Abstract
Kinesins are motor-based transport proteins that play diverse roles in various cellular processes. The trypanosome genome lacks the homologs of many conserved mitotic kinesins, but encodes a number of trypanosome-specific kinesins with unknown function. Here, we report the biochemical and functional characterization of TbKIN-C, a trypanosome-specific kinesin, which was initially identified through an RNAi screen for cytokinesis genes in T. brucei. TbKIN-C possesses in vitro ATPase activity and associates with cytoskeletal tubulin microtubules in vivo. It is distributed throughout the cytoskeleton with a focal enrichment at the posterior end of the cell during early cell cycle stages. RNAi of TbKIN-C resulted in distorted cell shape with an elongated posterior filled with tyrosinated tubulin microtubules. Silencing of TbKIN-C impaired the segregation of organelles and cytoskeletal structures and led to detachment of the new flagellum and a small portion of the cytoplasm. We also show that RNAi of TbKIN-C compromised cytokinesis and abolished the trans-localization of TbCPC1, a subunit of the chromosomal passenger complex, from the central spindle to the initiation site of cytokinesis. Our results suggest an essential role of TbKIN-C in maintaining cell morphology, likely through regulating microtubule dynamics at the posterior end of the cell.
Keywords: Trypanosoma brucei, kinesin, morphogenesis, tubulin, motor protein
Introduction
The trypanosomatids are a major family of parasites that cause several important human diseases, including African sleeping sickness (Trypanosoma brucei), Chagas disease in Central and South America (Trypanosoma cruzi), and Leishmaniasis (Leishmania spp.) in tropical and subtropical regions. The typical cell shape of a trypanosome cell is maintained by an array of subpellicular microtubules that are cross-linked to each other and to the plasma membrane (Gull, 1999). The subpellicular microtubules have the same intrinsic polarity with the fast-growing, dynamic ends located at the posterior end of the cell where cell elongation takes place during the growth phase of the cell cycle (Sherwin & Gull, 1989, Robinson et al., 1995). The microtubule cytoskeleton is crucial for maintenance of cell shape, cell polarity, and organelle distribution during the cell cycle of Trypanosoma brucei.
Trypanosomes possess a single flagellum that exits the cell body through the flagellar pocket, a specialized secretary organelle near the posterior end, and extends along the cell body towards the anterior end (Sherwin & Gull, 1989). The flagellum is, for most of its length, attached to the cell body via a specialized zone of the cortex, called the flagellum attachment zone (FAZ). The FAZ is made up of a single filament and a specialized set of four microtubules associated with the smooth endoplasmic reticulum (Kohl et al., 1999). The basal body of the flagellum in a G1-phase cell is positioned at the posterior end and is linked to the kinetoplast, the unique mitochondrial DNA network in trypanosomatids (Gull, 1999). The position, replication, and segregation of organelles and cytoskeletal structures such as the nucleus, kinetoplast, flagellum, Golgi, and basal body are tightly regulated and well coordinated within the cell cycle in trypanosomes (Woodward & Gull, 1990, Kohl & Gull, 1998). Cytokinesis is initiated from the anterior tip of the new FAZ, and the cleavage furrow ingresses unidirectionally along the long axis from the anterior towards the posterior end of the cell (Kohl et al., 2003, Vaughan & Gull, 2008, Vaughan et al., 2008). Strikingly, there is no evidence for the existence of an actomyosin contractile ring, the well established component of the cleavage furrow in yeasts and animals (Glotzer, 2005), at the cleavage furrow in trypanosomes (Garcia-Salcedo et al., 2004), suggesting that the cleavage furrow in this early branched eukaryote may possess an unusual structure with novel components.
Kinesins represent a superfamily of evolutionarily conserved microtubule-based motor proteins that perform diverse cellular functions, such as the regulation of microtubule dynamics and the transport of vesicles, organelles, chromosomes, and protein complexes (Hirokawa et al., 1998). The trypanosome genome encodes a large number (>40) of kinesin-like proteins, among which 13 are kinetoplastid-specific kinesin-like proteins with unknown functions (Wickstead & Gull, 2006). Additionally, homologs of many mitotic kinesins such as the spindle motor BimC, the central spindle kinesin MKLP1/Pavarotti/ZEN-4, and the chromokinesin KLP3A are also missing in the genome (Wickstead & Gull, 2006). Two orphan kinesins, TbKIN-A and TbKIN-B, co-immunoprecipitate with the Aurora kinase TbAUK1 and both are required for spindle assembly and chromosome segregation in T. brucei (Li et al., 2008a, Li et al., 2008b). Among the seven kinesin-13 family kinesins, only TbKif13-1 is nuclear and regulates spindle disassembly in T. brucei (Chan et al., 2010, Wickstead et al., 2010). The Leishmania ortholog of TbKif13-1, LmjKIN13-1, likely also plays a mitotic role, but unlike TbKif13-1, expression of LmjKIN13-1 is regulated during the cell cycle, probably by proteasome-mediated degradation (Dubessay et al., 2006). Another kinesin-13 protein, TbKif13-2, localizes to the tip of the flagellum (Chan et al., 2010, Wickstead et al., 2010) and appears to regulate flagellar length (Chan & Ersfeld, 2010). However, the effect of TbKif13-2 deletion or overexpression on flagellar length is limited (Chan & Ersfeld, 2010). This is in striking contrast to its Leishmania ortholog, LmjKIN13-2, which appears to play an essential role in flagellar length control (Blaineau et al., 2007). Other kinesins that have been functionally characterized include a novel C-terminal kinesin required for maintaining functional acidocalsisomes (Dutoya et al., 2001), and a novel farnesylated kinesin that is essential for viability (Engelson et al., 2011).
In this paper, we report the biochemical and functional characterization of TbKIN-C, a kinetoplastid-specific kinesin (Wickstead & Gull, 2006), in the procyclic form of T. brucei. TbKIN-C was originally identified through a small-scale genomic RNAi screen that aimed to identify genes required for cell division. We demonstrate that TbKIN-C associates with tubulin microtubules in vivo and likely regulates tubulin dynamics at the posterior end of the cell. We also show that RNAi of TbKIN-C led to cytokinesis defects and impaired the trans-localization of TbCPC1, a subunit of the chromosomal passenger complex (CPC) and a key regulator of cytokinesis in T. brucei (Li et al., 2008a), from the central spindle to the anterior tip of the new FAZ. TbKIN-C is the first trypanosome protein found to be enriched at the posterior end of the cell and is the first kinetoplastid-specific kinesin to be functionally characterized.
Results
Identification of TbKIN-C by a genomic RNAi screen
To identify genes that are involved in regulating cell division in the procyclic form of T. brucei, we carried out a small scale genomic RNAi screen following a previously published procedure (Morris et al., 2004). Cell lines with severe growth defects were chosen for further analysis (Supplemental Fig. 1A). One clone exhibited polyploidy and a drastic morphology change upon RNAi induction for three days (Supplemental Fig. 1B). To identify the gene that was knocked down in this RNAi mutant, the insert in the pZJM vector was amplified from purified genomic DNA and sequenced. BLAST search of the trypanosome genome database identified Tb927.8.2630, which was previously classified as a kinetoplastid-specific kinesin (group 2) of unknown function (Wickstead & Gull, 2006). To confirm that the observed phenotype of the RNAi mutant is indeed attributed to the knockdown of this novel kinesin, we cloned a different fragment of this kinesin gene into the pZJM vector and transfected the resulting construct into the 29–13 cell line. Upon RNAi induction, the phenotype of the new RNAi cell line was exactly the same as the mutant obtained in the initial screen, thus confirming that the RNAi mutant identified is indeed targeting this novel kinesin. Since this kinesin is kinetoplastid-specific and cannot be grouped into any known kinesin subgroups (Wickstead & Gull, 2006), we named this kinesin TbKIN-C after the two novel kinesins, TbKIN-A and TbKIN-B, identified in our previous report (Li et al., 2008a).
