Mitochondrial metabolism provides precursors to build macromolecules in growing cancer cells1,2. In normally-functioning tumor cell mitochondria, oxidative metabolism of glucose- and glutamine-derived carbon produces citrate and acetyl-CoA for lipid synthesis, which is required for tumorigenesis3. Yet some tumors harbor mutations in the citric acid cycle (CAC) or electron transport chain (ETC) that disable normal oxidative mitochondrial function4–7, and it is unknown how cells from such tumors generate precursors for macromolecular synthesis. Here we show that tumor cells with defective mitochondria use glutamine-dependent reductive carboxylation rather than oxidative metabolism as the major pathway of citrate formation. This pathway uses mitochondrial and cytosolic isoforms of NADP+/NADPH-dependent isocitrate dehydrogenase, and subsequent metabolism of glutamine-derived citrate provides both the acetyl-CoA for lipid synthesis and the 4-carbon intermediates needed to produce remaining CAC metabolites and related macromolecular precursors. This reductive, glutamine-dependent pathway is the dominant mode of metabolism in rapidly-growing malignant cells containing mutations in complex I or complex III of the ETC, in patient-derived renal carcinoma cells with mutations in fumarate hydratase (FH), and in cells with normal mitochondria subjected to acute pharmacological ETC inhibition. Our findings reveal the novel induction of a versatile glutamine-dependent pathway that reverses many of the reactions of the canonical CAC, supports tumor cell growth, and explains how cells generate pools of CAC intermediates in the face of impaired mitochondrial metabolism.
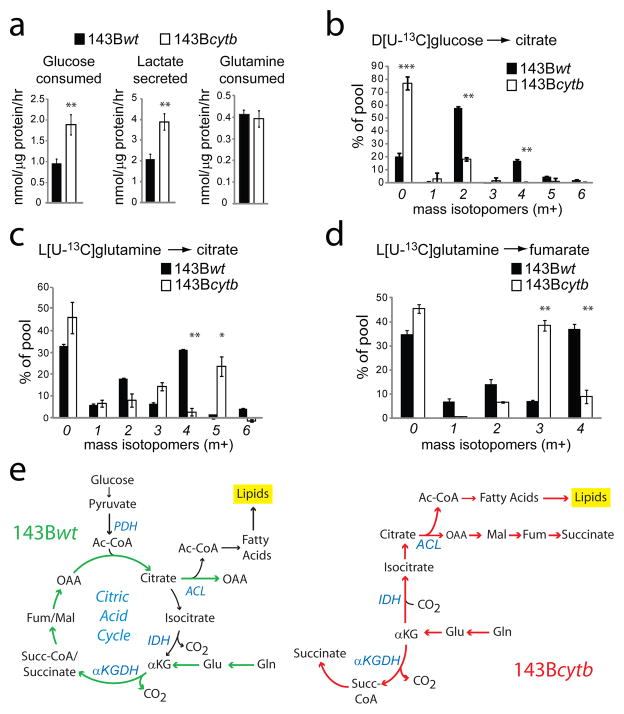
We first studied the metabolism of isogenic 143B human osteosarcoma cells that contained or lacked a loss-of-function mutation in ETC complex III (cytochrome b-c1 complex). These cell lines were generated by depleting 143B mitochondrial DNA (mtDNA) and repopulating with either wild-type mtDNA or mtDNA containing a frameshift mutation in the gene encoding cytochrome b (cytb), an essential complex III component8. Despite lack of respiration and complex III function in the mutants8, both wild-type (143Bwt) and cytb-mutant (143Bcytb) cells form colonies in soft agar9 and proliferate at comparable rates (Supplementary Fig. 1a), making these cells a good model to study growth during mitochondrial dysfunction. Both cell lines had detectable CAC intermediates, although citrate was less abundant and succinate was significantly more abundant in the 143Bcytb cells (Supplementary Fig. 1b). As expected for cells with defective oxidative phosphorylation, 143Bcytb cells had higher glucose consumption and lactate production than 143Bwt cells, indicating a metabolic shift towards aerobic glycolysis (Fig. 1a). To determine the effects of cytb mutation on the metabolic fates of glucose, we cultured both cell lines in medium containing D[U-13C]glucose and measured 13C enrichment of intracellular metabolites by mass spectrometry. In 143Bwt cells, most citrate molecules contained glucose-derived 13C (Fig. 1b). Citrate m+2 results from oxidative decarboxylation of glucose-derived pyruvate by pyruvate dehydrogenase (PDH) to form [1,2-13C]acetyl-coA, followed by condensation with an unlabeled oxaloacetate (OAA). Processing of citrate m+2 around one turn of the CAC produces citrate m+4 (Supplementary Fig. 2). In 143Bcytb cells, most citrate contained no glucose carbon (m+0), indicating suppressed PDH contribution to acetyl-CoA (Fig. 1b). Fumarate and malate m+2 were also decreased (Supplementary Fig. 1c,d).
