Abstract
Eukaryotic cytochrome c oxidase (COX) is the terminal enzyme of the mitochondrial respiratory chain. COX is a multimeric enzyme formed by subunits of dual genetic origin which assembly is intricate and highly regulated. The COX catalytic core is formed by three mitochondrial DNA encoded subunits, Cox1, Cox2 and Cox3, conserved in the bacterial enzyme. Their biogenesis requires the action of messenger-specific and subunit-specific factors which facilitate the synthesis, membrane insertion, maturation or assembly of the core subunits. The study of yeast strains and human cell lines from patients carrying mutations in structural subunits and COX assembly factors has been invaluable to identify these ancillary factors. Here we review the current state of knowledge of the biogenesis and assembly of the eukaryotic COX catalytic core and discuss the degree of conservation of the players and mechanisms operating from yeast to human.
Keywords: Mitochondria, cytochrome c oxidase, COX catalytic core, COX assembly
1. More than fifty years of eukaryotic cytochrome c oxidase assembly
Mitochondrial cytochrome c oxidase (COX) is a multimeric copper-heme A terminal oxidase that functions as an electron-driven proton pump and plays fundamental roles in eukaryotic cell respiration and aerobic energy production. COX catalyzes the transfer of electrons from ferrocytochrome c to molecular oxygen via the four redox active metal cofactors present in its catalytic core. Electrons enter COX through a mixed valence dinuclear copper center, the CuA site, located in subunit 2. Electrons are transferred from CuA to a low spin heme a located in subunit 1 and are subsequently transferred intra-molecularly to the active site where a high spin heme a3 and CuBform a binuclear center for oxygen binding (reviewed in [1-3]). The electron transfer reaction is coupled to the transfer of protons from the matrix to the intermembrane space ([4] and [5]) thus contributing to the generation of the proton gradient which is subsequently used by the F1F0 mitochondrial ATPase to drive ATP synthesis.
From a historical perspective [6], the identification of the subunit and prosthetic group components as well as the investigation of their assembly into a functional enzyme started early in the XX century and remains the center of ongoing intensive studies nowadays [7-12]. The metal content of COX was already known in the mid 1950s. However, the isolation of functionally intact COX, achieved approximately 50 years ago [13], opened the way for a detailed biochemical and biophysical analyses of the enzyme. It was soon recognized that COX was a protein complex and the number of component polypeptides was subsequently settled [14]. The current knowledge of the COX subunit composition and assembly of the functional enzyme has its foundations in work performed in the yeast Saccharomyces cerevisiae. In the late sixties and early 1970s, emphasis was placed on studying the biosynthetic origin of COX. Yeast cytoplasmic and mitochondrial protein synthesis was differentiated in vivo with antibiotics, thus providing the first clues that respiratory complexes, including COX, are contributed by two independent protein synthesis machineries [15]. It was then recognized that the three largest COX subunit polypeptides are translated by mitochondrial ribosomes [16] and that they have physical and chemical properties different from the subunits synthesized in cytoplasmic ribosomes [17]. The sequence of the corresponding yeast genes was soon disclosed [18, 19]. The later publication of the full sequence and organization of the human mitochondrial genome with the location of the COXI, COXII and COXIII genes confirmed that with a few exceptions in plants [20], the three core subunits are indeed encoded in the mitochondrial DNA. In fact, eukaryotic COX is formed by 11-13 subunits (11 in the yeast Saccharomyces cerevisiae and 13 in mammals) of dual genetic origin. The three-subunit core is surrounded by a set of nuclear-encoded small subunits that are important for both the assembly and function of the enzyme as well as its dimerization (reviewed in [8, 9, 21]). These subunits also serve to modulate the catalytic activity of the enzyme and to protect the core from oxidative damage. A list of COX homologue subunits in yeast and mammals is shown in Table 1.
Table 1.
Homologue COX subunits and COX assembly factors in the yeast Saccharomyces cerevisiae and human.
| YEAST | HUMAN | ROLE | ||
|---|---|---|---|---|
| GENE | PROTEIN | GENE | PROTEIN | |
| Catalytic core (mtDNA encoded structural subunits) | ||||
| COX1 | Cox1 | MTCOXI | COX1 | Catalytic core subunits |
| COX2 | Cox2 | MTCOXII | COX2 | |
| COX3 | Cox3 | MTCOXIII | COX3 | |
| Core protective shield (nDNA encoded structural subunits) | ||||
| COX4 | Cox4 | COXVb | COX5b | Subunits required for COX assembly and function |
| COX5a | Cox5a | COXIV-1 | COX4-1 | |
| COX6 | Cox6 | COXVa | COX5a | |
| COX7 | Cox7 | COXVIIa | COX7a | |
| COX8 | Cox8 | COXVIIc | COX7c | |
| COX9 | Cox7a | COXVIc | COX6c | |
| - | - | COXVIIb | COX7b | |
| -- | -- | COXVIII | COX8 | |
| COX12 | Cox9 | COXVIb | COX6b | Non-essential subunits |
| COX13 | Cox10 | COXVIa | COX6a | |
| Expression of catalytic core subunits | ||||
| MSS116 | Mss116 | - | - | Helicase involved in splicing of all COX1 and COB introns |
| Component of the RNA degradosome | ||||
| SUV3 | Suv3 | - | - | Helicase involved in COX1 aI5β intron splicing |
| Stability of intron-containing COX1 and COB transcripts Component of the RNA degradosome |
||||
| MRS1 | Mrs1 | - | - | Required for COX1 aI5β intron splicing Required for excision of the COB bI3 intron |
| MNE1 | Mne1 | - | - | Required for COX1 aI5β intron splicing |
| MSS18 | Mss18 | - | - | Required for COX1 aI5β intron splicing |
| Additional unidentified function | ||||
| COX24 | Cox24 | - | - | Required for splicing of aI2 and aI3 COX1 introns |
| Required for COX1 mRNA translation | ||||
| NAM2 | Nam2 | - | - | Required for COX1 aI4 intron splicing |
| CCM1 | Ccm1 | - | - | Required for COX1 aI4 intron splicing |
| PET309 | Pet309 | Yeast: Translational activator of COX1 mRNA | ||
| LRPPRC | LRPPRC | Human: mitochondrial mRNA stability | ||
| MSS51 | Mss51 | - | - | Translational activator of COX1 mRNA |
| Cox1 chaperone required for its stability/ maturation/assembly |
||||
| YGR021w | Ygr021w | Yeast: No role on COX biogenesis | ||
| TACO1 | TACO1 | Human: COX1 mRNA translational activator | ||
| PET111 | Pet111 | - | - | Translational activator of COX2 mRNA |
| PET54 | Pet54 | - | - | Translational activator of COX3 mRNA |
| Required for COX1 aI5β intron splicing and translation | ||||
| PET122 | Pet122 | - | - | Translational activator of COX3 mRNA |
| PET494 | Pet494 | - | - | Translational activator of COX3 mRNA |
| Membrane insertion and processing of catalytic core subunits | ||||
| OXA1 | Oxa1 | OXA1 | OXA1 | Membrane insertion of COX subunits, cytochrome b and ATPase proteolipid |
| COX20 | Cox20 | COX20 | COX20 | Cox2 chaperone. Presentation of Cox2-precursor to the IMP complex |
| COX18 | Cox18 | COX18 | COX18 | Export of the Cox2 C-terminus tail |
| MSS2 | Mss2 | -- | -- | Export of the Cox2 C-terminus tail |
| PNT1 | Pnt1 | -- | -- | Export of the Cox2 C-terminus tail |
| IMP1 | Imp1 | -- | -- | Responsible for the maturation of precursor Cox2 |
| IMP2 | Imp2 | IMMP2L | IMMP2L | Required for the stability and activity of Imp1 |
| SOM1 | Som1 | -- | -- | Third component of the yeast IMP complex. It could play a role in substrate recognition |
| Copper Metabolism and Insertion into catalytic core subunits | ||||
| COX17 | Cox17 | COX17 | COX17 | Delivery of copper to Sco1 and Cox11 |
| SCO1 | Sco1 | SCO1 | SCO1 | Transfer of copper to COX and/or reduction of cysteine |
| SCO2 | SCO2 | residues in subunit 2 | ||
| COX11 | Cox11 | COX11 | COX11 | Stable formation of the CuB and Mg centers |
| COX19 | Cox19 | COX19 | COX19 | CX9C proteins. They could play roles in redox control and copper trafficking in the intermembrane space |
| COX23 | Cox23 | COX23 | COX23 | |
| PET191 | Pet191 | PET191 | PET191 | |
| CMC1 | Cmc1 | CMC1 | CMC1 | |
| CMC2 | Cmc1 | CMC2 | CMC2 | |
| Heme A Biosynthesis | ||||
| COX10 | Cox10 | COX10 | COX10 | Farnesylation of protoheme |
| COX15 | Cox15 | COX15 | COX15 | Conversion of heme o to heme a |
| YAH1 | Yah1 | FDX2 | FDX2 | Collaborates with Cox15 in heme o hydroxylation |
| ARH1 | Arh1 | ADR | ADR | Collaborates with Cox15 in heme o hydroxylation |
| Assembly / Unknown | ||||
| COX16 | Cox16 | COX16 | COX16 | Unknown function |
| PET117 | Pet117 | - | -- | Unknown function |
| PET100 | Pet100 | -- | -- | Formation of assembly intermediates containing Cox7, Cox8, and Cox9 |
| SHY1 | Shy1 | SURF1 | SURF1 | Catalyzes an assembly step involving Cox1 |
| COX14 | Cox14 | - | - | Binds Cox1 and is required for its stability/maturation/assembly |
| COA1 | Coa1 | -- | -- | Binds Cox1 and is required for its stability/maturation/assembly |
| COA2 | Coa2 | -- | -- | Required for Cox1 stability/maturation/assembly |
|
COA3/ COX25 |
Coa3 Cox25 |
-- | -- | Binds Cox1 and is required for its stability/maturation/assembly |
|
CMC3/ COA4 |
Cmc3 Coa4 |
-- | -- | CX9C protein involved in late stages of COX assembly |
In another major breakthrough in eukaryotic COX research, the structure of COX from bovine heart mitochondria, determined at ~2.8 Å resolution [22-24], was revealed in the late 1990s. This represented a major milestone in the history of oxidative phosphorylation since COX was the first respiratory chain complex to have its high resolution crystal structure resolved. Structural studies on the simpler bacterial COX had given clues about the association of catalytic metal centers with specific homologue core subunits [25, 26] that were then confirmed for the mitochondrial enzyme. The structures allowed also to precisely determine the interface contacts among all subunit components and to answer questions concerning the coupling between electron transport and proton pumping at the atomic level. From these studies it is also known that non-catalytic metals such as zinc, magnesium and sodium are bound to the enzyme, although despite recent progress [27, 28] their roles remain largely unknown.
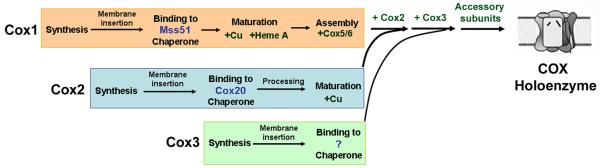
The process of COX assembly has been the subject of intense investigations over the last 50 years using different approaches and it still actively investigated nowadays. COX biogenesis is thought to be a linear process, with the different subunits and cofactors being added in an ordered manner. The concept of an assembly pathway characterized by the sequential incorporation of COX subunits was developed in the early 1980s from data obtained from studies that used rat liver mitochondria and followed the incorporation of radiolabeled subunits into the COX holocomplex [29]. The model was later confirmed by analyses of the human enzyme performed by Blue-Native electrophoresis. By analyzing the formation of assembly intermediates, it was concluded that assembly initiates around a seed formed by subunit 1 and proceeds with the formation of several discrete assembly intermediates probably representing rate-limiting steps in the process [30]. The study of assembly intermediates in yeast has been less productive since they do not seem to accumulate in detectable amounts in most COX defective strains [31], probably due to the marked downregulation of Cox1 synthesis in the absence of fully assembled COX [32]. However, subassemblies have been detected in yeast cox2 [33] and pet100 mutants [34] and are similar to those observed in mammalian cells. Adding to the concept of linear assembly, it is known that the biogenesis of the membrane forms of at least the two major catalytic core subunits 1 and 2 follow relatively independent lines with the participation of subunit-specific chaperones (Figure 1 and reviewed in [8, 10],). Furthermore, it has been proposed that COX subassemblies might interact with other respiratory chain components at early stages in the process of supercomplex assembly [35] and that newly imported nuclear DNA-encoded subunits can integrate not only into the COX holoenzyme by associating with pre-existing subunits but also into supercomplex forms by associating with intermediate assembly complexes [36].