TbKIN-C possesses in vitro ATPase activity and associates with tubulin microtubules in vivo
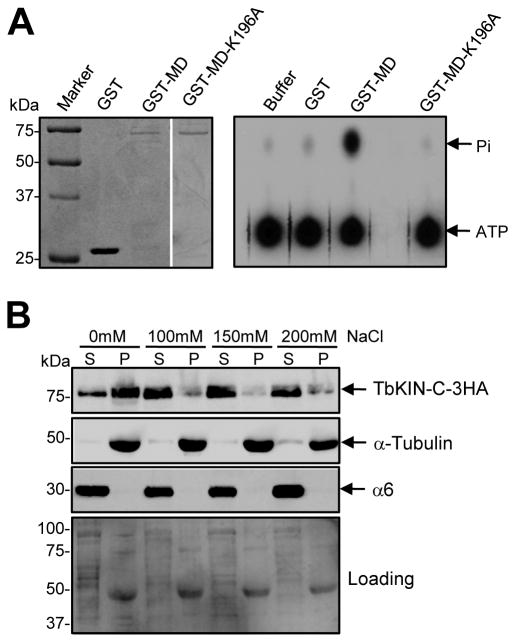
TbKIN-C encodes a putative kinesin with an N-terminal motor domain and three coiled-coil motifs in the C-terminus (Supplemental Fig. 1C, D). The motor domain of TbKIN-C contains the well conserved nucleotide- and microtubule-binding motifs (Supplemental Fig. 1C), indicating that TbKIN-C may possess ATPase activity and may be capable of binding to tubulin microtubules. To test whether TbKIN-C has ATPase activity, we expressed the full-length TbKIN-C in E. coli, but it was insoluble. We then expressed its motor domain fused with GST in bacteria, purified the recombinant protein to near homogeneity (Fig. 1A, left panel), and carried out in vitro ATPase activity assays (Fig. 1A, right panel). We found that the motor domain of TbKIN-C is capable of hydrolyzing ATP, as measured by phosphate release (Fig. 1A, right and lower panels). However, mutation of the well conserved lysine residue (Lys196; Supplemental Fig. 1C, arrowhead) in the ATP-binding motif of TbKIN-C to alanine completely disrupted ATPase activity (Fig. 1A), indicating that the activity associated with the wild-type TbKIN-C motor domain was not due to contamination from bacterial proteins.
Figure 1. TbKIN-C possesses in vitro ATPase activity and associates with tubulin microtubules in vivo.
(A). In vitro ATPase activity assay of the motor domain of TbKIN-C. The motor domain of TbKIN-C (MDTbKIN-C) and its K196A mutant (MD-KATbKIN-C) were purified as GST-fusion proteins from E. coli (left panel) and used for in vitro ATPase assays (right panel). Purified GST protein was included as a control. (B). In vivo association of TbKIN-C with tubulin microtubules in trypanosomes. 3HA-tagged TbKIN-C in soluble fraction and insoluble cytoskeleton fraction of trypanosome cells were detected by Western blot with anti-HA antibody. The same blot was also probed with anti-α-tubulin antibody and anti-α6 protein, a subunit of the 26S proteasome. S: supernatant; P: pellet.
The trypanosome cytoskeleton is characterized by a subpellicular corset of microtubules (Gull, 1999), and microtubule-associated proteins always co-precipitate with the cytoskeleton (Vedrenne et al., 2002). To examine whether TbKIN-C binds to tubulin microtubules in vivo, we tagged the endogenous TbKIN-C with a triple HA epitope at the C-terminus and prepared soluble and cytoskeleton (insoluble) fractions of T. brucei cells. Western blot indicated that TbKIN-C was present in both fractions (Fig. 1B). TbKIN-C protein in the insoluble fraction appeared to associate with tubulin microtubules that were mainly in the insoluble fraction as detected by Western blot with anti-α-tubulin antibody (Fig. 1B). However, when additional salt was added to the cytoskeleton preparation buffer, TbKIN-C protein in the cytoskeleton fraction was solubilized, and the majority of the protein (>95%) was detected in the soluble fraction (Fig. 1B). These results suggest that the majority of TbKIN-C associates with cytoskeletal microtubules and the remainder is localized in the cytoplasm in a soluble form.
Subcellular distribution of TbKIN-C during the cell cycle in the procyclic form of T. brucei
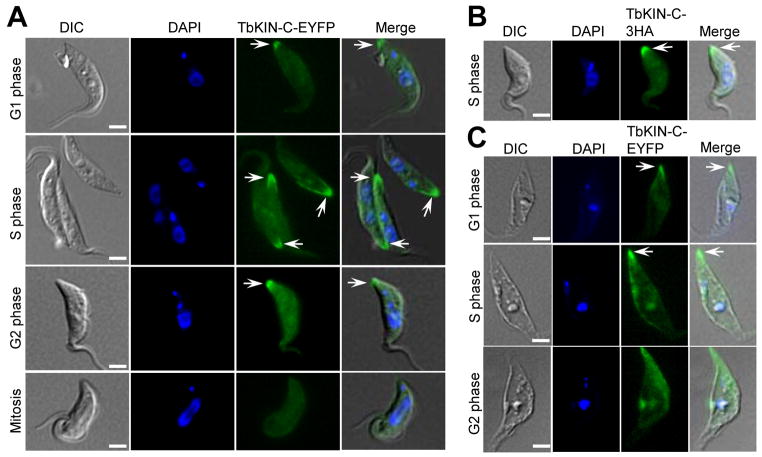
Localization of TbKIN-C during the cell cycle was monitored by immunofluorescence and fluorescence microscopic analysis of the endogenous TbKIN-C tagged with either a triple HA epitope or EYFP at either the N-terminus or the C-terminus. We found that TbKIN-C tagged with either epitope at either terminus displayed identical localization patterns (Fig. 2A, B). During G1 phase, TbKIN-C appeared to spread throughout the cell, but exhibited a focal enrichment at the posterior tip of the cell (Fig. 2A, arrow). During S and G2 phases, TbKIN-C was further concentrated at the posterior tip of the cell, albeit its overall cellular distribution was also enhanced (Fig. 2A, B). During mitosis, however, TbKIN-C signal at the posterior tip of the cell diminished, and its overall cellular level also appeared to decrease significantly (Fig. 2A). To test whether TbKIN-C associates with the cytoskeleton, we prepared the cytoskeleton of trypanosome cells and monitored TbKIN-C-EYFP localization. Similar to the pattern observed in intact cells, TbKIN-C-EYFP was concentrated at the posterior tip of G1 and S cells in addition to the overall distribution in the entire cell body (Fig. 2C). This result further indicates that TbKIN-C associates with cytoskeletal microtubules. Since newly synthesized tubulin microtubules are located at the posterior end of the cell during S phase and G2 phase in trypanosomes (Sherwin et al., 1987), enrichment of TbKIN-C at the posterior tip of the cell suggests that TbKIN-C may regulate the dynamics of newly synthesized tubulin microtubules (see below).
Figure 2. Subcellular localization of TbKIN-C in the procyclic form of T. brucei.
(A). Subcellular localization of TbKIN-C-EYFP during the cell cycle in intact procyclic cells. (B). Localization of TbKIN-C-3HA in intact procyclic cell. (C). Association of TbKIN-C-EYFP with the cytoskeleton of procyclic cells. Bars: 5 μm.
