Abstract
Nucleolin is an abundant multifunctional nucleolar protein with defined roles in ribosomal RNA processing, RNA polymerase I-catalyzed transcription, and the regulation of apoptosis. Earlier we reported that human nucleolin binds to the p53-antagonist Hdm2 as determined by reciprocal co-immunoprecipitation assays using cell lysates. We also demonstrated that nucleolin antagonizes Hdm2-mediated degradation of p53. Here, we identify specific domains of nucleolin and Hdm2 proteins that support mutual interaction and investigated the implications of complex formation on p53-ubiquitination and protein levels. Our data indicate that the nucleolin N-terminus as well as the central RNA-binding domain (RBD) are predominantly involved in binding to Hdm2. The nucleolin RBD robustly bound to the NLS/NES (nuclear localization and export signals) domain of Hdm2 in vitro, while N-terminus of nucleolin preferentially associated with the Hdm2 RING domain expressed in cells. We further demonstrate that the C-terminal GAR (Glycine-Arginine Rich) domain of nucleolin serves as the predominant binding domain for direct interaction with p53. While over-expression of nucleolin or its various domains had no significant effect on Hdm2 auto-ubiquitination, the nucleolin RBD antagonized the Hdm2 E3 ligase activity against p53, leading to p53 stabilization. Conversely, the adjacent GAR domain of nucleolin interacted with p53 causing a modest stimulatory effect on p53 ubiquitination. These data suggest that changes in nucleolin conformation can alter the availabilities of such domains in vivo to modulate the overall impact of nucleolin on Hdm2 activity and hence on p53 stability.
Keywords/Indexing: Nucleolin, MDM2, p53, Ubiquitination, Cancer therapy
Introduction
The Hdm2 oncoprotein (with non-human homologs referred to as Mdm2) is a well-studied E3 ubiquitin ligase that targets the p53 tumor suppressor protein for proteasomal degradation. Under normal physiological conditions, Hdm2-mediated p53 ubiquitination maintains low levels of p53 protein by triggering its degradation. Under conditions of cellular stress such as DNA damage or persistent oncogenic growth signals, the activity of Hdm2 towards p53 is reduced, raising p53 levels. This in turn causes growth arrest, the induction of apoptosis or senescence, dependent on the cellular milieu, in order to combat the stress conditions. Because the Hdm2-p53 connection is involved in life-and-death decisions for the cell, the interplay between these two factors is not surprisingly subject to regulation from various factors. A number of such factors are involved in ribosome structure or biogenesis including the large ribosomal subunits L5, L11 and L23 [1–6] as well as non-ribosomal proteins such as nucleophosmin/B23 [7–10], and nucleolin [11,12].
Nucleolin is an abundant nucleolar protein that stimulates the initial step of ribosomal RNA (rRNA) processing [11], and exerts both positive and negative effects on rDNA transcription, possibly through the FACT-like histone chaperone activity of nucleolin [13]. Over-expression of nucleolin is seen in many tumor types including leukemias (in which 25-fold higher nucleolin protein levels also stabilize Bcl-2 mRNA, increasing the resistance to apoptosis) [14], hepatocellular carcinomas [15], and breast tumors [16] (reviewed in [17]). Release of nucleolar stress factors (e.g. B23, ARF-Alternate Reading Frame protein and nucleolin) into the nucleoplasm upon various cellular stresses including DNA damage leads to p53-stabilization [8,18–22]. In contrast, binding of ribosomal protein L26 (RPL26) and nucleolin to the 5' untranslated region (UTR) of p53 mRNA, controls p53 translation and induction upon DNA damage [23]. Nucleolin levels can increase up to 4-fold in response to c-Myc or a heightened cellular proliferation rate [24,25]. We reported earlier that analogous 2- to 4-fold alterations of nucleolin protein levels cause a similar increase in p53 protein levels [12]. These findings have led to the proposal that heightened nucleolin levels found in hyper-proliferating cells provide a feedback mechanism to stimulate p53 and hence slow growth.
Although nucleolin and p53 physically interact [18], the increase in p53 levels occurs through nucleolin binding and inhibiting Hdm2 activity towards p53 [12]. The molecular basis for this effect remains dimly understood. In this report, we examined the molecular interactions between nucleolin and Hdm2 in both cell-free systems and in vivo. Here we demonstrate that specific domains of nucleolin and Hdm2 proteins are involved in mutual interactions. The results show novel associations between nucleolin-Hdm2 as well as nucleolin-p53, and add a unique mechanism into the regulation of p53 stability that is fine-tuned by availability of specific nucleolin domains through possible modifications in nucleolin conformation.
Results
In our earlier study we reported interactions of full-length nucleolin and Hdm2 proteins in vivo using reciprocal co-immunoprecipitation assays [12]. In order to map the interacting domains of nucleolin and Hdm2, we generated various constructs that expressed nucleolin truncation mutants (Fig. 1A). These constructs also allow the expression of GST- or GFP-tagged nucleolin in either a yeast or mammalian expression system. Similarly, GST-tagged Hdm2 FL and Hdm2-truncation mutants shown schematically in Fig. 1B, were produced using a prokaryotic expression system [8].
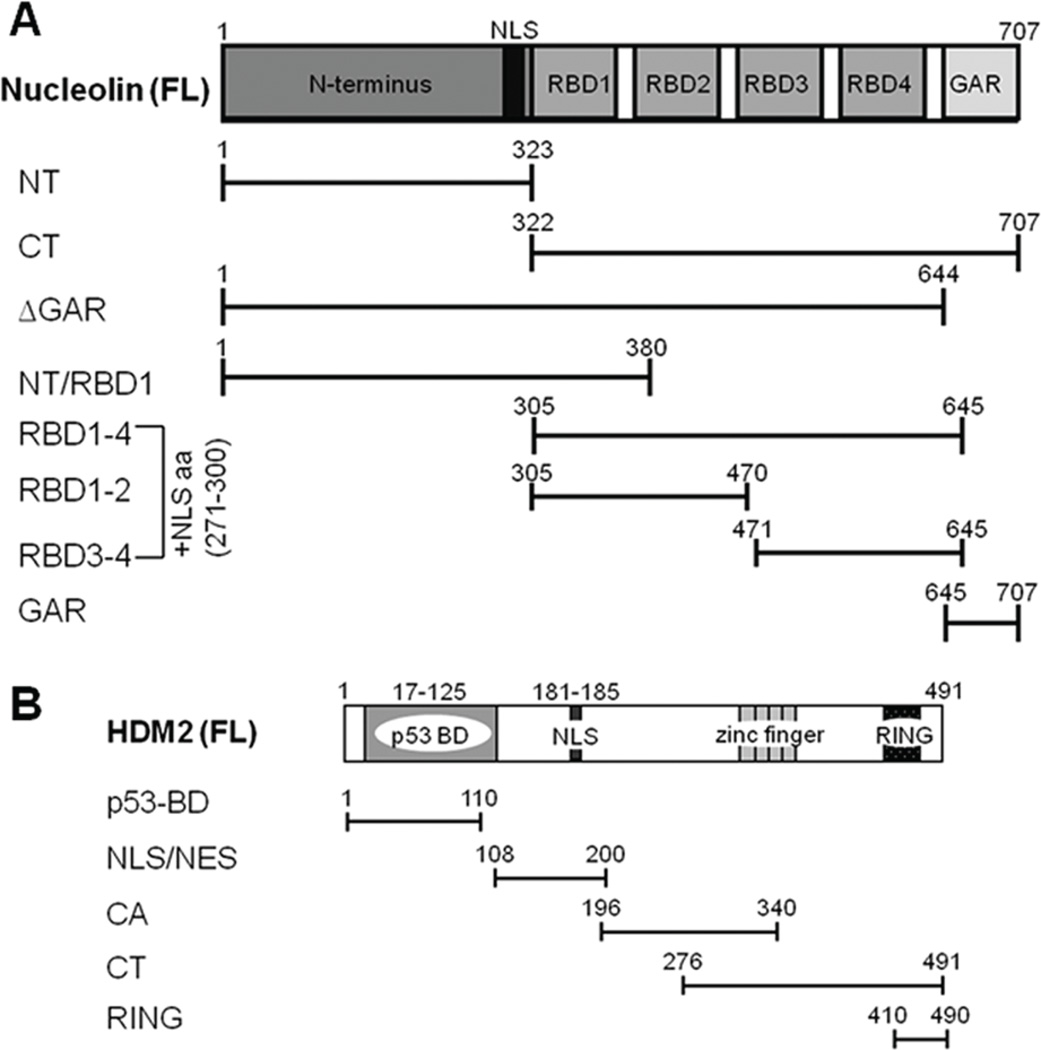
Fig. 1. Schematics of nucleolin and Hdm2 full-length (FL) and their domains.
A, The nucleolin FL and truncation mutant constructs that were used in these studies. These constructs express either a GST- or GFP tag (not shown) at the N-terminus. To support nuclear expression of RBD constructs, a nuclear localization signal (NLS) sequence was added to the N-terminus of the RBD sequence. B, Schematic showing the Hdm2 constructs used in these studies. For in vitro studies, all Hdm2 variants contained a GST-tag at the N-terminus (not shown).