Fig. 1. Simplified model for the process of COX assembly.
General chaperones and RNA-specific translational activators (not depicted here but see explanation in the text) are required for synthesis of the mtDNA-encoded subunits forming the core. Following their insertion into the inner membrane, Cox1 and Cox2 are matured by addition of metal cofactors. At some point, substrate-specific chaperones bind Cox1 and Cox2 to maintain them in an assembly-competent state. A predicted Cox3 chaperone has not been yet identified. Following Cox1 maturation, the nuclear DNA-encoded Cox5 and 6 subunits are added to Cox1 prior incorporation of the other core subunits and the rest of the accessory subunits to form the holoenzyme.
COX assembly is a protein assisted process. The systematic analysis of yeast mutants defective in COX assembly has been an invaluable strategy to identify the non-structural ancillary factors involved in COX assembly and subsequently attempt the reconstruction of the different steps of the assembly pathway. Screens of nuclear respiratory deficient mutants was an innovative strategy in the early 1970s [37, 38] and have revealed the existence of a large number of nuclear genes coding for accessory factors that selectively affect COX expression in yeast [39, 40]. Their functions, required for all steps of the process and significantly conserved from yeast to humans, are summarized in Table 1 and have been previously reviewed [7-10, 41, 42].
Over the last 20 years, mutant screen strategies have been also used in humans, since defective COX biogenesis results in devastating human mitochondrial diseases frequently involving brain, skeletal muscle and heart tissues (reviewed in [11, 21, 43-45]). To date, with the double exception of an infantile encephalomyopathy caused by a mutation in the nuclear encoded subunit COX6B1 [46] and an exocrine pancreatic insufficiency caused by a mutation in the COX4I2 gene [47], all Mendelian disorders presenting COX deficiency have been assigned to mutations in ancillary factors. Specifically, mutations have been found in SURF1, required for the formation of early assembly intermediates [48, 49], SCO1 and SCO2, required for COX copper metallation [50-54], COX10 and COX15, essential for heme A biosynthesis [55-57], and finally in LRPPRC [58] and TACO1 [59], required for the expression of COX subunits. Mutant fibroblast cell lines from patients suffering from these disorders have been used to study the accumulation of subassembly intermediates in the absence of specific COX assembly factors and obtain information concerning the assembly step either catalyzed or affected by the mutated factor [31, 60, 61]. These studies have also provided information concerning mammalian specific COX biogenetic factors, such as TACO1, an evolutionary conserved protein although it functions as a mitochondrial COX1 translational activator specifically in mammals [59].
Over the last ten years, particular attention has been devoted to the assembly of the COX catalytic core, which is the focus of this review. New players and regulatory pathways have been identified disclosing increasing levels of complexity. Our aim here is to summarize the current understanding of the biogenesis and assembly of the eukaryotic cytochrome c oxidase catalytic core in yeast and will include comparative notes of the process as it occurs in human cells.
2. Expression of mitochondrial DNA encoded COX core subunits
2.1. Mitochondrial COX gene transcription and mRNA processing in Saccharomyces cerevisiae
In S. cerevisiae, the COX2 and COX3 genes for subunits 2 and 3, respectively, have no introns and the primary transcripts are individually transcribed and matured by the general mitochondrial transcription and 3′ processing machinery. General mitochondrial transcription has been recently reviewed elsewhere [62].
In contrast, the COX1 gene is transcribed as a polycistronic precursor RNA that includes the genes COX1, ATP8, ATP6 and ENS2. This transcript is processed between the COX1 and ATP8 cistrons to release COX1 mRNA (reviewed in [63]). Additionally, the COX1 gene contains multiple introns of two different types, groups I (introns aI3, aI4, aI5α, and aI5β) and II (introns aI1, aI2, and aI5γ) [18]. Maturation of the subunit 1 pre-mRNA depends on proteins referred to as maturases (related to DNA endonucleases for group I introns and reverse transcriptases for group II introns [64]) whose genes are located in the COX1 introns [65, 66]. In addition, an increasing number of nuclear gene products have been described that are also essential for maturation of the mRNA (see full list in Table I). This includes the Mss116 RNA helicase [67], which is a general mitochondrial splicing factor that acts in facilitating the RNA folding reaction required for self-splicing [68, 69] and the Suv3 RNA helicase, which is required for the processing of intron aI5β by recycling the intron-splicing factor Mrs1 [70]. At least two proteins are required for COX1 aI4 intron splicing, Ccm1 [71] and Nam2 [72] . Other proteins are involved, which specific functions in most cases remain to be fully understood such as the COX3 mRNA translational activator Pet54 [73, 74], Mrs1 [75], Mne1 [76] and Mss18 [73], four proteins required for the processing of COX1 aI5β intron, and the recently identified Cox24, required for splicing of COX1 aI2 and aI3 introns [77]. Several nuclear gene products implicated in mitochondrial splicing seem to have multiple functions as suggested by the fact that yeast strains with a null mutation in one of these genes and devoid of mitochondrial introns do not recover full respiratory competency. That is the case of Cox24 [77] and Pet54 [78], which also seem to be required for efficient Cox1 synthesis. Nam2 is the yeast mitochondrial leucyl-tRNA synthetase and is additionally required for mitochondrial DNA maintenance [79]. Mss116 and Suv3 were found to be part of the degradosome, an enzymatic complex that takes part in turnover of mitochondrial RNAs [80]. Also Mss18 performs a second role which remains to be identified [73]. In contrast, both Δmrs1 and Δmne1 yeast strains carrying intronless mtDNA recover respiratory competency indicating that Mrs1 and Mne1 are only required for mitochondrial pre-mRNA splicing [76, 81].
As expected, defective COX1 mRNA processing and splicing result in null or poor translation and a subsequent COX assembly defect.
2.2. Mitochondrial COX messenger-specific translational factors in Saccharomyces cerevisiae
In S. cerevisiae, translation of each mitochondrial COX mRNA depends on one or more nuclear encoded translational activators (for review, see [82]). These mRNA-specific translational factors are either integral or peripheral inner membrane proteins that recognize the 5′-untranslated region (UTR) of their target transcripts. At least two proteins, Mss51 [83] and Pet309 [84], are required for synthesis of Cox1; Pet111 is required for Cox2 synthesis [85, 86]; and three gene products, Pet54, Pet122 and Pet494, govern the synthesis of Cox3 [87-89].
Most of the understanding of the interaction of these translational activators with the RNA leaders has been elucidated by genetic studies [87, 88, 90]. Although they show specificity for their target RNA sequence, in most cases it remains to be explored whether the interaction is physical and direct, or if they rather function through interactions with the mitoribosome and/or unidentified proteins. Current speculations suggest that translational activators play a role in the spatial and temporal organization of mitochondrial protein synthesis and thus may serve to couple translation to insertion of the newly synthesized hydrophobic products into the membrane near or at the site of their assembly into multisubunit complexes [91, 92]. The expression and stoichiometry of these translational activators is important to maintain a balanced accumulation of the mitochondrial encoded COX subunits. With the exception of Mss51 (see below), these factors have been found to interact with each other forming high molecular complexes [89, 91], suggesting some level of co-regulation in the expression of the core-forming subunits of COX [93]. The stability of these complexes and the function of the different factors can be affected if the expression of one of the components of the complex is altered [91], possibly by sequestering other proteins in a non functional state. For example, overexpression of Pet111 attenuates Cox1 synthesis and COX assembly and limits the respiratory growth of yeast, an effect that can be complemented by concomitant overexpression of Mss51 and Pet309 [93]. Interactions have also been noted between a mitochondrial transcription factor, Nam1, and translational activators, including Pet111, Pet309, and Pet494 [91, 94] raising the possibility that mitochondrial transcription may be coupled to translation.
2.3. Mitochondrial COX gene expression in human
Most of the yeast genes specifically involved in the expression of the three mtDNA encoded proteins forming the core of the enzyme seem to be absent in humans. This can be explained by qualitative differences between the two species in their mitochondrial DNA and mRNA. Splicing factors are not required in humans because COX1, as any other gene in the human mitochondrial genome lacks introns. Transcription is polycistronic and the structural genes are flanked by tRNAs which serve as marks for processing by specific RNAses to free the individual transcripts. In mammalian mitochondria, the mRNAs are subsequently polyadenylated. Detailed information about mammalian mitochondrial transcription can be found in recent reviews [95, 96]
Concerning translation, the existence of mRNA-specific translational factors in mammalian mitochondria has long been a subject of speculation. Mammalian mitochondrial mRNAs lack 5′UTRs entirely. Thus, a Shine/Dalgarno interaction between the mRNA and the 12S rRNA is not used during mitochondrial translation. The yeast mitochondrial mRNAs also lack a typical Shine/Dalgarno element. However, in yeast the mRNA-specific translational activators could be involved in the localization of the small ribosomal subunit near the translational start codon [97]. Since most aspects of COX biogenesis are functionally conserved phylogenetically, it has been suggested that such factors, if they exist in mammals, interact with the coding regions of mammalian mitochondrial mRNAs. Unfortunately, these translation factors are poorly conserved at the primary sequence level even among fungi, making their orthologues difficult to be identified in mammalian genomes. A breakthrough in this area has recently been made, showing that homologues with poorly conserve sequences can still play related functions. This is the case of the yeast nuclear gene PET309 [84] and its human homologue LRPPRC. Mutations in LRPPRC are responsible for the Canadian form of Leigh syndrome [58]. Although the sequence similarity between their products is very weak (26%), the reciprocal BLAST best match of LRPPRC found in yeast is PET309 [58]. LRPPRC plays roles in stabilizing and handling all mRNAs, particularly those for the COX core subunits [98]. This protein also interacts with SLIRP, a stem-loop RNA binding protein, in a high molecular weight complex that contains mature mitochondrial mRNAs. However, it has been shown that LRPPRC does not participate directly in translation [98].
TACO1, a specific translational activator of human COXI, was recently identified by genome-wide linkage analysis in a patient affected by Leigh syndrome associated with an isolated COX deficiency [59]. TACO1 is a soluble matrix protein which could act by securing an accurate start of COXI mRNA translation or by stabilizing the elongating polypeptide and ensuring completion of its translation [59]. Deletion of the yeast homologue, YGR021W, does not produce any respiratory deficient phenotype and its role is unclear. The identification of mammalian-specific mitochondrial COX gene translational activators and the characterization of their mechanism of action are expected to provide crucial information concerning how translation of mammalian mitochondrial mRNAs is activated.
3. Biogenesis and assembly of yeast COX Subunit 1
3.1. Cox1 synthesis, membrane insertion and stability
3.1.1. Exclusive properties of COX1 mRNA -specific translational activators
The two translational activators identified to be essential for the translation of the COX1 mRNA in S. cerevisiae, Pet309 and Mss51, have structural and functional properties worthy to be highlighted in this section.
Pet309 is the only mitochondrial translational activator identified in S. cerevisiae that contains PPR motifs. Proteins that contain these sequence repeats in other organisms are usually involved in specific steps of RNA metabolism. They are implicated in precursor transcript stability and processing, as well as translation [99-102]. The small number of PPR proteins in S. cerevisiae contrasts with the number of family members in S. pombe, in which nine PPR proteins were recently reported to regulate mitochondrial gene expression, among them Ppr4, a putative homologue of Pet309, specifically affecting Cox1 translation [101]. S. cerevisiae Pet309 is predicted to contain at least seven PPR motifs located in the central portion of the protein. Studies involving these motifs showed that they are necessary for Pet309 translation of COX1 mRNA, but are not required for the stability of the transcript [103]. Although Pet309 has been established as a specific Cox1 translational activator, also necessary for the stability of the mRNA transcript, it remains to be explored whether Pet309 interacts directly with the 5′UTR of COX1 mRNA, and what the role of the PPR motifs would be in this predicted interaction.