Figure 1. A reductive pathway of glutamine metabolism in cancer cells lacking activity of ETC complex III.
a, Glucose utilization, lactate secretion and glutamine utilization in 143Bwt and 143Bcytb cells. b, Mass isotopomer analysis of citrate in cells cultured with D[U-13C]glucose and unlabeled glutamine. c,d, Mass isotopomer analysis of citrate and fumarate in cells cultured with L[U-13C]glutamine and unlabeled glucose. Data are the average ± S.D. for three independent cultures. * p<0.05; ** p<0.005, Student’s t-test. e, Schematic of glutamine metabolism in 143Bwt and 143Bcytb cells. Colored arrows follow the paths of glutamine-derived carbon. Abbreviations: Ac-CoA, acetyl-CoA; OAA, oxaloacetate; Gln, glutamine; Glu, glutamate; αKG, α-ketoglutarate; Succ-CoA, succinyl-CoA; Fum, fumarate; Mal, malate; PDH, pyruvate dehydrogenase; ACL, ATP-citrate lyase; IDH, isocitrate dehydrogenase; αKGDH, α-ketoglutarate dehydrogenase.
Glutamine is a major respiratory substrate in cancer cells, providing energy and anaplerotic carbon for growth3,10. Both 143Bwt and 143Bcytb cells require glutamine for colony formation, implying a respiration-independent function for glutamine in cell growth9. 143Bcytb and 143Bwt cells consumed glutamine at similar rates (Fig. 1a). We cultured both cell lines with L[U-13C]glutamine to define the metabolic fates of glutamine. Similar to other glutamine-dependent cancer cells11, 143Bwt cells used glutamine as the major anaplerotic precursor, resulting in a large amount of fumarate, malate and citrate m+4 (Fig. 1c,d; Supplementary Figs. 1e and 3, top). In contrast, 143Bcytb cells produced only trace quantities of citrate m+4. Instead, they produced citrate m+5 through reductive carboxylation of glutamine-derived α-ketoglutarate (Fig. 1c). This reaction involves addition of an unlabeled carbon by isocitrate dehydrogenase (IDH) acting in reverse relative to the canonical oxidative CAC12. Cleavage of citrate m+5 by ATP-citrate lyase then produces acetyl-CoA m+2 and OAA m+3, with the unlabeled carbon retained on OAA (Supplementary Fig. 3, bottom). Examination of other CAC metabolites in 143Bcytb cells revealed that they were formed downstream of citrate m+5 and OAA m+3. These cells contained abundant malate and fumarate m+3, which was essentially absent from 143Bwt cells (Fig. 1d and Supplementary Fig. 1e). Thus these metabolites are generated from reductive, rather than oxidative glutamine metabolism (Fig. 1e). Succinate was formed through both reductive and oxidative glutamine metabolism in 143Bcytb cells (Supplementary Fig. 1f). Glutamine-dependent reductive carboxylation was also observed in 143Bcytb cells cultured in medium containing a full complement of amino acids (Supplementary Fig. 4), and in cells with a genetic defect in ETC complex I (Supplementary Fig. 5).