RNAi of TbKIN-C in the procyclic form of T. brucei led to growth defect and cell death
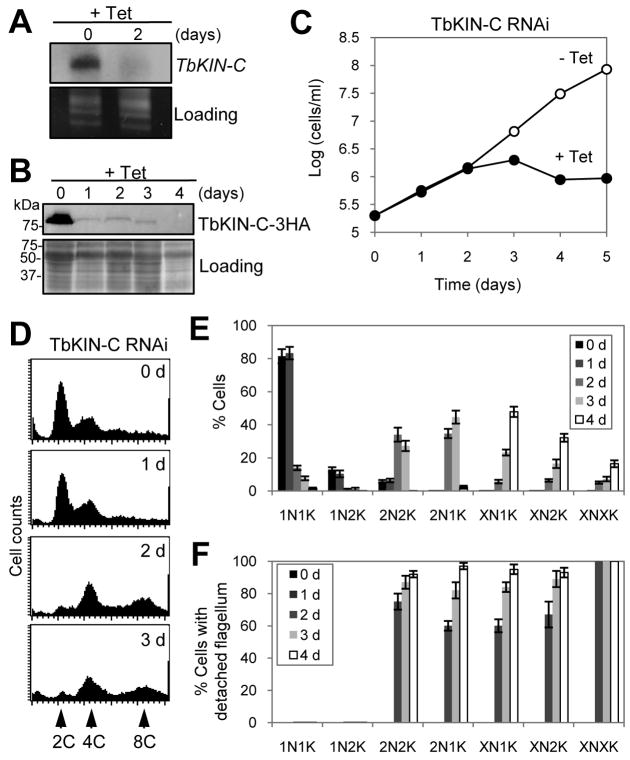
To understand the function of TbKIN-C in T. brucei, we further characterized TbKIN-C RNAi cells. Northern blot showed that TbKIN-C mRNA was decreased to less than 5% after RNAi induction for two days (Fig. 3A). To monitor the protein level of TbKIN-C, the endogenous TbKIN-C was tagged with a triple HA epitope at the C-terminus in the RNAi cell line, and Western blot with anti-HA antibody showed that upon RNAi induction for one day TbKIN-C-3HA was depleted from the cells (Fig. 3B). This led to significant growth inhibition and eventual cell death after four days (Fig. 3C), indicating that TbKIN-C is essential for cell viability in the procyclic form. Further, flow cytometry analysis showed that RNAi of TbKIN-C resulted in the accumulation of cells with 4C DNA content and 8C DNA content after RNAi induction for 2–3 days (Fig. 3D), suggesting the accumulation of polyploid cells. Similar results were obtained with three different clones (data not shown).
Figure 3. RNAi of TbKIN-C in the procyclic form of T. brucei resulted in growth inhibition and eventual cell death.
(A). TbKIN-C mRNA level in control and RNAi cells detected by Northern blot. (B). TbKIN-C protein level in control and RNAi cells detected by Western blot. (C). RNAi of TbKIN-C resulted in growth inhibition and eventual cell death. (D). Flow cytometry analysis of TbKIN-C RNAi cells. (E). Tabulation of cells with different number of nucleus (N) and kinetoplast (K) upon TbKIN-C knockdown. (F). Percentage of TbKIN-C RNAi cells with detached flagellum.
To further characterize the effect of TbKIN-C RNAi on cell division, control and RNAi cells were stained with DAPI for nuclear and kinetoplast DNA, and the number of cells with different number of nuclei and kinetoplasts was counted. After RNAi induction for 2–3 days, the number of cells with one nucleus and one kinetoplast (1N1K) and 1N2K was reduced from around 80% to less than 5%, which was accompanied by a gradual increase of 2N2K cells from about 10% to 30% of the total population (Fig. 3E). In addition to 2N2K cells, 2N1K cells emerged to around 40% of the total population after RNAi for three days (Fig. 3E and Supplemental Fig. 2). Moreover, cells with multiple nuclei and one kinetoplast (XN1K) and XN2K and XNXK also emerged (Supplemental Fig. 2), which, after RNAi induction for four days, constituted about 50%, 30%, and 20% of the total population, respectively (Fig. 3E). The 2N1K and XN1K cells appeared to contain an enlarged kinetoplast, whereas the two kinetoplasts in the 2N2K and XN2K cells either associated with each other or were very closely apposed (Supplemental Fig. 2). These observations suggest that kinetoplast segregation was inhibited. No zoid (0N1K) cells were observed in TbKIN-C RNAi populations, suggesting that cytokinesis was completely inhibited.
To examine whether there was any defect in mitosis in TbKIN-C RNAi cells, we monitored the progression of mitosis in both the control and TbKIN-C RNAi cells. Immunofluorescence assays were carried out using anti-H3K76me2 antibody, which only stains mitotic cells in trypanosomes (Janzen et al., 2006) and therefore can serve as a marker for mitotic progression in trypanosomes. We found that most (>90%) of the 2N cells and all the XN cells were positively stained by anti-H3K76me2 antibody (Supplemental Fig. 3), indicating that mitotic progression is not impaired by TbKIN-C RNAi. However, since anti-H3K76me2 antibody cannot be used to check whether mitotic exit is normal, it remains unclear whether mitotic exit in TbKIN-C RNAi cells was affected. Further, a close examination of the 2N and XN cells deficient in TbKIN-C did not identify any ingressing cleavage furrows (Supplemental Figs. 2 and 3). Collectively, these results suggest that TbKIN-C RNAi inhibited cell division without compromising mitosis.
Effect of TbKIN-C RNAi on the synthesis of the new flagellum and flagellum attachment zone
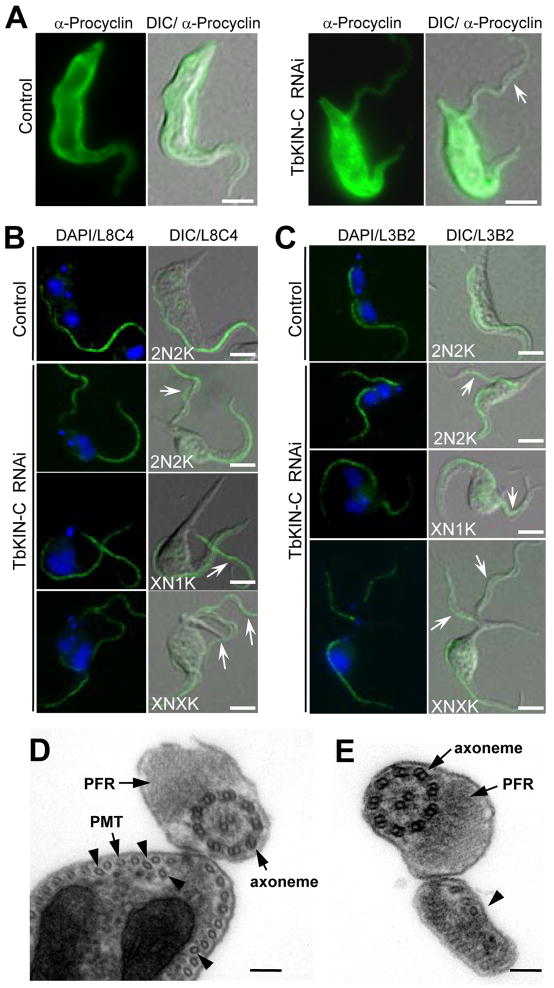
A striking phenotype caused by TbKIN-C RNAi is the detachment of flagellum, which was observed in more than 90% of the 2N2K, 2N1K, XN1K, and XN2K cells and in all the XNXK cells (Fig. 3F and Supplemental Fig. 2). Intriguingly, a close examination of the detached flagellum in TbKIN-C RNAi cells indicated that it appeared to contain the membrane and a small portion of the cytoplasm (Supplemental Fig. 2, arrows), and the former was confirmed via immunostaining with anti-Procyclin antibody (Fig. 4A, arrow). Immunofluorescence assays with L8C4 antibody, which specifically stains the flagellum in trypanosomes, showed that all the 2N cells and all the XN1K and XN2K cells contained two full-length flagella with one of them detached from the cell body, whereas all the XNXK cells possessed multiple full-length flagella with only one of them attached to the cell body (Fig. 4B). These data suggest that TbKIN-C deficiency did not affect flagellum synthesis and that only the newly formed flagella were detached upon TbKIN-C RNAi.