Mss51 is unique among the COX-related translational activators because in addition of its requirement for the synthesis of Cox1, it plays additional roles in the stability of the newly synthesized peptide and its incorporation into early COX intermediates [32, 104]. Genetic studies have shown an interaction of Mss51 with the 5′UTR of COX1 mRNA [104]. However, when the 5′UTR of COX1 mRNA was swapped for the same region of the COX2 mRNA, the translation of the chimeric COX1 mRNA proceeded in the absence of Pet309, but not in cells that lacked Mss51. These results suggested that Mss51 has a second role in Cox1 biogenesis beyond its function in translational activation. Besides its interaction with the COX1 message, Mss51 has been found to interact with newly synthesized Cox1 [32, 104]. It has been proposed that such an interaction could serve to facilitate Cox1 elongation during synthesis [104, 105]. We have shown that Mss51 acts to stabilize Cox1 and maintain it in an assembly competent state before it proceeds to downstream assembly steps [32, 104, 106]. The double function of Mss51 provides the basis for a regulatory mechanism that coordinates Cox1 synthesis with COX assembly as explained below.
Despite efforts in several labs to identify the precise role of Mss51 in Cox1 translation, several basic questions remain to be answered. For example, it has not been directly established whether and how a physical interaction of Mss51 with the 5′UTR of COX1 mRNA occurs in vivo. So far, only yeast-three-hybrid system studies have provided some evidence for a physical interaction between a hydrophilic region located in the N-terminal 177 residues of the mature Mss51 protein and a target in the 5′-UTR of COX1 mRNA within 245 nucleotides upstream of the initiation codon [105].
Noticeably, we have reported evidence for aberrant translation of COX1 mRNA species resulting in a polypeptide termed mp15, in the absence of Mss51 but not in the absence of Pet309 [105]. Most probably, mp15 is a truncated translation product of a partially processed COX1 mRNA [105]. These results suggest that binding of Mss51 to the 5′-UTR of COX1 mRNA could be necessary for optimal initiation of translation by Pet309, whereas the interaction of Mss51 with newly synthesized Cox1 may regulate elongation of the nascent polypeptide [105] as previously proposed [104].
3.1.2. Translational regulation of Cox1 synthesis, Cox1 chaperones and Cox1 pre-assembly complexes
COX assembly requires the accumulation of its constitutive subunits in a defined stoichiometric ratio. Studies over the last decade have led to the notion of two mechanisms responsible for the concerted accumulation of COX subunits in yeast mitochondria. First, most unassembled COX subunit 1 and the other highly hydrophobic core subunits 2 and 3 are very efficiently posttranslationally degraded by the ATP-dependent AAA proteases of the inner mitochondrial membrane [107]. Additional proteases have been recently found to specifically prevent the accumulation of immature Cox1, including the conserved metallopeptidase Oma1 [108]. Active degradation will avoid the accumulation of unassembled proteins that could have a tendency to aggregate and disturb membrane homeostasis or to form pro-oxidant species as discussed below.
Second, Cox1 is subjected to assembly-controlled translational auto-regulation [32, 78, 104-106, 109-111]. This kind of translational regulation was initially found to operate in the assembly of photosynthetic complexes in chloroplasts from the alga Chlamydomonas reinardthii [112, 113] and in higher plants [114] and termed control by epistasis of synthesis. A distinctive characteristic of these organellar translational auto-regulatory systems is the involvement of ternary factors, mRNA-specific translational activators, whose availability would be regulated by the specific gene products. In the case of yeast COX, the ternary factor is Mss51 [32, 78, 104-106, 109, 110].
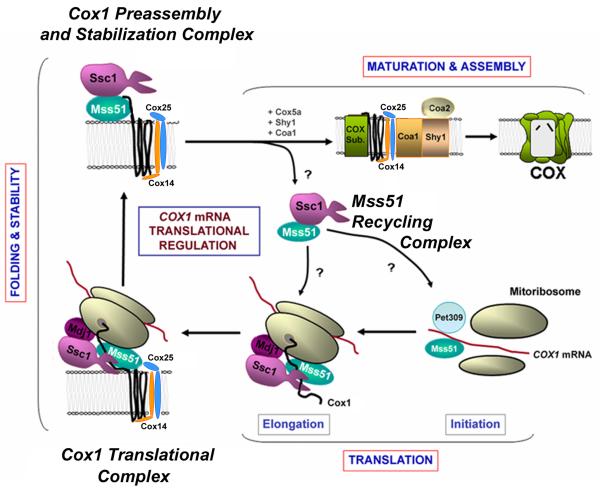
Mss51 dynamically interacts with Cox1 and several components of the COX biogenetic process. During Cox1 synthesis on the mitoribosomes, Mss51 and newly synthesized Cox1 form a transient complex [32, 104] that is stabilized by the COX assembly factors Cox14 [32] and Cox25 (also termed Coa3 [110, 111]) and additionally contain the mitochondrial Hsp70 chaperone Ssc1, and its co-chaperone Mdj1 [106]. Ssc1 and its co-chaperone Mdj1 were shown to form a complex with nascent polypeptide chains on mitochondrial ribosomes probably to facilitate their proper folding during translation [115]. The presence of all these proteins in a COX1 translational complex points towards cooperation of general and specific chaperones in the folding and stabilization of newly synthesized Cox1.
Cox1 is a highly hydrophobic protein and it spans 12 trans-membrane domains in the inner mitochondrial membrane, connected by short hydrophilic loops that protrude either to the inter-membrane mitochondrial space or the matrix. As Cox1 is being synthesized, it is co-translationally inserted into the inner mitochondrial membrane with the aid of the Oxa1 machinery [116], which interacts with the mitoribosome. Cox14 and Cox25/Coa3 are small single transmembrane proteins with a hydrophilic C terminus. While this region of Cox14 resides in the intermembrane space and is glutamine rich, the C terminus of Cox25 resides in the matrix milieu and contains a positively charged lysine rich terminal domain [110]. The topologies of Cox14 and Cox25 suggest these proteins could interact with Cox1 transmembrane domains and direct their insertion into the inner membrane. Definitely, they serve to promote the stability of the Cox1-Mss51-Ssc1 complex by interacting with Cox1 and holding the complex from both the intermembrane space and the matrix sides of the inner membrane. In fact, in the absence of either of these proteins, newly synthesized Cox1 is rapidly degraded [32, 106, 111].
Following Cox1 synthesis and membrane insertion, a Ssc1-Mss51-Cox1-Cox14-Cox25 pre-assembly complex remains stable until Cox1 proceeds to downstream assembly steps. This complex, abundant in wild-type cells, could represent a Cox1-containing complex serving as a reservoir of stable Cox1 ready to be matured and/or to progress in the COX assembly process when required. We have postulated that the Mss51 interactions within the translational and pre-assembly complexes down-regulate Cox1 synthesis when COX assembly is impaired by trapping Mss51 and limiting its availability for COX1 mRNA translation [106, 110] (Figure 2). The C-terminal residues of Cox1 have recently been shown to be essential for Mss51 sequestration and to stabilize the Ssc1-Mss51-Cox14-Cox25 interaction [78].
Fig. 2. Coordination of Cox1 synthesis with COX assembly.
The model depicts the roles of Mss51, Cox14, Cox25 and the mtHsp70 chaperone Ssc1 on translational regulation of COX biogenesis (see explanation in the text).
According to the translational regulation model (Figure 2), the release of Mss51-Ssc1 from the pre-assembly complex and Mss51 availability for Cox1 synthesis [106] probably occur when Cox1 acquires its prosthetic groups or interacts with other COX subunits, a step possibly catalyzed by Shy1 (SURF1 yeast homologue), a protein involved in maturation and/or assembly of Cox1 [32, 35, 117]. Coa1, another COX assembly factor, could also participate in Cox1 maturation and stabilize the Cox1-Ssc1-Mss51-Cox14-Cox25 complex prior to its interaction with Shy1. Once Mss51 is released from the Cox1 pre-assembly complex, Cox14 and Cox25 remain interacting with increasingly matured COX assembly intermediates [35, 111, 118]. Coa1 and Shy1 are subsequently incorporated to interact with Cox1-containing subassemblies downstream from the roles of Mss51 in COX biogenesis. In the absence of Coa1, Cox1 synthesis proceeds normally as in the case of cox14 or cox25 mutants. However, the Cox1 pre-assembly complex is formed in the absence of Coa1 [106] and it is now generally accepted that this protein is not part of Mss51-containing complexes [106, 111, 119]. Coa1 could play roles in Cox1 maturation perhaps in collaboration with Shy1[35, 118], an step that could be synchronized with the incorporation of nuclear encoded subunits Cox5a and Cox6 into early Cox1 assembly intermediates and coupled to the release of Mss51 from the Cox1 pre-assembly complex [78, 117].
The presence of Ssc1 in the pre-assembly complex could serve to facilitate the proper Cox1 folding during membrane insertion as mentioned earlier or to present it to matrix-localized proteolytic systems in the case of unproductive folding that would prevent membrane insertion. Alternatively or additionally, it could play a role in the coordination of the recycling of Mss51 from its posttranslational function to become available for COX1 mRNA translation [106]. In fact, we have proposed that when Mss51 is released from the pre-assembly complex, it forms a very stable heterodimeric complex with Ssc1 [106]. In the absence of COX1 mRNA and/or Cox1, all Mss51 is bound to Ssc1 in this binary complex which seems to be the reservoir of Mss51 when not engaged in its functions in Cox1 biogenesis. This heterodimer may be the source of Mss51 that is competent for translation [106]. Supporting this possibility, overexpression of Mss51 or null mutations in cox14 and cox25/coa3, results in wild-type levels of Cox1 synthesis, although the protein is rapidly degraded and Mss51 fully accumulates into the heterodimeric complex with Ssc1, thus preventing the trap into higher molecular weight complexes, and is available for subsequent rounds of translation [106, 110]. Among the many open questions remaining, it seems important to know whether Mss51 is released from the pre-assembly complex bound to Ssc1, or a new Ssc1 molecule binds Mss51 following its release. Similarly, exploring whether Mss51 plays a role in COX1 mRNA translational activation alone or complexed to Ssc1 requires further investigation.
Additionally, whether the COX1 translational regulatory system is conserved in higher eukaryotes remains unknown. Cox1 synthesis seems to proceed normally in cells from patients carrying mutations that compromise COX assembly. It can be claimed that in these cases, the mutant factors always retain some residual activity which could prevent the activation of the regulatory mechanism. However, Cox1 synthesis was not altered in mouse cells carrying a null allele of cox10 [120] .
3.2. Maturation of Cox1 by incorporation of its metal prosthetic groups
Cox1 contains two metal centers, heme a and the binuclear center formed by CuB and the high spin heme a3, which are essential for the catalytic activity of the enzyme. The incorporation of these prosthetic groups is necessary for the maturation and correct folding of the Cox1 polypeptide. When and how the metal groups are incorporated into Cox1 is of high interest in the field but remain to be fully understood.
3.2.1. Heme a biosynthesis, incorporation into Cox1 and coupling to COX assembly
Heme a is a prosthetic group present in eukaryotic and some bacterial COX. Most bacterial oxidases contain heme b, a heme a precursor species. COX is the only enzyme in mitochondria that requires heme a as a cofactor. Heme a differs from protoheme (heme b) in that the C2 vinyl side chain is replaced by a hydroxyfarnesyl and the methyl group is oxidized into a formyl group [121]. Heme a biogenesis from its ancestral heme b form is a stepwise process and has been reviewed elsewhere [21, 122]. The first reaction is catalyzed by Cox10, the heme A:farnesyltransferase, and involves the formation of a heme o intermediate that carries a hydroxyfarnesyl group at the C-2 position. The conversion of heme o to heme a requires the oxidation of the methyl substituent in position C-8 to a formyl group which could occur in two discrete monooxygenase steps [121]. Several models have been proposed to explain the conversion of heme o to heme a. In a model, the heme o methyl group is first hydroxylated by the oxygenase Cox15, which acts in concert with ferredoxin Yah1 and the ferredoxin reductase Arh1 [123], while the subsequent oxidation of the C-8 methanol to a formyl group would be catalyzed by an enzyme yet to be identified [123]. On another model, Cox15 may utilize two successive monooxygenase reactions to generate a geminal diol (which could then spontaneously dehydrate) [124]. On a third, perhaps less likely alternative, a peroxidase-type mechanism can be invoked to generate an aldehyde directly from a methyl group, or a peroxidase could oxidize an alcohol intermediate [124]. Studies in Bacillus subtillis have provided strong evidence supporting that Cox15 oxidizes heme o to heme a via successive monooxygenase reactions [124]. On a functional note, it has been proposed that the farnesylation of heme may be important for Cox1 folding and packing [122], which may account for the necessity to contain heme a over heme b.
