Abstract
The total synthesis of [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8) and its unique AgOAc-promoted single-step conversion to [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7), conducted on a fully deprotected substrate, are disclosed. The synthetic approach not only permits access to 7, but it also allows late stage access to related residue 4 derivatives, alternative access to [Ψ[CH2NH]Tpg4]vancomycin aglycon (6) from a common late stage intermediate, and provides authentic residue 4 thioamide and amidine derivatives of the vancomycin aglycon that will facilitate ongoing efforts on their semisynthetic preparation. In addition to early stage residue 4 thioamide introduction, allowing differentiation of one of seven amide bonds central to the vancomycin core structure, the approach relied on two aromatic nucleophilic substitution reactions for formation of the 16-membered diaryl ethers in the CD/DE ring systems, an effective macrolactamization for closure of the 12-membered biaryl AB ring system, and the defined order of CD, AB, and DE ring closures. This order of ring closures follows their increasing ease of thermal atropisomer equilibration, permitting the recycling of any newly generated unnatural atropisomer under progressively milder thermal conditions where the atropoisomer stereochemistry already set is not impacted. Full details of the evaluation of 7 and 8 along with several related key synthetic compounds containing the core residue 4 amidine and thioamide modifications are reported. The binding affinity of compounds containing the residue 4 amidine with the model d-Ala-d-Ala ligand 2 was found to be only 2–3 times less than the vancomycin aglycon (5) and this binding affinity is maintained with the model d-Ala-d-Lac ligand 4, representing a nearly 600-fold increase in affinity relative to the vancomycin aglycon. Importantly, the amidines display effective dual, balanced binding affinity for both ligands (Ka 2/4 = 0.9–1.05) and they exhibit potent antimicrobial activity against VanA resistant bacteria (E. faecalis, VanA VRE) at a level accurately reflecting these binding characteristics (MIC = 0.3–0.6 µg/mL), charting a rational approach forward in the development of antibiotics for the treatment of vancomycin-resistant bacterial infections. In sharp contrast, 8 and related residue 4 thioamides failed to bind either 2 or 4 to any appreciable extent, do not exhibit antimicrobial activity, and serve to further underscore the remarkable behavior of the residue 4 amidines.
Introduction
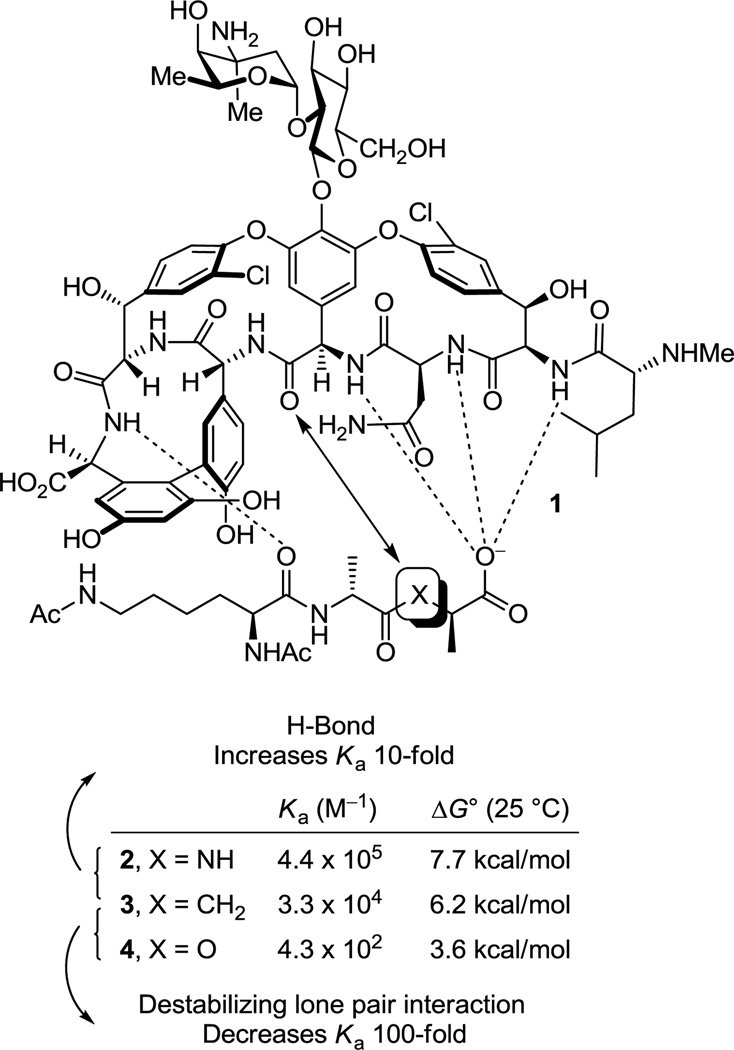
Vancomycin (1) is the most widely recognized member of a large family of glycopeptide antibiotics that are used for the treatment of resistant bacterial infections. Vancomycin was first disclosed over 50 years ago (1955) by Eli Lilly1 and its structure was determined 25–30 years later (1983),2 long after its clinical introduction in 1958. Clinical uses of vancomycin include the treatment of patients on dialysis, patients allergic to β-lactam antibiotics, and patients undergoing cancer chemotherapy.3 However, the most widespread clinical use of vancomycin is the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections, for which it is regarded as the drug of last resort.4 The prevalence of MRSA in intensive care units (ICU, 60% of SA infections in the US are MRSA, 2003)5 and the movement of MRSA from a hospital-acquired to a community-acquired infection has intensified the need to treat such resistant bacterial infections. It is estimated that MRSA is responsible for 19,000 deaths and invasively afflicts 94,000 annually in the US (2005) at a cost of $3–4 billion per year.5c,d In addition, vancomycin resistant strains of other bacteria are also on the rise with US ICU clinical isolates of vancomycin-resistant Enterococcus faecalis (VRE) approaching 30% (2003),5 albeit in strains presently sensitive to other antibiotics. Even more significant and feared by all is the recent emergence of MRSA strains now resistant or insensitive to vancomycin (VRSA and VISA). This poses a major health problem and has stimulated efforts to develop vancomycin analogues6 or alternative antibiotics to combat this resistance.7
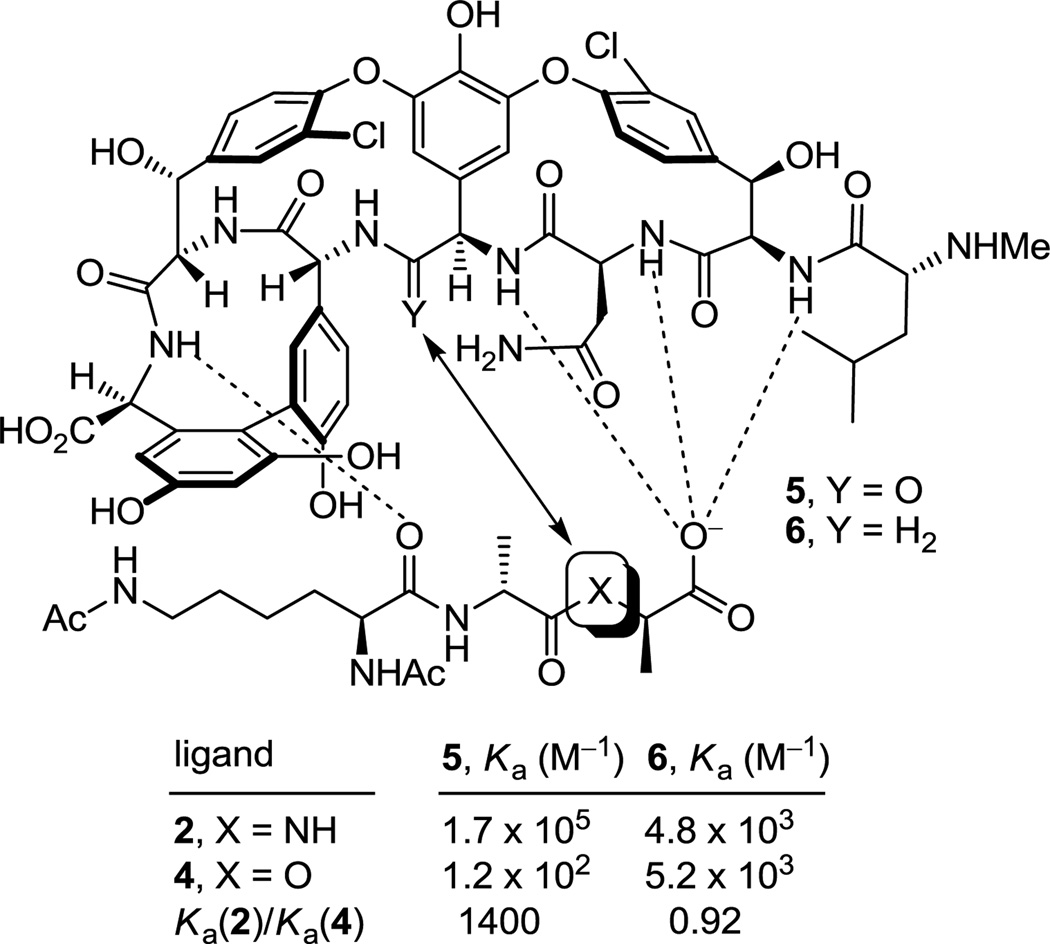
Vancomycin inhibits bacterial cell wall synthesis, binding to the peptidoglycan peptide terminus N-acyl-d-Ala-d-Ala found in cell wall precursors,8 by sequestering the substrate from transpeptidase and inhibiting cell wall cross-linking. Shortly after the disclosure of the structure of vancomycin, Williams provided the NMR structure of a N-acyl-d-Ala-d-Ala complex with the antibiotic that was found to be stabilized by an extensive array of hydrophobic van der Waals contacts within the vancomycin binding pocket and five key H-bonds lining the pocket and this has since been confirmed in X-ray studies of such model complexes (Figure 1).9 In the most commonly encountered vancomycin-resistant bacterial strains (VanA and VanB), a vancomycin challenge is sensed10 and subsequently induces a remodeling of the precursor peptidoglycan terminus from d-Ala-d-Ala to d-Ala-d-Lac.11 Normal d-Ala-d-Ala production continues despite the presence of vancomycin, but now a late-stage remodeling to d-Ala-d-Lac ensues to avoid the action of the antibiotic. The substitution of a linking ester for an amide with the exchange of a single atom (NH→O) reduces the binding to vancomycin 1000-fold and accounts for the 1000-fold higher MICs seen against vancomycin-resistant bacteria.11 A subtle but important feature that emerged from this understanding is the recognition that efforts to rationally redesign vancomycin to directly treat such resistant bacteria, which arise from this single atom change in a bacterial cell wall precursor, should strive to devise compounds that not only bind d-Ala-d-Lac, but that also maintain binding to d-Ala-d-Ala.
Figure 1.
Structure of vancomycin (1), schematic representation of interaction with model ligands 2–4, and measured binding data.
The altered complex of vancomycin with N-acyl-d-Ala-d-Lac lacks the central H-bond of the d-Ala-d-Ala complex and suffers a repulsive lone pair interaction between the vancomycin residue 4 carbonyl and the d-Ala-d-Lac ester oxygens (Figure 1). We provided an experimental estimation of the magnitude of these two effects by examining the model ligands 2–4 that indicated it is the repulsive lone pair interaction (100-fold), not the H-bond loss (10-fold), that is responsible for the largest share of the reduced binding affinity (1000-fold).12 Not only were the estimates consistent with intuitive expectations, but their combined magnitude matched expectations resulting from the stabilizing binding energy of an amide H-bond (0.0–1.5 kcal/mol) and a destabilizing lone pair/lone pair interaction (1.6–2.7 kcal/mol).13 These observations have significant ramifications on the reengineering of vancomycin to bind d-Ala-d-Lac, suggesting that the design could focus principally on removing the destabilizing lone pair interaction rather than reintroduction of a H-bond and that this may be sufficient to compensate for most of binding affinity lost with d-Ala-d-Lac.
The resurgence of interest in vancomycin and the identification of the molecular origin of the bacterial resistance emerged as the first total syntheses of its family members were completed.14 In our efforts complementary to those reported by Evans15 and Nicolaou,16 an initial total synthesis of the vancomycin aglycon17 was extended to the more complex tetracyclic teicoplanin18 and ristocetin19 aglycons, as well as the structurally related chloropeptins,20 in which the synthetic approach was continuously refined.21 In the midst of these studies and concurrent with efforts probing modifications to vancomycin itself,22 we initiated efforts on the redesign of vancomycin to bind d-Ala-d-Lac, completing the total synthesis and evaluation of [Ψ[CH2NH]Tpg4]vancomycin aglycon (6)23 in which the residue 4 amide carbonyl was replaced with a methylene group. Confirming the conclusions drawn from the binding studies summarized in Figure 1 and relative to vancomycin aglycon (5), 6 exhibited a 40-fold increase in affinity for d-Ala-d-Lac (4, Ka = 5.2 × 103 M−1) and a 35-fold reduction in affinity for d-Ala-d-Ala (2, Ka = 4.8 × 103 M−1), providing the first modified glycopeptide with dual, balanced binding properties. Accurately reflecting these binding properties, 6 exhibited improved, but modest, antimicrobial activity against vancomycin-resistant bacteria (VanA VRE MIC = 31 µg/mL). Thus, this removal of a single atom from the antibiotic countered bacterial resistance that is derived from the single atom change in the cell wall peptidoglycan precursor, provided the first reengineered vancomycin rationally designed to exhibit dual d-Ala-d-Ala and d-Ala-d-Lac binding, and established the successful foundation on which our continuing studies are based.