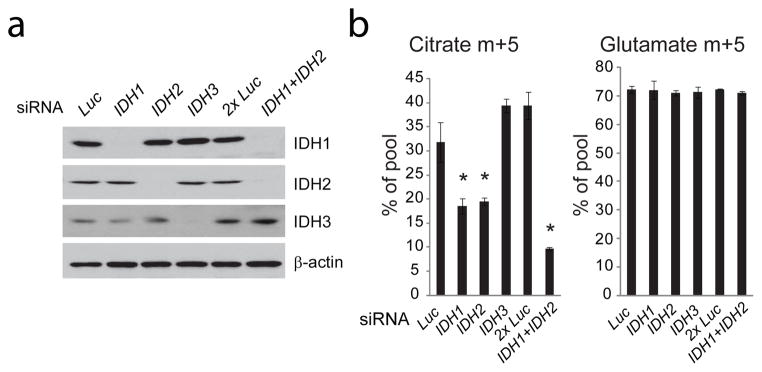
To determine which of the three IDH isoforms contributed to reductive carboxylation, we used RNA interference (RNAi) to silence expression of IDH1, IDH2 and IDH3 in 143Bcytb cells and measured glutamine-derived 13C labeling in citrate. Silencing either of the NADP+/NADPH-dependent IDH isoforms, IDH1 or IDH2, reduced citrate m+5 in cells cultured with L[U-13C]glutamine and silencing both at once further reduced citrate m+5 (Fig. 2a,b). Conversion of glutamine to glutamate was unaffected (Fig. 2b). Silencing IDH3, the mitochondrial NAD+/NADH-dependent isoform, had no effect on labeling (Fig. 2a,b). Thus reductive carboxylation in 143Bcytb cells involves IDH1 and IDH2, but not IDH3. Because IDH1 and IDH2 proteins are localized primarily in the cytoplasm and mitochondria, respectively, of both cell lines (Supplementary Fig. 6), the data suggest that reductive carboxylation in 143Bcytb cells may occur in either compartment.
Figure 2. NADP+/NADPH-dependent isoforms of isocitrate dehydrogenase contribute to reductive carboxylation.
a, Transient silencing of isocitrate dehydrogenase (IDH) proteins in 143Bcytb cells with short interfering RNAs (siRNAs) directed against IDH1, IDH2 or IDH3. An siRNA directed against luciferase (Luc) was used as a negative control. When IDH1 and IDH2 siRNAs were transfected concurrently (IDH1+IDH2), a transfection with the same nanomolar amount of Luc siRNA (2xLuc) was used as a negative control. b, Mass isotopomer analysis of citrate and glutamate in 143Bcytb cells cultured with L[U-13C]glutamine after silencing of IDH isoforms. Data are the average ± S.D. of three independent cultures. *p<0.05, Student’s t-test.
To determine whether IDH1 and IDH2 are required for growth, we stably silenced each enzyme using lentiviral RNAi. In 143Bwt cells, silencing either isoform reduced cell growth (Supplementary Fig. 7), consistent with findings in other cancer cells with normal mitochondria13. In 143Bcytb cells, a similar growth reduction occurred when either isoform was silenced (Supplementary Fig. 7). Thus both IDH1 and IDH2 support growth in cells that use glutamine-dependent reductive carboxylation.