While Cox1 hemylation is essential for COX assembly, little is known about the players and mechanism of heme a insertion into Cox1. Due to the reactive nature of the heme a moiety, it is likely that a heme-binding protein assists the transfer of heme a from its synthesis site to Cox1. Cox10 and Cox15 are both integral proteins in the inner mitochondrial membrane [123, 125], which could possibly give proximity of heme a synthesis to Cox1 insertion. Due to the requirement of heme a incorporation into Cox1 for its stability and folding, it has been suggested that this event could occur either co-translationally or during Cox1 insertion into the membrane. However, it has been recently shown that the two heme a cofactor sites in Cox1 form downstream of Mss51- and Coa1-containing Cox1 preassembly and stabilization intermediates [119]. These Cox1 intermediates form normally in cells defective in heme a biosynthesis or in cox1 mutant strains with heme a axial His mutations [119]. Additionally, analyses of purified Mss51-containing Cox1 preassembly and stabilization complex have failed to detect any traces of heme a (our unpublished results), thus indicating that heme insertion clearly occurs at a post-translational stage. In contrast, the Mss51-free, Shy1-containing Cox1 assembly intermediate is perturbed in the absence of heme a, thus suggesting that the incorporation of heme a into Cox1 occurs within this subassembly [119].
Shy1 is the yeast homolog of Surf1, a mammalian gene identified in the COX deficiency leading to Leigh syndrome in humans. Shy1 has been proposed to be involved in either the formation or the stabilization of the heme a3 site. This hypothesis was initially based on the observation that in a Rhodobacter sphaeroides surf1 null mutant the heme a3 site is not fully populated [126]. Noticeably, in this mutant the heme a site was found to be formed. This result brings the question of the possible existence of two heme a insertases. Although there has not been any report of other candidate proteins involved in heme delivery to Cox1, it is not likely that Shy1/Surf1 is the only protein involved in this process due to the 10-15% residual COX activity detected in the yeast null mutant and mutant surf1 human cells. Recent data on Paracoccus denitrificans Surf1 isoforms, have shown that when co-expressed in Escherichia coli together with enzymes for heme a synthesis, they have the ability to bind heme a in a 1:1 stoichiometry with Kd values in the submicromolar range [127]. Nonetheless, these findings have yet to be confirmed in vivo and in eukaryotes. The bacterial study also identified a conserved histidine as a residue crucial for heme binding [127]. COX10 is a weak multicopy suppressor of yeast Δshy1 cells, thus connecting Shy1 to heme biosynthesis [117, 118]. However, mutations of either of the two conserved His residues in yeast Shy1 did not significantly affect its function [128]. Alternatively, Shy1 function may enhance the stabilization of the heme a3 site rather than playing a direct role in heme a delivery. A role of Shy1 in incorporation of additional COX subunits into early Cox1 subassemblies has not been fully discarded. For example, overexpression of Cox5a and Cox6 significantly suppresses the respiratory defect of Δshy1 cells [117, 118]. Enhanced levels of these subunits may stabilize Cox1 in Δshy1 cells, enabling progression to later stages of COX assembly. We could speculate that, for example, addition of Cox5a, which transmembrane helix is tightly packed against Cox1 could contribute to the stabilization of the metal centers in Cox1. However, studies of COX assembly intermediates in fibroblasts from human patients carrying mutations in SURF1, have disclosed the accumulation of Cox1 alone or in an early intermediate containing human subunit IV and V (yeast subunits 5a and 6), which has suggested that SURF1 could play a role in the incorporation of subunit II into these nascent intermediates [129]. In any case, heme a insertion into the binuclear center of Cox1 seems to occur prior to addition of Cox2 as the farnesyl group of heme a3 is located at the interface between the two subunits [22].
Due to the high reactivity of the heme a moiety, it seems logical that its synthesis would be coordinated to its incorporation into Cox1. In fact, the biosynthesis of heme a has been shown to be regulated by downstream events in the COX assembly process [130]. In most yeast COX mutants there is a drastic reduction of steady-state levels of heme a [130]. The overexpression of COX15, particularly when co-overexpressed with the ferredoxin YAH1 acted as a suppressor of the heme a accumulation defect in COX mutants including mutants in which Cox1 was not synthesized. This observation suggested that the absence of heme a in the mutants is not due to a rapid turnover of the cofactor in the absence of COX subunit 1, but rather to a feedback regulation of the heme a synthesis when the COX assembly process is blocked [130]. These results suggest that heme a synthesis can proceed even in the absence of stable Cox1 peptide. In addition to low heme a synthesis, COX mutants with the obvious exception of cox10, also show an accumulation of heme o, indicating that this compound is stable. Heme o levels were also very low in a cox15 null mutant, a phenotype that was not rescued by the overexpression of COX10. This observation suggested that the first step of the heme a biosynthesis is also positively regulated in a Cox15 dependent manner [130]. It remains to be tested whether increased heme a levels by COX15 overexpression in the absence of COX assembly bypasses Cox1 downregulation. In that event, it could be proposed a direct connection between COX1 translation and heme a biosynthesis regulations. However, in a cox14 null mutant, in which Cox1 synthesis is not downregulated due to the instability of the Mss51:Cox1 early intermediate [32], heme a levels are low and there is a significant accumulation of the heme o intermediate, compared to a wild type strain [130], thus indicating that an increase in Cox1 synthesis does not stimulate heme a biogenesis.
The identity of the COX assembly intermediate that could operate in the regulation of heme a biosynthesis remains to be identified. Recent studies in yeast have identified a new COX assembly gene, COA2, essential for COX assembly, which absence impairs Cox1 maturation and induces a rapid degradation of newly synthesized Cox1 [108]. Coa2 was shown to transiently interact with Shy1 [131]. Coa2 has been connected to Cox1 hemylation because the respiratory deficiency of coa2Δ cells is suppressed by the presence of a catalytically active mutant allele of Cox10 (N196K). The suppressor activity of this Cox10 variant is actually dependent on its catalytic function and the presence of Cox15 [108]. Noticeably, Cox10 forms a high mass oligomeric complex which stability, enhanced by the N196K mutation, seems to depend on Coa2 [108]. Oligomerization of wild-type Cox10 appears to be dependent on the Coa1-containing Cox1 complex, but the N196K protein can oligomerize in the absence of Coa1. Consistently, the Cox10 oligomer is not formed in Δcox14 cells where Cox1 is synthesized but not assembled. However, it is unlikely that Cox10 oligomerization is necessary for its function as a heme o synthase, since the cox14 null mutant accumulates high levels of the heme o intermediate as mentioned earlier [130].
The pathways involved in heme a biosynthesis are highly conserved from yeast to human. The conservation of Cox10 and Cox15 along evolution is underlined by human COX10 and COX15 heterologous complementation, albeit poor in the former case, of yeast cox10 and cox15 null mutants, respectively [125, 132]. Mutations in human COX10 and COX15 genes have been associated with severe infantile cardiomyopathy and Leigh syndrome in human [55-57]. The analysis of COX assembly intermediates suggests that heme a is incorporated into COX1 at a very early stage of COX assembly, prior to the formation of the COX1-COX4-COX5a complex, that fail to accumulate in patient fibroblasts [56, 57, 60]. The heme a regulatory system could be different in mammalian cells. Analyses of mitochondrial heme content in COX15 deficient fibroblasts from a human patient suffering from hypertrophic cardiomyopathy showed levels of heme o significantly higher than in control fibroblasts [57].
3.2.2. Copper metabolism and insertion into Cox1
Despite being essential for life [133], the high redox activity of copper means that it can be extremely toxic and promote the formation of reactive oxygen species. Therefore, a network of transporters strictly controls the movement of copper in living systems. Most components of the copper homeostatic machinery and their mechanisms of action were elucidated in S. cerevisiae and have been extensively reviewed elsewhere [134]. In mitochondria, two enzymes, COX and mt-Sod1 receive copper within the IMS [12, 135]. This requires a specific copper transport pathway to this organelle. The mitochondrial matrix contains a pool of copper bound by a not yet fully characterized nonproteinaceous ligand [136]. This pool is the copper source for metallation of COX and mt-Sod1 [137]. The ligand has also been found in the cytoplasm and it has been suggested that it may recruit copper to mitochondria in place of a copper chaperone [137]. Exactly how copper makes its way from plasma membrane copper transporters to the mitochondrial matrix and IMS is of great interest to the field and remains to be understood.
COX copper metallation involves the IMS metallochaperone Cox17 [138], a small hydrophilic protein that contains a CCxC copper binding motif [139]. The last cysteine in the motif is the first in an overlapping twin Cx9C structural motif [140]. Once copper reaches Cox17 within the IMS, it is accepted that this protein transfers copper ions to two chaperones [141], Sco1 [142] and Cox11 [143, 144] that facilitate copper insertion into the Cox2-CuA and Cox1-CuB sites, respectively. How copper reaches Cox17 in the mitochondrial IMS is still an open question. In addition to Cox17, the IMS houses additional conserved Cx9C proteins which absence produce heterogeneous effects in COX assembly and function [145]. Several pieces of information obtained in S. cerevisiae suggest that at least some of these proteins could be part of a copper transfer pathway towards COX [135]. These proteins include Cox19 [146], Cox23 [147], Pet191 [148], Cmc1 [149] and Cmc2 [150]. We have hypothesized that copper could be transferred from the matrix pool across the inner membrane by an uncharacterized transporter to the membrane-bound Cmc1/2 proteins [135]. In a daisy chain transfer mechanism, copper would be successively transferred to the soluble Cox19, Cox23 and ultimately to Cox17 for final delivery to Cox11 and Sco1. This mechanism would be expected to involve transfer of both reducing potential and Cu(I) to generate reduced Cu(I)-bound proteins. Such a mechanism could be useful to accumulate a small copper pool in the IMS available for regulated transfer to Cox17 [135]. Alternatively, the small CX9C proteins could play a role in copper transfer towards Cox17 by modulating the local redox environment within the IMS. Interestingly, Cmc1 and Cmc2 are not only required for full expression of COX but indirectly modulate copper delivery to mt-Sod1, thus suggesting a connection between the two pathways [149, 150]. These aspects of mitochondrial copper metabolism remain speculative and have been extensively reviewed recently [135, 151].
Focusing on copper metallation of Cox1, the CuB site is formed by one copper ion coordinated by three histidine ligands and present in close proximity to the heme a3 moiety. The Cox11 metallochaperone is formed by a transmembrane domain and a copper binding globular domain facing the intermembrane space where copper transfer occurs [143, 144]. The soluble C-terminal domain of Cox11 forms a dimer that coordinates one Cu(I) per monomer. The two Cu(I) ions in the dimer exist in a binuclear cluster and appear to be coordinated by three conserved cysteine residues [144]. The mechanism by which Cox11 transfers copper to the CuB site remains to be elucidated.
Cox1 copper metallation was initially envisioned to occur co-translationally due to the positioning of the CuB site, deeply buried below the inner mitochondrial membrane. Such a hypothesis was supported by studies that show a small portion of Cox11 can interact with mitochondrial ribosomes [152]. However, functional analysis of the domains of yeast Cox11 suggests that the weak interaction with the ribosomes must be mediated by another protein, as the Cox11 matrix domain is non-essential [12, 153]. On the other hand, in Schizosaccharomyces pombe Cox11 exists as a fusion protein with Rsm22, a component of the small subunit of mitochondrial ribosomes [153] and in S. cerevisiae, the synthetic non-cleavable fusion of Cox11 with Rsm22 is fully functional [153]. It is tempting to speculate that the transient interaction with the ribosomes could serve to recruit Cox11 to the place of Cox1 synthesis to regulate its copper metallation once Cox1 has been membrane inserted and stabilized by binding to the Mss51, Cox14 and Cox25 chaperones.
In fact, more recent investigations have established that the CuB site formation does not actually occur while newly synthesized Cox1 is interacting with Mss51, but near or at the Shy1-containing Cox1 assembly intermediate [119], perhaps simultaneously to the incorporation of the heme a3 moiety into the heterobimetallic center. This conclusion was supported by the detection of a transient interaction between Cox11 and Shy1 and by the marked attenuation of the Shy1-containing Cox1 in Δcox11 which was partially restored with a nonfunctional Cox11 mutant [119].