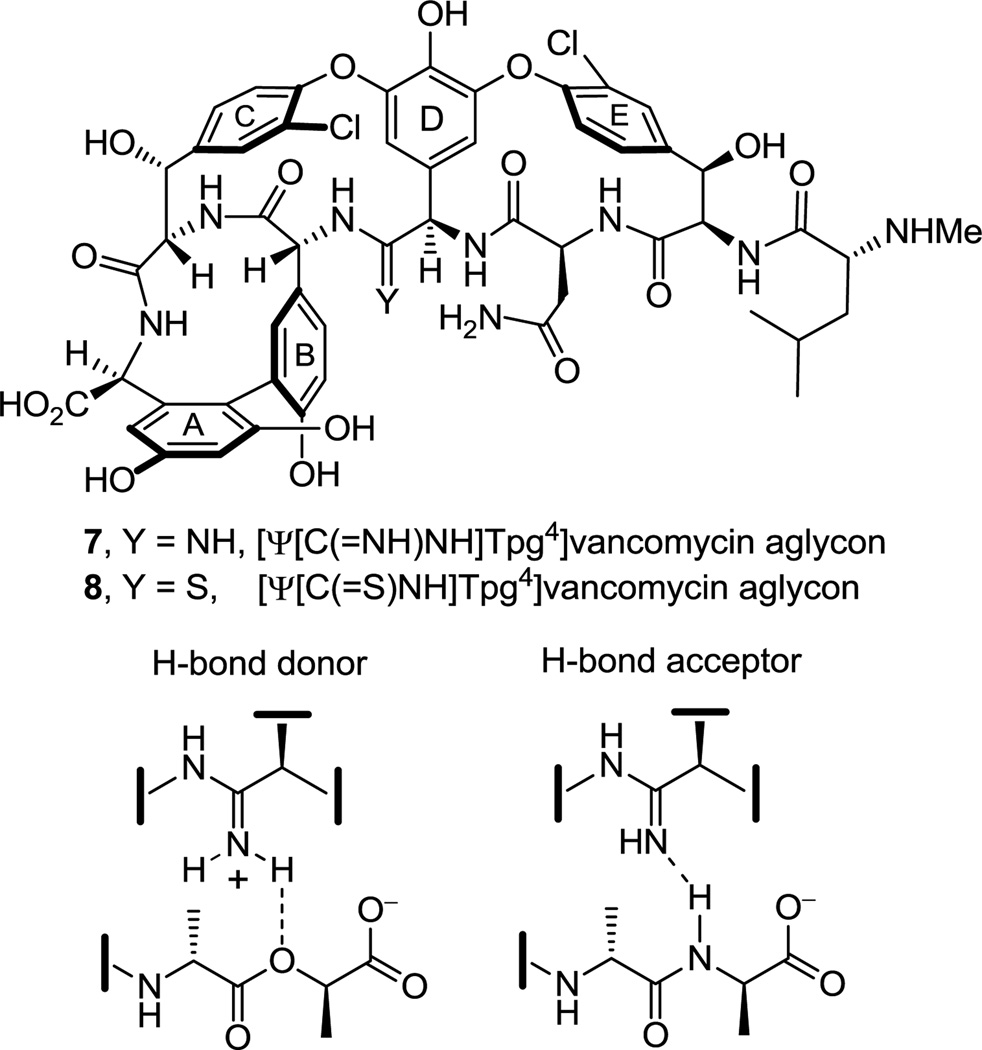
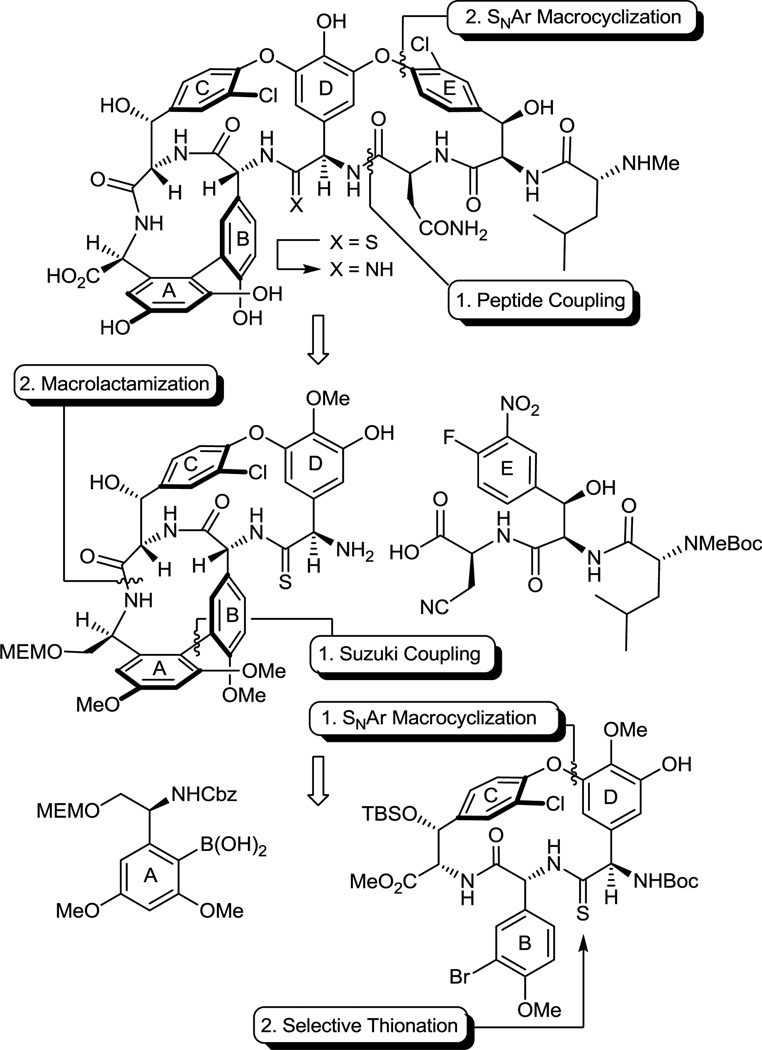
Having first addressed the vancomycin analogue 6 for which we were most confident in the projected binding characteristics, we turned our attention to an additional key member in the series [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7),23 replacing the residue 4 amide with the corresponding amidine (Figure 3). Although the methylene derivative 6 exhibited the desired dual binding and the expected enhancement in d-Ala-d-Lac affinity, it also displayed an expected reduced d-Ala-d-Ala affinity. The key feature to be addressed with [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7) was whether the incorporation of the residue 4 amidine could accommodate or further enhance d-Ala-d-Lac binding by removing the destabilizing lone pair repulsion and perhaps by serving as a H-bond donor, while simultaneously maintaining the required vancomycin affinity for d-Ala-d-Ala by virtue of serving as an alternative H-bond acceptor (Figure 3). Such binding characteristics of 7 were not easy to anticipate as it is not clear whether the ester oxygen of d-Ala-d-Lac could actually serve as a H-bond acceptor,24 or whether an amidine, that is likely protonated, might remain a good H-bond acceptor for d-Ala-d-Ala. Because the projected behavior of 7 was uncertain and since use of amidines as isosteres of amides in cyclic peptides has been largely unexplored,25,26 the preparation of 7 was slated to follow that of 6 in our efforts. Key to the approach to 7 pursued herein is the use of the residue 4 thioamide 8 for amidine introduction in the last step, conducted on a fully functionalized and fully deprotected vancomycin aglycon. This strategy not only permits access to 7, but it also allows late stage access to a series of related key analogues including 8 itself, alternative access to our methylene derivative 6 from a common late stage intermediate (8), and provides authentic samples of residue 4 thioamide and amidine derivatives of vancomycin aglycon that will facilitate ongoing efforts on the semisynthetic preparation of either 6 or 7. These latter two features of the work ensured that our investment in the efforts would be valuable even if the properties of 7 had proved disappointing. Herein, we report the total synthesis of [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8), its direct single-step conversion to [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7) by use of a unique AgOAc-promoted reaction, the preparation of a series of related key compounds that also contain the core residue 4 amidine and thioamide modifications, and details of their evaluation alongside the initial results we communicated for 7.27
Figure 3.
Structures of [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7), [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8), and the potential dual binding behavior of the amidine in 7.
Results and Discussion
Thionation survey
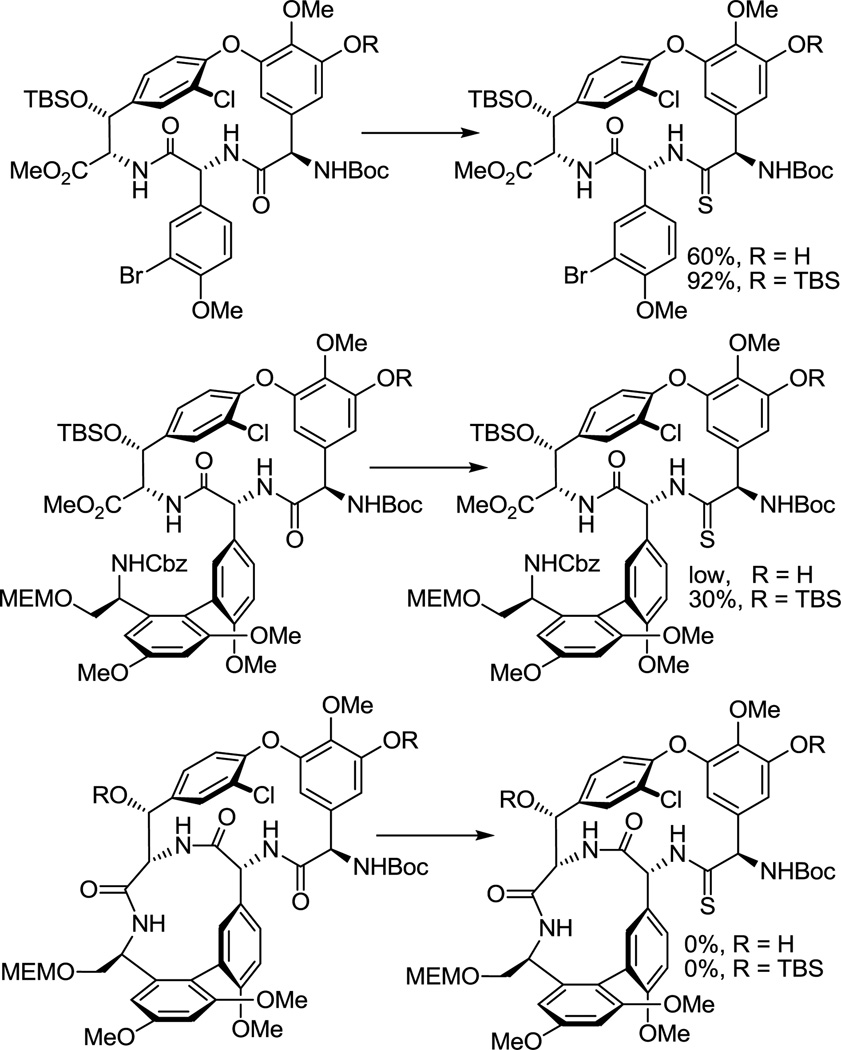
Although not yet comprehensively examined, efforts to selectively convert the amide linking residues 4 and 5 to a thioamide in a vancomycin aglycon derivative have not yet been successful in our hands. Consequently, we surveyed progressively simpler partial structures available from our vancomycin aglycon synthesis. At the initiation of our efforts, we found that the residue 4 amide could be selectively converted to the corresponding thioamide using Lawesson’s reagent within the isolated CD ring system where it is the sterically more accessible of only two possible amides, and that this could be further improved by conducting the reaction on the t-butyldimethylsilyl (TBS) ether protected phenol, providing a remarkably selective and high yielding stage for thionation (Figure 4). At this early stage, the CD ring system thioamide emerged as our key precursor from which 7 and 8 were to be prepared. No efforts were made to extensively profile the less successful reactions and it is likely that each would benefit from a detailed study.
Figure 4.
Thionation survey using Lawesson’s reagent in toluene.
With a thionation stage established and following a strategy first introduced in our vancomycin aglycon synthesis, the overall approach to 7 and 8 was anticipated to rely on two key aromatic nucleophilic substitution reactions for formation of the 16-membered diaryl ethers in the sequential CD and DE ring formations, a macrolactamization for closure of the 12-membered biaryl AB ring system, and the defined order of CD, AB, and DE ring closures to permit selective thermal atropoisomerization of the newly formed ring systems or their immediate precursors. Thus, in addition to any kinetic atroposelectivity that may be achieved in each macrocyclization, the order of ring closures follows their increasing ease of thermal atropisomer equilibration (Ea for CD, AB, and DE ring systems = 30.4, 25.1, and 18.7 kcal/mol, respectively),21 permitting the recycling of any newly generated unnatural atropisomer under progressively milder thermal conditions where any atropisomer stereochemistry already in place is not impacted. This additional indirect control of the atropisomer stereochemistry allows all synthetic material to be funneled into the single atropdiastereomer found in the natural product.