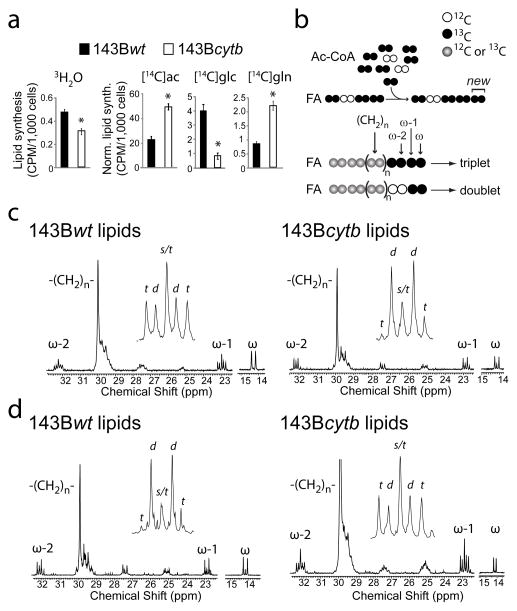
We next determined whether reductive glutamine metabolism contributed to lipogenesis, a biosynthetic activity required for cancer cell growth. Both 143Bwt and 143Bcytb cells produced fatty acids de novo, as indicated by incorporation of 3H2O into lipids (Fig. 3a). The two cell lines were then cultured with 14C-labeled lipogenic substrates to examine carbon preferences for lipogenesis (Fig. 3a). Both cell lines had an intact pathway of lipogenesis from exogenously-supplied acetate. The enhanced labeling in 143Bcytb cells likely resulted from an increased specific activity of the intracellular 2-carbon pool because of suppressed PDH. Lipids derived from 143Bcytb cells contained little radioactivity from glucose, but abundant radioactivity from glutamine, indicating a switch in the extent to which these nutrients supplied lipogenesis. To quantify the contribution of glucose and glutamine to lipogenic acetyl-CoA, cells were cultured in medium containing either D[U-13C]glucose and unlabeled glutamine, or L[U-13C]glutamine and unlabeled glucose. Extracted lipids were analyzed by 13C NMR. Because fatty acids are synthesized by the sequential addition of 2-carbon units, analyzing 13C-13C coupling between the ω-1 and ω-2 carbons – which enter the fatty acyl chain on different 2-carbon units – enables calculation of 13C enrichment in lipogenic acetyl-CoA3. In the ω-1 multiplet, 13C-13C coupling produces a triplet when two 13C-labeled 2-carbon units are added in succession, whereas one labeled and one unlabeled unit produce a doublet (Fig. 3b). Therefore 13C labeling in lipogenic acetyl-CoA is proportional to the contribution of the triplet to the overall area of the ω-1 multiplet. In 143Bwt cells cultured with D[U-13C]glucose, the ω-1 resonance was dominated by the triplet, which accounted for 65% of the total area (Fig. 3c, left). Glutamine was a minor source of fatty acyl carbon, as the triplet accounted for only 15% of the area when the cells were cultured with L[U-13C]glutamine (Fig. 3d, left). This pattern was reversed in the 143Bcytb cells, with glutamine providing 67% of the lipogenic carbon (Fig. 3c,d, right; Supplementary Fig. 8a). Thus, reductive glutamine metabolism was the major pathway of fatty acid production in newly-synthesized lipids during 143Bcytb growth.
Figure 3. Glutamine is the major lipogenic precursor in cells lacking oxidative CAC function.
a, Cells were cultured in medium containing 3H2O (21% volume), or 14C tracers of acetate (ac, 5 μCi), glucose (glc, 10 μCi) or glutamine (gln, 10 μCi). Lipids were analyzed for 3H or 14C content by scintillation counting. In the 14C experiments, raw counts per thousand cells were normalized to the absolute rate of fatty acid synthesis established using 3H2O. *p<0.05, Student’s t-test. b, Schematic outlining synthesis of fatty acids (FA) from acetyl-CoA (Ac-CoA) and the source of the triplet and doublet at ω-1 in 13C NMR spectroscopy. c,d, 13C NMR of lipids labeled with D[U-13C]glucose (c) or L[U-13C]glutamine (d). Insets are expansions of the ω-1 resonance. Abbreviations: s, singlet; d, doublet; t, triplet.
