig. 5a; Supplementary Schemes 4 and 5). As a source of Smo protein, detergent extracts were prepared from membranes isolated from smo−/−fibroblasts stably expressing functional, YFP-tagged Smo (smo−/−:YFP-Smo cells)39. Consistent with a physical interaction between nat-20(S)-OHC and Smo, YFP-Smo was captured on nat-20(S)-yne bearing magnetic beads (Fig. 5c). Binding was proportional to the amount of YFP-Smo added (Supplementary Fig. 8b) and control beads (Supplementary Scheme 5) captured 10-fold lower amounts of YFP-Smo (Fig. 5c). To provide more precise relative quantitation of YFP-Smo captured in these affinity chromatography experiments, all immunoblots shown were developed and quantitated using the LiCor infrared detection system rather than chemiluminescence.
Figure 5. Smo binds to a nat-20(S)-OHC analog immobilized on beads.(a).
Chemical structures of nat-20(S)-yne (4), containing a terminal alkyne compatible with click chemistry, a nat-20(S)-yne derivative (5) which is 4 coupled to a PEG linker, and 5 immobilized on magnetic beads. (b) A Gli-luciferase reporter assay demonstrates that nat-20(S)-yne (EC50 ~390 nM) is 8-fold more potent at activation of Hh signaling compared to nat-20(S)-OHC. (c) An immunoblot, depicted at two exposures, showing the amount of YFP-Smo precipitated by nat-20(S)-yne-beads or control beads from membrane extracts made from either smo−/−:YFP-Smo cells or smo−/− cells. The complete immunoblot is included in Supplementary Fig. 10. (d) Control beads or nat-20(S)-yne beads were incubated with membrane extracts made from 293T cells expressing YFP-Smo, a GFP-tagged somatostatin receptor (SSTR3-GFP) or a GFP-tagged serotonin receptor (HTR6-GFP). The amount of YFP/GFP-tagged receptor precipitated in each case was measured from the immunoblot shown in Supplementary Fig. 8c and plotted. (e) Free nat-20(S)-yne can inhibit the binding of YFP-Smo to nat-20(S)-yne beads (IC50=1.3μM). Error bars represent the SEM from three independent experiments. (f) nat-20(S)-yne beads incubated with YFP-Smo containing extracts were washed and subjected to ligand-elution with the indicated sterols (500μM each). The amount of YFP-Smo eluted in each case was measured by immunoblotting (Supplementary Figs. 8d and 10).
Several controls were performed to establish the specificity of this interaction. A band at the position corresponding to YFP-Smo was not observed on immunoblots when affinity chromatography was performed using smo−/− cells (lacking YFP-Smo), confirming the specificity of the signal detected with the anti-GFP antibody (Fig. 5c). Notably, the pattern of background bands captured from smo−/− extracts by control beads and nat-20(S)-yne beads was similar (Fig. 5c), highlighting the relevance of the selective interaction between nat-20(S)-yne beads and YFP-Smo. In addition, two other 7-pass transmembrane receptors that that localize in primary cilia did not show a selective interaction with nat-20(S)-yne beads (Fig. 5d, Supplementary Fig. 8c).
The binding of YFP-Smo to nat-20(S)-yne beads could be inhibited by free nat-20(S)-yne added to the extract in a dose-dependent fashion (IC50 = 1.3 μM), demonstrating that this interaction was mediated through the sterol itself and not through the PEG linker (Fig. 5e). Three Smo ligands, cyclopamine, SANT-1, and itraconazole, that inhibit nat-20(S)-OHC induced Hh signaling (Fig. 3) did not inhibit the interaction between YFP-Smo and nat-20(S)-yne beads when used at concentrations up to 5-fold higher than their respective IC50 values (Supplementary Fig. 9). Finally, to demonstrate the enantioselectivity of the interaction between Smo and nat-20(S)-OHC, we tested the ability of various ligands to elute YFP-Smo captured on nat-20(S)-yne beads. Both nat-20(S)-yne and nat-20(S)-OHC could elute YFP-Smo from the beads. However, ent-20(S)-OHC and an inactive, structurally related oxysterol, 22(S)-OHC, were both ineffective (Fig. 5f, Supplementary Fig. 8d). Since nat- and ent-20(S)-OHC have identical hydrophobic characteristics, evident from their identical interactions with lipid bilayers (Fig. 2d), the selective elution with one enantiomer strongly supports a specific interaction between nat-20(S)-OHC and Smo. Taken together, these biochemical data corroborate the pharmacological data and demonstrate a physical interaction between nat-20(S)-OHC and Smo.