CD Ring system synthesis
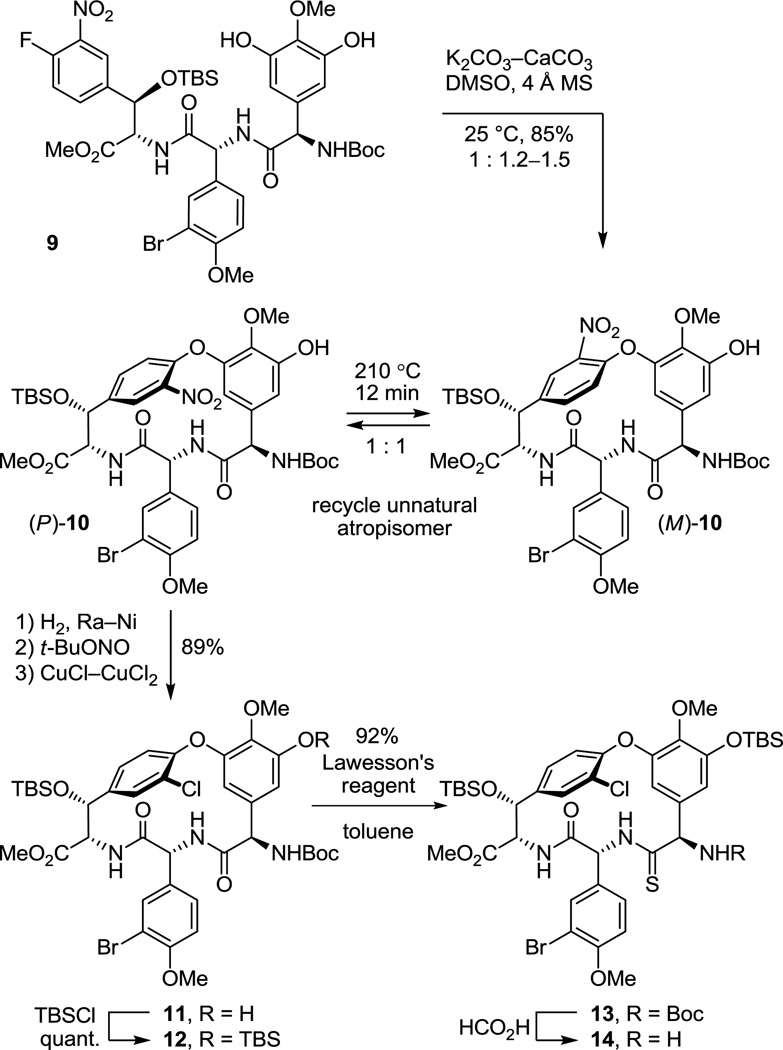
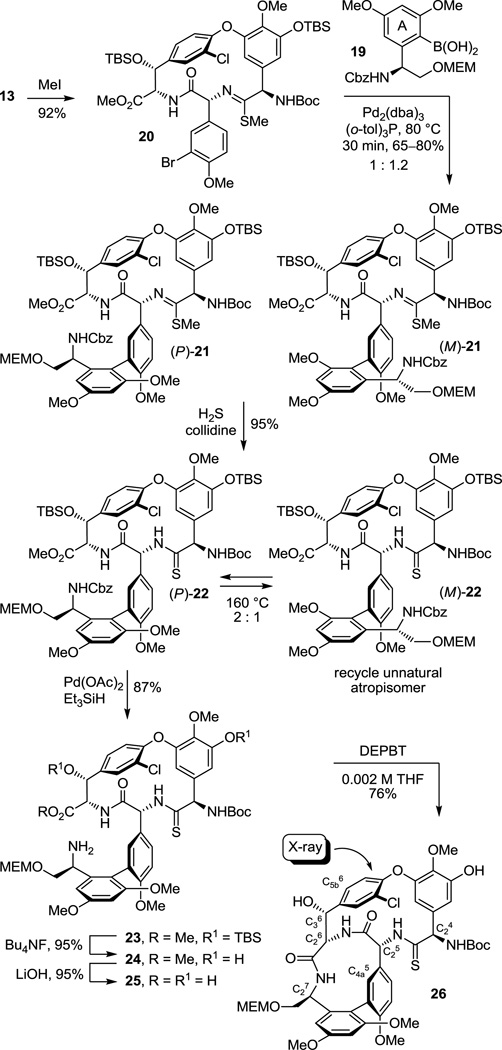
As a result of these studies, we were presented the opportunity to reexamine the CD macrocyclization reaction enroute to the vancomycin aglycon 5, potentially improving our original synthesis and benefiting from the experience gained with such aromatic nucleophilic substitution reactions since its disclosure. The acyclic tripeptide 9 was prepared in three steps (72% overall) from the constituent amino acid precursors as previously detailed (Scheme 1).17 Treatment of 9 with K2CO3/CaCO3 (5 equiv each, 4 Å MS, 0.005 M DMF, 45 °C, 12 h) provided 10 in good yield (50–60%) as a separable 1:1 mixture of atropisomers mirroring the results originally disclosed.17 Increasing the amount of CaCO3 (10 vs 5 equiv), which serves to scavenge liberated fluoride as insoluble CaF2 preventing competitive silyl ether deprotection and subsequent base-catalyzed retro aldol cleavage of the newly formed CD ring system, slightly improved the conversions (65%). Consequently, a reexamination of the impact of base (Li2CO3, Na2CO3, K2CO3, Cs2CO3, Rb2CO3), additive (CaCO3), and solvent (THF, DMF, DMSO) on the reaction was conducted, enlisting progressively larger amounts of reagents (Supporting Information Table S1). From these studies, K2CO3/CaCO3 reemerged as the most effective combination and with the increased reagent amounts (20 equiv each) now provided 10 as an approximate 1:1 mixture of atropisomers in yields as high as 68%. The most significant improvements were observed by exploring the impact of solvent while enlisting the increased reagent amounts. As anticipated, the facility of the cyclization increased substantially with DMSO > DMF > THF such that effective rates of ring closure were observed at 25 °C (DMSO), 45 °C (DMF), or 75 °C (THF). With the increased amounts of reagents and especially the with the larger amounts of CaCO3, the reaction in anhydrous DMSO (0.005 M, 6–8 h, 25 °C) was now unusually clean affording 10 as a separable 1:1.2 mixture of atropisomers in excellent yield (75–85%) free of apparent silyl ether deprotection, retro aldol cleavage, or inadvertent epimerization. Although the scale up of this optimized reaction intermittently suffered progressively lower conversions with the reemergence of competitive desilylation and ester hydrolysis, these were effectively addressed using either finely milled nano-CaCO3 (avg size = 0.34 µm vs 26 µm diameter) or carefully flame-dried reagents (20 equiv of K2CO3, 20 equiv of CaCO3, 3 wt equiv 4 Å MS, 0.005 M DMSO, 10 h, 25 °C). The isolated unnatural atropisomer (M)-10 was thermally equilibrated, regenerating a 1:1 mixture of M:P atropisomers from which additional amounts of the natural (P)-atropisomer were isolated, and this was conducted as previously disclosed (o-Cl2C6H4, 150 °C, 48 h, 85% recovery)17 or under improved microwave conditions (o-Cl2C6H4, 210 °C, 12 min, 89% recovery). Thus, the isolation and re-equilibration of the unnatural atropisomer was used to funnel all material into the synthetic sequence.
Scheme 1.
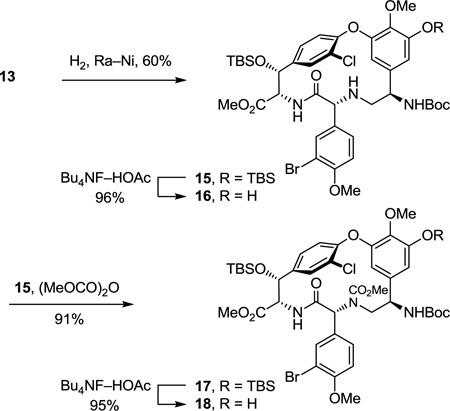
The cyclized material (P)-10 was converted to the aryl chloride 11 (82–89%) by reduction of the nitro group, diazotization of the resulting aniline, and Sandmeyer substitution following procedures previously disclosed.17 Although 11 could be converted to the corresponding thioamide upon treatment with Lawesson’s reagent (1 equiv, toluene, 80 °C, 8 h, 54–63%), protection of the phenol as its TBS ether 12 (quant.), preventing its competitive reaction with the reagent, and subsequent thionation with Lawesson’s reagent (1 equiv, toluene, 55–60 °C, 2–3 h) provided 13 in superb conversions (85–92%) with complete selectivity for the residue 4 amide. One of the challenges in characterizing 13 in early studies was the presence of carbamate rotamers (1:1 to 1:8 depending on the solvent) that complicated assessment of the reaction selectivity. Although these rotamers did not readily coalesce with variable temperature NMR at modest temperatures (< 60 °C) in a variety of solvents, N-Boc removal from 13 (HCO2H, CHCl3, 0 °C) provided the corresponding free amine 14 as a single compound. Additionally and without optimization, reduction of the thioamide 13 (H2, Ra–Ni, MeOH, –20 to 0 °C, 3 h, 60%) in the presence of formamidine acetate to prevent competitive aryl dechlorination and subsequent selective phenol TBS ether deprotection of 15 (1 equiv of Bu4NF, 5 equiv of HOAc, THF, 25 °C, 0.5 h, 96%) provided 16 (equation 1).23 Alternatively, treatment of 15 with a large excess of dimethyl dicarbonate (4 N aq NaOH–THF (1:2), 25 °C, 30 min, 91%) prior to subsequent selective phenol TBS ether deprotection of 17 (2 equiv of Bu4NF, 5 equiv of HOAc, THF, 25 °C, 1 h, 95%) provided 18, identical in all respects to authentic material prepared in our earlier efforts.23 Not only does this provide an alternative synthesis of the key intermediate 18 employed in our synthesis of [Ψ[CH2NH]Tpg4]vancomycin aglycon23 (6) and provide precedent for a late-stage conversion of 8 to 6, but the conversion of 13 to 18 also served to confirm the thionation site and macrocycle stereochemistry.
 |
(1) |
ABCD Ring system synthesis
Completion of the ABCD ring system required introduction of the 12-membered AB ring system and was expected to follow protocols we first introduced with vancomycin itself.17 Although direct Suzuki coupling of 13 with the A-ring boronic acid 19 proceeded to give the biaryl product 22 in good yield (69–79%) in early efforts, the reaction proved difficult to dependably reproduce (Scheme 2). Initially, sulfur byproducts from the thionation reaction were suspected to shut down this reaction, but even careful purification of 13 did not seem to alter this reaction irreproducibility. Rather, it appeared to be directly related to the presence of the substrate thioamide itself. Converting thioamide 13 to the corresponding methyl thioimidate 20 (2 equiv of MeI, 4 equiv of K2CO3, acetone, 25 °C, 3 h, 92%), protecting the coordinating thioamide as a less nucleophilic thioimidate, provided a substrate that cleanly and reproducibly participated in the Suzuki coupling reaction. Thus, coupling of 20 with the boronic acid 19 (2.5 equiv) catalyzed by Pd2(dba)3 (0.3 equiv) in the presence of the ligand (o-tol)3P (1.5 equiv) in toluene/MeOH/1 M aq NaHCO3 (3/1/0.5; 0.1 M in substrate) proceeds in 15–30 min at 80 °C providing 21 as a mixture of atropisomers (typically 1:1.2) in good yield (65–80%). Like the substrate bearing a residue 4 amide for which we first developed these reaction conditions,17 the Suzuki coupling of 20 proceeds at remarkable rates and conversions considering the hindered nature of the boronic acid and the electron rich character of the hindered aryl bromide. Liberation of the thioamide (H2S, 2 equiv of collidine, MeOH, 25 °C, 2 h, 95%) provided (P)-22 and its separable unnatural atropisomer. The unnatural atropisomer (M)-22 was thermally equilibrated under conditions that do not affect the CD atropisomer stereochemistry (o-Cl2C6H4, 120 °C, 10–24 h or microwave at 160 °C, 30 min, 2.6–1.8:1 P:M 22 with 83–99% recovery) to provide the thermodynamically preferred (P)-22, funneling all material into the synthetic sequence. The use of Na2CO3 (vs NaHCO3) in the Suzuki coupling led to lower conversions and detectable epimerization, the inclusion of collidine in the thioamide liberation minimized trace racemization, attempts to equilibrate the separable thioimidate atropisomers were not effective and led to the generation of a detectable new diastereomer, and the overall conversions were higher if the intermediate thioimidate atropisomers were not separated prior to thioamide liberation where the preparative separation was simpler and the product stability greater.
Scheme 2.