Identification of multiple domains of nucleolin that support association with Hdm2
First, in order to establish direct nucleolin and Hdm2 interactions, we used an in vitro Far-Western approach. Specific GST-tagged constructs that span all the different domains on nucleolin (e.g. NT, CT and GAR domains) were selected and expressed in yeast (see Materials and Methods). The purified proteins were subjected to SDS-PAGE and transferred to nitrocellulose. For detecting direct binding, membrane-bound nucleolin variants (Fig. 1A) were incubated with purified GST-Hdm2-FL protein. Following extensive washing, the associated Hdm2 was detected using anti-Hdm2 antibodies (Fig. 2A). Hdm2 was seen to have robust association with nucleolin-FL (lane 2) and p53 (lane 8), the latter factor being a positive control. Although lane 8 indicates a cleaved p53, we have identified positive interactions between Hdm2 and p53 in multiple experiments with GST-tagged p53 (Fig. S1). Hdm2 also showed a strong interaction with the individual N-terminal (NT; lane 3) and C-terminal (CT; lane 4) truncation products and with larger constructs containing the NT and RBD regions (e.g., NT/RBD1; lane 5 and ΔGAR; lane 6). Only the extreme C-terminal nucleolin-GAR domain showed minimal interaction with Hdm2 (lane 7), as did the GST (lane 1) and BSA (lane 9) negative controls. The amounts of loaded GST-nucleolin fusion proteins were visualized on a parallel blot and probing directly with anti-GST antibodies (Fig. 2B). These data indicate that both the nucleolin NT and central RBD domains can independently bind Hdm2.
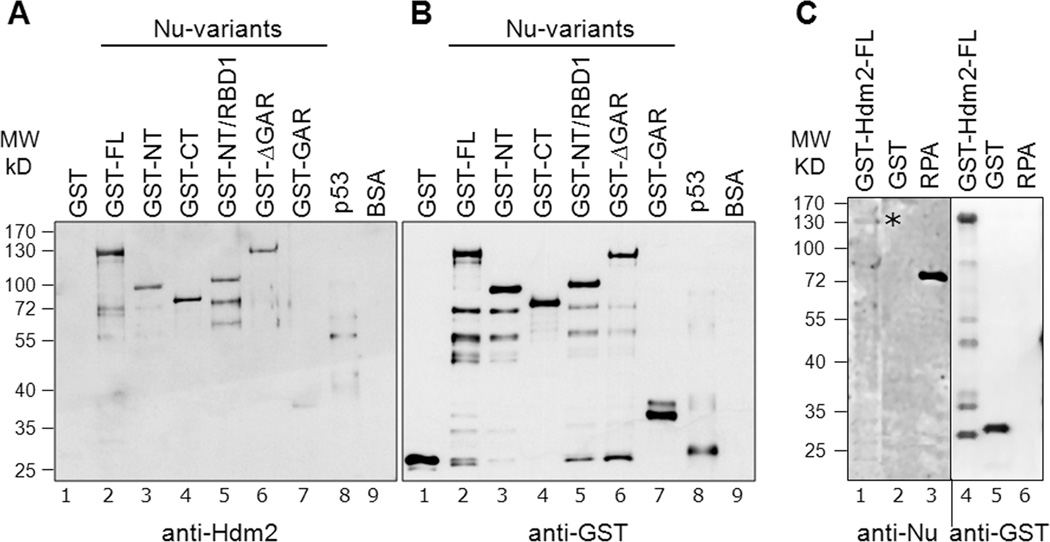
Fig. 2. Elucidation of direct nucleolin-Hdm2 interactions and identification of Hdm2-interacting domains on nucleolin.
A, Equivalent amounts (400 ng) of GST-tagged nucleolin FL (lane 2) or nucleolin domains (lanes 3 to 7) were subjected to SDS-PAGE. As negative controls for the nucleolin-Hdm2 interaction, GST alone (lane 1) and BSA (lane 9) were used. Cleaved p53 (after removal of GST-tag, lane 8) served as a positive control. The separated proteins were then electrotransferred to a nitrocellulose membrane for Far-Western analysis. The membrane was probed with purified Hdm2 protein (0.2 µg/ml), as described in Materials and Methods, and the bound Hdm2 detected by a monoclonal anti-Hdm2 antibody (SMP14). B, Equivalent loading of the GST-nucleolin fusion proteins was confirmed by running a parallel blot and probing directly with anti-GST antibodies. The multiple lower molecule bands visualized could be attributed to self-cleaving properties of nucleolin at the C-terminus [42,43]. These data indicate that both the nucleolin NT and RBD domains can independently bind Hdm2. C, Nucleolin interaction with Hdm2 in reciprocal Far-Western. Equivalent amounts (400 ng) of GST-tagged Hdm2 FL (lane 1), GST (lane 2), and non-GST-tagged RPA (lane 3) were subjected to SDS-PAGE and then electrotransferred to a nitrocellulose membrane for subsequent Far-Western analysis. The membrane was probed with purified nucleolin FL (isolated from yeast with a GST tag), and the bound nucleolin was then detected using an anti-nucleolin antibody (MS-3). The location of bound nucleolin is denoted with an asterisk. Equal loading of GST and GST-Hdm2 FL was confirmed by running a parallel blot, and probing directly for anti-GST antibodies (RPA is not GST-tagged and hence not recognized by anti-GST). Although each three lane group was originally present on the same blot, they were spliced adjacent to each other for this figure.
In a reciprocal Far-Western blot, we then assayed the ability of nucleolin-FL to physically interact with Hdm2. After bacterial expression, we isolated GST-tagged Hdm2 FL and other Hdm2 truncation mutants as shown schematically in Fig. 1B. The membrane-bound Hdm2 was then probed with nucleolin-FL isolated from yeast, and the interacting nucleolin subsequently detected by an anti-nucleolin antibody (Fig. 2C). A relatively weak nucleolin signal was observed associated with Hdm2-FL (lane 1), as well as the Hdm2 p53-binding domain (data not shown). As controls, nucleolin was found to bind replication protein A (RPA-lacking a GST tag, lane 3), as shown previously [26], but did not associate with GST alone (lane 2). Equal loading of GST and GST-Hdm2 FL was confirmed by running a parallel blot, and probing the membrane for GST (lanes 4–6).
Together, these Far-Western data substantiate our earlier co-immunoprecipitation data demonstrating that full-length nucleolin and Hdm2 interact in cells, with these new data suggesting that these interactions are direct. Because many of these domains are highly acidic (e.g., nucleolin-NT, Hdm2-central-acidic domain) or basic (e.g., nucleolin-GAR), we sought to provide further evidence for the interactions between specific domains, using purified Hdm2 domains on beads.
Elucidation of specific domains on nucleolin sufficient to interact with Hdm2
We next identified the specific interacting domains on nucleolin and Hdm2, examining the ability of nucleolin to associate with GST-tagged Hdm2 domains on beads. We created an additional construct that clearly defined the RBD1–4 element within the CT domain (Fig. 1A). Because the expression of nucleolin in yeast is poor (and bacterially-expressed nucleolin is insoluble; data not shown), we expressed GFP-tagged nucleolin variants in human H1299 cells (Fig. 3A). Representative GST-tagged Hdm2 variants utilized in the binding reactions were identified using anti-GST antibodies (Fig. 3B).