Discussion
In addition to their roles in regulating the local structure and dynamics of membranes, sterols can impact a broad range of physiological and pathological processes, ranging from lipid metabolism17–20 and atherosclerosis40, to apoptosis23, inflammation24, and cancer susceptibility41. In many of these cases, the specific mechanisms and molecular pathways by which sterols exert their effects on cells remain unknown. One intriguing possibility is that endogenous oxysterols function as second messengers in cellular signaling pathways. A prominent example supporting this idea is the Hh signaling pathway, which has long been postulated to be regulated by endogenous sterols12,42.
Although the molecular basis of this sterol dependency remains to be understood, oxysterols have recently emerged as candidates for regulatory small molecules in Hh signaling. nat-20(S)-OHC is an effective activator of Hh signaling, but is a particularly enigmatic and poorly understood molecule. While detectable in tissues43, biochemical pathways for the synthesis and degradation of this molecule remain unknown. Herein, we utilized a combination of chemical and pharmacological methods to understand the mechanism by which nat-20(S)-OHC influences Hh signaling.
A classical problem in the study of sterols (and most amphiphiles) comes from the difficulty in distinguishing their effects on membrane properties from their direct effects on proteins31. The use of enantiomers represents an incisive and generally applicable methodology to make this critical distinction, thus, we report here the first synthesis of ent-20(S)-OHC (3). This enantiomer was completely inactive in Hh pathway assays, suggesting that nat-20(S)-OHC activates Hh signaling by binding to a highly chiral protein binding pocket, rather than by incorporating into a dynamic lipid membrane wherein even “ordered” domains are likely rapidly assembling and disassembling, and are therefore insensitive to stereochemical changes31. In addition, we have also reported the improved synthesis of the C-20 epimer of nat-20(S)-OHC, nat-20(R)-OHC44 (2). nat-20(R)-OHC did not activate Hh signaling, further exemplifying the structural discrimination associated with a specific protein interaction.
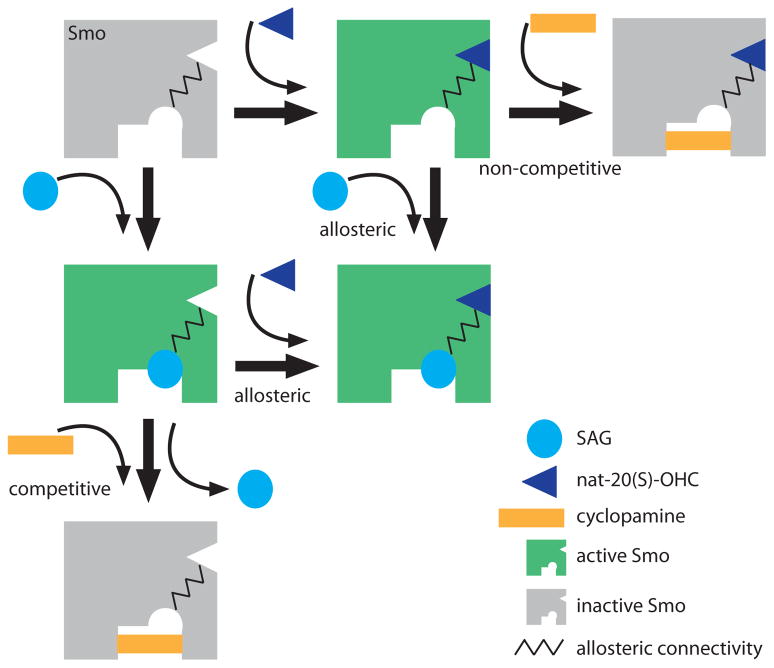
Our pharmacological and biochemical studies provide strong evidence for the direct effect of nat-20(S)-OHC on Smo, an oncoprotein and important cancer drug target. The high degree of synergy seen between nat-20(S)-OHC and the direct Smo agonist SAG is most parsimoniously explained by a positive allosteric interaction between the two molecules mediated through distinct sites on Smo. Since SAG and cyclopamine display a competitive interaction9 both in binding and activation assays, nat-20(S)-OHC most likely binds to a site distinct from the canonical cyclopamine binding site that has been the target for most anti-Smo drugs9–11. Taken together, our data is consistent with the presence to at least two binding sites on Smo, one that binds to nat-20(S)-OHC and a second that binds to SAG and cyclopamine (Fig. 6). SAG and nat-20(S)-OHC display a positive allosteric interaction, SAG and cyclopamine a competitive interaction and cyclopamine and nat-20(S)-OHC a non-competitive interaction (Fig. 6). Given the similarity of Smo to G-protein-coupled-receptors (GPCRs), our finding of a Smo-oxysterol interaction is reminiscent of recent evidence showing that metarhodopsin I45 and the β2-adrenergic receptor46 bind to cholesterol through their transmembrane regions and evidence showing that oxysterols can function in chemotaxis of leukocytes by acting on a GPCR21,22. Thus, it is likely that sterols represent a class of ligands that can play an important role in modulating the activation of many GPCR-initiated signaling pathways.