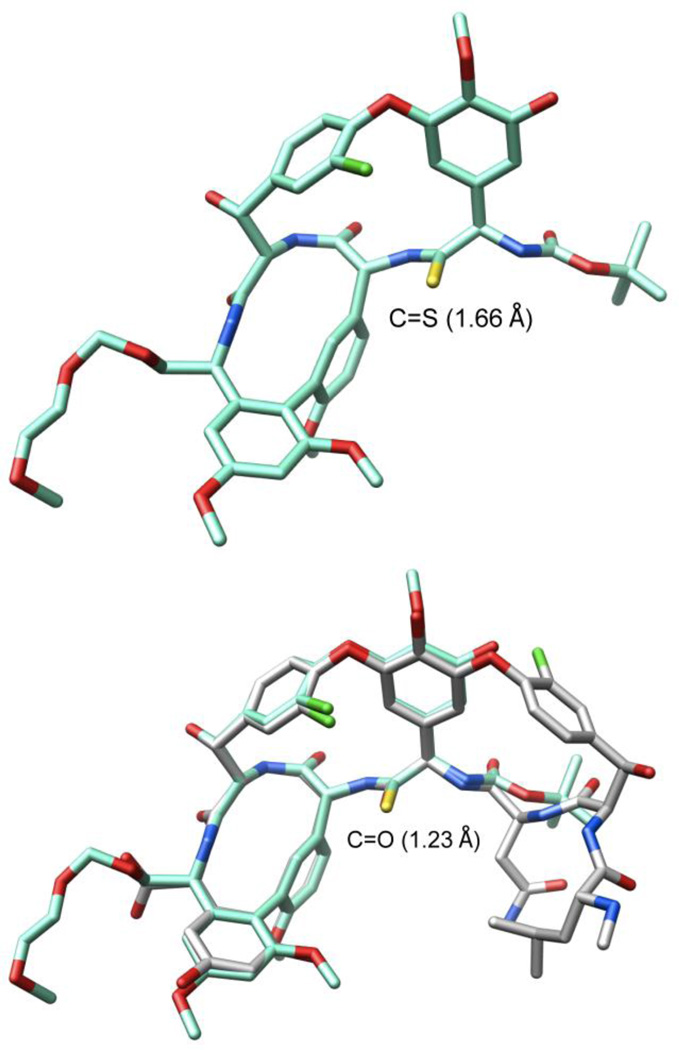
Ionic Pd-catalyzed benzyloxycarbonyl (Cbz) removal on (P)-22 (0.3–0.5 of equiv Pd(OAc)2, 10 equiv of Et3SiH, 1–1.5 equiv of NMM, CH2Cl2, 25 °C, 24 h, 87%),28,29 silyl ether deprotection of 23 (7 equiv of Bu4NF, 8 equiv of HOAc, THF, 25 °C, 3 h, 95%), hydrolysis of the resulting methyl ester 24 (2 equiv of LiOH, 2:1 t-BuOH–H2O, 0 °C, 2 h, 87–100%), and macrocyclization of the amino acid 25 (2–5 equiv of DEPBT,30 2–5 equiv of NMM, 0.002 M THF, 0–25 °C, 12–17 h, 70–79%) provided 26, the fully functionalized ABCD ring system bearing the core residue 4 thioamide. The use of ionic hydrogenolysis conditions for the Cbz deprotection, which provided 23 in yields as high as 97% (80–97%), avoided competitive thioamide reduction observed to be problematic with conventional reagents. Additionally, it provides an initial and unusually stable triethylsilyl (TES) carbamate29 that required subsequent deliberate deprotection to fully release the free amine (saturated aq. NH4Cl, 2 h). The silyl ether removal with Bu4NF–HOAc (vs Bu4NF) prior to methyl ester hydrolysis, rendering the methyl ester sterically more accessible, as well as careful optimization of the hydrolysis reaction conditions prevented competitive ester epimerization. Reversing the order of ester and carbamate deprotections led to competitive resilylation of the required free alcohol and phenol under the ionic hydrogenolysis reaction conditions. Additionally, a number of reagents were examined to effect the macrolactamization of the AB ring system, including EDCI–HOBt (42–50% and dr 2.5–3:1), FDPP (40–66% and dr 5:1), and PyBop (20% and dr 1:2), that suffered comparatively lower conversions and more extensive racemization than 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4-one (DEPBT),30 which was superb at minimizing racemization (71% and dr 7:1).31 More subtly, conducting the macrocyclization with the A-ring C-terminus MEM ether (vs methyl ester) accelerates the rate of macrocyclization, diminishing competitive epimerization of the activated carboxylic acid that accompanies ring closure, and improves the overall conversions.17 However, it necessitates a late stage alcohol deprotection and subsequent oxidation to provide the C-terminus carboxylic acid, a conversion viewed as potentially problematic in the presence of a residue 4 thioamide. Finally, the structure and conformation of 26 initially was established by NMR, exhibiting diagnostic NOE’s analogous to those observed with vancomycin itself, confirming the atropisomer stereochemistry, the cis amide between residues 5 and 6 central to the AB ring system, and the location of the thioamide [e.g., C4a5-H/C25-H, C4a5-H/C26-H, C25-H/C26-H (cis amide), C5a6-H/C36-H and C5a6-H/C26-H (atropisomer stereochemistry); vancomycin numbering]. These conformational properties of 26 were not surprising since earlier studies, including those describing the more flexible methylene analogue 6, showed that the AB ring system and its rigid conformation bearing a cis amide central to even its isolated structure defines the conformation characteristic of the vancomycin ABCD ring system.17,23 The structure, stereochemistry, and conformation of 26 were unambiguously established with a single-crystal X-ray structure determination32 conducted on crystals grown from acetone–hexane that overlays nearly precisely with the ABCD ring system of vancomycin with the exception that the C=S bond length (1.66 Å) of the thioamide 26 exceeds the C=O bond length (1.23 Å) of the vancomycin amide (Figure 5).33
Figure 5.
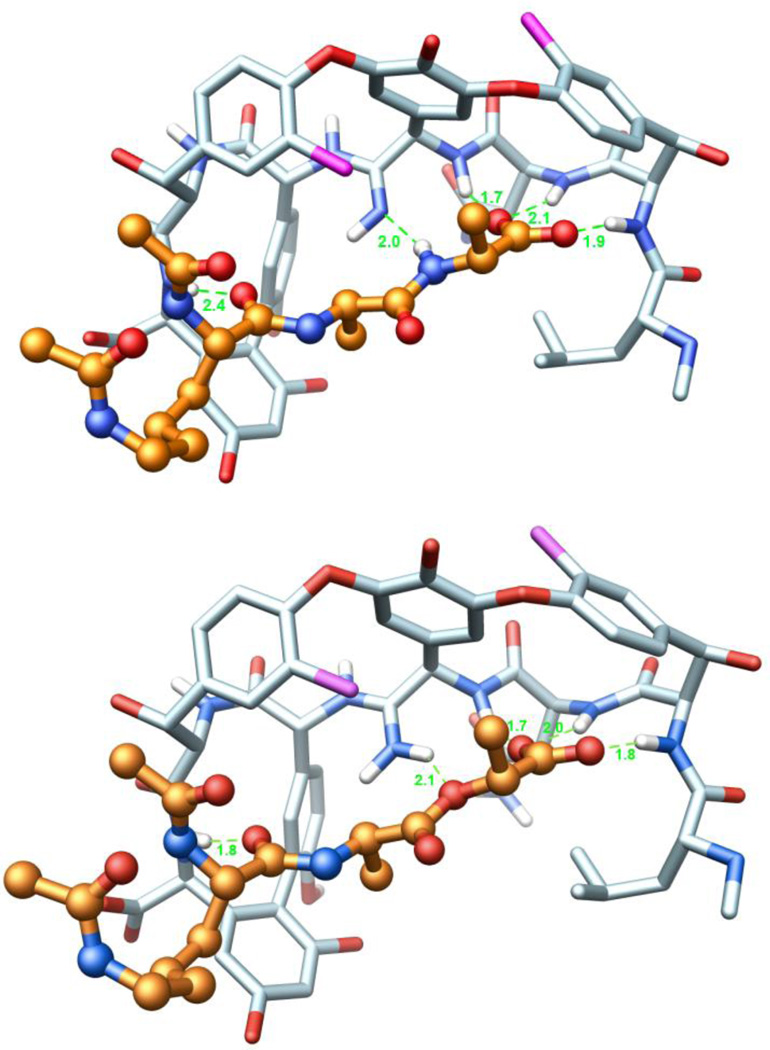
X-ray structure of 2632 and its overlay with an X-ray structure of vancomycin (1) taken from a bound complex with a model d-Ala-d-Ala ligand (PDB code 1FVM, 1.8 Å resolution).33 Superimposition performed with the molecular modeling program MOLOC34 and graphics generated using Chimera.35
Introduction of the DE ring system and synthesis of [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8)
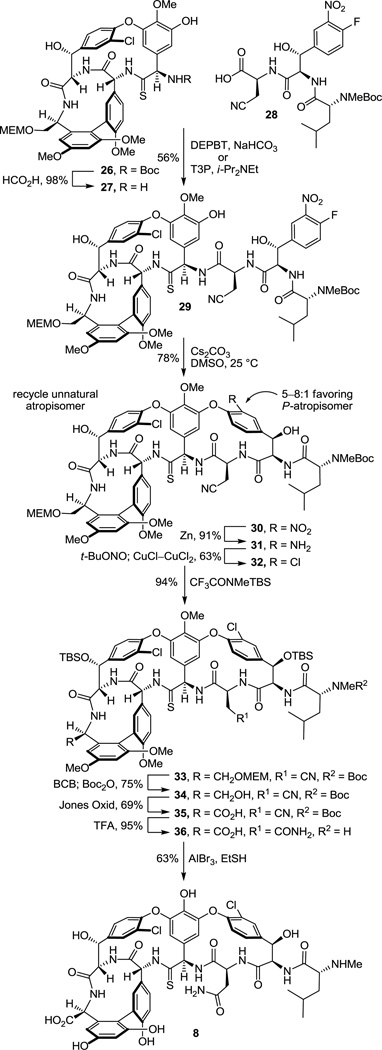
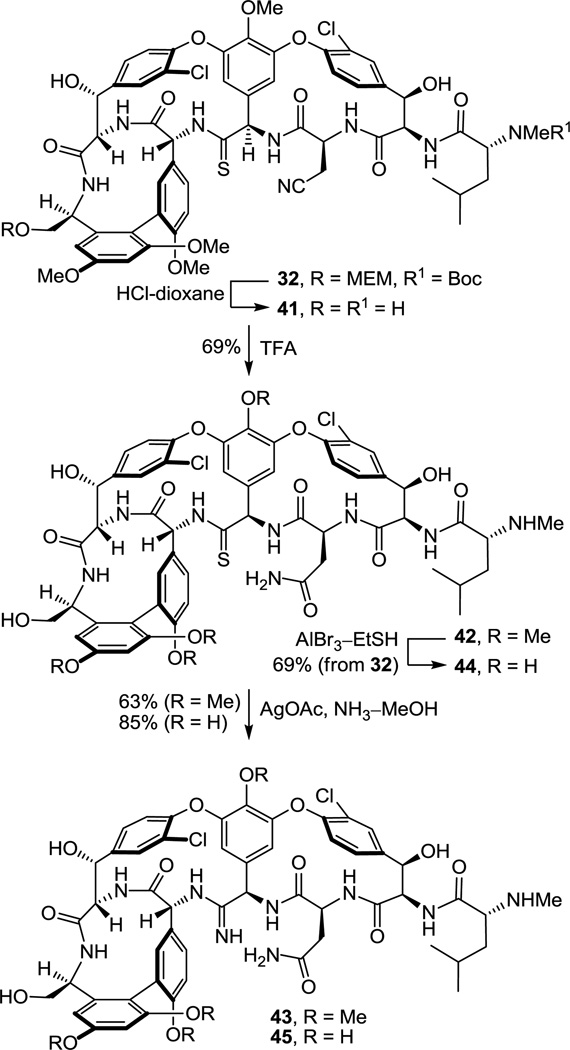
This set the stage for the introduction of the DE ring system. N-Boc deprotection of 26 (85–98%) and subsequent coupling of the amine 27 with 2817 provided substrate 29 for the final cyclization (Scheme 3). This coupling reaction is more challenging than it might appear with nearly all reagents providing substantial amounts of racemization (e.g., EDCI–HOAt, 0 °C, dr = 2:1, 60% 29). In earlier efforts on teicoplanin where competitive racemization is even more problematic,18 we found that DEPBT30 was the only reagent capable of promoting the analogous coupling without significant racemization and DEPBT (2 equiv) was also found to work well for promoting the coupling (4 equiv of NaHCO3, THF, 0 °C, 16 h) of 27 with 28 (68%, dr > 4.5:1, 56% 29). A continued survey of newer or alternative reagents also revealed that commercially available propylphosphonic anhydride (T3P, 2.4 equiv)36 performed equally well (4 equiv of i-Pr2NEt, THF, 0 °C, 0.5–1.5 h), providing 29 in essentially identical conversions (68%) and dr (4.5:1, 56% 29). In addition, the reaction promoted by T3P proceeded at a much faster rate (1 vs 16 h) and proved much easier to conduct because of the workup water soluble byproducts, suggesting that it represents an attractive alternative reagent with coupling characteristics similar to DEPBT. The final macrocyclization, an aromatic nucleophilic substitution reaction for formation of the diaryl ether and closure of the 16-membered DE ring system, was effected by treatment of 29 with Cs2CO3 (3–4 equiv, 0.005 M DMSO, 9–12 h) at room temperature to provide 30 and its DE atropisomer in superb yield (74–78%) as a separable 5–8:1 mixture of atropisomers favoring the natural (P)-isomer (58–66%). A range of reagents were found to be effective for promoting this cyclization (10 equiv of CsF, 50 equiv of K2CO3–CaCO3, or 50 equiv of K2CO3) under remarkably mild room temperature reaction conditions when conducted in DMSO, approaching or matching the conversions and atropodiastereoselectivity (4–7:1) observed with Cs2CO3 (5–8:1). The distinguishing feature of Cs2CO3 that led to its use in our efforts going forward was its ease of implementation (non-hygroscopic), as well as the reduced number of equivalents of reagent needed to promote the ring closure that simplified workup of the reaction mixture. The minor unnatural DE atropisomer was collected and, when accumulated in sufficient quantities, thermally equilibrated (1:1, o-Cl2C6H4, microwave at 170 °C, 30 min) without impacting the AB and CD atropisomer stereochemistry, funneling all material into the synthetic sequence. Conversion of the aryl nitro substituent to the corresponding chloride 32 (55–60%) proceeded smoothly under conditions that avoid use of excess oxidant (t-BuONO) and the resulting competitive thioamide oxidation. Compound 32 served as our storage point for the synthetic efforts from which material was carried forward to 7 and 8 only as needed, or diverted to allow the preparation of related key analogues (e.g., 42–45). Protection of the secondary alcohols as their TBS ethers (95%) and subsequent MEM ether deprotection using B-bromocatecholborane (BCB) followed by N-Boc reprotection provided the C-terminus primary alcohol 34 (75%), poised for oxidation to the corresponding carboxylic acid 35 (Scheme 3).
Scheme 3.