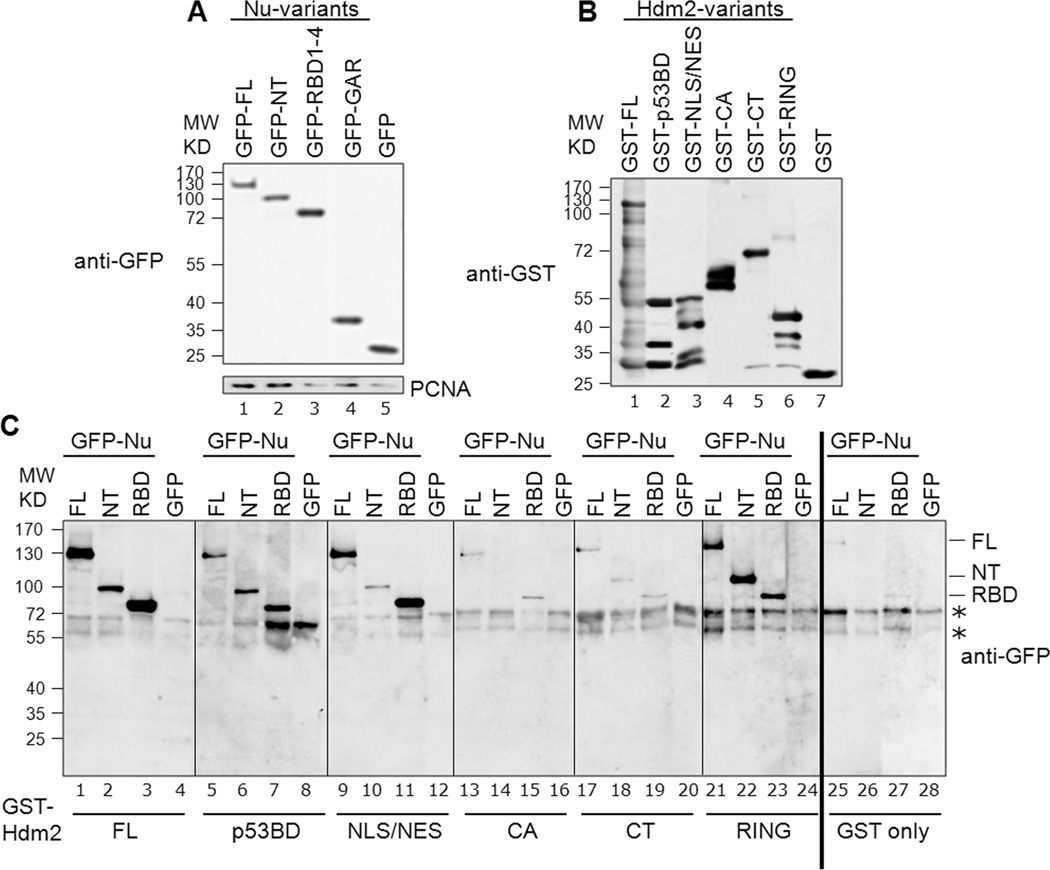
Fig. 3. Determination of specific domains involved in nucleolin-Hdm2 interactions using cell lysates.
A, Representative expression of the GFP-tagged nucleolin constructs: The various nucleolin constructs, as indicated, were transfected into H1299 cells. Lysates were prepared from these cells and examined by Western blotting for GFP. PCNA served as a loading control. B, Representative corresponding GST-tagged Hdm2 variants utilized in a given binding reaction: For a given binding reaction, the same blot was analyzed following stripping of anti-GFP and reprobing with anti-GST. Only one representative lane for each is shown and rearranged for easier display. C, Identification of interacting nucleolin and Hdm2 domains. Binding assays were performed using equivalent amounts of GST-tagged Hdm2 variants bound to glutathione-Sepharose beads. These constructs were Hdm2 FL (lanes 1–4), p53-BD (lanes 5–8), NLS/NES (lanes 9–12), CA (lanes 13–16), CT (lanes 17–20) and RING (lanes 21–24). GST alone was used as the negative control (lanes 25–28). The bead-bound Hdm2 variants were incubated with 1 mg of protein lysates from H1299 cells expressing GFP-tagged nucleolin FL, NT, RBD1–4 (RBD), or GFP alone, as indicated. The presence of nucleolin (full-length and truncation domains) was determined by subjecting the bead-bound material to Western analysis, probing for GFP. The non-specific signals seen in the GFP blot are indicated by asterisks. All the blots for this figure were exposed to film simultaneously and developed for identical time periods. After scanning, the particular sections were cropped and rearranged for easier display.
Cell lysates containing the nucleolin variants were incubated with the purified GST-Hdm2 domains on glutathione beads. The ability of nucleolin variants to associate with these domains was determined by subjecting the bead-bound material to SDS-PAGE and Western blotting using anti-GFP antibodies (Fig. 3C). First testing Hdm2-FL, significant association with nucleolin-FL (lane 1), and the NT (lane 2) and RBD1–4 (lane 3, labeled as RBD) domains was found, while no binding of GFP alone was detected (lane 4). The nucleolin GAR domain was found to associate with all Hdm2 constructs as well as with GST (data not shown). Even though there is apparent stronger association of nucleolin GAR with Hdm2 than with GST, we conclude the GAR binding to be artifactual. The nucleolin-FL, NT, and RBD1–4 variants were not seen to have any substantial interaction with GST alone (lanes 25–27), demonstrating the specificity of the interaction.
Testing individual Hdm2 domains, we observed strong interactions of nucleolin-FL with Hdm2 NLS/NES (lane 9) and RING (lane 21) domains, and weaker association with the p53BD (lane 5). Testing specific nucleolin domains, the RBD1–4 robustly interacted with Hdm2-NLS/NES (lane 11) and significantly associated with Hdm2-p53BD (lane 7). In contrast, nucleolin-NT had the strongest apparent interaction with Hdm2 RING domain (lane 22) and a marginal interaction with the Hdm2-p53BD (lane 6). We did not find appreciable association of nucleolin, or its truncation domains, to the Hdm2 central acidic (CA) domain (lanes 13–15), nor to the CT domain (lanes 17–19). Similarly, no binding of GFP alone was detected to the various Hdm2-domains (lanes 8, 12, 16, 20 and 24) or GST alone (lane 28). Note that we observed binding of nucleolin-NT to Hdm2-RING, even though nucleolin association with the CT domain was minimal. These data suggest that the conformation of the CT can inhibit nucleolin binding to the contained RING entity, perhaps by shielding interacting features of the RING domain from contact with nucleolin.
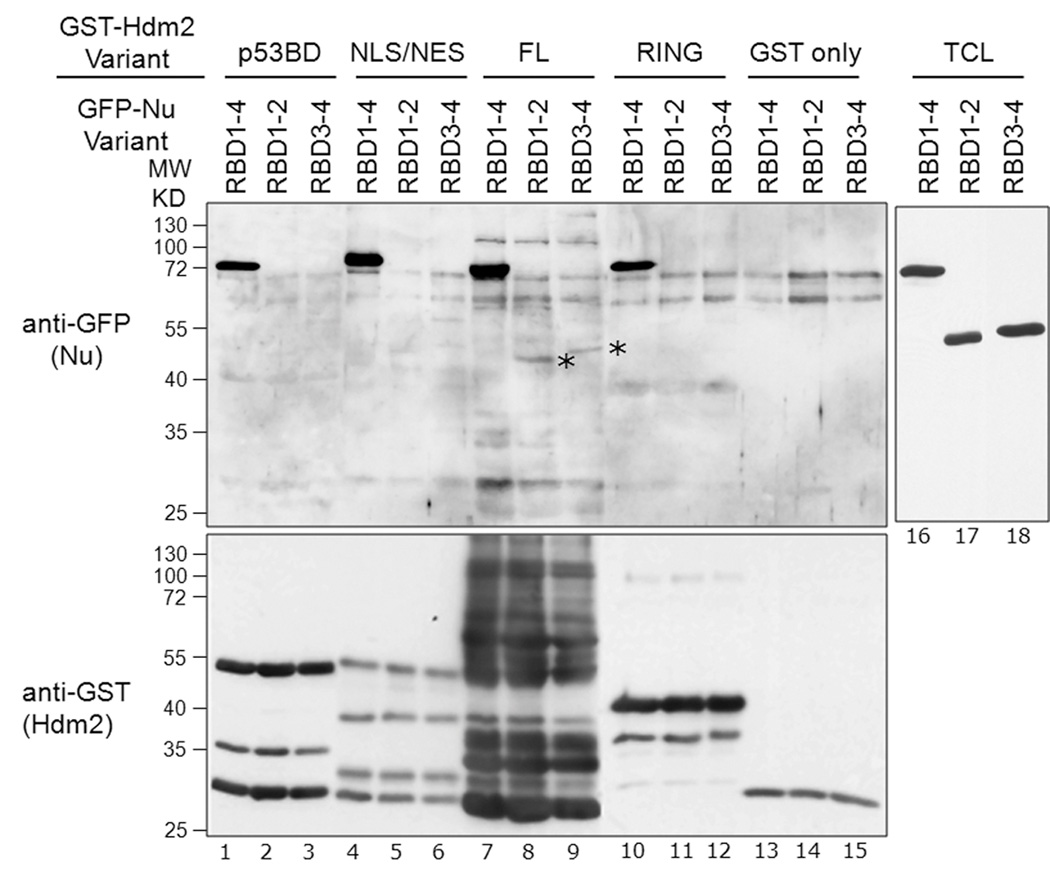
Because the nucleolin RBD contains four RNA-binding motifs, we determined if the RBD could be subdivided into smaller domains that were competent in binding Hdm2. The nucleolin RBD1–4 construct was therefore halved into two fragments that contained either RNA-binding motifs 1/2 (RBD1–2) or 3/4 (RBD3–4; see Fig. 1A). These subdomains, along with RBD1–4, were expressed as GFP-tagged proteins in H1299 cells and the resulting cell lysates were used for binding assays, as above. Surprisingly, the smaller RBD1–2 and RBD3–4 were unable to bind significantly to any of the Hdm2 constructs that were bound by RBD1–4, apart from a barely detectable interaction with FL Hdm2 (Fig. 4, lanes 8–9, denoted by asterisks). Thus, we did not identify a smaller RBD region capable of binding Hdm2. Because RNA-binding motifs do not generally function as protein-binding modules, it is possible that the presence of the inter-motif linker segments (e.g. between RBD2 and RBD3) serve as Hdm2-interacting elements.