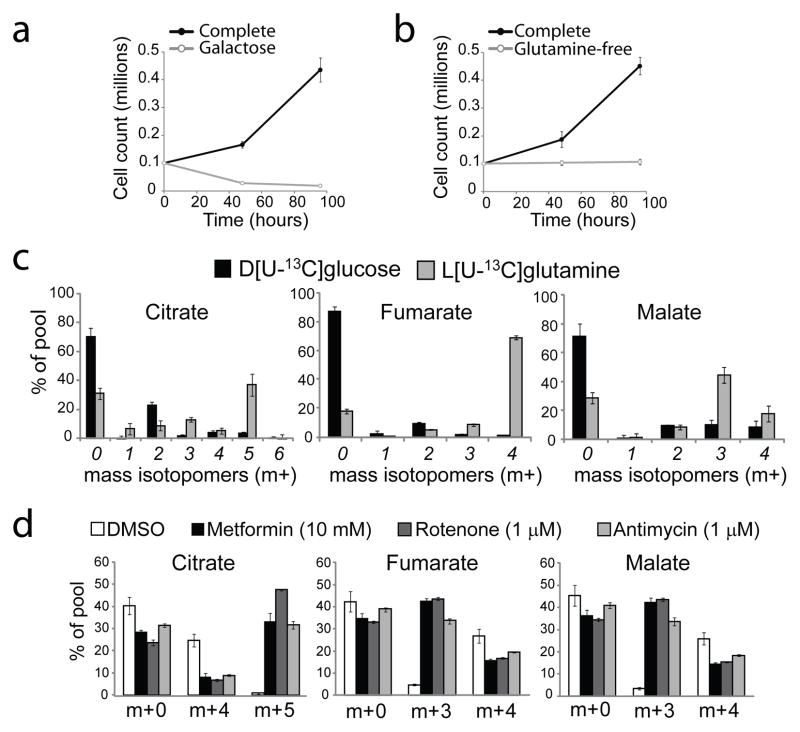
Although most cancer cells have functional mitochondria, a subset of human tumors harbors mutations that impair mitochondrial metabolism. Two major classes of mutations occur in genes required for function of the CAC enzymes succinate dehydrogenase (SDHA-D, SDHAF2) or fumarate hydratase (FH)14. To test whether glutamine-dependent reductive carboxylation occurs in tumor cells harboring these mutations, we studied UOK262 cells derived from a renal tumor in a patient with hereditary leiomyomatosis renal cell carcinoma syndrome. In this syndrome, affected individuals inherit one loss-of-function FH allele and their tumors display loss of heterozygosity for the FH locus15. UOK262 cells are defective in respiration and devoid of FH activity15. They failed to survive culture in glucose-free medium containing galactose and glutamine (Fig. 4a), a condition that forces cells to use mitochondrial metabolism to generate ATP16. Lack of FH activity precludes the use of glutamine as a carbon source in the conventional oxidative CAC. Yet the UOK262 cells required glutamine to proliferate (Fig. 4b). Culture of these cells with D[U-13C]glucose produced mass isotopomer data similar to 143Bcytb cells, with minimal formation of citrate m+2 (Fig. 4c, left). In cells cultured with L[U-13C]glutamine, the most abundant mass isotopomer of citrate was m+5 formed by reductive carboxylation (Fig. 4c, left). Fumarate was predominantly formed through oxidative glutamine metabolism, producing fumarate m+4 (Fig. 4c, middle). However, a large fraction of the malate pool (m+3) was formed via citrate cleavage and subsequent reductive metabolism (Fig. 4c, right). Lipid labeling using D[U-13C]glucose or L[U-13C]glutamine revealed that, like 143Bcytb cells, glutamine is the major carbon source for fatty acid synthesis (Supplementary Fig. 8b). Thus these cancer cells produce CAC intermediates using two distinct pathways to compensate for the loss of FH enzyme activity: oxidative metabolism of glutamine to fumarate, and glutamine-dependent reductive carboxylation to produce citrate, acetyl-CoA for lipogenesis, and remaining 4-carbon CAC intermediates.
Figure 4. Glutamine-dependent reductive carboxylation in renal carcinoma cells lacking fumarate hydratase and during ETC inhibition in tumor cells with normal mitochondria.