We approached this conversion with considerable concern, recognizing that most reagents previously employed in the vancomycin field would likely oxidize the residue 4 thioamide, promoting its conversion to an amide. As a result, a series of reagents were examined first with the primary alcohol derived from 26 in hopes of identifying one capable of finessing this conversion. These studies were largely unsuccessful, leading to competitive or preferential thioamide oxidation. With the benefit of results derived from concurrent efforts on the conversion of 34 to the corresponding amidine, we came to recognize that the thioamide of 34 is deeply embedded in the core structure, often sterically protected from reaction. As a result, we found that Jones oxidation37 (4 equiv CrO3, aq. H2SO4–acetone, 12–24 h) under controlled reaction conditions selectively and directly oxidizes the primary alcohol 34 to the corresponding carboxylic acid 35 (63–69%) in a single step with a manageable amount of competitive oxidative conversion of the thioamide to the amide (≥10:1). Somewhat larger amounts of the amide (5–7:1) were observed with use of less H2SO4 as an additive, but were further minimized with the use of a larger amount of the additive. Significantly, the amide byproduct produced proved identical in all respects to authentic material,17 confirming the structure and stereochemistry of the precursor and product thioamides 34 and 35. More popular oxidizing reagents and two-step procedures typically deployed on such occasions including Dess–Martin periodinane, Swern oxidation, SO3–Pyr, Bu4NRuO4 (TPAP), pyridinium dichromate (PDC),38 pyridinium chlorochromate (PCC),38 and 2-iodoxybenzoic acid (IBX) provided little or no desired reaction or complex mixtures of products even when employed in the presence of thiourea as a diversion additive, and 2,2,6,6-tetramethypiperidine-N-oxyl/(bisacetoxyiodo)benzene (TEMPO–BAIB) selectively provided what appeared to be the isolable thioamide S-oxide. The subsequent penultimate conversion of the nitrile found in 35 to the corresponding primary carboxamide 36 was not successful when we enlisted the mild conditions (H2O2, K2CO3, DMSO–H2O, 25 °C) developed for vancomycin itself due to competitive thioamide oxidation.17 Recognizing the robust stability of vancomycin to acidic conditions, the nitrile hydrolysis to the carboxamide was accomplished by exposure to TFA–H2SO439 (25 °C) or more simply trifluoroacetic acid39 (TFA, 25 °C, 12 h, 95%), which also served to remove the N-terminus Boc group. Finally, treatment of 36 with AlBr3 (250 equiv) in EtSH (25 °C, 12 h)17 served to globally remove the four aryl methyl ethers and two TBS ethers, providing [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8) in good conversions (63%). The physical properties and chromatographic behavior of 8, like that of the intermediate thioamides, proved similar to vancomycin aglycon or the corresponding residue 4 amides, albeit characteristically being slightly less polar in their behavior.
Synthesis of [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (7)
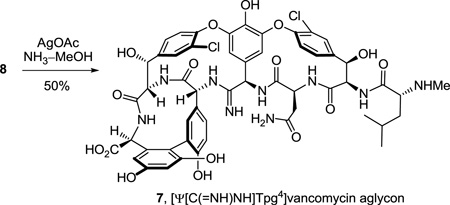
Key to the work and central to our objectives was the conversion of a thioamide to an amidine. Optimally, we felt this should be conducted on 8 itself requiring only one additional step for the preparation of 7, establishing it as a late stage penultimate intermediate from which additional deep-seated and site specific changes in this key region of the vancomycin core structure could be conducted. Thus, single-step treatment of the fully deprotected vancomycin aglycon thioamide 8 with AgOAc40 (10 equiv) in saturated NH3–MeOH at 25 °C (12 h) provided the amidine 7, [Ψ[C(=NH)NH]Tpg4]vancomycin aglycon (>50%), cleanly as a colorless solid that proved to be stable to extensive handling and purification (equation 2). Although there is no direct precedent that we are aware of detailing the use of AgOAc for promoting direct amidine generation upon reaction of ammonia with a thioamide, it was examined based on its well-recognized thiophilicity.40 AgOAc proved more effective than alternative, commonly employed reagents (e.g., HgCl2 or Hg(OAc)2)41 whose use in protic solvents with ammonia were found to be precluded by extensive salt precipitation. Beyond the homogeneous reaction conditions and central to the success with AgOAc is the ability to use a relatively poor nucleophile (NH3) in a protic solvent (MeOH) where the substrate is readily soluble, in a reaction conducted on the fully functionalized and deprotected vancomycin aglycon without the detection of reagent-derived competitive reactions. The product amidine 7 is considerably more polar than 5 or 8, most likely reflecting amidine protonation, is readily soluble in H2O or H2O–MeOH but insoluble in MeCN, and required addition of TFA to the sample before reverse-phase high performance liquid chromatography (HPLC) purification (5 µm C18, 0.46 × 15 cm, 30% CH3OH/H2O–0.07% TFA, 3 mL/min, Rt = 17.9 min or 5–20% MeCN/H2O–0.07% TFA, 3 mL/min, Rt = 21.6 min). Although our experience in handling 7 is still limited, the conversion of 8 to 7 is clean and complete (>85% by LCMS) although it is conservatively reported herein as 50%. We have occasionally experienced losses of material during HPLC purification that may be related to additional unappreciated features of its physical properties that are under continued examination.
 |
(2) |
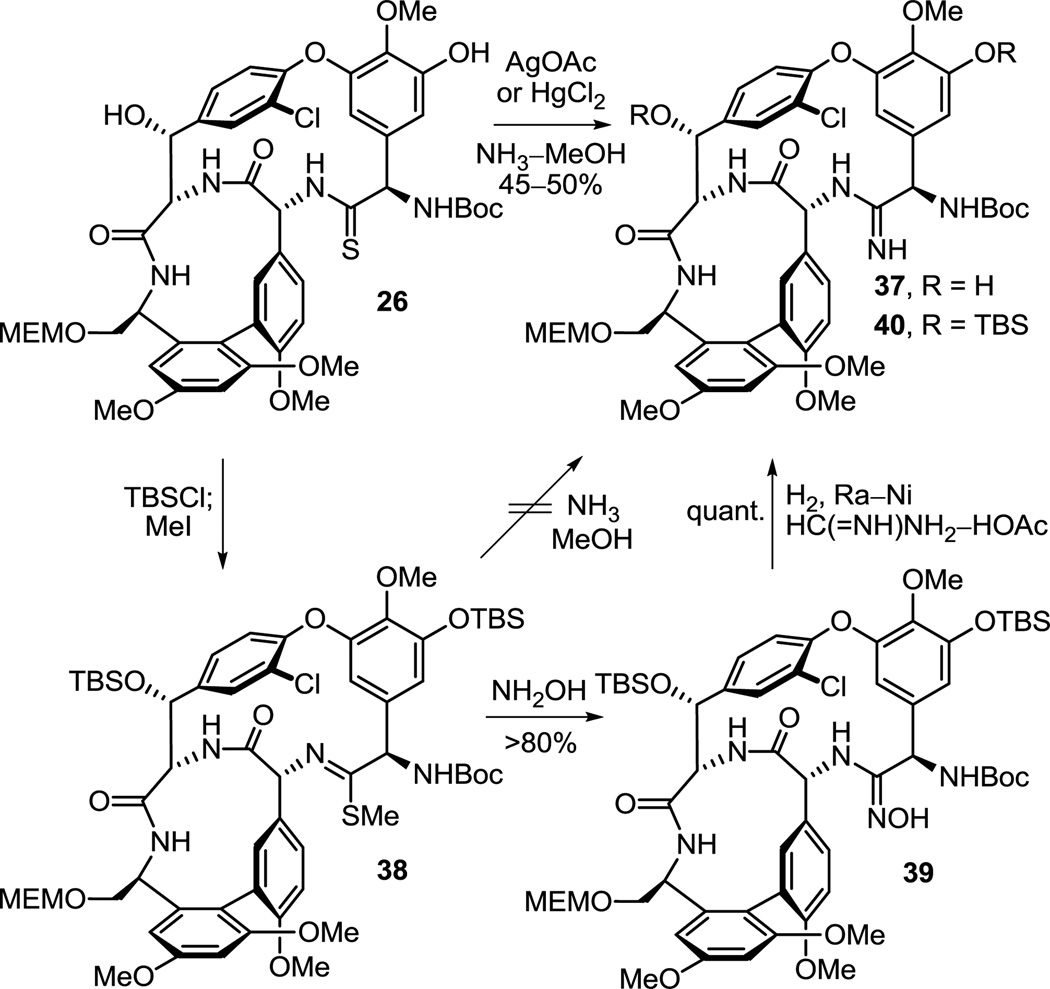
The simplicity of this transformation does not do justice to the efforts that went into its adoption. Throughout the course of our studies and many times in pursuit of establishing the viability of earlier stage amidine introductions in alternative routes to 7, the conversions of the thioamides in 13, 26, 32 and 34 to the corresponding amidines were examined. Although these proved instructive and provided additional key derivatives for comparative examination (e.g., 43 and 45), establishing the experience needed to conduct this transformation within the chemical and structural framework of a fully functionalized and fully deprotected vancomycin aglycon, they were not especially predictive of what approach might be successful when applied to 8 itself. In retrospect, we now understand that many of the subtleties of these observations reflect the protecting group status of the earlier stage intermediates rather than the reactions themselves, in particular the residue 3 nitrile that can also react with NH3. Nonetheless, the efforts with 26 proved most predictive and, without optimization, several direct conversions of its thioamide to the corresponding amidine were found to be effective of which the AgOAc (vs HgCl2 or Hg(OAc)2)41 promoted reaction (10 equiv, NH3–MeOH, 25 °C) proved to be the cleanest, most direct, and most effective (Scheme 4). The Hg(II)-promoted reactions, while often clean and more rapid, always suffered from poor recovery due in part to the precipitated salts that form in the reaction mixtures with NH3. Efforts to conduct the conversion in a stepwise fashion with NH3 displacement on the intermediate methyl thioimidate 38 were surprisingly ineffective, although more powerful nucleophiles including hydroxylamine cleanly react with 38. Reduction (H2, Ra–Ni or TiCl3) of the resulting hydroxyamidine (amidoxime) 39, also available directly through a AgOAc or HgCl241 promoted reaction of hydroxylamine with the thioamide, provided the corresponding amidine 40, but required controlled conditions for reduction with Ra–Ni to avoid competitive aryl dechlorination. Significantly, analogous thioamide functionalizations are planned with 8 itself, and the results of such studies will be disclosed in due course.
Scheme 4.
Additional residue 4 thioamide and amidine derivatives
Previous studies have shown that the C-terminus carboxylic acid of vancomycin does not have an impact on d-Ala-d-Ala binding and it has been explored extensively as a productive derivatization site, tolerating conversion to ester, amide, and alcohol derivatives. Similarly, phenol methylation is well tolerated, often is beneficial for imparting antimicrobial activity against vancomycin-resistant bacteria,22,42 and does not significantly impact model d-Ala-d-Ala or d-Ala-d-Lac binding. Consequently, earlier stage tricyclic thioamide and amidine derivatives were also prepared bearing a C-terminus hydroxymethyl group with and without the corresponding phenol methyl ether protection and made available for examination (Scheme 5). These earlier stage intermediates not only allowed the development of refined conditions for the amidine introduction, but their examination served to establish the generality of the amidine and thioamide biological properties. Concurrent C-terminus MEM ether and N-terminus Boc deprotection of 32 by treatment with 4 N HCl–dioxane (25 °C, 8 h) and subsequent residue 3 nitrile hydrolysis of 41, enlisting TFA treatment (25 °C, 12 h) followed by a workup with MeOH (25 °C, 1 h) to convert the intermediate trifluoroacetoxy imidate to the primary carboxamide, provided thioamide 42 (66%, 2 steps). We later found that omitting the HCl-catalyzed deprotections and simply treating 32 with TFA (25 °C, 16 h) followed by workup with MeOH (25 °C, 18 h) provided 42 directly (69%), effecting the MEM ether and Boc deprotections along with the nitrile conversion to the primary carboxamide. Single-step, direct conversion of 42 to the corresponding amidine was effected by treatment with AgOAc (10 equiv) in saturated NH3–MeOH (25 °C, 24 h) to provide 43 (63%), which proved surprisingly more challenging to work with43 than either 7 or 45 (below).
Scheme 5.
However, global deprotection of the four methyl ethers in the thioamide 42 (250 equiv of AlBr3, EtSH, 25 °C, 4 h) cleanly provided the free phenol 44 (61–69% from 32) without impacting the thioamide. Single-step, direct conversion of 44 to the corresponding amidine with AgOAc (10 equiv) in saturated NH3–MeOH (25 °C, 24 h) provided the well-behaved and stable amidine 45 (85%), which differs from the structure of 7 only at the C-terminus (hydroxymethyl group vs carboxylic acid). Interestingly, reverse-phase HPLC purification of 45 (5–20% MeCN/H2O–0.7%TFA) revealed that the compound is actually slightly more polar than the corresponding carboxylic acid 7. Finally and in the course of these efforts, we also established that treatment of the corresponding residue 4 amide 47 with an even larger excess of AgOAc (50 equiv) in NH3–MeOH for a more extended reaction time (48 h, 25 °C) provided only recovered starting material, indicating no side reactions at sites other than the thioamide might be expected upon exposure to the reaction conditions.
Binding to model d-Ala-d-Ala and d-Ala-d-Lac ligands
The binding assays were conducted at pH 5.1 in a citrate buffer as originally disclosed by Perkins.8,44 At this pH and regardless of any structural perturbation in a protonated amidine pKa (12.5), the residue 4 amidine would be expected to be fully protonated in the absence of a ligand, even more so than a simple amine (pKa = 11.5). Similarly, the presence of the excess citrate in the buffer insures nonspecific binding of a ligand carboxylic acid is not observed in the vancomycin binding pocket. Thus, although the incorporation of the amidine in the potentially hydrophobic vancomycin binding pocket might be expected to lower the protonated amidine pKa, and one might be concerned with whether the residue 4 amidine may nonspecifically bind a ligand carboxylic acid, the binding assay conditions mitigate such concerns, providing conditions under which to critically evaluate only the specific interaction of ligands with the candidate analogues.