Fig. 4. Subdomains of nucleolin RBD1–4 are unable to bind Hdm2.
A, Dissection of nucleolin RBD to identify subdomains sufficient to interact with Hdm2: GFP-tagged nucleolin RBD1–4, RBD1–2 and RBD3–4 constructs were transiently expressed in H1299 cells. Lysates were prepared and 1 mg protein was used in each binding assay containing equivalent amounts of purified GST-tagged Hdm2; p53-BD (lanes 1–3), NLS/NES (lanes 4–6), FL (lanes 7–9), RING (lanes 10–12), or GST alone (lanes 13–15), each bound to glutathione-Sepharose beads. After washing, the bead-bound material was separated by SDS-PAGE, and then analyzed by Western for GFP (upper panel) or GST (lower panel). Total cell lysates (TCL) were also Western blotted for GFP, showing equivalent expression of each RBD variant (lanes 16–18). These data indicate that only RBD1–4 showed robust binding to Hdm2-FL (lane 7) or the Hdm2 truncation domains (lanes 1, 4, and 10). Barely detectable binding of nucleolin-RBD1–2 and nucleolin-RBD3–4 domains to Hdm2-FL was seen (indicated by asterisks, lanes 8–9).
In conclusion, our data suggest that the central nucleolin RBD1–4 element, most strongly interacts with the Hdm2 NLS/NES region, in addition to making significant contacts to the p53-BD and RING domains of Hdm2. Furthermore, the nucleolin-NT associates strongly with the Hdm2 RING domain. These interaction experiments are summarized in Table 1.
Table 1.
Summary of nucleolin-Hdm2 interactions
| Hdm2 Construct |
Nucleolin | |||||
|---|---|---|---|---|---|---|
| FL | NT | RBD1–4 | RBD1–2 | RBD3–4 | GFP | |
| FL | +++ positive |
++ positive |
+++ positive |
+/− | +/− | − negative |
| p53-BD | ++ | + | ++ | − | − | − |
| NLS/NES | +++ | +/− | +++ positive |
− | − | − |
| CA | − | − | − | n.d. | n.d. | − |
| CT | − | − negative |
− | n.d. | n.d. | − |
| RING | ++ | +++ negative |
++ | − | − | − |
| GST | − negative |
− | − | − | − | − |
The strength of the apparent interactions, derived from GST-binding assays using cell lysates expressing GFP-nucleolin (Figure 3 and 4), is indicated as follows: − none; −/+ v. weak; + weak, ++ moderate, +++ strong. Interactions that were not determined are indicated as n.d. The results from in vitro binding assays with in vitro translated nucleolin domains with GST-tagged purified Hdm2 proteins (Fig. 6) are listed as “positive” or “negative” binding where indicated.
Nucleolin interacts with p53 predominantly by its CT-GAR domain
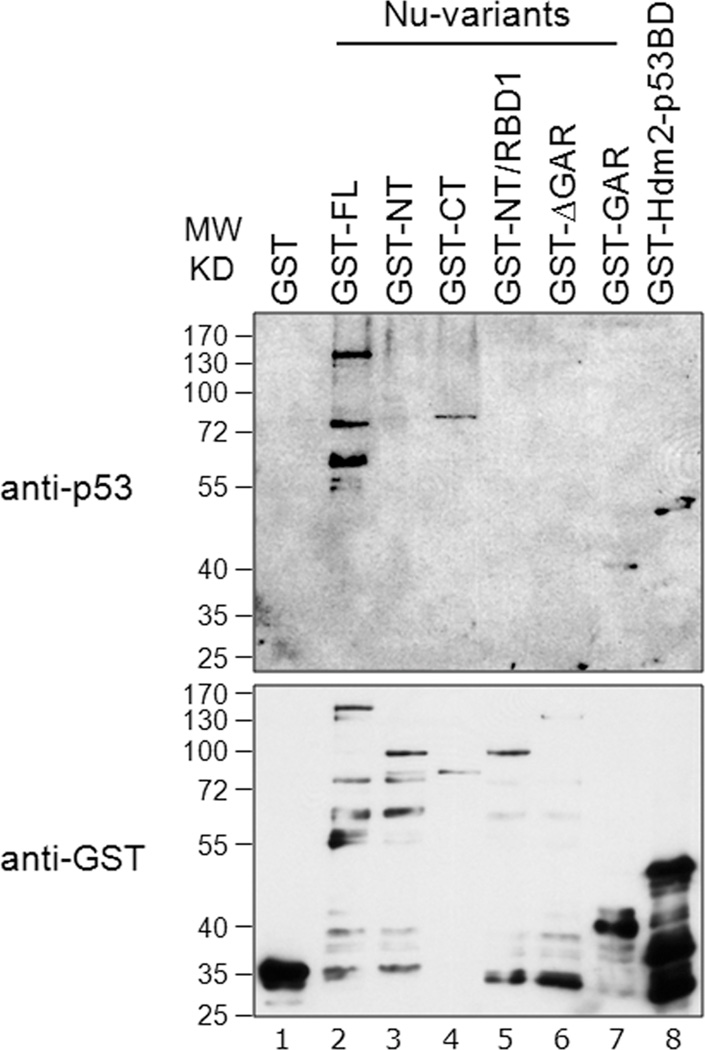
Because nucleolin interacts with p53 in addition to Hdm2 [18], we wished to identify the nucleolin domain that interacts with p53. We had previously observed that the extreme C-terminal 30 residues of p53 protein were necessary both for association with nucleolin, and to stimulate nucleolin mobilization from the nucleolus following genotoxic stress [18]. However, the nucleolin domain involved in p53 binding was not identified. Using Far-Western analysis, we electrotransferred various GST-tagged nucleolin domains onto a nitrocellulose membrane, and then incubated the membrane with purified p53 protein. Probing for the presence of bound p53, we observed positive signals with nucleolin-FL and the CT construct that includes the RBD1–4 and GAR domains (Fig. 5, lanes 2 and 4, respectively). The nucleolin NT (lane 3), NT/RBD1 (lane 5) and ΔGAR (lane 6) constructs, all lacking the GAR domain, failed to show any detectable p53 binding. In contrast, p53 was observed to bind the GAR domain alone (lane 7) and, as expected, to the p53 binding-domain of Hdm2 (lane 8). We therefore conclude that the nucleolin GAR domain is both necessary and sufficient to support interaction with p53 in vitro. Interestingly, our previous studies also found that overexpression of GAR reduced p53 protein levels [27]. This idea is further supported by our finding that GAR domain directly interacts with p53 and not through Hdm2 resulting in an increased Hdm2-mediated p53 ubiquitination in vitro (see Figs. 6B, lane 9; 7B, lane 9; and S2A, lane 7, below).
Fig. 5. Nucleolin GAR interacts with p53.
Equivalent amounts (200 ng) of GST-tagged nucleolin-FL (lane 2) or truncation domains (lanes 3 to 7) were subjected to SDS-PAGE. After transfer to a nitrocellulose membrane, the membrane was probed with purified GST-p53 protein (0.2 µg/ml) in a Far-Western assay (upper panel, lanes 1 to 8). The bound p53 was then visualized using a monoclonal anti-p53 antibody (DO-1). To visualize GST-tagged proteins, a parallel blot was run and probed directly for anti-GST antibodies (lower panel, lanes 1 to 8). GST alone (lane 1) and GST-tagged Hdm2 p53-BD (lane 8) were used as negative and positive binding controls, respectively.