It remains unclear in what specific order the CuB-heme a3 center is metallated. Studies in a Rhodobacter sphaeroides cox11 null mutant have shown that the heme a3 moiety can be delivered to this site in the absence of copper [143]. Studies in yeast have revealed that null cox11 mutants, as well as sco1 mutants, show higher sensitivity to hydrogen peroxide than a wild type strain, which was attributed to the presence of a highly reactive Cox1-heme a3 intermediate [154]. It has been proposed that independently of its copper binding function, Cox11 may bind and stabilize Cox1 through transient interactions in a conformer of Cox1 that has less solvent accessibility to the heme a3. However, a direct Cox11-Cox1 interaction has not been detected. Additionally, the amount of the Shy1-containing Cox1 intermediate was found markedly attenuated in Δcox11 as mentioned earlier [119], an observation that would suggest its accumulation could produce deleterious effects but at the same time makes intriguing the small threshold concentration of this putative intermediate that seems to be necessary to produce such an effect.
Human mitochondrial copper metabolism involves conserved elements. Homologue of the yeast genes COX11, COX17, SCO1, COX19, COX23, PET191, CMC1 and CMC2 have been identified but in most cases their functions remains poorly characterized [155]. The human COX17 homologue shares 48% sequence identity with its yeast counterpart. However depletion of COX17 by siRNA in HeLa cells causes the accumulation of a COX1-containing COX assembly intermediates devoid of COX2, suggesting that the role for human COX17 in the metallation of COX2 could be more essential than for COX1 (Oswald et al., 2009). However, the metal content of the stable COX1 was not reported. On another respect, a homozygous mutation in C2orf64, the human homologue of yeast PET191 gene, has been recently identified in two siblings affected by fatal neonatal cardiomyopathy associated with severe COX deficiency [156]. Accumulation of a small COX assembly intermediate containing COX1, but not COX2, COX4 or COX5b in patient fibroblasts suggests that PET191 is involved in an early step of COX assembly [156].
4. Biogenesis and assembly of COX Subunit 2
4.1. Cox2 synthesis, membrane insertion and stability
Cox2 synthesis requires the action of Pet111, a membrane bound COX2 mRNA-specific translational activator [85, 86]. Pet111 interacts with the 5′-UTR of COX2 to promote translation [85, 86] and is present in the mitochondrial membranes at a low concentration thus limiting the synthesis of Cox2. On a different type of regulation, translation of COX2 mRNA is known to be controlled by intrinsic antagonistic signals. The mRNA sequence of the first 14 COX2 codons, which specify the precursor Cox2 (pCox2) N-terminal leader peptide discussed below, contain a positively acting element required for downstream translation of COX2 mRNA [157]. Three additional sequences located in the Cox2 N-terminal region of the mature protein have been described to play an inhibitory role in the absence of the positive element [158]. Interplay among these signals during translation has been hypothesized it may ensure the coordination of Cox2 synthesis and subsequent assembly into COX.
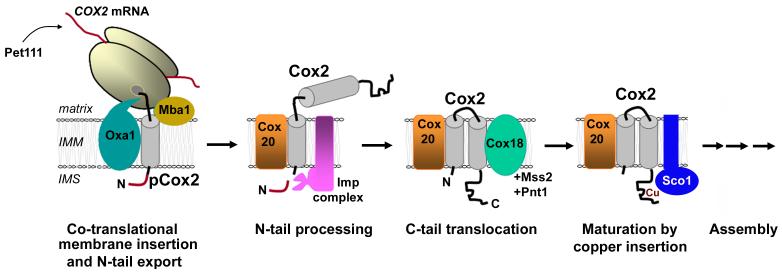
Cox2 is an integral membrane protein containing two transmembrane domains and presents an N-out C-out topology (both termini protruding into the IMS). Cox2 is synthesized as a precursor protein (pCox2) with a cleavable amino terminal extension (Fig. 3). Prior to the cleavage, pCox2 interacts with the Oxa1 machinery which facilitates membrane insertion of the first pCox2 transmembrane domain and concomitant export of its N-terminal domain across the inner membrane [159, 160]. This step is required for the export of the pCox2 C-terminal tail [161]. Oxa1 belongs to a conserved protein family known as YidC/Alb3/Oxa1 family, the bacterial, mitochondrial and plastid members of which assist the membrane insertion of proteins [162]. Oxa1 is an integral inner membrane protein that contains five transmembrane domains and presents N out-C in orientation. The C-terminus of Oxa1 interacts with the large ribosomal subunit within close proximity to the polypeptide exit tunnel, thus facilitating the co-translational insertion of the nascent hydrophobic peptides into the mitochondrial inner membrane [163, 164]. Oxa1 cooperates with the membrane protein Mba1 which acts as a ribosome receptor to recruit ribosomes to the inner membrane and helps Oxa1 in the orientation of the ribosome exit site towards the inner membrane insertion machinery [116, 165]. However, both ribosomal subunits remain bound to the inner membrane in the absence of Oxa1 and Mba1, indicating that other factors collaborate to the tethering of ribosomes to the membrane [165]. The function of Oxa1 is not limited to Cox2 insertion, but extends to the insertion of other polytopic proteins such as the mitochondrially encoded Cox1 and Cox3 proteins [116, 160] as mentioned earlier. Independently of the lack of cleavable N-terminus in mammalian Cox2, the human homolog of Oxa1 (OXA1L) is able to functionally replace the yeast protein leading to a correct COX assembly, suggesting that both proteins play a similar role [166].
Fig. 3. Biogenesis of Cox2.
The scheme depicts the several steps and proteins involved of synthesis, topogenesis, maturation and assembly of Cox2 (see explanation in the text). IMM, inner mitochondrial membrane; IMS, intermembrane space.
The export of the S. cerevisiae Cox2 C-terminus depends on the inner membrane potential, while export of the N-tail does not [159], which suggests that although the two processes depend on Oxa1, they involve distinct mechanisms [159, 161]. Downstream the role of Oxa1, the product of COX18 [167] in conjunction with two other inner membrane proteins, Mss2 and Pnt1, has been proposed to subsequently promote the insertion of the second, C-proximal, pCox2 transmembrane domain [159, 167-169]. Cox18 resembles some Oxa1 family members but its role is specific for the translocation of Cox2 [168]. However, a direct interaction between Cox2 and Cox18 has not yet been shown [168]. Cox18 physically interacts with Mss2 and Pnt1, suggesting that they form a translocon complex although the detailed molecular mechanisms involved in the export of the Cox2 C-terminal tail remains to be elucidated [168]. Interestingly, copper binding to Cox2 is not a requirement for C-tail translocation since missense substitutions of two of copper-binding residues in Cox2 did not prevent its export [170]. Despite the similarities between Oxa1 and Cox18, OXA1 overexpression is not able to suppress the respiratory deficiency of Δcox18 cells [171]. However, it does promote some translocation of the Cox2 C-terminal domain across the inner membrane and increased accumulation of Cox2, which remains unassembled. This observation has suggested that in addition to its role in C-tail translocation, Cox18 is required to deliver Cox2 to a state competent for COX assembly [171]. Interestingly, Δcox18 cells overexpressing Oxa1 recover respiratory competency after acquiring recessive mutations in Mgr1 and Mgr3, which products are known to be subunits of the inner membrane AAA protease supercomplex, where they associate with Yme1 and participate in membrane protein degradation [171]. However, the absence of Mgr1/3 does not act by stabilizing Cox2 and it has been proposed that Yme1 could probably chaperone the folding and/or assembly of Oxa1-exported Cox2 under these conditions [171].
Following export, pCox2 interacts with the Cox2-specific chaperone Cox20 [172] which presents the precursor protein to the three-subunit inner membrane peptidase (IMP)-complex [173, 174] that catalyzes the proteolytic removal of the pCox2 amino-terminal sequence. The Cox20-Cox2 interaction most probably occurs after export of the Cox2 N-tail domain and prior to the action of Cox18 because the leader sequence of pCox2 is cleaved in Δcox18 cells [175]. The IMP-complex is formed by three subunits; Imp1 and Imp2 have catalytic functions with different substrate specificities [176, 177] and Som1 stabilizes the complex [173]. Imp1 is responsible for the proteolytic cleavage of pCox2, but its function requires the stabilizing presence of the other two subunits [173, 176]. Based on sequence similarity, a putative human homolog of IMP2 has been reported [178], although considering the absence of experimental data about its function, it remains uncertain whether it is a true ortholog. In any case, it is unlikely that it could be related to Cox2 biogenesis considering that mammalian Cox2 lacks any cleavable amino terminal extension.
Once the pCox2 leader sequence is cleaved, Cox20 probably remains bound to mature Cox2, stabilizes the protein and facilitates its assembly into COX assembly intermediates [172]. The essential role of Cox20 as a Cox2 chaperone is supported by its conservation through evolution. A human homologue was recently identified by BLAST sequence analyses [10]. Although the protein localizes to mitochondria in HeLa cells (our unpublished results), its function as a Cox2 chaperone remains to be fully characterized.
2.2. Maturation of Cox2 by insertion of its copper prosthetic group
The CuA binuclear center in Cox2 is coordinated by a CxExCGx2Hx2M motif [22, 25] and exists as a [Cu2+/Cu1+] complex as revealed by EPR spectroscopy [179-181]. The metallochaperone for the formation of the CuA center of Cox2 is the product of SCO1 [142].
Sco1 is anchored to the mitochondrial inner membrane through a transmembrane α-helix and exposes the copper binding site in the intermembrane space where copper transfer occurs [139, 153]. In vitro experiments have shown that a soluble truncated form of Sco1 is able to bind copper donated from Cox17 [153, 182]. However, it remains unclear how the transfer of copper occurs because a physical interaction between Cox17 and Sco1 has not yet been detected probably due to its transient nature in vivo. Sco1 has a metal binding Cx3C motif analogous to the copper binding motif of Cox2, and this motif is essential for its function, as demonstrated by site-direct mutagenesis [183]. SCO1 was originally identified as a multicopy suppressor of a cox17 null mutant [138] and has been shown to directly interact with Cox2 [184]. In addition, since the CuA center is formed by a Cu(I) ion and a Cu(II) ion, it remains to be elucidated if Sco1 mediates the transfer of both different valent ions or, alternatively if two Cu(I) are inserted in Cox2 by Sco1 and the active site is successively oxidized. Sco1 is able to bind both Cu(I) and Cu(II) [185] and NMR studies have suggested that Cox17 transfers reducing potential and Cu(I) to enable Sco1 metallation [186]. Besides the role of Sco1 in copper insertion, its structural similarity with disulfide reductases has suggested that it could be involved in the reduction of cysteines in the Cox2 copper binding site thus facilitating copper incorporation [186-189]. Sco1 can form homodimeric complexes [184] which could facilitate the performance of both functions by the collaborative action of each monomer.
Yeast SCO1 has a highly conserved homologue, SCO2 [190]. Although its absence does not affect COX assembly [142], Sco2 overexpression partially rescues the respiratory defect of sco1 point mutant cells [142] as well as the defect of cox17 mutants when the growth media is supplemented with copper [142]. However, its precise function remains to be disclosed.
The CuA active site is located above the inner membrane surface, in a domain of Cox2 folded in a β-barrel and protruding into the mitochondrial intermembrane space. The Sco1-mediated insertion of copper into Cox2 probably takes place after the extrusion of the Cox2 copper binding domain in the intermembrane space and before the incorporation of this subunit to a COX assembly intermediate [136].
2.3. Roles of mammalian Cox2-specific copper insertion chaperones
Similar to yeast, humans have two Sco1 proteins, although both are homologues of yeast Sco1. They were also termed SCO1 and SCO2 [191]. Both are essential for COX assembly and mutations in SCO1 [50] and SCO2 [51-53] result in severe mitochondrial disorders. The functional differences among the two yeast and human isoforms is explained by the fact that the two genes probably originated from a duplication that occurred separately in the two organisms [51].