Growth of UOK262 renal carcinoma cells in complete medium, in medium in which glucose was replaced with galactose (a), and in medium lacking glutamine (b). c, Mass isotopomer analysis of citrate, fumarate and malate from UOK262 cells cultured in medium containing D[U-13C]glucose and unlabeled glutamine, or L[U-13C]glutamine and unlabeled glucose. d, Mass isotopomer analysis for citrate, fumarate and malate from 143Bwt cells cultured in medium containing unlabeled glucose and L[U-13C]glutamine. The labeled glutamine was introduced at the same time the cells were treated with vehicle (DMSO), metformin, rotenone or antimycin. Metabolites were extracted 6 hours later. In all experiments, data are the average ± S.D. of three independent cultures.
Finally, we tested whether glutamine-dependent reductive carboxylation was unique to cells with permanent abnormalities in mitochondrial metabolism. 143Bwt cells and transformed mouse embryonic fibroblasts (MEFs) were subjected to acute ETC inhibition. Exposure to inhibitors of complex III (antimycin) or complex I (rotenone) induced a switch from oxidative to reductive metabolism of glutamine (Fig. 4d, Supplementary Fig. 9a). Enhanced reductive carboxylation was evident one hour after ETC inhibition and was not eliminated by pre-treatment with the protein synthesis inhibitor cycloheximide (Supplementary Fig. 9b,c). Metformin, an antidiabetic agent that inhibits respiration through effects on complex I17, also induced glutamine-dependent reductive carboxylation at high doses (Fig. 4d, Supplementary Fig. 9a). Interestingly, doses that mimicked plasma concentrations in patients with acute metformin toxicity and lactic acidosis induced reductive glutamine metabolism (Supplementary Fig. 10). These data demonstrate that glutamine-dependent reductive carboxylation is a common cellular response to impaired mitochondrial metabolism.
Glutamine-dependent reductive carboxylation has been described previously as a minor source of isocitrate/citrate and lipogenic carbon in mammalian cells12,13,18–20. In Plasmodium falciparum, a unicellular protozoan with limited respiration, the pathway is a major source of citrate and acetyl units21. We show that this pathway can also function as the predominant metabolic strategy to produce lipids for cancer cell growth, and can be induced acutely during ETC inhibition. We speculate that reductive carboxylation is stimulated by a disturbance in the redox ratio of the mitochondria caused by ETC impairment, decreasing the NAD+/NADH ratio and rendering oxidative function of the CAC less efficient. This ratio could be dissipated in part by NAD(P)-transhydrogenases, which transfer reducing equivalents from NADH to NADPH, and this in turn may drive NADPH-dependent reductive carboxylation by IDH1 and IDH2. This redirection of the CAC extends the versatility of glutamine metabolism in cancer cell growth. In cells with normal mitochondria, glutamine oxidation is frequently the major source of anaplerosis, providing most of the carbon to pools of CAC intermediates and facilitating production of macromolecular precursors in the mitochondria22. Glutamine-dependent reductive carboxylation enables glutamine to retain this status as a growth-promoting nutrient even when mitochondrial metabolism is impaired. Therefore, during malignant transformation the acquisition of somatic mutations that impair mitochondrial function and stimulate aerobic glycolysis (the Warburg effect) does not diminish the importance of glutamine metabolism or of the mitochondria themselves for tumor cell proliferation.