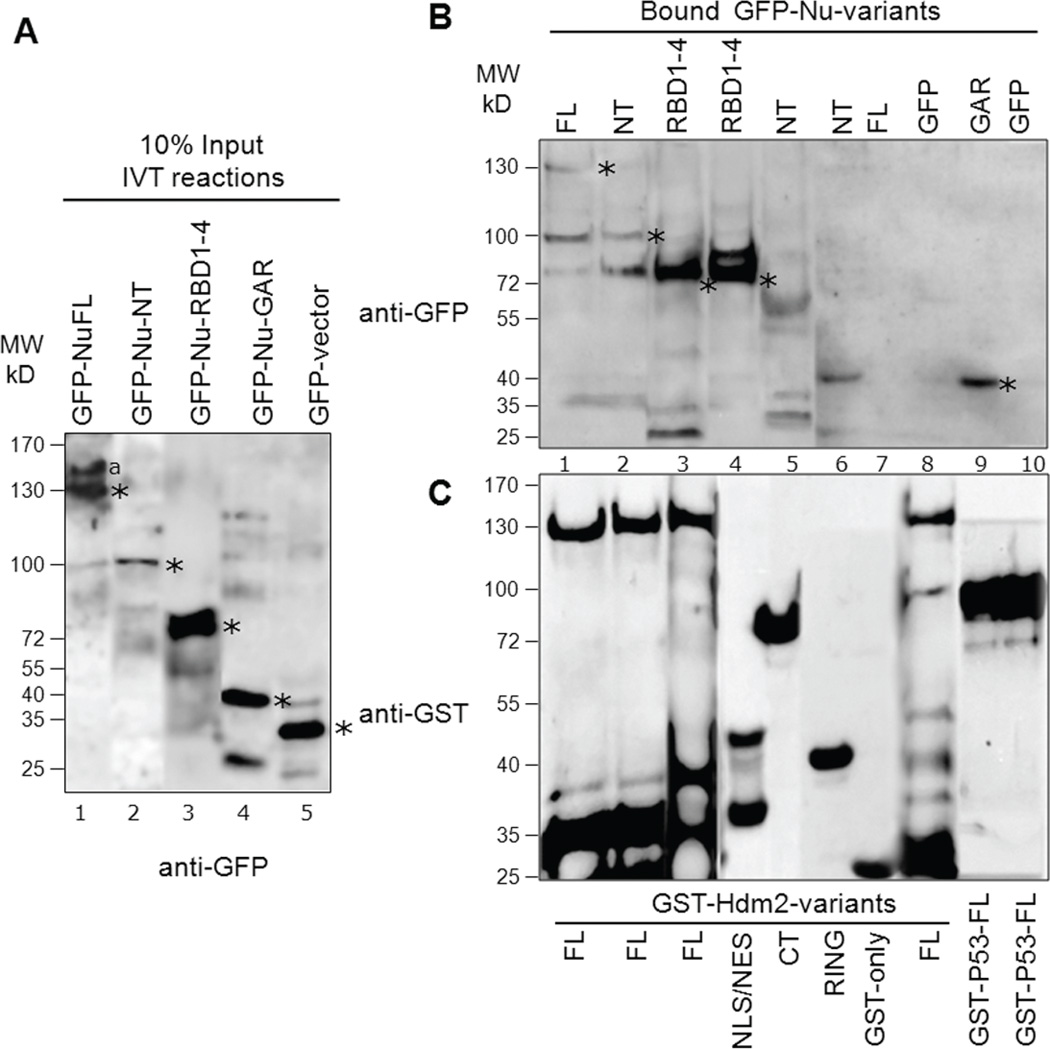
Fig. 6. Nucleolin directly interacts with Hdm2 and p53 using distinct domains in vitro.
In vitro translated (IVT) GFP-tagged proteins (nucleolin variants or GFP alone) were incubated with purified GST-tagged proteins (Hdm2, p53 or GST alone) to test association. A, A representative blot indicating the translated GFP-nucleolin variants: 10% of input levels used in binding reactions shown in panel B were analyzed by Western for GFP. We repeatedly observed lesser IVT product for nucleolin-NT (lane 2). The location of expected molecular size of each nucleolin-variant and GFP is denoted with an asterisk. The higher molecular weight product (lane 1, denoted by “a”) could be due to partial translational read-through of the normal mRNA stop codon during the IVT reaction. Lower molecular bands observed could be due to proteolysis of the translated product. B, Identification of nucleolin domains that interact with Hdm2 and p53: Bound fractions from binding reactions were analyzed by SDS-PAGE and Western blotting using anti-GFP. GFP-Nu-FL, NT and RBD1–4 directly interact with GST-Hdm2-FL (lanes 1–3). RBD1–4 also strongly interacts with GST-Hdm2 NLS/NES domain (lane 4) while nucleolin-NT domain does not interact with either Hdm2-CT or RING domain (lanes 5–6). As negative controls, the translated GFP-nucleolin-FL did not interact with GST alone (lane 7), nor did GFP significantly bind to GST-Hdm2-FL (lane 8). On the other hand, nucleolin-GAR domain strongly interacts with GST-p53-FL (lane 9) while no interaction was detected for the negative control GFP with p53 (lane 10). Detectable binding of nucleolin-variants to Hdm2-FL or p53-FL are indicated by asterisks. C, The blots were subsequently stripped and reprobed with anti-GST to reveal the loading of the purified GST-tagged proteins in the corresponding binding reaction. The lower molecular weight products observed indicate that some proteolysis of translated products (blot B) or GST-tagged constructs (blot C) occur, despite of the presence of protease inhibitor in the binding reactions. After scanning, the particular sections were cropped and rearranged for easier display.
Nucleolin directly interacts with Hdm2 and p53 by distinct domains in vitro
To rule out the possibility that an unknown factor associating with nucleolin in the H1299 cell lysates could significantly influence the detected nucleolin-Hdm2 interactions, we performed a more direct binding assay. We prepared GFP-tagged nucleolin variants using a coupled transcription/translation reaction, and tested the ability of these proteins to associate with purified GST-tagged Hdm2 proteins on beads. We achieved significant expression of each nucleolin variant using this cell-free system (Fig. 6A). We did however repeatedly observe less translation product for nucleolin-NT (Fig. 6A, lane 2), possibly due to the unstable nature of this protein domain. We tested each of the nucleolin species for their ability to associate with purified GST-tagged Hdm2-FL, and found that a significant level of interaction of nucleolin-FL, NT, and RBD1–4 with Hdm2 (Fig 6B, lanes 1–3). These data are in accordance with the aforementioned binding experiments using cell-lysates and provide compelling evidence that the nucleolin-Hdm2 interaction is direct.
We also tested a subset of the nucleolin truncation constructs for their ability to associate with Hdm2 domains. Most importantly, we observed that nucleolin-RBD1–4 preferentially binds to the NLS/NES domain of the Hdm2 (Fig. 6B, lanes 4), as found above. We however failed to observe any significant binding between nucleolin-NT and Hdm2 domains (e.g., CT and RING, Fig. 6B, lanes 5–6). This negative binding could be due to less nucleolin-NT input per reaction or because the translated NT domain is not stably folded to provide functional interactions in vitro. As negative controls, the translated GFP-nucleolin-FL did not interact with GST alone, nor did GFP significantly bind to GST-Hdm2-FL (Fig. 6B, lanes 7–8). As anticipated, we observed a significant interaction between the GFP-tagged nucleolin-GAR domain and GST-p53FL, while no interaction was detected for the negative control GFP with p53 (Fig. 6B, lanes 9–10). Reprobing the same blot with anti-GST antibodies revealed similar loading of each of the purified GST-tagged proteins in the corresponding binding reaction (Fig. 6C).
These direct binding assays cause us to conclude that nucleolin interacts with Hdm2 through its NT and RBD1–4 domains, with RBD1–4 strongly associating with the Hdm2 NLS/NES domain in vitro. The results from nucleolin-Hdm2 direct binding experiments are summarized in Table 1 as “positive” or “negative” interactions. Furthermore, nucleolin-GAR domain is sufficient for direct interaction with p53.
Nucleolin does not significantly alter Hdm2 auto-ubiquitination
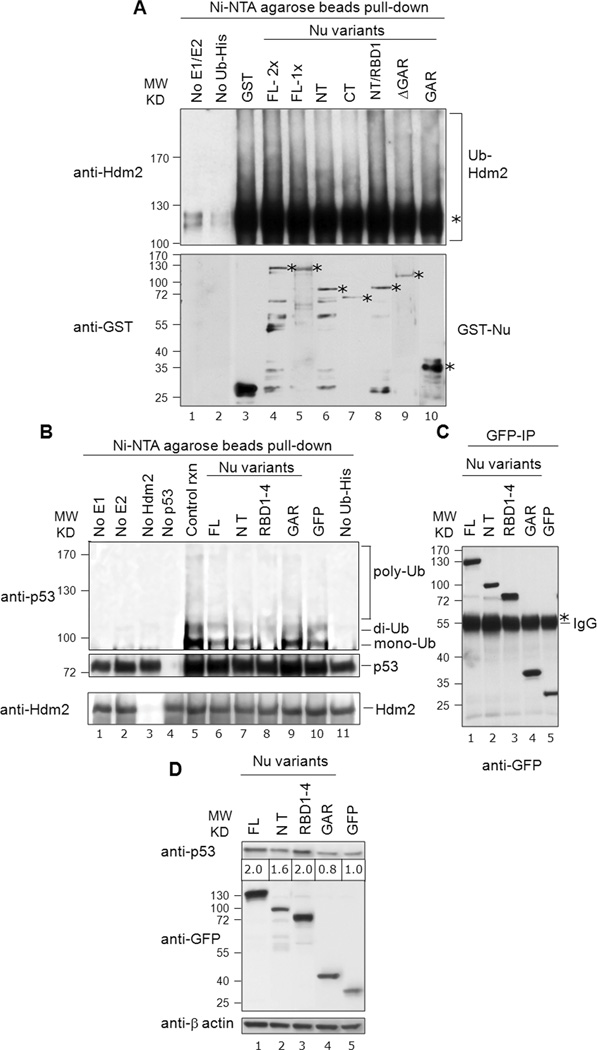
Because we identified multiple domains on both nucleolin and Hdm2 that could support association, we next tested the functional significance of these interactions. We focused on the Hdm2 E3 ubiquitin ligase activity, initially analyzing Hdm2 auto-ubiquitination in vitro. Purified Hdm2 was incubated with required reaction components (e.g., E1 and E2) and His-tagged ubiquitin (Ub-His). The ubiquitinated species were then purified by Ni2+-affinity chromatography and analyzed by Western for Hdm2 (Fig. 7A, upper panel). We observed robust formation of apparent mono-ubiquitinated Hdm2 (lane 3 to 10; marked with *), as well as higher-molecular weight poly-ubiquitinated species. Auto-ubiquitination was dependent on the presence of both E1 and E2 (lane 1) and Ub-His (lane 2). Addition of nucleolin-FL (lanes 4 and 5), as compared to the control reaction lacking nucleolin (lane 3) did not significantly alter the amount of poly-ubiquitinated Hdm2. Although slight changes to Hdm2 auto-ubiquitination were observed in the presence of various truncated nucleolin proteins (lanes 6 to 10), on the whole, no significant effects of any nucleolin variant on Hdm2 auto-ubiquitination were noted. The amounts of loaded GST-nucleolin fusion proteins were visualized on a parallel blot and found to be generally similar (Fig. 7A, bottom panel). These in vitro auto-ubiquitination results are consistent with the lack of effect on Hdm2 auto-ubiquitination caused by overexpression of nucleolin-FL in vivo, reported previously [12]. Thus, we conclude that nucleolin does not significantly alter Hdm2 auto-ubiquitination.