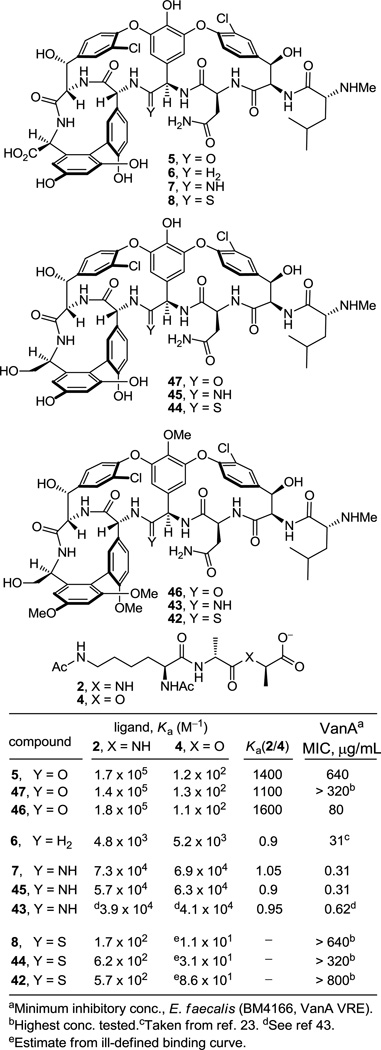
Although [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8) was prepared as the key immediate precursor to 7, it proved especially revealing to examine its binding characteristics with the model d-Ala-d-Ala and d-Ala-d-Lac ligands 2 and 4, respectively. Since thioamides are regarded as weaker H-bond acceptors than the corresponding amides, the affinity of 8 for 2 was anticipated to be reduced, whereas its potential behavior toward 4 was more difficult to predict. As reported earlier,27 its behavior and that of the related thioamides 42 and 44 proved stunning (Figure 6). All three thioamides failed to bind either the model d-Ala-d-Ala or d-Ala-d-Lac ligands 2 and 4 to any appreciable extent and all three failed to exhibit appreciable antimicrobial activity against sensitive or vancomycin-resistant bacteria. The 1000-fold loss in affinity for the d-Ala-d-Ala ligand 2 relative to the vancomycin aglycon (5) is both general (8, 42, and 44) and remarkable, indicating that this simple exchange in a single atom (O→S) is sufficient to completely disrupt binding. Although the weaker H-bonding capability of a thioamide thiocarbonyl may contribute to this lowered affinity, the magnitude of the loss indicates something more fundamental is responsible. As reported earlier,27 we suggest that both the increased bond length of the thiocarbonyl (1.66 vs 1.23 Å, see Figure 5) and the larger van der Waals radii of sulfur (1.80 vs 1.52 Å) are sufficient to sterically displace and completely disrupt the intricate binding of d-Ala-d-Ala. This behavior of all three residue 4 thioamides, which initially tempered our expectations of 7, serves as a sharp contrast to the remarkable behavior of the residue 4 amidines detailed below.
Figure 6.
Comparison of residue 4 amide, thioamide, and amidine derivatives.
The behavior of the residue 4 amidine derivatives proved truly remarkable (Figure 6). Both the bond length of an amidine (1.30 vs 1.23 Å) and the van der Waals radii of a nitrogen atom (1.55 vs 1.52 Å) closely approximate those of an amide carbonyl and oxygen atom, suggesting they may serve as effective amide isosteres in peptides. The binding affinity44 of 7 and the closely related amidine 45 with the model d-Ala-d-Ala ligand 2 was only approximately 2-fold less than that of the residue 4 amides including vancomycin aglycon itself and 15-fold higher than the corresponding methylene derivative 6, suggesting that the residue 4 amidine functions well as a H-bond acceptor for the amide NH in the model ligand. Moreover, this binding affinity of 7 and the closely related amidine 45 was maintained with the model d-Ala-d-Lac ligand 4, representing a nearly 600-fold increase in affinity relative to the residue 4 amides including the vancomycin aglycon (5) and a more than 10-fold increase in affinity relative to the methylene derivative 6. Significantly, 7 and the related residue 4 amidines display effective dual, balanced binding affinity for both model ligands (Ka 2/4 = 0.9–1.05) at a level that is within 2–3 fold that exhibited by vancomycin for d-Ala-d-Ala.
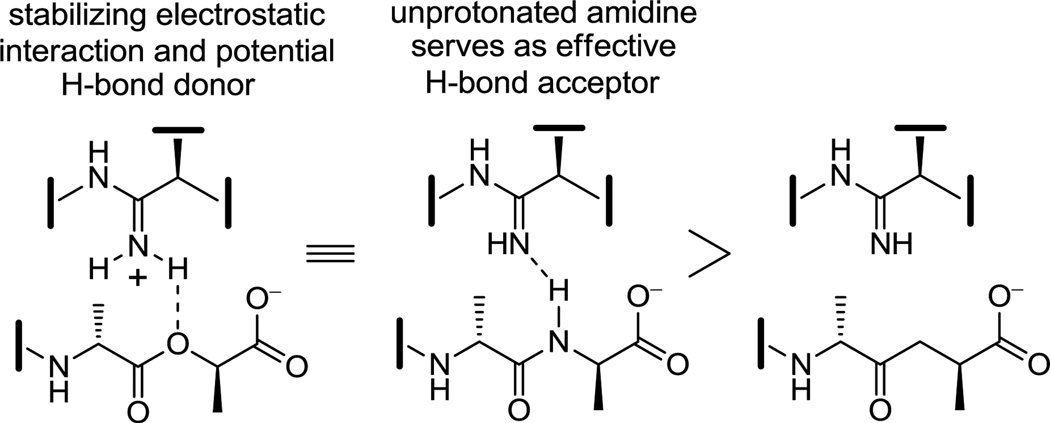
Although the behavior of 7 and the related residue 4 amidines toward the model d-Ala-d-Ala ligand 2 may not be too surprising on the surface, it requires a presumably ligand induced unprotonated (vs protonated) amidine to function effectively as a H-bond acceptor for the ligand amide NH. The behavior of the amidines toward the d-Ala-d-Lac ligand 4 is even more remarkable. There is no precedent on which to suggest that the residue 4 amidine could function as such a good H-bond donor to the ester oxygen of the d-Ala-d-Lac ligand to achieve this level of increased affinity. Rather, we have suggested that this is largely the result of a now stabilizing electrostatic interaction between the protonated amidine and the ester oxygen lone pairs27 (Figure 7). Thus, removing the vancomycin carbonyl oxygen atom and its destabilizing lone pair repulsion with the d-Ala-d-Lac ester oxygen atom and replacing it with a protonated amidine nitrogen and its now complementary stabilizing electrostatic interaction and H-bond donor capability reinstates essentially full binding affinity to the altered ligand. Although our studies do not experimentally distinguish whether the amidine is protonated or unprotonated when binding d-Ala-d-Lac and either is feasible, an unprotonated amidine would offer only the stabilization of a questionable H-bond and less or little in the way of additional electrostatic stabilization. As a result and because the amidine (pKa = 12.5) would be expected to be fully protonated in the absence of a ligand under the conditions of the assay (pH = 5.1), it is unnecessary to invoke and unlikely to involve binding of the unprotonated amidine. Beautifully, this represents a complementary single atom exchange in the antibiotic (O→NH) to counter a corresponding single atom exchange in the cell wall precursors of resistant bacteria (NH→O).
Figure 7.
Proposed dual binding behavior of amidine derivatives toward d-Ala-d-Lac and d-Ala-d-Ala.
To further support the unique binding behavior of the residue 4 amidines and that it is directly related to their proposed interaction with the linking heteroatoms of the d-Ala-d-Ala and d-Ala-d-Lac ligands 2 and 4, the binding of the amidines 7 and 45 was also examined with the ketone ligand 3,12 lacking a linking heteroatom. The amidines were found to bind the ketone ligand 3 with affinities of 7.4 × 103 M−1 (7) and 8.8 × 103 M−1 (45), representing binding approximately 10-fold less effective than their binding to either 2 or 4 and consistent with both qualitative and quantitative expectations of the behavior of an unprotonated amidine (Figure 7).
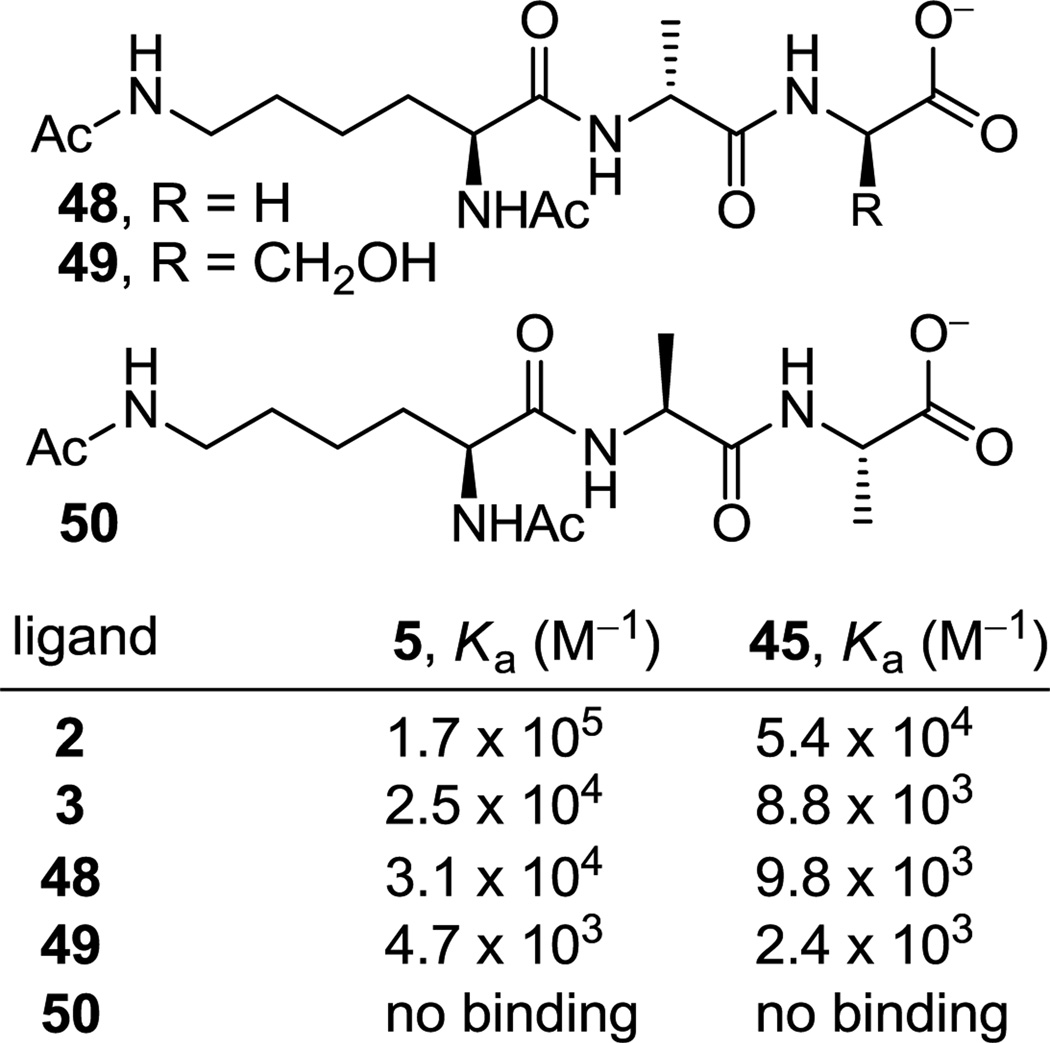
The binding of the amidine 45 with a series of additional ligands was also examined, and its comparison with vancomycin aglycon (5) demonstrate that they display identical trends (Figure 8). The ligands 48 and 49, which have been previously examined with vancomycin and related compounds,44 represent models of an alternative, less effective vancomycin binding site in the bacterial cell wall (48)44c and a modified peptidoglycan precursor found in a less prevalent form of vancomycin-resistant (VanC) bacteria (49).44e For both ligands and precisely in line with the differences observed with the d-Ala-d-Ala ligand 2 (2–3 fold), the amidine 45 binding was 2–3 fold lower the that of the vancomycin aglycon. Relative to the respective binding to d-Ala-d-Ala (2) and like 5, amidine 45 binding to the d-Ala-Gly ligand 48, lacking only the methyl group on the terminal amino acid, is reduced 5.5-fold (vs 5.5-fold for 5) and that of d-Ala-d-Ser 49, bearing the additional hydroxyl group, is lowered 23-fold (vs 36-fold for 5). Importantly and also like 5,44c no binding of amidine 45 with the l-Ala-l-Ala ligand 50 was observed, ruling out a nonspecific interaction with the examined ligands. This study further establishes that the amidines (1) bind such ligands in a manner identical to the residue 4 amides, (2) are subject to the same structural recognition features that dominate the vancomycin interaction with d-Ala-d-Ala and related ligands, and (3) eliminates the possibility that the amidines may be interacting with the ligands in a different or unique manner.
Figure 8.
Additional comparison ligand binding affinities of amidine 45 versus amide 5.
Additionally, the binding titration assay final mixture of 45 with the d-Ala-d-Lac ligand 4, containing the final 10-fold excess of ligand 4, was monitored for >2 months (room temperature) for evidence of either N-acylation of 45 by 4 or hydrolysis of 4 catalyzed by 45 that conceivably could contribute to the properties of the residue 4 amidines. However, both 45 and ligand 4 were unchanged under these conditions (pH 5.1) and no evidence of substrate acylation or ligand hydrolysis was detected (MS).
Models of amidine binding
Energy minimized models of the free base amidine binding to (Ac)2-l-Lys-d-Ala-d-Ala (2) and the protonated amidine binding to (Ac)2-l-Lys-d-Ala-d-Lac (4) are presented in Figure 9 along with the modeled H-bond distances. Most notably, those of the latter are nearly indistinguishable from those observed in the X-ray structure of vancomycin bound to the model (Ac)2-l-Lys-d-Ala-d-Ala ligand (2) illustrated in Figure 5. Analogous efforts to model the binding of the protonated amidine with the model (Ac)2-l-Lys-d-Ala-d-Ala ligand (2) did not provide as productive a complex that maintains the intricate interactions characterizing the natural product X-ray and NMR structures.