Methods Summary
143Bwt and 143Bcytb and UOK262 cells were cultured as described9,15. MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS, Hyclone), penicillin/streptomycin and 6mM L-glutamine. Cell growth was monitored in sub-confluent cultures by trypsinizing and counting with a hemacytometer. Glucose, lactate, and glutamine were measured as described23. Isotopic labeling was performed in DMEM with 10% dialyzed FBS supplemented with either 15mM D-[U-13C]glucose or 2mM L-[U-13C]glutamine. Aqueous metabolites were analyzed using an Agilent 6970 gas chromatograph and an Agilent 5973 mass selective detector. To determine relative metabolite abundance across samples, the area of the total ion current peak for the relevant metabolite was compared to that of an internal standard (2-oxobutyrate) and normalized for protein content. Analysis of 13C enrichment and mass isotopomer distribution was performed as published11,24. For radioisotope lipid synthesis assays, cells were cultured for 18 hours in complete medium supplemented with 3H2O or 14C tracers (Perkin Elmer). Lipids were extracted and analyzed as described3. For lipid synthesis experiments using NMR, cells were plated in 10-cm dishes and allowed to proliferate exponentially in medium containing D[U-13C]glucose and unlabeled glutamine, or unlabeled glucose and L[U-13C]glutamine until two confluent 15-cm dishes were obtained. Lipids were then extracted and analyzed by 13C NMR as described3. Gene silencing was performed using commercial small interfering RNAs (Thermo) directed against IDH1, IDH2 or IDH3. Cells were transfected with DharmaFECT transfection reagent (Thermo) and protein abundance was examined after 72 hours using western blots with commercial antibodies (Santa Cruz Biotechnology). Cells were incubated with L[U-13C]glutamine for two hours prior to extraction of metabolites for mass isotopomer distribution analysis. Stable silencing used lentiviral shRNAs from the Mission shRNA pLKO.1-puro library (Sigma). Additional details are in the online Methods.
Supplementary Material
Acknowledgments
Kosaku Uyeda and members of the DeBerardinis and Chandel laboratories provided critical assessment of the data, and Jessica Sudderth and Chendong Yang provided experimental assistance. We thank Carlos Moraes and I.F.M. de Coo for the 143Bwt and 143Bcytb cell lines, Immo E. Scheffler for CCL16-B2 cells and Takao Yagi for CCL16-NDI1 cells. This work was supported by grants to R.J.D. from the N.I.H. (K08DK072565, R01CA157996, and RR02584), the Cancer Prevention and Research Institute of Texas (CPRIT, HIRP100437) and the Robert A. Welch Foundation (I1733); to N.S.C. from the N.I.H. (R01CA123067), the LUNGevity Foundation and a Consortium of Independent Lung Health Organizations convened by Respiratory Health Association of Metropolitan Chicago; and to E.S.J. by an N.I.H. grant (DK078933). The work was also supported by the Intramural Research Program of the N.I.H., National Cancer Institute Center for Cancer Research. N.I.H. training grants supported A.R.M. (5T32GM083831), W.W.W. (T32CA009560) and L.B.S. (T32GM008061). T.C. was supported by a CPRIT training grant.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. A.R.M., W.W.W., N.S.C. and R.J.D. designed the research. A.R.M., W.W.W., L.B.S., E.S.J., T.C. and P-H.C. performed the experiments. A.R.M., W.W.W., L.B.S., E.S.J., P-S.C., T.C., N.S.C. and R.J.D. analyzed the data. Y.Y. and W.M.L. provided the FH-deficient (UOK262) cells. A.R.M., N.S.C. and R.J.D. wrote the paper.
None of the authors have competing financial interests to declare. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 6.Tomlinson IP, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 7.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rana M, de Coo I, Diaz F, Smeets H, Moraes CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann Neurol. 2000;48:774–781. [PubMed] [Google Scholar]
- 9.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 11.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Des Rosiers C, Fernandez CA, David F, Brunengraber H. Reversibility of the mitochondrial isocitrate dehydrogenase reaction in the perfused rat liver. Evidence from isotopomer analysis of citric acid cycle intermediates. J Biol Chem. 1994;269:27179–27182. [PubMed] [Google Scholar]
- 13.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J Mol Med. 2011;89:213–220. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, et al. UOK 262 cell line, fumarate hydratase deficient (FH-/FH-) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet. 2010;196:45–55. doi: 10.1016/j.cancergencyto.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 17.El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 18.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–20627. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol. 2002;283:H1505–1514. doi: 10.1152/ajpheart.00287.2002. [DOI] [PubMed] [Google Scholar]
- 20.Lemons JM, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olszewski KL, et al. Branched tricarboxylic acid metabolism in Plasmodium falciparum. Nature. 2010;466:774–778. doi: 10.1038/nature09301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.