Fig. 7. Nucleolin RBD1–4 selectively inhibits p53 ubiquitination in vitro, but not Hdm2 auto-ubiquitination.
A, Nucleolin does not significantly alter Hdm2 auto-ubiquitination: Nucleolin FL or truncation domains (150 ng; 1×) were analyzed for their effects on Hdm2 auto-ubiquitination (upper panel, lanes 4 to 10), using His-tagged ubiquitin. Reaction products were purified on Ni-NTA-agarose beads (i.e., to isolate the ubiquitinated products), and then subjected to SDS-PAGE and Western analysis for Hdm2. The mono-ubiquitinated Hdm2 is marked with an asterisk. Control reactions lacked the E1/E2 ubiquitination factors (lane 1) or Ub-His (lane 2). These data indicate that nucleolin does not significantly alter Hdm2 auto-ubiquitination. To visualize GST-tagged nucleolin proteins, a parallel blot was prepared and probed directly for anti-GST antibodies (lower panel). The location of expected molecular size of each nucleolin-variant and GST is denoted with an asterisk. B, Nucleolin RBD1–4 significantly inhibits p53-ubiquitination: Nucleolin FL (40 ng) or truncation domains were added to ubiquitination reactions including p53. The same blot subsequently stripped and reprobed with anti-Hdm2 to reveal the equivalent loading of the purified Hdm2 in the corresponding reaction (bottom panel). Various controls are also included, as indicated (lanes 1 to 5, 10 and 11). C, Immunoprecipitated GFP-nucleolin fusion proteins on IgG beads, utilized in corresponding in vitro p53-ubiquitination reactions were visualized on a parallel blot after probing with anti-GFP antibodies. The heavy band of IgG is also visible (marked with an asterisk). D, Effects of nucleolin domain expression on p53 levels: Constructs expressing various GFP-tagged nucleolin domains were transfected into U2OS cells. Post-transfection (36 h), endogenous p53 was detected by Western using an anti-p53 (DO-1) antibody. GFP blot indicated the expression levels of each nucleolin domain while actin served as a loading control. Note that the level of p53 protein (denoted below the p53 Western blot) was normalized to β-actin expression, after quantitation with Image J software (NIH). We observe a dose-dependent effect of expression of nucleolin protein and p53 stabilization with the range of 1.2 to 2.0-fold increase in p53 protein levels (compare with Fig. S3).
The nucleolin RBD1–4 domain significantly reduces p53-ubiquitination
We next tested the effect of nucleolin domains on Hdm2-mediated p53-ubiquitination in vitro, using purified proteins. The control reaction demonstrated the robust formation of ubiquitinated p53, primarily of putative mono- and di-ubiquitinated conjugates, but also of a modest amount of poly-ubiquitinated p53 (Fig. 7B, lane 5). The specificity of the reaction was demonstrated by the lack of ubiquitination in the absence of E1 (lane 1), E2 (lane 2), Hdm2 (lane 3), and Ub-His (lane 11). The presence of nucleolin-FL reproducibly caused a marked reduction in the amount of poly-ubiquitinated p53 (compare lane 6 vs. 5). Testing individual nucleolin domains, while the NT domain was not found to have any major effects on p53 modification (lane 7), the RBD1–4 construct caused nearly a complete loss of p53 mono-ubiquitination (lane 8). In contrast, addition of the nucleolin-GAR domain caused a modest stimulation of evident mono-ubiquitinated p53 (lane 9). Even though each of the nucleolin constructs was GFP-tagged, the presence of GFP was not seen to have any detectable effect on p53 ubiquitination (lane 10). Reprobing the same blot demonstrated equivalent loading of the added Hdm2 in each reaction (Fig. 7B, bottom panel). Immunoprecipitated GFP-nucleolin fusion proteins, utilized in the corresponding p53-ubiquitination reactions in vitro, were visualized on a parallel blot using anti-GFP antibodies and also found to be present at similar levels (Fig. 7C). Similar p53-ubiquitination assays were performed using purified GST-tagged nucleolin and yielded comparable results (Fig. S2). Along with corroborating our previous p53 ubiquitination studies testing overexpression of nucleolin-FL in vivo [12], these data demonstrate that both full-length nucleolin and individual nucleolin domains can modulate Hdm2-mediated p53-ubiquitination.
Effects of nucleolin domains on p53 expression in vivo
We determined the effect of expression of the various nucleolin domains on p53 protein levels in U2OS cells (Fig. 7D), normalizing the amounts of p53 to those of β-actin. The p53 protein levels increased 2.0-fold higher in the presence of nucleolin-FL (lane 1) and as compared to the GFP control (lane 5); this finding is in line with the effects of nucleolin on p53 expression we described earlier [12]. Expression of nucleolin-RBD1–4 reproducibly increased p53 expression levels 2.0-fold (lane 3), while the GAR domain caused a slight decrease in p53 levels (lane 4). Interestingly, the NT domain that has apparent strong interactions with the Hdm2-RING domain in cells but failed to interact in vitro, had a lesser effect on p53 stabilization (lane 2). We repeated these experiments on a different p53-WT cell-line, SJSA, and found similar effects on p53 levels (Fig. S3). Combined, both experiments indicated that there is a dose-dependent effect of nucleolin protein expression and p53 stabilization. Depending upon the expression levels of various nucleolin constructs (e.g. FL, NT or RBD1–4), p53 levels were increased from 1.2 to 2.0-fold. Importantly, each of these effects was consistent with the modulatory effects of these same domains on p53 ubiquitination in vitro.
Discussion
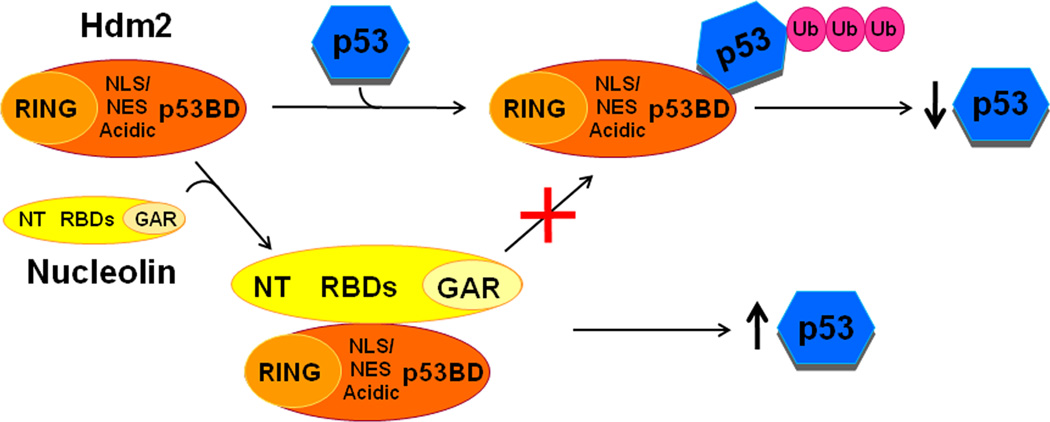
Hdm2 is a key E3 ubiquitin ligase that is critical for the regulation of p53 stability and thereby p53 activity in cells. Significant control over Hdm2 activity has been demonstrated to be exerted by factors involved in ribosomal RNA processing (nucleolin, nucleophosmin/B23), or by particular ribosome subunits (L5, L11 and L23). Here, our examination of the nucleolin-Hdm2 association finds two distinct physical interactions between nucleolin and Hdm2 that are mediated by multiple domains on each factor (schematically depicted in Fig. 8). Nucleolin RBD was found to bind directly with the Hdm2 p53-BD, NLS/NES, and RING, while nucleolin NT associate with the Hdm2 p53-BD and RING. Furthermore, these interactions can yield both positive and negative alterations in Hdm2 activity, and similar effects on p53 levels. Although the expression of the full-length nucleolin increases p53 protein levels under our growth conditions (this report; [12]), our results clearly raise the potential for nucleolin to have both positive and negative effects on Hdm2 activity, dependent on nucleolin conformation and/or modification state that enable availability of specific domains.