Figure 6. A model for the allosteric regulation of Smo by small molecules.
The key postulate of this model is the presence of two distinct binding sites on Smo, one for SAG and one for nat-20(S)-OHC, that show a positive allosteric interaction, denoted by the zig-zag line. Cyclopamine binds to a site that overlaps with SAG and thus is an orthosteric competitive antagonist of SAG. However, the effect of cyclopamine is “agonist-specific” because it acts as a non-competitive antagonist of nat-20(S)-OHC. When cyclopamine binds to Smo, nat-20(S)-OHC cannot activate the protein even at saturating concentrations. The active conformation of Smo, one that is capable of transducing the Hh signal, is colored green and the inactive conformation is colored gray.
Like GPCRs, Smo likely adopts a range of conformations with different signaling properties39. Different Smo ligands clearly stabilize distinct ensembles of these conformations. One striking example comes from the differences between SANT-1 and cyclopamine, two Smo ligands that show a competitive binding interaction. While both molecules inhibit Hh target gene transcription, SANT-1 inhibits Smo accumutation in cilia while cyclopamine drives Smo accumulation in cilia, proving that they stabilize distinct conformations39. This is consistent with our finding that nat-20(S)-OHC displays a non-competitive interaction with cyclopamine but a competitive interaction with SANT-1. From a therapeutic perspective, Smo may be susceptible to allosteric regulation and future drug discovery efforts should focus on targeting of such allosteric sites. In addition, it will be important to determine whether different Smo ligands can favor the coupling of Smo to distinct sets of downstream signaling complexes.
While our results provide evidence for the allosteric activation of Smo by nat-20(S)-OHC, further studies are needed to address the effects of endogenous nat-20(S)-OHC on Hh pathway activity in cells and animals. The EC50 of nat-20(S)-OHC for Hh target gene induction (~3 μM) is in the same range as EC50 values (4–7 μM) reported for the activation of LXRαreceptors by the endogenous ligands 24-OHC and 22-OHC19,20. While the concentration of nat-20(S)-OHC in Hh-responsive cells or embryos has not been carefully measured, concentrations of oxysterols in tissues have been estimated to be in the 0.1–10μM range47,48. While this seems lower than our measured EC50 for nat-20(S)-OHC, it is important to note that the affinity constants (Kd values) for oxysterol-protein interaction are often more than an order of magnitude lower than EC50s, presumably because much of the oxysterol is not available to the receptor when its added to cell culture19. In addition, the local concentration of a lipophilic molecule in a cellular compartment can be substantially higher than the bulk concentration measured in a tissue. Future progress in this area will require new methods to reliably measure nat-20(S)-OHC levels in cells and an understanding of how this molecule is synthesized, transported and degraded such that its levels can be perturbed. nat-20(S)-yne (4), a potent, click-chemistry compatible analog of nat-20(S)-OHC, provides an invaluable bio-orthogonal reporter to dissect nat-20(S)-OHC function in cells and animals. A rigorous understanding of how sterols influence Smo and other GPCRs is certain to provide novel avenues for the modulation of these key therapeutic targets in a variety of human diseases.