Figure 4. Effect of TbKIN-C RNAi on the synthesis of flagellum and flagellum attachment zone.
(A). Immunostaining of control and TbKIN-C RNAi cells with anti-Procyclin antibody. The arrow indicates the detached flagellum that contains the membrane and cytoplasm. Bars: 5 μm. (B). Effect of TbKIN-C RNAi on the growth of the new flagellum. Cells were fixed in cold methanol, stained with L8C4 antibody for flagella, and counterstained with DAPI for DNA. Arrows point to the detached flagella. Bars: 5 μm. (C). Effect of TbKIN-C RNAi on growth of the new flagellum attachment zone. Cells were fixed in cold methanol, stained with L3B2 antibody for FAZ, and counterstained with DAPI for DNA. Arrows point to the detached flagellum that contains full-length FAZ. Bars: 5 μm. (D, E). Cross-section through the attached flagellum (D) and the detached flagellum (E) of TbKIN-C RNAi cells. The flagellar axoneme, paraflaegllar rod (PFR), and the subpellicular microtubules (PMT) are indicated. The arrowheads in panel D indicate the disorganized PMT, and the arrowhead in panel E points to the portion of cytoplasm detached together with the flagellum. Bars: 100 nm.
The flagellum of a trypanosome cell is attached to the cell body through the FAZ, and it is known that defects in FAZ structure lead to flagellum detachment (Vaughan et al., 2008). To investigate whether there was any defect in the FAZ contributing to flagellum detachment in TbKIN-C RNAi cells, we stained the cells with the L3B2 antibody, a marker for the FAZ (Kohl et al., 1999), and found that the detached flagellum contained a full-length FAZ (Fig. 4C). These observations indicated that flagellum detachment in TbKIN-C RNAi cells was not due to defects in the FAZ. Transmission electron microscopic analysis of TbKIN-C RNAi cells showed that both the attached old flagellum and the detached new flagellum possessed normal axoneme and paraflagellar rod (PFR) structure (Fig. 4D, E). However, the subpellicular microtubule corset (PMT) appeared to be disorganized in TbKIN-C RNAi cells with some microtubules placed further away from the membrane (Fig. 4D, arrowheads). Additionally, we were unable to detect the specialized microtubule quartet underneath the FAZ in all the TbKIN-C RNAi cells and in many uninduced control cells. Therefore, it is not clear whether this is a direct effect of TbKIN-C RNAi. Together, these data suggest that microtubule structure in the flagellum was not disrupted by TbKIN-C depletion, but organization of the subpellicular microtubule corset appeared to be slightly affected.
RNAi of TbKIN-C impaired the segregation of organelles and cytoskeletal structures
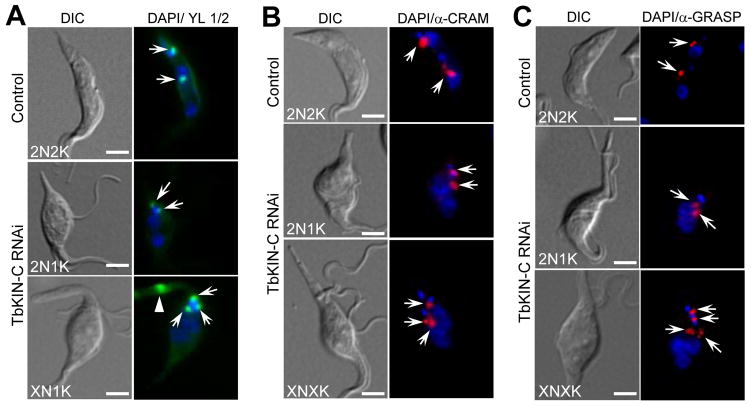
Kinetoplast segregation apparently was inhibited in TbKIN-C RNAi cells (Supplemental Fig. 2). Since kinetoplast segregation is mediated by basal body segregation (Robinson & Gull, 1991), we asked whether basal body replication and/or segregation were impaired by TbKIN-C knockdown. Immunostaining with YL 1/2 antibody showed that the two basal bodies in 2N1K cells were very close (<2μm) to each other and that the multiple basal bodies in XN1K cells formed a cluster around the enlarged kinetoplast (Fig. 5A, arrows). This is in striking contrast to the two well-segregated basal bodies in control 2N2K cells, which are generally separated ~ 5μm apart during cell cycle progression (Absalon et al., 2007). This indicates that segregation, but not replication, of the basal bodies was abrogated in TbKIN-C RNAi cells. To examine whether replication and/or segregation of other organelles and cytoskeletal structures were inhibited in TbKIN-C RNAi cells, we stained the control and TbKIN-C RNAi cells with anti-CRAM and anti-GRASP antibodies to detect the flagellar pocket and Golgi body, respectively. Similar to the basal body, the flagellar pockets and Golgi bodies were not well separated in TbKIN-C RNAi cells (Fig. 5B, C), suggesting defects in segregation of these organelles/structures.
Figure 5. Effect of TbKIN-C RNAi on replication and segregation of basal body, Golgi, and flagellar pocket.
(A). Effect of TbKIN-C RNAi on basal body replication and segregation. Basal bodies (arrows) were stained by YL 1/2 antibody. The arrowhead indicates the accumulation of tyrosinated α-tubulin in the elongated posterior of the cell (see Fig. 6D below). (B). Effect of TbKIN-C RNAi on the replication and segregation of flagellar pocket (arrows) as monitored by anti-CRAM antibody. (C). Effect of TbKIN-C RNAi on Golgi replication and segregation. Golgi (arrows) was immunolabeled by anti-GRASP antibody. Bars: 5 μm.
Depletion of TbKIN-C led to the elongation of the posterior that is filled with tyrosinated α-tubulin microtubules
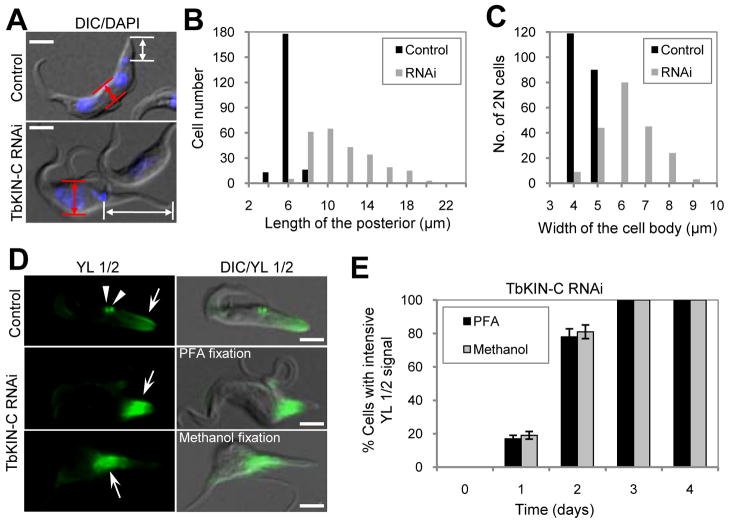
One prominent effect of TbKIN-C RNAi, other than flagellum detachment, is the gross distortion of cell shape with the middle portion of the cell becoming gradually rounded up and the posterior portion of the cell becoming slender and elongated (Fig. 4 and Supplemental Figs. 2 and 3). We measured the distance between the kinetoplast and the posterior end in both the control and TbKIN-C RNAi cells (Fig. 6A, white arrows). The length of the posterior in ~90% of the control cells was between 5–6 μm with the average length of the posterior calculated to be 5.5 μm (Fig. 6B). In contrast, the average length of the posterior in TbKIN-C RNAi cells was about 11.6 μm, and in some extreme cases the posterior was elongated to more than 20 μm in length (Fig. 6B). We also measured the width of the middle portion of the cells containing two nuclei (2N) during the first two days of RNAi (Fig. 6A, red arrows) because the morphology change could be due to the direct effect of TbKIN-C RNAi. The average width of control 2N cells was about 4.9 μm, but the average width of TbKIN-C-deficient 2N cells was increased to 6.7 μm with a few cells becoming almost 10 μm wide in the middle portion (Fig. 6C).