Fig. 8. Schematic indicating identified nucleolin:Hdm2 domains interactions and its biological significance.
Nucleolin-Hdm2 interactions by the RBD1–4 and NT domains of nucleolin presumably inhibit Hdm2-p53 interaction via its p53BD and central domain that contained the NLS/NES region. This antagonism results in decreased Hdm2-mediated p53-ubiquitination, stabilizing cellular p53 protein levels.
Past work has found that the association of nucleolin with other factors is altered both by genotoxic stress and the nucleolin phosphorylation state, presumably by causing changes to nucleolin conformation [27]. It had also been observed that the conformation of nucleolin, as examined by the sensitivity to protease digestion, was altered by the presence of poly (G) RNA [28]. These results suggest that the nucleolin conformation is sensitive to environmental conditions, and to the loss of its pre-rRNA target caused by stress. Thus, the ability of multiple domains in both nucleolin and Hdm2 to interact suggests that alterations in nucleolin conformation, which increase or decrease the exposure of particular nucleolin domains, can fine-tune the activity of Hdm2 towards p53.
The L5, L11 and L23 ribosome subunits have each been found to associate with the central acidic domain of Hdm2 [1,2,5]. These interactions also lead to loss of activity of Hdm2 towards p53, and thus p53 stabilization. Binding of the L5 and L11 proteins was also found to be dependent on the structural integrity of the C4 zinc finger region that is contained within the Hdm2 central acidic domain [29,30]. In contrast, nucleolin interacted with the strongest apparent affinity to the Hdm2 NLS/NES and RING entities, and to a lesser extent with the p53-binding domain. We did not observe any noticeable association of nucleolin with the Hdm2 central acidic domain, demonstrating that nucleolin inhibits Hdm2-mediated p53 ubiquitination using a mechanism distinct from that used by the three ribosomal factors.
Because nucleolin, or any of its isolated domains, did not detectably affect Hdm2 auto-ubiquitination in vitro, the ability of nucleolin to inhibit p53 ubiquitination does not appear to result from a general inhibition of Hdm2 activity. Instead the effect of nucleolin on Hdm2 is specific to ubiquitination of p53. Expression of the nucleolin RBD had the strongest effect on p53 stabilization, but the mechanism of Hdm2 inhibition is made complicated by the ability of RBD to associate with the Hdm2 NLS/NES, RING, and p53-BD entities. Which of these interactions causes an inhibition of Hdm2-mediated p53 ubiquitination? First considering the Hdm2 NLS/NES sequence, it has been shown that Hdm2 association with p53 can induce nuclear export, an event required for p53 degradation [31]. However, this re-localization is dependent on the p53, but not the Hdm2, NES. The significance of nucleolin association with the Hdm2 NLS/NES is therefore as yet unclear. In contrast, the association of the RBD with the p53-binding domain could be easily envisioned to yield a specific effect on p53 ubiquitination by preventing p53 association with Hdm2. Nutlin-1, a small molecule antagonist of Mdm2, functions by binding to the p53-binding pocket in the p53-BD, destabilizing the Hdm2-p53 complex [32]. Lastly, it is also possible that the association of the nucleolin RBD (or NT) domain with the Hdm2 RING is responsible for the observed inhibition of Hdm2 activity. The RING domain is an element essential for p53 ubiquitination, and past work has shown that acetylation of Mdm2 primarily in the RING domain inhibits p53 degradation [33]. Although nucleolin does not efficiently bind with the Hdm2-CT constructs containing RING domain in binding assays with cell lysates and nucleolin-NT does not interact with Hdm2-RING domain in vitro, it can be envisioned that binding of nucleolin-RBD to the NLS/NES or p53-BD may expose the RING domain for interaction with nucleolin-NT in vivo. More work will be needed to further decipher the roles of the nucleolin RBD and NT on Hdm2 activity, particularly in the context of the full-length nucleolin protein.
Our study provides further demonstration of the importance of factors involved in ribosome biogenesis in the regulation of the Hdm2-p53 axis. In addition, it is now clear that the modulation of Hdm2 activity can provide a potential route for anti-cancer therapy [34–37]. Dissecting nucleolin domains that are inhibitory to Hdm2 can thus provide valuable data for the development of alternative anti-Hdm2 regulators to aid cancer therapeutics.
Materials and Methods
Expression vectors
The expression constructs for human nucleolin (Nu, NCBI: NP_005372.2 or UniProt: P19338); full-length (FL, aa 1–707), the N-terminal acidic domain (NT), the central RNA-binding region, and C-terminal Gly/Arg-rich (GAR) element, containing an N-terminal GFP-tag, were described previously [12,27]. Additional constructs that allow expression of specific subsets of the four nucleolin RNA-binding domains (RBD) were also generated (see Figure 1A). Because the nucleolin nuclear localization signal (NLS) is located in the NT, this NLS sequence was added to the N-terminus of the RBD sequence to support nuclear expression of these constructs. Constructs expressing GST-tagged Hdm2, (UniProt:Q00987, full-length and truncation mutants; [8]) were obtained from Dr. David Meek (U. Dundee, UK). The construct expressing GST-tagged human p53 (UniProt:P04637) was obtained from Dr. A. Levine (Institute for Advanced Study, Princeton, NJ).
Antibodies and other biochemicals
The primary antibodies used for Western blotting were as follows: GFP, rabbit polyclonal (Molecular Probes, Invitrogen Corp. Carlsbad, CA, USA); GST, mouse monoclonal; β-actin, mouse monoclonal (all from Sigma-Aldrich, St Louis, MO, USA); nucleolin, mouse monoclonal MS-3; Hdm2, mouse monoclonal SMP14; and p53 mouse monoclonal DO-1; PCNA, rabbit polyclonal (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA). The secondary antibodies used were anti-mouse and anti-rabbit HRP-conjugated antibodies (GE Healthcare Bio-Sciences Corp. Piscataway, NJ, USA). Various reagents for protein purification and biochemical assays were purchased commercially including Ni-NTA agarose beads (Qiagen, Valencia, CA, USA) and glutathione Sepharose beads (GE Healthcare, USA).
Cell culture and transfection
All human cell lines were obtained from ATCC. H1299, a p53-null lung cancer cell line, was used primarily for expression and purification of various nucleolin domains. Wild-type p53-expressing cell lines, such as U2OS and SJSA, were used for analyzing expression of p53. Plasmid transfections were performed using Effectene transfection reagent (Qiagen).
Western blot analysis
For Western blotting of total cellular proteins, pelleted cells were lysed by incubation (30 min) on ice in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% (v/v) NP-40, 2 mM Dithiothreitol [DTT], and 2 mM phenylmethylsulfonyl fluoride [PMSF]) and total cell lysate (TCL) was prepared. Following SDS-PAGE and transfer onto a nitrocellulose membrane, membranes were stained with Ponceau S to visualize transferred proteins. After blocking in non-fat dry milk and incubation with primary antibodies, the membranes were incubated with anti-mouse or -rabbit HRP-conjugated antibody, and visualized by ECL-Plus reagent (Perkin-Elmer, Wellesley, MA, USA).
Protein expression and purification
GST-tagged nucleolin-FL and various nucleolin truncation domains were purified from yeast as described earlier [27,38], following induction of nucleolin expression with 2% galactose (2 to 4 h at 30°C). GST-Hdm2 and GST-p53 were purified from E. coli BL21 cells, following a 3 to 4 h induction (30°C) with 0.2 to 0.4 mM IPTG [39]. Protease inhibitors (Roche Diagnostics Corporation, Indianapolis, IN, USA) were used during the entire process of lysis and purification. Bound proteins were then eluted from the glutathione-Sepharose beads (GE Healthcare) using 10 mM reduced glutathione, and dialyzed against 50 mM Tris, pH 7.5, 100 mM NaCl and 20% glycerol. GST-tagged nucleolin constructs were utilized in Far-western, Hdm2 auto-ubiquitination and some p53-ubiquitination assays.
Because the yield of nucleolin protein purified from yeast was low, we utilized human H1299 cells as an alternate source from which to purify nucleolin. Following transfection of the appropriate GFP-tagged nucleolin-expression construct (full-length or mutant derivatives), expressed proteins were immunoprecipitated using polyclonal anti-GFP antibodies (Invitrogen) and purified using Protein A/G plus beads (Santa Cruz Biotechnology). In all cases, the quality of proteins was assayed by SDS-PAGE, using Coomassie Blue or silver staining. Proteins were quantified using Bradford protein reagent (Bio-Rad) or by silver staining (ProteoSilver Plus kit, Sigma), employing BSA as a standard. Immunoprecipitated GFP-tagged nucleolin on beads were typically used in p53-ubiquitination assays in vitro.