In contrasts to the yeast Sco proteins, human SCO1 and SCO2 have been shown to perform independent, cooperative functions in COX assembly [192, 193]. Sco proteins contain a CX3C copper-binding motif, shown to be essential for their function in COX2 biogenesis [185, 193]. Furthermore both SCO1 and SCO2 proteins have an affinity for copper higher than COX17 (Banci et al, 2010), which allow for the quantitative transfer of Cu(I) from COX17 to SCO1 and SCO2 [194, 195]. SCO1 protein exists as a mixed population of oxidized and reduced thiols, the proportion of which depends upon the presence of a functional SCO2 protein [193]. These data, together with the observation that Sco proteins contain a highly conserved thioredoxin domain [189], has brought Leary and co-authors to suggest a thiol-disulphide oxidoreductase function for SCO2 protein [193]. Following the proposed model, after COX2 metallation by SCO2 and SCO1-dependent simultaneous or sequential copper insertion, SCO2 re-oxidizes SCO1 cysteines, a reaction that allows to reset both proteins for further rounds of COX2 biogenesis [193]. While both Sco proteins are required for copper transfer to COX2, SCO2 is additionally necessary for COX2 synthesis, since SCO2 depletion decreases the accumulation of newly synthesized COX2 in culture cells [193]. These stage-specific functions of each Sco protein during Cox2 synthesis and formation of the CuA site probably serve to coordinate both processes during COX assembly.
Finally, in a process independent of COX biogenesis, both SCO1 and SCO2 play additional roles in the maintenance of cellular copper homeostasis cooperating to regulate copper efflux under conditions of excessive cellular copper [196].
5. Biogenesis and assembly of COX Subunit 3
5.1 Cox3 synthesis, membrane insertion and stability
Cox3, the third mitochondrial DNA encoded COX subunit, completes the COX catalytic core. Similar to Cox1 and Cox2, Cox3 is a highly hydrophobic protein with 7 transmembrane helixes embedded in the inner mitochondrial membrane. In contrast, Cox3 does not contain prosthetic groups and its function remains unclear. It has been proposed that Cox3 could be involved in the assembly and/or stability of subunits 1 and 2 or in the modulation of oxygen access to the binuclear center [197]. However, studies performed on bacterial COX, formed exclusively of the three core subunits, have suggested that Cox3 could play a role in modulating proton transfer through subunit 1 and 2 [198, 199].
In S. cerevisiae, Cox3 synthesis requires the activity of three nuclear encoded translational activators: Pet54, Pet122 and Pet494 [87, 200]. Deletion of any of these proteins severely reduces Cox3 synthesis, thus underlying their essential non-redundant role in Cox3 biogenesis [89]. The COX3 mRNA translational activators interact among them to form a COX3 specific activator complex [89], which, through Pet54, directly binds the 5′-UTR of COX3 mRNA to promote translation [74, 88, 90]. Pet54 plays multiple roles in COX biogenesis. It does not only act as a COX3 mRNA translational activator but it is also necessary for the splicing of the aI5β group I intron in the COX1 pre-mRNA [201]. In particular, it has been shown that the COX3 5′-UTR mRNA and the aI5β group I intron bind to the same or overlapping surface on Pet54. Moreover, the Pet54 binding sites in the COX3 5′-UTR mRNA and in the aI5β group I intron present 56% sequence similarity. Ultimately, the COX3 5′-UTR mRNA and the aI5β group I intron can compete for Pet54 binding [74]. Recently, based on the observation that Pet54 is required for Cox1 synthesis even in a yeast strain carrying intronless mtDNA, it has been proposed an additional yet undefined role of Pet54 in Cox1 biogenesis [78]. Taken together, these data suggest a further level of co-regulation of Cox1 and Cox3 expression.
Pet122 and Pet494 are integral inner membrane proteins, while Pet54 is a peripheral protein associated to the inner mitochondrial membrane [202, 203]. It is believed that, similarly to Cox1 and Cox2 biogenesis, the tethering of the translational apparatus to the inner mitochondrial membrane by these translational activators could facilitate the Oxa1-dependent co-translational membrane insertion of the newly synthesized highly hydrophobic Cox3 subunit.
In yeast, unassembled Cox3 is subjected to a rapid proteolytic turnover. Cox3, similar to the other core subunits, is a substrate of the matrix-AAA protease complex [107] that it is likely to recognize the Cox3 N-terminus protruding into the mitochondrial matrix.
5.2 Assembly of Cox3
Little is know about the addition of Cox3 into COX assembly intermediates. Although it is expected to be a protein assisted process, the identity of the putative Cox3-specific chaperone remains to be disclosed. It is believed, however, that Cox3 assembly occurs once the Cox1-Cox5a-Cox6 complex has been formed and Cox2 has been already incorporated. The addition of Cox3 stabilizes the core because in the absence of this subunit, Cox1 and Cox2 fail to accumulate in the mitochondrial membranes [32].
In human, a few mutations have been described in COX3-associated COX deficiency [43]. The brief list includes a 15-bp microdeletion in a patient with recurrent myoglobinuria [204], a mutation that also prevented COX assembly when introduced in yeast [205]. In both cases, Cox1 and Cox2 are rapidly degraded in the absence of Cox3. Also in Chlamydomonas, where Cox3 is encoded by a nuclear gene and presents a lower hydrophobicity than other Cox3 mitochondria-encoded proteins [20], the lack of this subunit leads to the absence of COX [206]. These results show that, independently of its genetic origin, the essential role of Cox3 in the assembly or stability of the catalytic core of COX is conserved though evolution.
6. Concluding remarks and open questions
Despite continuous efforts to fully understand the biogenesis and assembly of mitochondrial COX catalytic core subunits in several laboratories and the recent advances made in our understanding of the mechanisms involved, sorting out its complexity and how COX biogenesis is regulated remains a remarkable challenge. Whereas the number of COX ancillary factors identified continues increasing; the specific functions of most of them are only partially characterized. Specific functions such as the protein-assisted assembly of Cox3, the dehydrogenase activity required for the last step of heme a biosynthesis, or the Cox1 heme insertion chaperone, among many others, wait to have a protein performer identified and/or assigned. Regulatory pathways coordinating COX core subunit synthesis, cofactor availability, maturation and assembly also remain to be fully characterized. A final challenge will involve the identification of COX assembly factors and regulatory pathways conserved from yeast to human and those that evolved to adapt to the tissue-specific requirements of multicellular organisms. This has a great biomedical relevance since lesions affecting the expression and assembly of COX catalytic core subunits result in severe human mitochondrial encephalomyopathies.
Highlights.
-
-
Cytochrome c oxidase (COX) is the terminal mitochondrial respiratory chain enzyme.
-
-
The catalytic core of COX is formed by three mitochondrial DNA encoded subunits.
-
-
A large number of nuclear encoded ancillary factors are necessary for COX biogenesis.
-
-
Core subunit synthesis and maturation are key regulatory points during COX assembly.
Acknowledgements
We thank Dr. Myriam Bourens for critically reading the manuscript. Our research is supported by NIH Grant GM071775A (to AB), MDA Research Grant (to AB), MDA Development Grant (to FF) and NIH F31 fellowship GM081975 (to IS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hill BC. Modeling the sequence of electron transfer reactions in the single turnover of reduced, mammalian cytochrome c oxidase with oxygen. J. Biol. Chem. 1994;269:2419–2425. [PubMed] [Google Scholar]
- [2].Brunori M, Giuffre A, Malatesta F, Sarti P. Investigating the mechanism of electron transfer to the binuclear center in Cu-heme oxidases. J. Bioenerg. Biomembr. 1998;30:41–45. doi: 10.1023/a:1020503410377. [DOI] [PubMed] [Google Scholar]
- [3].Brunori M, Giuffre A, Sarti P. Cytochrome c oxidase, ligands and electrons. J. Inorg. Biochem. 2005;99:324–336. doi: 10.1016/j.jinorgbio.2004.10.011. [DOI] [PubMed] [Google Scholar]
- [4].Belevich I, Verkhovsky MI, Wikstrom M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- [5].Yoshikawa S, Muramoto K, Shinzawa-Itoh K, Aoyama H, Tsukihara T, Shimokata K, Katayama Y, Shimada H. Proton pumping mechanism of bovine heart cytochrome c oxidase. Biochim. Biophys. Acta. 2006;1757:1110–1116. doi: 10.1016/j.bbabio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [6].Scheffler IE. In: Mitochondria. Wiley-Liss, editor. New York: 1999. [Google Scholar]
- [7].Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006;291:C1129–1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- [9].Barrientos A, Barros MH, Valnot I, Rotig A, Rustin P, Tzagoloff A. Cytochrome oxidase in health and disease. Gene. 2002;286:53–63. doi: 10.1016/s0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- [10].Herrmann JM, Funes S. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- [11].Shoubridge EA. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- [12].Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- [13].Fowler LR, Richardson SH, Hatefi Y. A rapid method for the preparation of highly purified cytochrome oxidase. Biochim. Biophys. Acta. 1962;64:170–173. doi: 10.1016/0006-3002(62)90770-9. [DOI] [PubMed] [Google Scholar]
- [14].Rubin MS, Tzagoloff A. Assembly of the mitochondrial membrane system. IX. Purification, characterization, and subunit structure of yeast and beef cytochrome oxidase. J. Biol. Chem. 1973;248:4269–4274. [PubMed] [Google Scholar]
- [15].Clark-Walker GD, Linnane AW. In vivo differentiation of yeast cytoplasmic and mitochondrial protein synthesis with antibiotics. Biochem. Biophys. Res. Commun. 1966;25:8–13. doi: 10.1016/0006-291x(66)90631-0. [DOI] [PubMed] [Google Scholar]
- [16].Rubin MS, Tzagoloff A. Assembly of the mitochondrial membrane system. X. Mitochondrial synthesis of three of the subunit proteins of yeast cytochrome oxidase. J. Biol. Chem. 1973;248:4275–4279. [PubMed] [Google Scholar]
- [17].Poyton RO, Schatz G. Cytochrome c oxidase from bakers’ yeast. III. Physical characterization of isolated subunits and chemical evidence for two different classes of polypeptides. J. Biol. Chem. 1975;250:752–761. [PubMed] [Google Scholar]
- [18].Bonitz SG, Coruzzi G, Thalenfeld BE, Tzagoloff A, Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J. Biol. Chem. 1980;255:11927–11941. [PubMed] [Google Scholar]
- [19].Coruzzi G, Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence of subunit 2 of yeast cytochrome oxidase. J. Biol. Chem. 1979;254:9324–9330. [PubMed] [Google Scholar]
- [20].Perez-Martinez X, Funes S, Tolkunova E, Davidson E, King MP, Gonzalez-Halphen D. Structure of nuclear-localized cox3 genes in Chlamydomonas reinhardtii and in its colorless close relative Polytomella sp. Curr. Genet. 2002;40:399–404. doi: 10.1007/s00294-002-0270-6. [DOI] [PubMed] [Google Scholar]
- [21].Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: Insights into the COX assembly process. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- [23].Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- [24].Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- [25].Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- [26].Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Coyne HJ, 3rd, Ciofi-Baffoni S, Banci L, Bertini I, Zhang L, George GN, Winge DR. The characterization and role of zinc binding in yeast Cox4. J. Biol. Chem. 2007;282:8926–8934. doi: 10.1074/jbc.M610303200. [DOI] [PubMed] [Google Scholar]
- [28].Schmidt B, McCracken J, Ferguson-Miller S. A discrete water exit pathway in the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15539–15542. doi: 10.1073/pnas.2633243100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wielburski A, Nelson BD. Evidence for the sequential assembly of cytochrome oxidase subunits in rat liver mitochondria. Biochem. J. 1983;212:829–834. doi: 10.1042/bj2120829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 1998;254:389–394. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- [31].Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 2001;498:46–51. doi: 10.1016/s0014-5793(01)02447-4. [DOI] [PubMed] [Google Scholar]
- [32].Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Horan S, Bourges I, Taanman JW, Meunier B. Analysis of COX2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem J. 2005;390:703–708. doi: 10.1042/BJ20050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Church C, Goehring B, Forsha D, Wazny P, Poyton RO. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J. Biol. Chem. 2005;280:1854–1863. doi: 10.1074/jbc.M410726200. [DOI] [PubMed] [Google Scholar]
- [35].Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- [37].Ebner E, Mason TL, Schatz G. Mitochondrial assembly in respiration-deficient mutants of Saccharomyces cerevisiae. II. Effect of nuclear and extrachromosomal mutations on the formation of cytochrome c oxidase. J. Biol. Chem. 1973;248:5369–5378. [PubMed] [Google Scholar]