Figure 6. Effect of TbKIN-C RNAi on the distribution of tyrosinated α-tubulin.
(A). TbKIN-C RNAi resulted in extension of the posterior of the cell. Arrows show the distance between the kinetoplast and the posterior end of the cell. Bars: 5 μm. (B) The length of the posterior, from the kinetoplast to the posterior end of the cell, was measured by ImageJ software and plotted. A total of 200 cells were measured for both the control and RNAi cells. (C). The width of the middle portion of 2N cells (~200) was measured by ImageJ and plotted. The middle portion of the cell was defined as the middle between the two nuclei. (D). Cellular distribution of tyrosinated α-tubulin in TbKIN-C RNAi cells. Cells were fixed either with paraformaldehyde (PFA) or with cold methanol and stained with YL 1/2 antibody, which stains tyrosinated α-tubulin. Arrows point to YL 1/2 signal and arrowheads indicated the basal bodies in the control cell. Bars: 5 μm. (E). Percentage of TbKIN-C RNAi cells with intensive YL 1/2 staining in the posterior end of the cell.
The unusual enrichment of TbKIN-C at the posterior end of the cell (Fig. 2) suggests that TbKIN-C might regulate the dynamics of newly synthesized tubulin microtubules that are known to be also enriched at the posterior end of the cell (Sherwin et al., 1987, Robinson et al., 1995). To examine whether the elongated posterior was filled with newly synthesized tubulin microtubules, control and TbKIN-C RNAi cells were stained with YL 1/2 antibody (Kilmartin et al., 1982), which is known to specifically stain tyrosinated α-tubulin proteins that are newly assembled at the plus end of the microtubules in trypanosome cells (Sherwin et al., 1987). Similar to previous reports (Robinson et al., 1995, Sherwin et al., 1987, Sherwin & Gull, 1989), we found that the posterior of some control cells was stained by this antibody (Fig. 6D, arrow). Basal bodies were also stained by YL 1/2 (Fig. 6D, arrowheads). These cells are mostly G1 and S phase cells that are actively growing towards the posterior end through extension of microtubules (Sherwin et al., 1987). In TbKIN-C RNAi cells, however, the posterior of the cell, but not the tip of the posterior, was stained very intensively by YL 1/2 antibody (Fig. 6D, arrows), and in most cells the basal bodies were not readily identifiable, likely because they were masked by the massive amount of tyrosinated α-tubulins accumulated nearby. Immunostaining of methanol-fixed cells showed patterns similar to those observed in paraformaldehyde-fixed cells (Fig. 6D). The number of TbKIN-C RNAi cells with intensive YL 1/2 staining increased significantly to over 80% of the total population after two days of RNAi, but none of the control cells exhibited intensive YL 1/2 staining (Fig. 6E), suggesting accumulation of newly synthesized tubulin microtubules at these locations after RNAi depletion of TbKIN-C, which likely contributed to the observed elongation of the posterior of the cell.
RNAi of TbKIN-C abolishes migration of TbCPC1, a subunit of the chromosomal passenger complex, from the central spindle to the anterior tip of the new FAZ
It is known that during the transition from mitosis to cytokinesis the chromosomal passenger complex in trypanosomes is trans-localized from the central spindle to the anterior tip of the new FAZ where it promotes cytokinesis initiation (Li et al., 2008a, Li et al., 2009). During late anaphase, the elongated spindle appears to bend towards the FAZ (Li et al., 2009, Li et al., 2010), and, subsequently, the CPC travels to the anterior tip of the FAZ. During cytokinesis, the CPC moves along the cleavage furrow, which is initiated from the anterior tip of the FAZ, from the anterior towards the posterior end of the cell (Li et al., 2009). Since TbKIN-C RNAi resulted in defects in cytokinesis and the CPC is known to promote cytokinesis, we asked whether RNAi of TbKIN-C affected the localization of the CPC to the anterior tip of the FAZ. To test this, we examined the subcellular localization of EYFP-tagged TbCPC1 in TbKIN-C RNAi cells. In the un-induced control cells, TbCPC1-EYFP was detected at structures reminiscent of metaphase kinetochores, in an area between the two segregated nuclei consistent with it being located at the central spindle, and at the anterior tip of the new FAZ during telophase (Fig. 7A, arrowheads), similar to our previous observations (Li et al., 2008a, Li et al., 2008b, Li et al., 2009). Among the 2N2K control cells that expressed TbCPC1-EYFP, TbCPC1-EYFP was enriched on the central spindle in about 85% of the 2N2K cells or at the anterior tip of the new FAZ in the remaining 2N2K cells (Fig. 7B). In TbKIN-C RNAi cells, however, TbCPC1-EYFP was detected at structures reminiscent of kinetochores in about 60% of the 2N1K and 2N2K cells and in an area between the segregated nuclei in the remaining 2N1K and 2N2K cells (Fig. 7, arrows). Strikingly, none of the TbKIN-C-deficient cells contained TbCPC1-EYFP at the anterior tip of the new FAZ (Fig. 7B). These results suggest that localization of TbCPC1 to the anterior tip of the new FAZ was apparently impaired in TbKIN-C RNAi cells, which resulted in the re-distribution of the TbCPC1 back to the two segregated nuclei. Similar results were obtained when the kinase activity of TbAUK1 was inhibited during anaphase-cytokinesis transition in trypanosomes (Li et al., 2009), suggesting that when cytokinesis initiation is inhibited, CPC proteins always re-distribute back to the two nuclei. However, the underlying mechanism governing this behavior is unknown.
Figure 7. TbKIN-C RNAi abolishes the trans-localization of the chromosomal passenger protein from the central spindle to the cytokinesis initiation site.
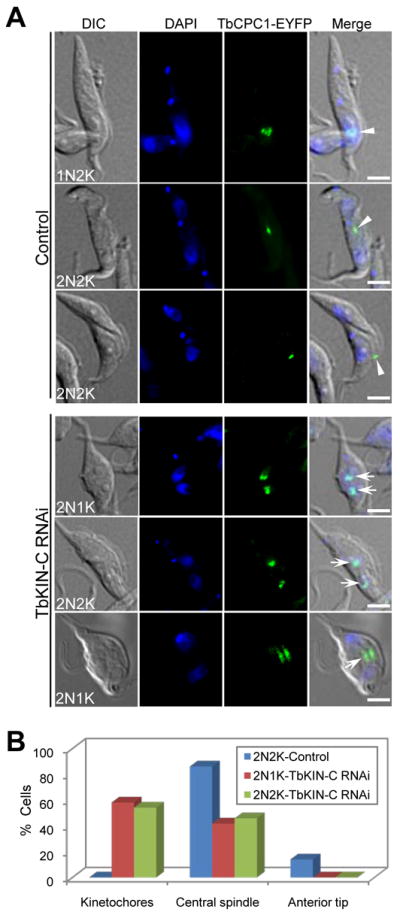
(A). Effect of TbKIN-C RNAi on localization of the chromosomal passenger protein TbCPC1 in the procyclic form. Control and TbKIN-C RNAi cells were fixed and stained with DAPI for DNA. Cells were examined for localization of EYFP-tagged TbCPC1. Arrowheads in control cells and arrows in TbKIN-C RNAi cells indicate the spots of TbCPC1-EYFP protein. Bars: 5 μm. (B). Tabulation of TbCPC1-EYFP localization in control cells and TbKIN-C RNAi cells (2N1K and 2N2K). About 200 cells were counted, and the data represent the average of three independent experiments.