Far-Western analysis
The detailed procedure for Far-Western blotting is described elsewhere [27,40]. Briefly, following SDS-PAGE of the purified GST-nucleolin variants (400 ng), proteins were transferred onto a nitrocellulose membrane and then subjected to a renaturation protocol. The membrane was subsequently incubated with purified Hdm2 (0.2 µg/ml) in PBS-T (phosphate buffered saline with 0.1% Tween 20) containing 0.25% non-fat dry milk, 1 mM DTT, and 2.5 mM PMSF for 2 h at RT. After washing the membrane, Hdm2 binding was detected using the SMP-14 monoclonal antibody and HRP-conjugated secondary antibodies; and visualized using ECL-Plus. The interactions of nucleolin with membrane-bound Hdm2 domains, and association of p53 with nucleolin domains, were similarly identified using monoclonal antibodies against nucleolin and p53, respectively.
Binding assays with cell lysates
To assay nucleolin-Hdm2 binding in vitro, the individual GST-Hdm2 constructs, previously purified on glutathione-Sepharose beads, were mixed with lysates (1 mg) from H1299 cells transfected with the specific GFP-nucleolin expression construct. An equal amount of bead-bound GST-Hdm2 proteins were used (as analyzed with anti-GST antibodies). After an overnight incubation at 4°C, beads were washed extensively with 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 20% (v/v) glycerol, 1 mM PMSF, 1 mM DTT and 1× protease inhibitor cocktail (Roche), resuspended in SDS-PAGE lysis buffer, and analyzed by Western blot.
In vitro direct binding assays
GFP-tagged nucleolin variants were prepared in vitro using the TNT Quick Coupled Transcription/Translation System (Promega Corporation, Madison, WI, USA). Because the GFP-tagged nucleolin constructs were contained within the pEGFP vector (Clontech Laboratories Inc., Mountain View, CA, USA) that lacks the T7 RNA polymerase promoter sequence, we first generated PCR products with T7 promoter site. Each GFP-nucleolin variant was PCR-amplified using the following primer set: forward, TCGAAATTAATACGACTCACTATAGGGTCGACCACCATGGTGAGCAAGGGCGAGGAGCT; reverse, TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTACAAATGTGGTATGGCTGA. These PCR-fragments were then transcribed using T7 RNA polymerase and translated using the TNT kit with the manufacturer’s protocol. The in vitro translated (IVT) products (40 µl) were then incubated overnight with 3 µg of purified GST-Hdm2, GST-p53 or GST proteins in BC100 reaction buffer [20mM Tris-HCl pH 7.3, 100mM NaCl, 10%glycerol, 1mM DTT, 0.2% BSA, 0.5mM PMSF and 1× protease inhibitors (Roche)] on a rotator at 4°C. To the reaction mixture was added glutathione-Sepharose beads, and the beads then washed six times with BC100. The bound proteins were eluted with 40 µl of 2× SDS sample buffer and boiled for 5 min. The presence of GFP-tagged protein was detected by SDS-PAGE and Western blotting.
In vitro ubiquitination assays
Hdm2 auto-ubiquitination and Hdm2-mediated p53-ubiquitination reactions were performed as described by Li et al. 2002 [41] with some modifications. Standard Hdm2 auto-ubiquitination reaction mixtures (25 µl) contained purified components including 20 ng E1 (Sigma-Aldrich), 200 ng E2 (UbcH5a; Sigma-Aldrich), 400 ng GST-Hdm2, 10 µg His-ubiquitin (Boston Biochem), and 150 ng nucleolin (1×) in reaction buffer (40 mM Tris, pH 7.6, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 2.4 µM ubiquitin aldehyde). For Hdm2-mediated p53-ubiquitination, purified GST-p53 (250 ng), and the amount of nucleolin was reduced to 40–100 ng. Reactions were carried out for 60 min at 37°C and His-tagged (i.e., ubiquitinated) products were either first purified using Ni-NTA agarose beads (Qiagen), or directly subjected to SDS-PAGE and analyzed by Western blotting using either anti-p53 or -Hdm2 antibodies.
Supplementary Material
Acknowledgements
We thank Kyung Kim and Checo Rorie for their technical assistance. This study was supported by NIH grant R01 GM083185, the New York University Cancer Institute, the Rita J. and Stanley Kaplan Comprehensive Cancer Center, and National Cancer Institute Grant P30CA16087 (JAB) and PSC-CUNY 60040-39 40 and 63776-0041 grants (AS).
List of abbreviation used in the manuscript
- BC100
Buffer C with 100 mM NaCl
- CT
C-terminus
- DTT
Dithiothreitol
- ECL
Enhanced Chemiluminescence
- FL
Full-length
- GAR
Glycine-Arginine rich
- Hdm2
Human homolog of Mdm2
- HRP-conjugated
Horseradish peroxidase
- IPTG
Isopropyl beta-D-1-thiogalactopyranoside
- IVT
In-vitro translated
- Mdm2
Mouse double minute2
- NLS/NES
Nuclear Localization Signal/Nuclear Export Signal
- NT
N-terminus
- p53-BD
p53-binding domain
- PMSF
phenylmethylsulfonyl fluoride
- RBD
RNA-binding domain
- rDNA
ribosomal DNA
- RING
Really Interesting New Gene
- RPA
replication protein A
- RPL26
ribosomal protein
- rRNA
ribosomal RNA
- TCL
total cell lysate
- UTR
untranslated region
Footnotes
Conflict of Interest
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
Supplementary Material
Refer to Web version on PubMed Central for supplementary material.
Fig. S1, Far-Western analyses of direct nucleolin-Hdm2 interactions.
Fig. S2, p53 ubiquitination using GST-nucleolin domains in vitro.
S3, Effects of nucleolin domain expression on p53 levels.
References
- 1.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat KP, Itahana K, Jin A, Zhang Y. Embo J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai MS, Lu H. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 5.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin A, Itahana K, O'Keefe K, Zhang Y. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 8.Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 9.Bertwistle D, Sugimoto M, Sherr CJ. Mol. Cell. Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Mol. Cell. Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginisty H, Amalric F, Bouvet P. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- 13.Angelov D, et al. Embo J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otake Y, et al. Blood. 2007;109:3069–3075. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trere D, Derenzini M, Sirri V, Montanaro L, Grigioni W, Faa G, Columbano GM, Columbano A. Hepatology. 1996;24:1269–1273. doi: 10.1002/hep.510240547. [DOI] [PubMed] [Google Scholar]
- 16.Ceccarelli C, Trere D, Santini D, Taffurelli M, Chieco P, Derenzini M. Micron. 2000;31:143–149. doi: 10.1016/s0968-4328(99)00071-2. [DOI] [PubMed] [Google Scholar]
- 17.Derenzini M. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- 18.Daniely Y, Dimitrova DD, Borowiec JA. Mol. Cell. Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 20.Rubbi CP, Milner J. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson MO. Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 22.Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. Cancer Res. 2005;65:9834–9842. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
- 23.Takagi M, Absalon MJ, McLure KG, Kastan MB. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 24.Sirri V, Roussel P, Gendron MC, Hernandez-Verdun D. Cytometry. 1997;28:147–156. [PubMed] [Google Scholar]
- 25.Greasley PJ, Bonnard C, Amati B. Nucl. Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniely Y, Borowiec JA. J. Cell Biol. 2000;149:799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M, Borowiec JA. Mol Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson MO, Kirstein MN, Wallace MO. Biochemistry. 1990;29:5682–5686. doi: 10.1021/bi00476a006. [DOI] [PubMed] [Google Scholar]
- 29.Gilkes DM, Chen L, Chen J. Embo J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstrom MS, Deisenroth C, Zhang Y. Cell Cycle. 2007;6:434–437. doi: 10.4161/cc.6.4.3861. [DOI] [PubMed] [Google Scholar]
- 31.Boyd SD, Tsai KY, Jacks T. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 32.Vassilev LT, et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Taplick J, Geva N, Oren M. FEBS Lett. 2004;561:195–201. doi: 10.1016/S0014-5793(04)00168-1. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y. Cancer Biol Ther. 2003;2:623–629. [PubMed] [Google Scholar]
- 35.Vassilev LT. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Shangary S, Wang S. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiman KG. Cell Death Differ. 2006;13:921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 38.Haluska P, Jr, Saleem A, Edwards TK, Rubin EH. Nucleic Acids Res. 1998;26:1841–1847. doi: 10.1093/nar/26.7.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxena A, Morozov P, Frank D, Musalo R, Lemmon MA, Skolnik EY, Tycko B. J Biol Chem. 2002;277:49935–49944. doi: 10.1074/jbc.M206497200. [DOI] [PubMed] [Google Scholar]
- 40.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 42.Fang SH, Yeh NH. Exp Cell Res. 1993;208:48–53. doi: 10.1006/excr.1993.1221. [DOI] [PubMed] [Google Scholar]
- 43.Tuteja R, Tuteja N. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.