Figure 9.
(Top) Ball-and-stick representation of a energy minimized structure (MOLOC34) of 7 (C-atoms: light blue) and (Ac)2-l-Lys-d-Ala-d-Ala (C-atoms: salmon). (Bottom) Ball-and-stick representation of a energy minimized structure of 7 (C-atoms: light blue) and (Ac)2-l-Lys-d-Ala-d-Lac (C-atoms: salmon). Hydrogen atoms of all amides are depicted in white, H-bonds are depicted as green-dotted lines, and H-bonding distances are indicated. The corresponding H-bond distances for vancomycin binding to (Ac)2-l-Lys-d-Ala-d-Ala (see Figure 5) in the X-ray structure are 1.9, 1.9, 1.8, 2.0, and 1.7 Å, respectively. Structures obtained by modification of a co-crystal structure of vancomycin aglycon and (Ac)2-l-Lys-d-Ala-d-Ala (PDB code: 1FVM).
Antimicrobial activity
Accurately reflecting their binding properties, each of the residue 4 amidines exhibited potent and comparable antimicrobial activity (MIC = 0.3–0.6 µg/mL) against VanA resistant bacteria (E. faecalis, VanA VRE), the most stringent of vancomycin-resistant bacteria, being roughly 1000-fold more potent than vancomycin (1) and vancomycin aglycon (5), Figure 7, and equipotent to the activity they display against sensitive bacterial strains (MIC = 0.3–2 µg/mL). By contrast, but also consistent with their binding properties, the residue 4 thioamides were found to be inactive against VanA VRE (Figure 7) as well as sensitive bacteria. The impressive antimicrobial activity of 7 and its generality observed with the related amidines 43 and 45 suggest that the clinical impact of such redesigned residue 4 amidine glycopeptide antibiotics is likely to be important, charting a rational approach forward in the development of antibiotics for the treatment of vancomycin-resistant bacterial infections.
Conclusions
The induced remodeling of the dipeptide terminus of peptidoglycan cell wall precursors from d-Ala-d-Ala to d-Ala-d-Lac in resistant bacteria with the exchange of a single atom reduces vancomycin binding affinity for its biological target by 1000-fold, leading to a loss in biological activity. Herein, we reported the total synthesis45 of a key vancomycin analogue designed to exhibit dual, balanced binding to both d-Ala-d-Ala to d-Ala-d-Lac, incorporating a complementary single atom exchange in the residue 4 amide, [Ψ[C(=NH)NH]Tpg4]-vancomycin aglycon (7). The approach, enlisting a unique and optimal AgOAc-promoted late stage single-step conversion from [Ψ[C(=S)NH]Tpg4]vancomycin aglycon (8), not only permitted access to 7 and a series of earlier stage amidines, but it now also allows late stage access to related analogues and alternative access to [Ψ[CH2NH]Tpg4]vancomycin aglycon (6) from a common late stage intermediate. The approach relied on two aromatic nucleophilic substitution reactions of o-nitrofluoroaromatics for formation of the 16-membered CD/DE diaryl ether ring systems, an effective macrolactamization for closure of the 12-membered biaryl AB ring system, and the defined order of CD, AB, and DE ring closures (Figure 10). This order of ring closures follows their increasing ease of thermal atropisomer equilibration, permitting the recycling of any newly generated unnatural atropisomer under progressively milder thermal conditions where the atropisomer stereochemistry already set is not impacted. This indirect control of the atropisomer stereochemistry, in addition to any kinetic atroposelectivity achieved in the cyclizations, allowed all synthetic material to be funneled into the single atropdiastereomer characterizing the natural product.
Figure 10.
Summary of synthetic strategy.
The binding affinity of 7 and the closely related residue 4 amidine 45 with the model d-Ala-d-Ala ligand 2 was found to be only 2-fold less than that of the vancomycin aglycon (5), indicating that the amidine functions well as a H-bond acceptor for the amide NH in the model ligand. More remarkable, this binding affinity was maintained with the model d-Ala-d-Lac ligand 4, representing a nearly 600-fold increase relative to the vancomycin aglycon (5). Thus, removing the vancomycin carbonyl oxygen atom and its destabilizing electrostatic interaction with the d-Ala-d-Lac ester oxygen atom and replacing it with a protonated amidine nitrogen and its complementary stabilizing electrostatic interaction and H-bond donor capability reinstates essentially full binding affinity to the altered ligand. Significantly, 7 and the related amidine 45 display dual, balanced binding affinity for both ligands (Ka 2/4 = 0.9–1.05) that is within 2–3 fold that of vancomycin for d-Ala-d-Ala. Accurately reflecting these binding properties, each of the residue 4 amidines exhibited potent antimicrobial activity (MIC = 0.3–0.6 µg/mL) against VanA resistant bacteria (E. faecalis, VanA VRE), being 1000-fold more potent than vancomycin (1) and vancomycin aglycon (5) and equipotent to the activity they display against sensitive bacterial strains (MIC = 0.3–2 µg/mL). Beautifully, this represents a single atom exchange in the antibiotic (O→NH) to counter a corresponding single atom exchange in the cell wall precursors of resistant bacteria (NH→O), charting a rational path forward in the development of antibiotics for the treatment of vancomycin-resistant bacterial infections. Further residue 4 amide modifications building off these synthetic accomplishments and the remarkable behavior of 7 enlisting 8 as a key intermediate are under investigation, efforts directed at the development of alternative approaches to the preparation of 7 or 8 are in progress, and the impact of peripheral modification of the key amidines for further enhancing their potency are being examined and the results of such studies will be disclosed in due time.46
Supplementary Material
Figure 2.
Dual, balanced binding properties of [Ψ[CH2NH]Tpg4]vancomycin aglycon (6).
Acknowledgments
We are especially grateful for the financial support of the National Institutes of Health (CA041101) and the Skaggs Institute for Chemical Biology. We wish to thank Dr. D. B. Horne for initial improvements in the macrocyclization of 9 and the initial successful selective thionation studies on 11, Dr. Y. Zhang for extending these studies to the thionation of 12, Dr. J. Elsner for substantial supplies of early intermediates prepared at mid stage of the efforts, and Dr. Raj Chadha for the X-ray crystal structure of 26. The authors would like to thank Dr. H. Gukasyan (Pfizer) for the generation and characterization of the nano-suspension of CaCO3 in DMSO. J.G.P. is the recipient of a NIH postdoctoral fellowship (CA144333, 2009–2011), A.O. is the recipient of a JSPS fellowship (2010–2011), R.C.J. is a Skaggs Fellow (2006–2011), and S.S. was the recipient of a Swiss NSF postdoctoral fellowship (SNSF PBZH2-115560, 2006–2007).
Footnotes
Supporting Information Available. Full experimental details and full citations 5c and 26a are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McCormick MH, Stark WM, Pittenger GE, Pittenger RC, McGuire JM. Antibiot. Annu. 1955–1956:606. [PubMed] [Google Scholar]
- 2. Harris CM, Kopecka H, Harris TM. J. Am. Chem. Soc. 1983;105:6915. See also: Harris CM, Harris TM. J. Am. Chem. Soc. 1982;104:4293. Williamson MP, Williams DH. J. Am. Chem. Soc. 1981;103:6580. Sheldrick GM, Jones PG, Kennard O, Williams DH, Smith GA. Nature. 1978;271:223. doi: 10.1038/271223a0. Williams DH, Kalman JR. J. Am. Chem. Soc. 1977;99:2768. doi: 10.1021/ja00450a058.
- 3.Nagarajan R, editor. Glycopeptide Antibiotics. New York: Marcel Dekker; 1994. [Google Scholar]
- 4.Reviews: Kahne D, Leimkuhler C, Lu W, Walsh CT. Chem. Rev. 2005;105:425. doi: 10.1021/cr030103a. Hubbard BK, Walsh CT. Angew. Chem. Int. Ed. 2003;42:730. doi: 10.1002/anie.200390202.
- 5.(a) CDC (2003) National Nosocomial Infections Surveillance (NNIS) System Report, Data Summary from January 1992 Through June 2004, Issued October 2004. Am. J. Infect. Control. 2004;32:470. doi: 10.1016/S0196655304005425. [DOI] [PubMed]; (b) Laxminarayan R. Antibiotic Resistance: The Unfolding Crisis. In Extending the Cure, Policy Responses to the Growing Treat of Antibiotic Resistance. In: Laxminarayan R, Malani A, Howard D, Smith DL, editors. Resources for the Future. Chapter 1. 2007. pp. 25–37. [Google Scholar]; (c) Klevens RM, et al. J. Am. Med. Assoc. 2007;298:1763. [Google Scholar]; (d) Walsh CT, Fischbach MA. Sci. Am. 2009;301(1):44. doi: 10.1038/scientificamerican0709-44. [DOI] [PubMed] [Google Scholar]
- 6.Reviews: Malabarba A, Nicas TI, Thompson RC. Med. Res. Rev. 1997;17:69. doi: 10.1002/(sici)1098-1128(199701)17:1<69::aid-med3>3.0.co;2-r. Malabarba A, Ciabatti R. Curr. Med. Chem. 2001;8:1759. doi: 10.2174/0929867013371716. Van Bambeke FV, Laethem YV, Courvalin P, Tulkens PM. Drugs. 2004;64:913. doi: 10.2165/00003495-200464090-00001.
- 7.(a) Süssmuth RD. ChemBioChem. 2002;3:295. doi: 10.1002/1439-7633(20020402)3:4<295::AID-CBIC295>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]; (b) Gao Y. Nat. Prod. Rep. 2002;19:100. doi: 10.1039/b100912p. [DOI] [PubMed] [Google Scholar]; (c) Walker S, Chen L, Helm J, Hu Y, Rew Y, Shin D, Boger DL. Chem. Rev. 2005;105:449. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]; (d) von Nussbaum F, Brands M, Hinzen B, Weigand S, Häbich D. Angew. Chem. Int. Ed. 2006;45:5072. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- 8. Nieto M, Perkins HR. Biochem. J. 1971;123:789. doi: 10.1042/bj1230789. Review: Perkins HR. Pharmacol. Ther. 1982;16:181. doi: 10.1016/0163-7258(82)90053-5.
- 9. Williams DH, Williamson MP, Butcher DW, Hammond SJ. J. Am. Chem. Soc. 1983;105:1332. Review: Williams DH, Bardsley B. Angew. Chem. Int. Ed. 1999;38:1172. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1172::AID-ANIE1172>3.0.CO;2-C. X-ray: Schaefer M, Schneider TR, Sheldrick GM. Structure. 1996;4:1509. doi: 10.1016/s0969-2126(96)00156-6.
- 10.For lead references, see: Koteva K, Hong H–J, Wang XD, Nazi I, Hughes D, Naldrett MJ, Buttner MJ, Wright GD. Nat. Chem. Biol. 2010;6:327. doi: 10.1038/nchembio.350. Hong H–J, Hutchings MI, Buttner MJ. Adv. Exp. Med. Biol. 2008;631:200. doi: 10.1007/978-0-387-78885-2_14.
- 11. Bugg TDH, Wright GD, Dutka–Malen S, Arthur M, Courvalin P, Walsh CT. Biochemistry. 1991;30:10408. doi: 10.1021/bi00107a007. Reviews: Walsh CT. Science. 1993;261:308. doi: 10.1126/science.8392747. Walsh CT, Fisher SL, Park I–S, Prahalad M, Wu Z. Chem. Biol. 1996;3:21. doi: 10.1016/s1074-5521(96)90079-4. Lessard IA, Walsh CT. Proc. Natl. Acad. Sci. USA. 1999;96:11028. doi: 10.1073/pnas.96.20.11028. Healy VL, Lessard IA, Roper DI, Knox JR, Walsh CT. Chem. Biol. 2000;7:R109. doi: 10.1016/s1074-5521(00)00116-2.
- 12.McComas CC, Crowley BM, Boger DL. J. Am. Chem. Soc. 2003;125:9314. doi: 10.1021/ja035901x. [DOI] [PubMed] [Google Scholar]
- 13. Morgan BP, Scholtz JM, Ballinger MD, Zipkin ID, Bartlett PA. J. Am. Chem. Soc. 1991;113:297. and refs. cited therein.
- 14.Reviews: Evans DA, DeVries KM. Drugs Pharm. Sci. 1994;63:63. Nicolaou KC, Boddy CNC, Brase S, Winssinger N. Angew. Chem. Int. Ed. 1999;38:2096. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. Boger DL. Med. Res. Rev. 2001;21:356. doi: 10.1002/med.1014.
- 15. Evans DA, Wood MR, Trotter BW, Richardson TI, Barrow JC, Katz JL. Angew. Chem. Int. Ed. 1998;37:2700. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2700::AID-ANIE2700>3.0.CO;2-P. Evans DA, Dinsmore CJ, Watson PS, Wood MR, Richardson TI, Trotter BW, Katz JL. Angew. Chem. Int. Ed. 1998;37:2704. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2704::AID-ANIE2704>3.0.CO;2-1. See also: Evans DA, Barrow JC, Watson PS, Ratz AM, Dinsmore CJ, Evrard DA, DeVries KM, Ellman JA, Rychnovsky SD, Lacour J. J. Am. Chem. Soc. 1997;119:3419. Evans DA, Dinsmore CJ, Ratz AM, Evrard DA, Barrow JC. J. Am. Chem. Soc. 1997;119:3417.