Discussion
In this report, we describe the functional characterization of TbKIN-C, a kinetoplastid-specific kinesin protein in T. brucei. TbKIN-C was initially identified through an RNAi screen for genes essential for cell division in the procyclic form of T. brucei. TbKIN-C exhibits an interesting subcellular localization pattern during the cell cycle, with enrichment at the posterior end of the cell during G1/S/G2 phases in addition to its distribution throughout the cell body (Fig. 2). Given that TbKIN-C is precipitated with the cytoskeleton (Fig. 1), it may associate with tubulin microtubules in the trypanosome cell. In a previous report, TbKIN-C was identified in the flagellum proteome (Broadhead et al., 2006), but our data indicate that it was not enriched in the flagellum at any cell cycle stages (Fig. 2), although we cannot rule out that some TbKIN-C proteins do associate with the flagellar microtubules. The enrichment of TbKIN-C at the posterior end of the cell resembles the distribution of tyrosinated α-tubulin, a marker for newly assembled microtubules (Sherwin et al., 1987), implying that this focal concentration of TbKIN-C may correlate with growth of the cell that is known to involve the extension of microtubules towards the posterior end during the early cell cycle in trypanosomes. Notably, we found that RNAi of TbKIN-C led to a massive accumulation of tyrosinated α-tubulin at the posterior of the cell (Fig. 6D, E), suggesting that TbKIN-C likely regulates microtubule dynamics by inhibiting the assembly of tubulin microtubules and consequently inhibiting the extension of the microtubule corset at the posterior end during G1/S/G2 phases. In support of this idea, the posterior of the TbKIN-C RNAi cell was apparently elongated (Fig. 6A, B).
An unusual phenotype caused by TbKIN-C RNAi is the detachment of the flagellum along with a portion of the cytoplasm (Fig, 4 and Supplemental Fig. 2). The flagellum in a trypanosome cell is attached to the cell body via the FAZ (Kohl et al., 1999), and it has been shown that defects in FAZ structure or synthesis result in flagellum detachment and defective cytokinesis (Li & Wang, 2008, Vaughan et al., 2008). Similarly, depletion of a flagellum adhesion glycoprotein (FLA1) also resulted in flagellum detachment, but it is not clear whether FAZ structure was compromised in FLA1 RNAi cells (LaCount et al., 2002). However, synthesis of the new FAZ appeared to be normal in TbKIN-C-deficient cells (Fig. 4). At present, it remains unclear how the new flagellum of the TbKIN-C RNAi cell is detached from the cell body. Given that the detached flagellum also contains a portion of the cytoplasm, it may indicate that the cell has undergone an asymmetrical cytokinesis event due to mis-positioning of the cleavage plane between the new and old flagella. However, we consider this unlikely because the CPC, the essential player for cytokinesis initiation (Li et al., 2008a, Li et al., 2009), was not targeted to the site of cytokinesis initiation during the mitosis-cytokinesis transition (Fig. 7, see discussion below). Therefore, without the CPC at the initiation site, cytokinesis will not be initiated, as we have demonstrated previously (Li et al., 2009). On the other hand, since TbKIN-C RNAi cells lost their regular shape before the new flagellum was detached, it is likely that the cell failed to maintain the new flagellum in the right position due to distorted morphology.
Depletion of TbKIN-C was found to inhibit cytokinesis (Fig. 3). Cytokinesis initiation and completion in trypanosomes is unique in that the cleavage furrow ingresses longitudinally from the anterior tip of the new FAZ towards the posterior end of the cell. Trypanosomes express a novel chromosomal passenger complex that migrates from the central spindle to the anterior tip of the new FAZ to initiate cytokinesis (Li et al., 2008a). Intriguingly, localization of the CPC to the anterior tip of the FAZ was apparently abolished in TbKIN-C RNAi cells (Fig. 7). Since the new FAZ was detached along with the new flagellum from the cell body in TbKIN-C RNAi cells (Fig. 4), it suggests that localization of TbCPC1 to the anterior tip of the FAZ requires the attachment of the FAZ to the cell body and/or connection of the new flagellum to the old flagellum. Most importantly, this result suggests that the cytokinesis defect caused by TbKIN-C RNAi could be attributed, at least partially, to the failure of targeting TbCPC 1 to the anterior tip of the FAZ where the TbCPC1 is known to promote cytokinesis initiation (Li et al., 2008a, Li et al., 2009). However, given that the defect in basal body segregation also inhibits cytokinesis (Pradel et al., 2006, Li & Wang, 2008, Selvapandiyan et al., 2007) and the two basal bodies were not well segregated in TbKIN-C RNAi cells (Fig. 5A), it is likely that the cytokinesis defect in TbKIN-C RNAi cells could also be attributed to the defect in basal body segregation. At present, we cannot ascertain which factor is the main cause for the defective cytokinesis, but since cytokinesis initiation requires coordination of organelle segregation and cellular morphogenesis, the cytokinesis defect in TbKIN-C RNAi cells could reflect the pleiotropic effect of the morphology change caused by altered microtubule dynamics.
Experimental Procedures
Trypanosome cell culture
The procyclic form of T. brucei strain 427 was cultured at 27°C in SDM-79 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals). Procyclic 29–13 cell line (Wirtz et al., 1999) was cultivated in SDM-79 medium containing 10% fetal bovine serum, 15 μg/ml G418 and 50 μg/ml hygromycin. Cells were routinely diluted when the density reaches 5 × 106/ml.
RNAi screen for trypanosome mutants defective in cell division
A genomic RNAi library with an average insert size of about 600-bp was constructed according to the published procedures (Morris et al., 2004, Morris et al., 2002, Drew et al., 2003). A small-scale RNAi screen was performed by carrying out 10 transfections in the procyclic 29–13 cell line, estimating to cover the trypanosome genome about one time (Morris et al., 2004). After electroporation, the parasites were pooled in SDM-79 medium containing 10% FBS, 15 μg/ml G418, and 50 μg/ml hygromycin and cultured at 27°C. After 24 hours, cells were spun down, resuspended in SDM-79 medium containing 2.5 μg/ml Phleomycin in addition to G418 and hygromycin, and cultured in 50 96-well plates at 27°C. About 200 cell lines were obtained and all of them were induced with 1.0 μg/ml tetracycline for defect in cell growth. Only two cell lines showed significant growth defect and were chosen for further study. The two cell lines were further cloned in 96-well plates and 3–5 clones for each cell line were obtained. DNA insert in the pZJM vector from each of these clones was amplified by PCR and sequenced. Identity of the gene fragment was determined by BLAST search of the trypanosome genome sequence.
RNA interference
To make a new RNAi construct against TbKIN-C gene, a 500-bp DNA fragment corresponding to the N-terminal coding region was cloned into pZJM and transfected into 29–13 cell line. This DNA fragment is different from the DNA insert in the pZJM obtained from RNAi screen. Successful transfectants were selected under 2.5 μg/ml Phleomycin and cloned by limiting dilution. To induce RNAi, 1 μg/ml tetracycline was added to the culture medium. Cell growth was monitored daily by counting the number of cells with a hemacytometer and plotted against induction time.