Methods
Cells and reagents
NIH 3T3 and 293T cells were obtained from ATCC, Wnt-L cells from Ethan Lee (Vanderbilt), SAG (≥95%) from Enzo Life Sciences, cyclopamine from Toronto Research Chemicals (≥98%), itraconazole (≥98%) from Sigma, the SANTs (≥95%) from EMD and purmorphamine (≥98%) from Cayman Chemicals. All sterols except ent-20(S)-OHC, nat-20(R)-OHC and nat-20(S)-yne were purchased from Steraloids (purity ≥98%).
Chemical synthesis
Schemes for nat-20(R)-OHC, ent-20(S)-OHC, nat-20(S)-yne and nat-20(S)-yne beads are fully described in the Supplementary Methods (Schemes 1–5).
Hedgehog reporter assays
NIH 3T3 cells in a 10 cm plate were transfected with 8 μg of a 19:1 ratio of firefly luciferase reporter driven by a Gli-responsive promoter28 and a constitutive Renilla luciferase reporter. The next day, transfected cells were seeded into a 96-well plate, grown to confluence, and treated overnight with drugs diluted in media containing 0.5% fetal bovine serum (FBS). Activity of both reporters was measured using the Dual-Luciferase Reporter kit (Promega) and read on a Berthold LB 96 V luminometer or a Tecan Infinite M1000 plate reader. The Gli luciferase to Renilla luciferase ratio was taken as a metric for Hh signaling.
Immunoblotting
Im1munoblotting was performed as described previously39, and detailed methods are provided in the supplementary information.
Ligand affinity chromatography
Smo−/− cells and smo−/−:YFP-Smo cells39 were lysed by hypotonic lysis in SEAT buffer (250 mM sucrose, 1 mM EDTA, 10 mM acetic acid, 10 mM triethanolamine, 10 μg/mL eupeptin-pepstatin-chymostatin (LPC) protease inhibitor mix and the SigmaFast EDTA-Free protease cocktail). After removal of nuclei by centrifugation (500xg, 5 min), membranes were pelleted by centrifugation at 95,000xg for 30 min. Membranes were extracted in a n-dodecyl-β-D-maltopyranoside (DDM) extraction buffer (50 mM Tris pH 7.4, 500 mM NaCl, 10% v/v glycerol, 0.1% w/v DDM and the SigmaFast EDTA-Free protease cocktail) for 4 hours at 4°C, followed by removal of insoluble material by centrifugation (100,000xg, 30 min). This DDM extract was incubated with nat-20(S)-yne magnetic beads or control beads for 12 hours at 4°C to allow binding. In experiments where soluble ligands were included as competitors, the extracts were incubated with free ligand for one hour at 4°C prior to the addition of beads. In all experiments, the amount of solvent was carefully equalized in each sample. After extensive washing, proteins captured on the beads were eluted with free ligand or with reducing SDS sample buffer. The presence of YFP-Smo in these eluates was determined by quantitative immunoblotting with an anti-GFP antibody (Novus, NB600-308, 1:5000) and infrared imaging (Li-Cor Odyssey system).
Quantitative Real-time PCR (Q-PCR)
After drug treatment (described above), RNA was harvested from cells in TRIzol (Invitrogen) and converted to cDNA using the iScript cDNA synthesis kit (BioRad) with a mixture of oligo(dT) and random hexamer primers. This cDNA was used as a substrate for Q-RT-PCR using primers for Gli1 (For: CCAAGCCAACTTTATGTCAGGG, Rev: AGCCCGCTTCTTTGTTAATTTGA) and the loading control GAPDH (For: AGTGGCAAAGTGGAGATT, Rev: GTGGAGTCATACTGGAACA). Q-RT-PCR assays were performed with IQ SYBR Green Supermix (Bio-Rad) on a Sequence Detector 7900HT (Applied Biosystems).
Immunofluorescence, microscopy and image analysis
Immunofluorescence, microscopy and image analysis were performed as described previously15, and detailed methods are included in the supplementary information.
Data analysis
All statistical analysis and curve fitting was done in GraphPad Prism. For microscopy data, the Smo fluorescence for each cilium was individually plotted, generating a scatter plot that represents variability in the data. To compare Smo levels between different conditions, the mean and 95% confidence interval are provided (n ~35).