- 16.(a) Nicolaou KC, Takayanagi M, Jain NF, Natarajan S, Koumbis AE, Bando T, Ramanjulu JM. Angew. Chem. Int. Ed. 1998;37:2717. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2717::AID-ANIE2717>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (b) Nicolaou KC, Natarajan S, Li H, Jain NF, Hughes R, Solomon ME, Ramanjulu JM, Boddy CNC, Takayanagi M. Angew. Chem. Int. Ed. 1998;37:2708. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2708::AID-ANIE2708>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]; (c) Nicolaou KC, Jain NF, Natarajan S, Hughes R, Solomon ME, Li H, Ramanjulu JM, Takayanagi M, Koumbis AE, Bando T. Angew. Chem. Int. Ed. 1998;37:2714. doi: 10.1002/(SICI)1521-3773(19981016)37:19<2714::AID-ANIE2714>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]; (d) Nicolaou KC, Mitchell HJ, Jain NF, Winssinger N, Hughes R, Bando T. Angew. Chem. Int. Ed. 1999;38:240. [Google Scholar]
- 17.(a) Boger DL, Miyazaki S, Kim SH, Wu JH, Loiseleur O, Castle SL. J. Am. Chem. Soc. 1999;121:3226. [Google Scholar]; (b) Boger DL, Miyazaki S, Kim SH, Wu JH, Castle SL, Loiseleur O, Jin Q. J. Am. Chem. Soc. 1999;121:10004. [Google Scholar]
- 18.(a) Boger DL, Kim SH, Miyazaki S, Strittmatter H, Weng J–H, Mori Y, Rogel O, Castle SL, McAtee JJ. J. Am. Chem. Soc. 2000;122:7416. [Google Scholar]; (b) Boger DL, Kim SH, Mori Y, Weng J–H, Rogel O, Castle SL, McAtee JJ. J. Am. Chem. Soc. 2001;123:1862. doi: 10.1021/ja003835i. [DOI] [PubMed] [Google Scholar]
- 19.Crowley BM, Mori Y, McComas CC, Tang D, Boger DL. J. Am. Chem. Soc. 2004;126:4310. doi: 10.1021/ja039795a. [DOI] [PubMed] [Google Scholar]
- 20.(a) Garfunkle J, Kimball FS, Trzupek JD, Takazawa S, Shimamura H, Tomishima M, Boger DL. J. Am. Chem. Soc. 2009;131:16036. doi: 10.1021/ja907193b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shimamura H, Breazzano SP, Garfunkle J, Kimball FS, Trzupek JD, Boger DL. J. Am. Chem. Soc. 2010;132:7776. doi: 10.1021/ja102304p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Breazzano SP, Boger DL. J. Am. Chem. Soc. 2011;133:18495. doi: 10.1021/ja208570q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Boger DL, Weng J–H, Miyazaki S, McAtee JJ, Castle SL, Kim SH, Mori Y, Rogel O, Jin Q. J. Am. Chem. Soc. 2000;122:10047. doi: 10.1021/ja003835i. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Castle SL, Miyazaki S, Wu JH, Beresis RT, Loiseleur O. J. Org. Chem. 1999;64:70. doi: 10.1021/jo980880o. [DOI] [PubMed] [Google Scholar]; (c) Boger DL, Miyazaki S, Loiseleur O, Beresis RT, Castle SL, Wu JH, Jin Q. J. Am. Chem. Soc. 1998;120:8920. [Google Scholar]; (d) Boger DL, Beresis RT, Loiseleur O, Wu JH, Castle SL. Bioorg. Med. Chem. Lett. 1998;8:721. doi: 10.1016/s0960-894x(98)00110-3. [DOI] [PubMed] [Google Scholar]; (e) Boger DL, Borzilleri RM, Nukui S, Beresis RT. J. Org. Chem. 1997;62:4721. doi: 10.1021/jo961346o. [DOI] [PubMed] [Google Scholar]; (f) Boger DL, Loiseleur O, Castle SL, Beresis RT, Wu JH. Bioorg. Med. Chem. Lett. 1997;7:3199. doi: 10.1016/s0960-894x(98)00110-3. [DOI] [PubMed] [Google Scholar]; (g) Boger DL, Zhou J, Borzilleri RM, Nukui S, Castle SL. J. Org. Chem. 1997;62:2054. doi: 10.1021/jo961346o. [DOI] [PubMed] [Google Scholar]; (h) Boger DL, Borzilleri RM, Nukui S. J. Org. Chem. 1996;61:3561. doi: 10.1021/jo961346o. [DOI] [PubMed] [Google Scholar]; (i) Boger DL, Borzilleri RM, Nukui S. Bioorg. Med. Chem. Lett. 1995;5:3091. [Google Scholar]; (j) Boger DL, Borzilleri RM. Bioorg. Med. Chem. Lett. 1995;5:1187. [Google Scholar]; (k) Boger DL, Nomoto Y, Teegarden BR. J. Org. Chem. 1993;58:1425. [Google Scholar]; (l) Boger DL, Yohannes D. J. Org. Chem. 1990;55:6000. [Google Scholar]
- 22.(a) Crane CM, Pierce JG, Leung SSF, Tirado–Rives J, Jorgensen WL, Boger DL. J. Med. Chem. 2010;53:7229. doi: 10.1021/jm100946e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crane CM, Boger DL. J. Med. Chem. 2009;52:1471. doi: 10.1021/jm801549b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McComas CC, Crowley BM, Hwang I, Boger DL. Bioorg. Med. Chem. Lett. 2003;13:2933. doi: 10.1016/s0960-894x(03)00513-4. [DOI] [PubMed] [Google Scholar]; (d) McAtee JJ, Castle SL, Jin Q, Boger DL. Bioorg. Med. Chem. Lett. 2002;12:1319. doi: 10.1016/s0960-894x(02)00130-0. [DOI] [PubMed] [Google Scholar]
- 23.Crowley BM, Boger DL. J. Am. Chem. Soc. 2006;128:2885. doi: 10.1021/ja0572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molcanov K, Kojic–Prodic B, Raos N. Acta Cryst. 2004;B60:424. doi: 10.1107/S0108768104014442. [DOI] [PubMed] [Google Scholar]
- 25.(a) Moser H, Fliri A, Steiger A, Costello G, Schreiber J, Eschenmoser A. Helv. Chim. Acta. 1986;69:1224. [Google Scholar]; (b) Jones RCF, Ward GJ. Tetrahedron Lett. 1988;29:3853. [Google Scholar]
- 26.(a) Kamenecka TM, et al. Bioorg. Med. Chem. Lett. 2004;14:2323. doi: 10.1016/j.bmcl.2004.01.100. [DOI] [PubMed] [Google Scholar]; (b) Inokuchi E, Oishi S, Kubo T, Ohno H, Shimura K, Matsuoka M, Fujii N. ACS Med. Chem. Lett. 2011;2:477. doi: 10.1021/ml200047e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Inokuchi E, Yamada A, Hozumi K, Tomita K, Oishi S, Ohno H, Nomizu M, Fujii N. Org. Biomol. Chem. 2011;9:3421. doi: 10.1039/c0ob01193b. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Pierce JG, James RC, Okano A, Boger DL. J. Am. Chem. Soc. 2011;133:13946. doi: 10.1021/ja207142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman RS, Shah JA. Synthesis. 1999:1399. [Google Scholar]
- 29.Sakaitani M, Ohfune Y. J. Org. Chem. 1990;55:870. [Google Scholar]
- 30.(a) Fan C–X, Hao X–L, Ye Y–H. Synth. Commun. 1996;26:1455. [Google Scholar]; (b) Li H, Jiang X, Ye Y–H, Fan C, Romoff T, Goodman M. Org. Lett. 1999;1:91. doi: 10.1021/ol990573k. [DOI] [PubMed] [Google Scholar]
- 31.(a) Jiang W, Wanner J, Lee RJ, Bounaud P–Y, Boger DL. J. Am. Chem. Soc. 2002;124:5288. doi: 10.1021/ja020237q. [DOI] [PubMed] [Google Scholar]; (b) Jiang W, Wanner J, Lee RJ, Bounaud P–Y, Boger DL. J. Am. Chem. Soc. 2003;125:1877. doi: 10.1021/ja0212314. [DOI] [PubMed] [Google Scholar]; (c) Shin D, Rew Y, Boger DL. Proc. Natl. Acad. Sci. USA. 2004;101:11977. doi: 10.1073/pnas.0401419101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Nam J, Shin D, Rew Y, Boger DL. J. Am. Chem. Soc. 2007;129:8747. doi: 10.1021/ja068573k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Boger DL, Colletti SL, Honda T, Menezes RF. J. Am. Chem. Soc. 1994;116:5607. [Google Scholar]
- 32.The structure, stereochemistry, and conformation of 26 were established with a single-crystal X-ray structure determination (CCDC 837960).
- 33.Nitanai Y, Kikuchi T, Kakoi K, Hanamaki S, Fujisawa I, Aoki K. J. Mol. Biol. 2009;5:1422. doi: 10.1016/j.jmb.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Gerber PR, Mueller K. J. Comput.-Aided Mol. Des. 1995;9:251. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:160. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.(a) Wissmann H, Kleiner H–J. Angew. Chem. Int. Ed. 1980;19:133. [Google Scholar]; (b) Dunetz JA, Xiang Y, Baldwin A, Ringling J. Org. Lett. 2011;13:5048. doi: 10.1021/ol201875q. [DOI] [PubMed] [Google Scholar]
- 37.(a) Bowden K, Heilbron IM, Jones ERH, Weedon BCL. J. Chem. Soc. 1946:39. [Google Scholar]; (b) Bowers A, Halsall TG, Jones ERH, Lemin AJ. J. Chem. Soc. 1953:2548. [Google Scholar]
- 38.(a) Corey EJ, Suggs JW. Tetrahedron Lett. 1975:2647. [Google Scholar]; (b) Corey EJ, Boger DL. Tetrahedron Lett. 1978:2461. [Google Scholar]; (c) Corey EJ, Schmidt G. Tetrahedron Lett. 1979:399. [Google Scholar]
- 39.Moorthy JN, Singhal N. J. Org. Chem. 2005;70:1926. doi: 10.1021/jo048240a. [DOI] [PubMed] [Google Scholar]
- 40.(a) Shibuya I, Taguchi Y, Tsuchiya T, Oishi A, Katoh E. Bull. Chem. Soc. Jpn. 1994;67:3048. [Google Scholar]; (b) Cacchi S, La Torre F, Misiti D. Chem. Ind. 1978:669. [Google Scholar]; (c) Corey EJ, Boger DL. Tetrahedron Lett. 1978:5. [Google Scholar]
- 41.(a) Marchand–Brynaert J, Moya–Portuguez M, Huber I, Ghosez L. J. Chem. Soc. Chem. Commun. 1983:818. [Google Scholar]; (b) Sauve G, Rao VS, Lajoie G, Belleau B. Can. J. Chem. 1985;63:3089. [Google Scholar]; (c) Bock MG, DiPardo RM, Evans BE, Rittle KE, Freidinger RM, Chang RSL, Lotti VJ. J. Med. Chem. 1988;31:264. doi: 10.1021/jm00396a042. [DOI] [PubMed] [Google Scholar]
- 42.(a) Leung SSF, Tirado–Rives J, Jorgensen WL. Bioorg. Med. Chem. 2009;17:5874. doi: 10.1016/j.bmc.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Leung SSF, Tirado–Rives J, Jorgensen WL. Bioorg. Med. Chem. Lett. 2009;19:1236. doi: 10.1016/j.bmcl.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samples of 43, even after repeated purification, can be isolated pure, but re-equilibrate (< 1 h) to three easily separable (HPLC) interconverting components (2:1:1), each of which display the same molecular weight: ESI-TOF HRMS m/z 1184.39 (M+ + H, C57H63Cl2N9O15 requires 1184.39), see Supporting Information. Binding studies conducted with this equilibrating constitutional, configurational, or conformational isomer mixture exhibited Ka’s of 3.9 × 104 M−1 (2) and 4.1 × 104 M−1 (4).
- 44.UV-difference titration assays were run as previously described: Nieto M, Perkins HR. Biochem. J. 1971;124:845. doi: 10.1042/bj1240845. Nieto M, Perkins HR. Biochem. J. 1971;123:773. doi: 10.1042/bj1230773. For D-Ala-Gly and L-Ala-L-Ala, see: Nieto M, Perkins HR. Biochem. J. 1971;123:789. doi: 10.1042/bj1230789. Perkins HR. Biochem. J. 1969;111:195. doi: 10.1042/bj1110195. For D-Ala-D-Ser, see: van Wageningen AMA, Staroske T, Williams DH. Chem. Commun. 1998:1171.
- 45.Szpilman AM, Carreira E. Angew. Chem. Int. Ed. 2010;49:9592. doi: 10.1002/anie.200904761. [DOI] [PubMed] [Google Scholar]
- 46.Abbreviations: BCB, B-bromocatecholborane; Boc, t-butyloxycarbonyl; Cbz, benzyloxycarbonyl or carboxybenzyl; dba, dibenzylideneacetone; DEPBT, 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4-one; DMAP, 4-(dimethylamino)pyridine; DMF, N,N-dimethylformamide; DMSO, dimethylsulfoxide; EDCI, 1-[3-(dimethylamino)propyl]-3-ethylcarbodiimide hydrochloride; FDPP, pentafluorophenyl diphenylphosphinate; HATU, O-(7-azabenzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate; HOAt, 1-hydroxy-7-azabenzotriazole; HOBt, 1-hydroxybenzotriazole; IBX, 2-iodoxybenzoic acid; MS, molecular sieves; NMM, N-methylmorpholine; NMP, N-methylpyrrolidinone; PyBop, (benzotriazol-1-yl)tripyrrolidinophosphonium hexafluorophosphate; Pyr, pyridine; Ra–Ni, Raney nickel; T3P, propylphosphonic anhydride; TBS, t-butyldimethylsilyl; TEMPO, 2,2,6,6-tetramethypiperidine-1-oxyl; TES, triethylsilyl; TFA, trifluoroacetic acid; THF, tetrahydrofuran.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.