- [38].Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system: isolation of nuclear and cytoplasmic mutants of Saccharomyces cerevisiae with specific defects in mitochondrial functions. J. Bacteriol. 1975;122:826–831. doi: 10.1128/jb.122.3.826-831.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].McEwen JE, Ko C, Kloeckner-Gruissem B, Poyton RO. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J. Biol. Chem. 1986;261:11872–11879. [PubMed] [Google Scholar]
- [40].Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Khalimonchuk O, Rodel G. Biogenesis of cytochrome c oxidase. Mitochondrion. 2005;5:363–388. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- [42].Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60:557–568. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Solans A, Zambrano A, Barrientos A. Cytochrome c oxidase deficiency: from yeast to human. Preclinica. 2004;2:336–348. [Google Scholar]
- [44].Pecina P, Houstkova H, Hansikova H, Zeman J, Houstek J. Genetic defects of cytochrome c oxidase assembly. Physiol. Res. 2004;53:S213–223. [PubMed] [Google Scholar]
- [45].Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- [46].Massa V, Fernandez-Vizarra E, Alshahwan S, Bakhsh E, Goffrini P, Ferrero I, Mereghetti P, D’Adamo P, Gasparini P, Zeviani M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shteyer E, Saada A, Shaag A, Al-Hijawi FA, Kidess R, Revel-Vilk S, Elpeleg O. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am. J. Hum. Genet. 2009;84:412–417. doi: 10.1016/j.ajhg.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- [49].Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Valnot I, Osmond S, Gigarel N, Mehaye B, Amiel J, Cormier-Daire V, Munnich A, Bonnefont JP, Rustin P, Rotig A. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000;67:1104–1109. doi: 10.1016/s0002-9297(07)62940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Papadopoulou LC, Sue CM, Davidson MM, Tanji K, Nishino I, Sadlock JE, Krishna S, Walker W, Selby J, Glerum DM, Coster RV, Lyon G, Scalais E, Lebel R, Kaplan P, Shanske S, De Vivo DC, Bonilla E, Hirano M, DiMauro S, Schon EA. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- [52].Jaksch M, Ogilvie I, Yao J, Kortenhaus G, Bresser HG, Gerbitz KD, Shoubridge EA. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:795–801. doi: 10.1093/hmg/9.5.795. [DOI] [PubMed] [Google Scholar]
- [53].Sue CM, Karadimas C, Checcarelli N, Tanji K, Papadopoulou LC, Pallotti F, Guo FL, Shanske S, Hirano M, De Vivo DC, Van Coster R, Kaplan P, Bonilla E, DiMauro S. Differential features of patients with mutations in two COX assembly genes, SURF-1 and SCO2. Ann. Neurol. 2000;47:589–595. [PubMed] [Google Scholar]
- [54].Salviati L, Sacconi S, Rasalan MM, Kronn DF, Braun A, Canoll P, Davidson M, Shanske S, Bonilla E, Hays AP, Schon EA, DiMauro S. Cytochrome c oxidase deficiency due to a novel SCO2 mutation mimics Werdnig-Hoffmann disease. Arch. Neurol. 2002;59:862–865. doi: 10.1001/archneur.59.5.862. [DOI] [PubMed] [Google Scholar]
- [55].Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rotig A. A mutation in the human heme A:farnesyltransferase gene (COX10 ) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:1245–1249. doi: 10.1093/hmg/9.8.1245. [DOI] [PubMed] [Google Scholar]
- [56].Antonicka H, Leary SC, Guercin GH, Agar JN, Horvath R, Kennaway NG, Harding CO, Jaksch M, Shoubridge EA. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003;12:2693–2702. doi: 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- [57].Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2003;72:101–114. doi: 10.1086/345489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U. S. A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmuller H, Chevrette M, Kaufman BA, Horvath R, Shoubridge EA. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–837. doi: 10.1038/ng.390. [DOI] [PubMed] [Google Scholar]
- [60].Williams SL, Valnot I, Rustin P, Taanman JW. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- [61].Stiburek L, Vesela K, Hansikova H, Pecina P, Tesarova M, Cerna L, Houstek J, Zeman J. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 2005;392:625–632. doi: 10.1042/BJ20050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome--from genetics to evolution and systems biology. Biochim. Biophys. Acta. 1797:1086–1098. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- [63].Dieckmann CL, Staples RR. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int. Rev. Cytol. 1994;152:145–181. doi: 10.1016/s0074-7696(08)62556-5. [DOI] [PubMed] [Google Scholar]
- [64].Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- [65].Lazowska J, Jacq C, Slonimski PP. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980;22:333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- [66].Levra-Juillet E, Boulet A, Seraphin B, Simon M, Faye G. Mitochondrial introns aI1 and/or aI2 are needed for the in vivo deletion of intervening sequences. Mol. Gen. Genet. 1989;217:168–171. doi: 10.1007/BF00330957. [DOI] [PubMed] [Google Scholar]
- [67].Seraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- [68].Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U. S. A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, Rhoades E, Hackney DD, Pyle AM, De La Cruz EM. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J. Mol. Biol. 2011;409:399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Turk EM, Caprara MG. Splicing of yeast aI5beta group I intron requires SUV3 to recycle MRS1 via mitochondrial degradosome-promoted decay of excised intron ribonucleoprotein (RNP) J. Biol. Chem. 2010;285:8585–8594. doi: 10.1074/jbc.M109.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Moreno JI, Buie KS, Price RE, Piva MA. Ccm1p/Ygr150cp, a pentatricopeptide repeat protein, is essential to remove the fourth intron of both COB and COX1 pre-mRNAs in Saccharomyces cerevisiae. Curr. Genet. 2009;55:475–484. doi: 10.1007/s00294-009-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Labouesse M, Dujardin G, Slonimski PP. The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell. 1985;41:133–143. doi: 10.1016/0092-8674(85)90068-6. [DOI] [PubMed] [Google Scholar]
- [73].Seraphin B, Simon M, Faye G. MSS18, a yeast nuclear gene involved in the splicing of intron aI5 beta of the mitochondrial cox1 transcript. EMBO J. 1988;7:1455–1464. doi: 10.1002/j.1460-2075.1988.tb02963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kaspar BJ, Bifano AL, Caprara MG. A shared RNA-binding site in the Pet54 protein is required for translational activation and group I intron splicing in yeast mitochondria. Nucleic Acids Res. 2008;36:2958–2968. doi: 10.1093/nar/gkn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kreike J, Schulze M, Pillar T, Korte A, Rodel G. Cloning of a nuclear gene MRS1 involved in the excision of a single group I intron (bI3) from the mitochondrial COB transcript in S. cerevisiae. Curr. Genet. 1986;11:185–191. doi: 10.1007/BF00420605. [DOI] [PubMed] [Google Scholar]
- [76].Watts T, Khalimonchuk O, Wolf RZ, Turk EM, Mohr G, Winge DR. Mne1 is a novel component of the mitochondrial splicing apparatus responsible for processing of a COX1 group I intron in yeast. J. Biol. Chem. 2011;286:10137–10146. doi: 10.1074/jbc.M110.205625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Barros MH, Myers AM, Van Driesche S, Tzagoloff A. COX24 codes for a mitochondrial protein required for processing of the COX1 transcript. J. Biol. Chem. 2005 doi: 10.1074/jbc.M510778200. [DOI] [PubMed] [Google Scholar]
- [78].Shingu-Vazquez M, Camacho-Villasana Y, Sandoval-Romero L, Butler CA, Fox TD, Perez-Martinez X. The carboxyl-terminal end of Cox1 is required for feedback-assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 2010;285:34382–34389. doi: 10.1074/jbc.M110.161976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990;224:209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- [80].Minczuk M, Dmochowska A, Palczewska M, Stepien PP. Overexpressed yeast mitochondrial putative RNA helicase Mss116 partially restores proper mtRNA metabolism in strains lacking the Suv3 mtRNA helicase. Yeast. 2002;19:1285–1293. doi: 10.1002/yea.906. [DOI] [PubMed] [Google Scholar]
- [81].Bousquet I, Dujardin G, Poyton RO, Slonimski PP. Two group I mitochondrial introns in the cob-box and coxI genes require the same MRS1/PET157 nuclear gene product for splicing. Curr. Genet. 1990;18:117–124. doi: 10.1007/BF00312599. [DOI] [PubMed] [Google Scholar]
- [82].Towpik J. Regulation of mitochondrial translation in yeast. Cell Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- [83].Decoster E, Simon M, Hatat D, Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet. 1990;224:111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- [84].Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mulero JJ, Fox TD. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Costanzo MC, Seaver EC, Fox TD. At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 1986;5:3637–3641. doi: 10.1002/j.1460-2075.1986.tb04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Costanzo MC, Fox TD. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Brown NG, Costanzo MC, Fox TD. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Costanzo MC, Fox TD. A point mutation in the 5′-untranslated leader that affects translational activation of the mitochondrial COX3 mRNA. Curr. Genet. 1995;28:60–66. doi: 10.1007/BF00311882. [DOI] [PubMed] [Google Scholar]
- [91].Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767–775. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- [93].Fiori A, Perez-Martinez X, Fox TD. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol. Microbiol. 2005;56:1689–1704. doi: 10.1111/j.1365-2958.2005.04658.x. [DOI] [PubMed] [Google Scholar]
- [94].Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- [96].Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- [97].Fox TD, Hershey JWB, M BM, Sonenberg N. Translational Control. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1996. pp. 733–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Manthey GM, Przybyla-Zawislak BD, McEwen JE. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- [100].Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- [101].Kuhl I, Dujeancourt L, Gaisne M, Herbert CJ, Bonnefoy N. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 2011 Jul 3; doi: 10.1093/nar/gkr511. e-published ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tavares-Carreon F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingu-Vazquez M, Torres-Larios A, Perez-Martinez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 2008;283:1472–1479. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- [104].Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:523–535. doi: 10.1091/mbc.E06-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Langer T, Kaser M, Klanner C, Leonhard K. AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem. Soc. Trans. 2001;29:431–436. doi: 10.1042/bst0290431. [DOI] [PubMed] [Google Scholar]
- [108].Bestwick M, Khalimonchuk O, Pierrel F, Winge DR. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell Biol. 2010;30:172–185. doi: 10.1128/MCB.00869-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wollman FA, Minai L, Nechushtai R. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim Biophys Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- [113].Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA. Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans. 2001;29:421–426. doi: 10.1042/bst0290421. [DOI] [PubMed] [Google Scholar]
- [114].Wostrikoff K, Stern D. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6466–6471. doi: 10.1073/pnas.0610586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hell K, Neupert W, Stuart RA. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Fontanesi F, Jin C, Tzagoloff A, Barrientos A. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 2008;17:775–788. doi: 10.1093/hmg/ddm349. [DOI] [PubMed] [Google Scholar]
- [118].Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Caughey WS, Smythe GA, O’Keeffe DH, Maskasky JE, Smith MI. Heme A of cytochrome c oxidase. Structure and properties: comparisons with hemes B, C, and S and derivatives. J. Biol. Chem. 1975;250:7602–7622. [PubMed] [Google Scholar]
- [122].Carr HS, Winge DR. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- [123].Barros MH, Carlson CG, Glerum DM, Tzagoloff A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001;492:133–138. doi: 10.1016/s0014-5793(01)02249-9. [DOI] [PubMed] [Google Scholar]
- [124].Brown KR, Allan BM, Do P, Hegg EL. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry. 2002;41:10906–10913. doi: 10.1021/bi0203536. [DOI] [PubMed] [Google Scholar]
- [125].Glerum DM, Tzagoloff A. Isolation of a human cDNA for heme A:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8452–8456. doi: 10.1073/pnas.91.18.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- [127].Bundschuh FA, Hannappel A, Anderka O, Ludwig B. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 2009;284:25735–25741. doi: 10.1074/jbc.M109.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bestwick M, Jeong MY, Khalimonchuk O, Kim H, Winge DR. Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol. Cell Biol. 2010;30:4480–4491. doi: 10.1128/MCB.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 1999;8:2533–2540. doi: 10.1093/hmg/8.13.2533. [DOI] [PubMed] [Google Scholar]
- [130].Barros MH, Tzagoloff A. Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 2002;516:119–123. doi: 10.1016/s0014-5793(02)02514-0. [DOI] [PubMed] [Google Scholar]