For Hh reporter assays, each point represents the mean of triplicate wells, with error bars representing the standard error of the mean (SEM). Each result in the paper was repeated at least three times with similar outcomes. Relative luciferase activity was calculated by dividing Gli luciferase by Renilla luciferase luminescence. Fold-change in reporter activity was calculated by dividing each replicate by the mean reporter activity of the solvent-treated control. Normalized (% of Max) Hh reporter activity was calculated by setting the minimum and maximum values of a curve to 0% and 100% respectively using the “normalize” function of GraphPad Prism. For the binding inhibition curves, percent specific binding refers to the amount of binding detected above non-specific background binding, defined as the binding seen even with maximal concentrations of the inhibitor and subtracted from all values. This background binding was set as 0% and the maximum binding detected in the absence of any inhibitor set to 100%. In all graphs, dotted lines are straight connectors between points, and solid lines represent non-linear curve fits of the data to a sigmoidal (variable slope) equation. Curve fits were calculated in GraphPad Prism using the log(agonist/inhibitor) versus response or normalized response as appropriate.
Bliss independence analysis
For Bliss independence analysis38, we calculated the fractional activation (F) produced by each drug dose by dividing its reporter activity by the maximal Hh reporter activity obtained with that drug. The Bliss score for a combination of two drugs (A and B) was calculated with the equation FA+FB(1−FA), representing the predicted activation if the two drugs act independently. This score was compared to the observed response when the two drugs were combined at the same doses. The Bliss analysis presented in Fig. 4f uses data from Figs. 4a and 4e, and the analysis in Fig. 4h uses data from Fig. 4g.
Liposome expansion assay
Carboxyfluorescein (CF)-loaded unilamellar liposomes were prepared and used in the dequenching assay as described previously33, and detailed methods are outlined in the supplementary information.
Supplementary Material
Acknowledgments
We thank members of the Rohatgi Lab for helpful discussions, G. Luchetti for help with ligand affinity chromatography, P. Niewiadomski and A. Lebensohn for critical reading of the manuscript, Ethan Lee for Wnt-L cells, Kirk Mykytyn for the SSTR3-GFP and HTR6-GFP constructs, Alejandro Sweet-Cordero for use of a Li-Cor Odyssey imager and Matthew Scott for support and use of a confocal microscope. Mass spectrometry analysis was conducted at the NIH/NCRR Mass Spectrometry Facility at Washington University, supported by the NIH (RR00954, DK020579, DK056341). This work was supported by a Pew Scholar Award and an Innovation Research Grant from the Stand Up to Cancer Foundation to R.R., by NIH grants to D.F.C (GM47969 and HL67773) and P.S. (HL67773), and by NIH training grants to S.N. (5 T32 GM007276) and L.K.M (5 T32 HL007275).
Footnotes
Author Contributions
L.K.M., K.K. and D.F.C. designed and synthesized the oxysterol analogs. S.N. performed cellular experiments with oxysterols. J.R. and P.S. designed and performed vesicle expansion experiments. All authors analyzed the data and contributed to the manuscript. S.N. and R.R. wrote the paper with input from L.K.M., D.F.C, J.R. and P.S.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 2.ClinicalTrials.gov. 1993 May 19; 2008 [cited 2011 February 5]; Available from: cilincaltrials.gov.
- 3.Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 4.Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 5.Marigo V, et al. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 6.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JK, et al. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heretsch P, Tzagkaroulaki L, Giannis A. Cyclopamine and hedgehog signaling: chemistry, biology, medical perspectives. Angew Chem Int Ed. 2010;49:3418–3427. doi: 10.1002/anie.200906967. [DOI] [PubMed] [Google Scholar]
- 9.Chen JK, et al. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha S, Chen JK. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol. 2006;2:29–30. doi: 10.1038/nchembio753. [DOI] [PubMed] [Google Scholar]