Northern blot
Total RNA was purified from T. brucei cells with the TRIzol reagent (Invitrogen). Northern blot was performed as previously described (Li et al., 2003). Briefly, total RNA was blotted onto nitrocellulose membrane in a 20× SSC (150mM NaCl and 0.15mM sodium citrate) solution. Northern hybridization was carried out overnight at 42°C in 50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt’s solution with 0.1mg/ml salmon sperm DNA. The membrane was washed with 2× SSC and 0.1% SDS for 30 minutes, 1× SSC and 0.1% SDS for 30 minutes, and 0.5× SSC and 0.1% SDS for 30 minutes before exposing to the X-ray film. The intensity of the bands was quantified by the ImageJ software.
Purification of GST fusion protein and in vitro ATPase activity assay
Wild-type TbKIN-C and the mutant TbKIN-C bearing K196A mutation in its ATP-binding motif were each cloned cloned into pGEX-4T-3 vector for expression of recombinant proteins in E. coli. The motor domain (MD) of TbKIN-C and its K196A mutant were also cloned into pGEX-4T-3 vector. Recombinant proteins were expressed in E. coli BL21 cells and purified through a column of glutathione Sepharose 4B beads. Purified recombinant proteins were dialyzed against 50mM Tris-Cl and 50 mM NaCl. Only GST-MDTbKIN-C and GST-MD-K196A TbKIN-C were soluble and used for in vitro ATPase activitiy assay.
In vitro ATP hydrolysis catalyzed by GST-MDTbKIN-C and its K196A mutant was assayed by measuring the release of radiolabelled Pi from γ-32P-ATP according to our previous procedure (Dang & Li, 2011). Recombinant GST-MDTbKIN-C or its K196R mutant was incubated with 20 mM Tris-Cl, pH8.0, 1 mM MgCl2, 30 mM KCl, 4%(w/v) sucrose, 80 μg/ml BSA, 1 mM ATP, 0.2 μCi γ-32P-ATP for 2 hrs at 37°C. Reaction was stopped by chilling on ice, and 1.0μl of the reaction solution was spotted onto a polyethyleneimine (PEI) cellulose thin-layer plate. Ascending chromatography was performed in the solution containing 0.5M LiCl and 1M formic acid at room temperature for 20 min. The plate was air dried and exposed to X-film. Spots of phosphate and ATP area were excised from the plate and counted in a liquid scintillation counter.
Epitope tagging of endogenous TbKIN-C in the procyclic form of T. brucei
TbKIN-C was cloned into the pC-EYFP-Neo vector and the pC-3HA-Bla vector, respectively. Both vectors were obtained by modifying the pC-PTP-Neo and the pC-HA-Bla vectors (Schimanski et al., 2005). The resulting constructs were linearized and electroporated into the wild-type 427 cell line. The pC-TbKIN-C-3HA-Bla construct was also transfected into pZJM-TbKIN-C cell line. Correct in situ tagging of one of the two alleles was confirmed by PCR and subsequent sequencing. Stable transfectants were selected under 40 μg/ml G418 or 10 μg/ml Blasticidin and cloned by limiting dilution.
Preparation of trypanosome cytoskeleton
Cytoskeleton preparation of procyclic cells was performed according to previous procedures (Robinson et al., 1991). Cells expressing TbKIN-C-3HA were collected, washed 3 times with PBS, and incubated in PEME buffer (0.1 M PIPES, pH6.9, 2 mM EGTA, 1 mM Magnesium sulfate, 0.1 mM EDTA) containing 1% Nonidet P-40 at room temperature for 5 min. Various concentrations of NaCl were added to the cytoskeleton preparation buffer to solubilize any cytoskeleton-associated proteins. Cells were then spun down, and soluble fraction and insoluble pellet fraction were separated. Both fractions were boiled in 1× SDS-PAGE sampling buffer for 5 min and analyzed by Western blot with anti-HA mAb, anti-α-tubulin mAb (Sigma-Aldrich), and anti-α6 pAb for cytoplasm marker (Li et al., 2002).
Western blot
To prepare trypanosome cell lysate for Western blot, procyclic cells expressing 3HA-tagged proteins were spun down, re-suspended in SDS-PAGE sampling buffer, boiled for 5 min at 100°C, and cleared by centrifugation. The lysate wasfractionated in SDS-PAGE, transferred onto PVDF membrane, and immuno-blotted with anti-HA mAb as described (Li & Wang, 2008). The membrane was stained with coomassie blue to serve as the loading control.
Flow cytometry
Flow cytometry analysis of propidium iodide-stained trypanosome cells was carried out as previously described (Li et al., 2006). Briefly, T.brucei cells were washed once in PBS and re-suspended in 0.1 ml PBS. The cells were fixed by adding 0.2 ml 10% ethanol, 0.2 ml 50% ethanol and 1.0 ml 70% ethanol (all in PBS with 5% glycerol), and incubated at 4°C. The cells were washed once with PBS and re-suspended in PBS. DNase-freeRNase (10 μg/ml) and propidium iodide (20 μg/ml)were added before flow cytometry analysis. The DNA content of propidium iodide-stained cells was analyzed with a fluorescence-activatedcell sorting scan (FACScan) analytical flow cytometer (BD Biosciences).
Immunofluorescence microscopy
Cells were harvested by centrifugation, washed once in PBS, and fixed in 4% paraformaldehyde or in cold methanol. The fixed cells were washed with PBS, suspended in PBS, and adhered to poly-L-Lysine treated cover-slips. Cells were treated with blocking buffer (1% BSA and 0.1% Triton X-100 in PBS) for 1 hr at room temperature. Cells were then incubated with primary antibodies diluted in PBS containing 1% BSA for 1 hr at room temperature. After three washes with wash buffer (0.1% Triton X-100 in PBS), cells were incubated with FITC-conjugated anti-mouse IgG or anti-rabbit IgG diluted in PBS containing 1% BSA for 1 hr at room temperature. The following antibodies were used: YL 1/2 mAb for detecting tyrosinated α-tubulin used at 1:400 dilution (Kilmartin et al., 1982); α-CRAM pAb for flagellar pocket used at 1:400 dilution (Liu et al., 2000); α-GRASP pAb for Golgi used at 1: 1,000 dilution (He et al., 2004); α-H3K76Me2 pAb for detecting mitotic cells used at 1:50 dilution (Janzen et al., 2006); L8C4 mAb for detecting flagellum used at 1:50 dilution (Kohl et al., 1999); L3B2 for detecting the FAZ used at 1:50 dilution (Kohl et al., 1999); and anti-Procyclin mAb for detecting the membrane of procyclic cells used at 1:200 dilution (Li et al., 2003). After three washes with wash buffer, the slides were mounted in VectaShield mounting medium (Vector Labs) containing DAPI, and examined using an inverted microscope (model IX71, Olympus) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60× 1.42-NA DIC objective. Images were acquired and processed using the Slidebook5 software (Intelligent Imaging Innovations, Inc).
Transmission electron microscopy
Trypanosome cells were fixed in glutaraldehyde, and then treated with Millonig’s buffer for 10 min. Gently draw off the Millonig’s and layer 2% OsO4 for 60 min at 4°C. Cells were washed with water, dehydrated with ethanol (50% ethanol for 5min, 70% ethanol for 10min, 95% ethanol for 10min, and finally treated three times with 100% ethanol for 10min) and then embedded in resin (treated two times with 100% propylene oxide for 15 min, 50% LX-112 resin/50% propylene oxide for 2 hrs, and finally with 100% LX-112 resin for 2 hrs). Embed the fixed cells in BEEM capsules, and polymerize overnight at 70 °C. Subsequently, the 120nm thin sections were cut using a Leica Ultracut-R microtome and a diamond knife (Daitome-U.S.), placed on 150 mesh copper grids (EMS), and stained 15 min with 2% uranyl acetate. The thin sections were then rinsed with water and incubated with Renold’s lead citrate for 5 min. Grids were imaged using a JEOL 1400 TEM at 60kv and captured with a Gatan CCD camera.