- [131].Pierrel F, Khalimonchuk O, Cobine PA, Bestwick M, Winge DR. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol. Cell Biol. 2008;28:4927–4939. doi: 10.1128/MCB.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Bureik M, Schiffler B, Hiraoka Y, Vogel F, Bernhardt R. Functional expression of human mitochondrial CYP11B2 in fission yeast and identification of a new internal electron transfer protein, etp1. Biochemistry. 2002;41:2311–2321. doi: 10.1021/bi0157870. [DOI] [PubMed] [Google Scholar]
- [133].Pena MM, Lee J, Thiele DJ. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 1999;129:1251–1260. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- [134].Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- [135].Horn D, Barrientos A. Mitochondrial copper metabolism and delivery to cytochrome c oxidase. IUBMB Life. 2008;60:421–429. doi: 10.1002/iub.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Cobine PA, Ojeda LD, Rigby KM, Winge DR. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 2004;279:14447–14455. doi: 10.1074/jbc.M312693200. [DOI] [PubMed] [Google Scholar]
- [137].Cobine PA, Pierrel F, Bestwick ML, Winge DR. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 2006;281:36552–36559. doi: 10.1074/jbc.M606839200. [DOI] [PubMed] [Google Scholar]
- [138].Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- [139].Beers J, Glerum DM, Tzagoloff A. Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J. Biol. Chem. 1997;272:33191–33196. doi: 10.1074/jbc.272.52.33191. [DOI] [PubMed] [Google Scholar]
- [140].Heaton D, Nittis T, Srinivasan C, Winge DR. Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 2000;275:37582–37587. doi: 10.1074/jbc.M006639200. [DOI] [PubMed] [Google Scholar]
- [141].Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- [142].Glerum DM, Shtanko A, Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:20531–20535. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- [143].Hiser L, Di Valentin M, Hamer AG, Hosler JP. Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 2000;275:619–623. doi: 10.1074/jbc.275.1.619. [DOI] [PubMed] [Google Scholar]
- [144].Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 2002;277:31237–31242. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- [145].Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. Systematic analysis of the twin cx(9)c protein family. J. Mol. Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- [146].Rigby K, Zhang L, Cobine PA, George GN, Winge DR. Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:10233–10242. doi: 10.1074/jbc.M610082200. [DOI] [PubMed] [Google Scholar]
- [147].Barros MH, Johnson A, Tzagoloff A. COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 2004;279:31943–31947. doi: 10.1074/jbc.M405014200. [DOI] [PubMed] [Google Scholar]
- [148].McEwen JE, Hong KH, Park S, Preciado GT. Sequence and chromosomal localization of two PET genes required for cytochrome c oxidase assembly in Saccharomyces cerevisiae. Curr. Genet. 1993;23:9–14. doi: 10.1007/BF00336742. [DOI] [PubMed] [Google Scholar]
- [149].Horn D, Al-Ali H, Barrientos A. Cmc1p is a conserved mitochondrial twin Cx9C protein involved in cytochrome c oxidase biogenesis. Mol. Cell. Biol. 2008;28:4354–4364. doi: 10.1128/MCB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Horn D, Zhou W, Trevisson E, Al-Ali H, Harris TK, Salviati L, Barrientos A. The conserved mitochondrial twin CX9C protein Cmc2 is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J. Biol. Chem. 2010;285:15088–15099. doi: 10.1074/jbc.M110.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Khalimonchuk O, Winge DR. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim Biophys Acta. 2007;1783:618–628. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Khalimonchuk O, Ostermann K, Rodel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr. Genet. 2005;47:223–233. doi: 10.1007/s00294-005-0569-1. [DOI] [PubMed] [Google Scholar]
- [153].Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J. Biol. Chem. 2005;280:22664–22669. doi: 10.1074/jbc.M414077200. [DOI] [PubMed] [Google Scholar]
- [154].Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- [155].Horn D, Fontanesi F, Barrientos A, W SN. Yeast as a Model for Human Disease. Transworld Research Network; Trivandrum. India: 2009. pp. 41–61. Chapter 3. [Google Scholar]
- [156].Huigsloot M, Nijtmans LG, Szklarczyk R, Baars MJ, van den Brand MA, Hendriksfranssen MG, van den Heuvel LP, Smeitink JA, Huynen MA, Rodenburg RJ. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 2011;88:488–493. doi: 10.1016/j.ajhg.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bonnefoy N, Bsat N, Fox TD. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell Biol. 2001;21:2359–2372. doi: 10.1128/MCB.21.7.2359-2372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Williams EH, Fox TD. Antagonistic signals within the COX2 mRNA coding sequence control its translation in Saccharomyces cerevisiae mitochondria. RNA. 2003;9:419–431. doi: 10.1261/rna.2182903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol. Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- [162].Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Bonnefoy N, Kermorgant M, Groudinsky O, Minet M, Slonimski PP, Dujardin G. Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1-mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11978–11982. doi: 10.1073/pnas.91.25.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Souza RL, Green-Willms NS, Fox TD, Tzagoloff A, Nobrega FG. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- [168].Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell. 2002;13:1122–1131. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Broadley SA, Demlow CM, Fox TD. Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol. Cell Biol. 2001;21:7663–7672. doi: 10.1128/MCB.21.22.7663-7672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Fiumera HL, Broadley SA, Fox TD. Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol. Cell Biol. 2007;27:4664–4673. doi: 10.1128/MCB.01955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Fiumera HL, Dunham MJ, Saracco SA, Butler CA, Kelly JA, Fox TD. Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18. Genetics. 2009;182:519–528. doi: 10.1534/genetics.109.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Hell K, Tzagoloff A, Neupert W, Stuart RA. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 2000;275:4571–4578. doi: 10.1074/jbc.275.7.4571. [DOI] [PubMed] [Google Scholar]
- [173].Jan PS, Esser K, Pratje E, Michaelis G. Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol. Gen. Genet. 2000;263:483–491. doi: 10.1007/s004380051192. [DOI] [PubMed] [Google Scholar]
- [174].Graef M, Seewald G, Langer T. Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell Biol. 2007;27:2476–2485. doi: 10.1128/MCB.01721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Carlson CG, Barrientos A, Tzagoloff A, Glerum DM. COX16 encodes a novel protein required for the assembly of cytochrome oxidase in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:3770–3775. doi: 10.1074/jbc.M209893200. [DOI] [PubMed] [Google Scholar]
- [176].Nunnari J, Fox TD, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- [177].Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- [178].Petek E, Windpassinger C, Vincent JB, Cheung J, Boright AP, Scherer SW, Kroisel PM, Wagner K. Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am. J. Hum. Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Kroneck PM, Antholine WA, Riester J, Zumft WG. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II),Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 1988;242:70–74. doi: 10.1016/0014-5793(88)80987-6. [DOI] [PubMed] [Google Scholar]
- [180].Malmstrom BG, Aasa R. The nature of the CuA center in cytochrome c oxidase. FEBS Lett. 1993;325:49–52. doi: 10.1016/0014-5793(93)81411-r. [DOI] [PubMed] [Google Scholar]
- [181].Lappalainen P, Aasa R, Malmstrom BG, Saraste M. Soluble CuA-binding domain from the Paracoccus cytochrome c oxidase. J. Biol. Chem. 1993;268:26416–26421. [PubMed] [Google Scholar]
- [182].Nittis T, George GN, Winge DR. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 2001;276:42520–42526. doi: 10.1074/jbc.M107077200. [DOI] [PubMed] [Google Scholar]
- [183].Rentzsch A, Krummeck-Weiss G, Hofer A, Bartuschka A, Ostermann K, Rodel G. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr. Genet. 1999;35:103–108. doi: 10.1007/s002940050438. [DOI] [PubMed] [Google Scholar]
- [184].Lode A, Kuschel M, Paret C, Rodel G. Mitochondrial copper metabolism in yeast: interaction between Sco1p and Cox2p. FEBS Lett. 2000;485:19–24. doi: 10.1016/s0014-5793(00)02176-1. [DOI] [PubMed] [Google Scholar]
- [185].Horng YC, Leary SC, Cobine PA, Young FB, George GN, Shoubridge EA, Winge DR. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 2005;280:34113–34122. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- [186].Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Martinelli M, Palumaa P, Wang S. A hint for the function of human Sco1 from different structures. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8595–8600. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Ye Q, Imriskova-Sosova I, Hill BC, Jia Z. Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins. Biochemistry. 2005;44:2934–2942. doi: 10.1021/bi0480537. [DOI] [PubMed] [Google Scholar]
- [188].Abajian C, Rosenzweig AC. Crystal structure of yeast Sco1. J. Biol. Inorg. Chem. 2006;11:459–446. doi: 10.1007/s00775-006-0096-7. [DOI] [PubMed] [Google Scholar]
- [189].Chinenov YV. Cytochrome c oxidase assembly factors with a thioredoxin fold are conserved among prokaryotes and eukaryotes. J. Mol. Med. 2000;78:239–242. doi: 10.1007/s001090000110. [DOI] [PubMed] [Google Scholar]
- [190].Smits PH, De Haan M, Maat C, Grivell LA. The complete sequence of a 33 kb fragment on the right arm of chromosome II from Saccharomyces cerevisiae reveals 16 open reading frames, including ten new open reading frames, five previously identified genes and a homologue of the SCO1 gene. Yeast. 1994;10:S75–80. doi: 10.1002/yea.320100010. [DOI] [PubMed] [Google Scholar]
- [191].Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M. Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics. 1998;54:494–504. doi: 10.1006/geno.1998.5580. [DOI] [PubMed] [Google Scholar]
- [192].Leary SC, Kaufman BA, Pellecchia G, Guercin GH, Mattman A, Jaksch M, Shoubridge EA. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004;13:1839–1848. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- [193].Leary SC, Sasarman F, Nishimura T, Shoubridge EA. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- [194].Banci L, Bertini I, Ciofi-Baffoni S, Gerothanassis IP, Leontari I, Martinelli M, Wang S. A structural characterization of human SCO2. Structure. 2007;15:1132–1140. doi: 10.1016/j.str.2007.07.011. [DOI] [PubMed] [Google Scholar]
- [195].Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6803–6808. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- [197].Brunori M, Antonini G, Malatesta F, Sarti P, Wilson MT. Cytochrome-c oxidase. Subunit structure and proton pumping. Eur. J. Biochem. 1987;169:1–8. doi: 10.1111/j.1432-1033.1987.tb13572.x. [DOI] [PubMed] [Google Scholar]
- [198].Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: proton transfer through the respiratory complexes. Annu. Rev. Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Hosler JP. The influence of subunit III of cytochrome c oxidase on the D pathway, the proton exit pathway and mechanism-based inactivation in subunit I. Biochim. Biophys. Acta. 2004;1655:332–339. doi: 10.1016/j.bbabio.2003.06.009. [DOI] [PubMed] [Google Scholar]
- [200].Kloeckener-Gruissem B, McEwen JE, Poyton RO. Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene is Saccharomyces cerevisiae. J. Bacteriol. 1988;170:1399–1402. doi: 10.1128/jb.170.3.1399-1402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Valencik ML, Kloeckener-Gruissem B, Poyton RO, McEwen JE. Disruption of the yeast nuclear PET54 gene blocks excision of mitochondrial intron aI5 beta from pre-mRNA for cytochrome c oxidase subunit I. EMBO J. 1989;8:3899–3904. doi: 10.1002/j.1460-2075.1989.tb08569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Costanzo MC, Seaver EC, Fox TD. The PET54 gene of Saccharomyces cerevisiae: characterization of a nuclear gene encoding a mitochondrial translational activator and subcellular localization of its product. Genetics. 1989;122:297–305. doi: 10.1093/genetics/122.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].McMullin TW, Fox TD. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:11737–11741. [PubMed] [Google Scholar]
- [204].Keightley JA, Hoffbuhr KC, Burton MD, Salas VM, Johnston WS, Penn AM, Buist NR, Kennaway NG. A microdeletion in cytochrome c oxidase (COX) subunit III associated with COX deficiency and recurrent myoglobinuria. Nat. Genet. 1996;12:410–416. doi: 10.1038/ng0496-410. [DOI] [PubMed] [Google Scholar]
- [205].Meunier B. Site-directed mutations in the mitochondrially encoded subunits I and III of yeast cytochrome oxidase. Biochem. J. 2001;354:407–412. doi: 10.1042/0264-6021:3540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Remacle C, Coosemans N, Jans F, Hanikenne M, Motte P, Cardol P. Knock-down of the COX3 and COX17 gene expression of cytochrome c oxidase in the unicellular green alga Chlamydomonas reinhardtii. Plant Biol. 2010;74:223–233. doi: 10.1007/s11103-010-9668-6. [DOI] [PubMed] [Google Scholar]