- 11.Romer JT, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran RB, Scott MP. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci USA. 2006;103:8408–8413. doi: 10.1073/pnas.0602852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwyer JR, et al. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JS, et al. Novel oxysterols have pro-osteogenic and anti-adipogenic effects in vitro and induce spinal fusion in vivo. J Cell Biochem. 2011;112:1673–1684. doi: 10.1002/jcb.23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 16.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 17.LeBlanc MA, McMaster CR. Lipid binding requirements for oxysterol-binding protein Kes1 inhibition of autophagy and endosome-trans-Golgi trafficking pathways. J Biol Chem. 2010;285:33875–33884. doi: 10.1074/jbc.M110.147264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan A, et al. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janowski BA, et al. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, et al. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 2007;5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannedouche S, et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, et al. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–523. doi: 10.1038/nature10226. [DOI] [PubMed] [Google Scholar]
- 23.Panini SR, Sinensky MS. Mechanisms of oxysterol-induced apoptosis. Curr Opin Lipidol. 2001;12:529–533. doi: 10.1097/00041433-200110000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Park K, Scott AL. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J Leukoc Biol. 2010;88:1081–1087. doi: 10.1189/jlb.0610318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theunissen JJ, et al. Membrane properties of oxysterols. Interfacial orientation, influence on membrane permeability and redistribution between membranes. Biochim Biophys Acta. 1986;860:66–74. doi: 10.1016/0005-2736(86)90499-2. [DOI] [PubMed] [Google Scholar]
- 26.Rentero C, et al. Functional implications of plasma membrane condensation for T cell activation. PLoS One. 2008;3:e2262. doi: 10.1371/journal.pone.0002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olkkonen VM, Hynynen R. Interactions of oxysterols with membranes and proteins. Mol Aspects Med. 2009;30:123–133. doi: 10.1016/j.mam.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki H, et al. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 29.Infante RE, et al. Purified NPC1 protein. I Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 30.Massey JB, Pownall HJ. Structures of biologically active oxysterols determine their differential effects on phospholipid membranes. Biochemistry. 2006;45:10747–10758. doi: 10.1021/bi060540u. [DOI] [PubMed] [Google Scholar]
- 31.Covey DF. ent-Steroids: novel tools for studies of signaling pathways. Steroids. 2009;74:577–585. doi: 10.1016/j.steroids.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannock DA, et al. Effects of natural and enantiomeric cholesterol on the thermotropic phase behavior and structure of egg sphingomyelin bilayer membranes. Biophys J. 2003;84(2 Pt 1):1038–1046. doi: 10.1016/S0006-3495(03)74920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westover EJ, et al. Cholesterol depletion results in site-specific increases in epidermal growth factor receptor phosphorylation due to membrane level effects. Studies with cholesterol enantiomers. J Biol Chem. 2003;278:51125–51133. doi: 10.1074/jbc.M304332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gale SE, et al. Side chain oxygenated cholesterol regulates cellular cholesterol homeostasis through direct sterol-membrane interactions. J Biol Chem. 2009;284:1755–1764. doi: 10.1074/jbc.M807210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rominger CM, et al. Evidence for allosteric interactions of antagonist binding to the smoothened receptor. J Pharmacol Exp Ther. 2009;329:995–1005. doi: 10.1124/jpet.109.152090. [DOI] [PubMed] [Google Scholar]
- 36.Kenakin TP. A Pharmacology Primer. 3. Elsevier Inc; 2009. [Google Scholar]
- 37.Kim J, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzgerald JB, et al. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 39.Rohatgi R, et al. Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torocsik D, Szanto A, Nagy L. Oxysterol signaling links cholesterol metabolism and inflammation via the liver X receptor in macrophages. Mol Aspects Med. 2009;30:134–152. doi: 10.1016/j.mam.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Brown AJ. Cholesterol, statins and cancer. Clin Exp Pharmacol Physiol. 2007;34:135–141. doi: 10.1111/j.1440-1681.2007.04565.x. [DOI] [PubMed] [Google Scholar]
- 42.Cooper MK, et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 43.Lin YY, Welch M, Lieberman S. The detection of 20S-hydroxycholesterol in extracts of rat brains and human placenta by a gas chromatograph/mass spectrometry technique. J Steroid Biochem Mol Biol. 2003;85:57–61. doi: 10.1016/s0960-0760(03)00137-7. [DOI] [PubMed] [Google Scholar]
- 44.Mijares A, et al. Studies on the C-20 epimers of 20-hydroxycholesterol. J Org Chem. 1967;32:810–812. doi: 10.1021/jo01278a066. [DOI] [PubMed] [Google Scholar]
- 45.Ruprecht JJ, et al. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts KD, Bandy L, Lieberman S. The occurrence and metabolism of 20 alpha-hydroxycholesterol in bovine adrenal preparations. Biochemistry. 1969;8:1259–1270. doi: 10.1021/bi00831a061. [DOI] [PubMed] [Google Scholar]
- 48.Lutjohann D, et al. Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci USA. 1996;93:9799–9804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.