Supplementary Material
Acknowledgments
We are grateful to Dr. George A. M. Cross of Rockefeller University for providing the 29–13 cell line and anti-H3K76me2 and anti-Procyclin antibodies, and to Dr. Paul Englund of Johns Hopkins School of Medicine for providing the pZJM vector. We also thank Dr. Arthur Günzl of the University of Connecticut Health Center for providing the pC-PTP-Neo and pC-HA-Bla vectors and Dr. Keith Gull of University of Oxford for providing the L8C4 and L3B2 antibodies. Anti-CRAM antibody and anti-GRASP antibody are generous gifts of Dr. Mary Lee from New York University and Dr. Graham Warren from Max F. Perutz Laboratories in Austria, respectively. We thank Drs. Steven Kolodziej and Julie Chang of University of Texas Medical School at Houston for preparing trypanosome thin sections and technical assistance. We are also grateful to Dr. Kevin Morano of the University of Texas Medical School at Houston for critical reading of the manuscript. This work was supported by the start-up funds from the University of Texas Medical School at Houston and, in part, by the NIH grant AI090070 to Z. L.
References
- Absalon S, Kohl L, Branche C, Blisnick T, Toutirais G, Rusconi F, Cosson J, Bonhivers M, Robinson D, Bastin P. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS ONE. 2007;2:e437. doi: 10.1371/journal.pone.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pages M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Chan KY, Ersfeld K. The role of the Kinesin-13 family protein TbKif13-2 in flagellar length control of Trypanosoma brucei. Mol Biochem Parasitol. 2010;174:137–140. doi: 10.1016/j.molbiopara.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Matthews KR, Ersfeld K. Functional characterisation and drug target validation of a mitotic kinesin-13 in Trypanosoma brucei. PLoS Pathog. 2010;6:e1001050. doi: 10.1371/journal.ppat.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang HQ, Li Z. The Cdc45/Mcm2-7/GINS complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J Biol Chem. 2011 doi: 10.1074/jbc.M111.240143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew ME, Morris JC, Wang Z, Wells L, Sanchez M, Landfear SM, Englund PT. The adenosine analog tubercidin inhibits glycolysis in Trypanosoma brucei as revealed by an RNA interference library. J Biol Chem. 2003;278:46596–46600. doi: 10.1074/jbc.M309320200. [DOI] [PubMed] [Google Scholar]
- Dubessay P, Blaineau C, Bastien P, Tasse L, Van Dijk J, Crobu L, Pages M. Cell cycle-dependent expression regulation by the proteasome pathway and characterization of the nuclear targeting signal of a Leishmania major Kin-13 kinesin. Mol Microbiol. 2006;59:1162–1174. doi: 10.1111/j.1365-2958.2005.05013.x. [DOI] [PubMed] [Google Scholar]
- Dutoya S, Gibert S, Lemercier G, Santarelli X, Baltz D, Baltz T, Bakalara N. A novel C-terminal kinesin is essential for maintaining functional acidocalcisomes in Trypanosoma brucei. J Biol Chem. 2001;276:49117–49124. doi: 10.1074/jbc.M105962200. [DOI] [PubMed] [Google Scholar]
- Engelson EJ, Buckner FS, Van Voorhis WC. An Essential Farnesylated Kinesin in Trypanosoma brucei. PLoS ONE. 2011;6:e26508. doi: 10.1371/journal.pone.0026508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, Nolan DP. A differential role for actin during the life cycle of Trypanosoma brucei. Embo J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gull K. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol. 1999;53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- He CY, Ho HH, Malsam J, Chalouni C, West CM, Ullu E, Toomre D, Warren G. Golgi duplication in Trypanosoma brucei. J Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Gull K. Molecular architecture of the trypanosome cytoskeleton. Mol Biochem Parasitol. 1998;93:1–9. doi: 10.1016/s0166-6851(98)00014-0. [DOI] [PubMed] [Google Scholar]
- Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. Embo J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Barrett B, Donelson JE. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J Biol Chem. 2002;277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- Li Y, Li Z, Wang CC. Differentiation of Trypanosoma brucei may be stage non-specific and does not require progression of cell cycle. Mol Microbiol. 2003;49:251–265. doi: 10.1046/j.1365-2958.2003.03575.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Lee JH, Chu F, Burlingame AL, Gunzl A, Wang CC. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS ONE. 2008a;3:e2354. doi: 10.1371/journal.pone.0002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tu X, Wang CC. Okadaic acid overcomes the blocked cell cycle caused by depleting Cdc2-related kinases in Trypanosoma brucei. Exp Cell Res. 2006;312:3504–3516. doi: 10.1016/j.yexcr.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Li Z, Umeyama T, Li Z, Wang CC. Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot Cell. 2010;9:705–716. doi: 10.1128/EC.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Umeyama T, Wang CC. The chromosomal passenger complex and a mitotic kinesin interact with the Tousled-like kinase in trypanosomes to regulate mitosis and cytokinesis. PLoS ONE. 2008b;3:e3814. doi: 10.1371/journal.pone.0003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Umeyama T, Wang CC. The Aurora Kinase in Trypanosoma brucei plays distinctive roles in metaphase-anaphase transition and cytokinetic initiation. PLoS Pathog. 2009;5:e1000575. doi: 10.1371/journal.ppat.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang CC. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot Cell. 2008;7:1941–1950. doi: 10.1128/EC.00249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zou CB, Yao Y, Hoyt MA, McDonough S, Mackey ZB, Coffino P, Wang CC. An easily dissociated 26 S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J Biol Chem. 2002;277:15486–15498. doi: 10.1074/jbc.M109029200. [DOI] [PubMed] [Google Scholar]
- Liu J, Qiao X, Du D, Lee MG. Receptor-mediated endocytosis in the procyclic form of Trypanosoma brucei. J Biol Chem. 2000;275:12032–12040. doi: 10.1074/jbc.275.16.12032. [DOI] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Drew ME, Englund PT. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. Embo J. 2002;21:4429–4438. doi: 10.1093/emboj/cdf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Motyka SA, Drew ME, Englund PT. An RNAi-based genomic library for forward genetics in the African trypanosome. In: Sohail M, editor. Gene Silencing by RNA Interference: Technology and Application. Boco Raton: CRC Press LLC; 2004. pp. 241–257. [Google Scholar]
- Pradel LC, Bonhivers M, Landrein N, Robinson DR. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J Cell Sci. 2006;119:1852–1863. doi: 10.1242/jcs.02900. [DOI] [PubMed] [Google Scholar]
- Robinson D, Beattie P, Sherwin T, Gull K. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 1991;196:285–299. doi: 10.1016/0076-6879(91)96027-o. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol Biol Cell. 2007;18:3290–3301. doi: 10.1091/mbc.E07-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell. 1989;57:211–221. doi: 10.1016/0092-8674(89)90959-8. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S, Gull K. The structural mechanics of cell division in Trypanosoma brucei. Biochem Soc Trans. 2008;36:421–424. doi: 10.1042/BST0360421. [DOI] [PubMed] [Google Scholar]
- Vaughan S, Kohl L, Ngai I, Wheeler RJ, Gull K. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist. 2008;159:127–136. doi: 10.1016/j.protis.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Vedrenne C, Giroud C, Robinson DR, Besteiro S, Bosc C, Bringaud F, Baltz T. Two related subpellicular cytoskeleton-associated proteins in Trypanosoma brucei stabilize microtubules. Mol Biol Cell. 2002;13:1058–1070. doi: 10.1091/mbc.01-06-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Carrington JT, Gluenz E, Gull K. The expanded Kinesin-13 repertoire of trypanosomes contains only one mitotic Kinesin indicating multiple extra-nuclear roles. PLoS ONE. 2010;5:e15020. doi: 10.1371/journal.pone.0015020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95(Pt 1):49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.