Summary
The chemical identity and integrity of the genome is challenged by the incorporation of ribonucleoside triphosphates (rNTPs) in place of deoxyribonucleoside triphosphates (dNTPs) during replication. Misincorporation is limited by the selectivity of DNA replicases. We show that accumulation of ribonucleoside monophosphates (rNMPs) in the genome causes replication stress and has toxic consequences, particularly in the absence of RNase H1 and RNase H2, which remove rNMPs. We demonstrate that postreplication repair (PRR) pathways—MMS2-dependent template switch and Pol ζ-dependent bypass—are crucial for tolerating the presence of rNMPs in the chromosomes; indeed, we show that Pol ζ efficiently replicates over 1–4 rNMPs. Moreover, cells lacking RNase H accumulate mono- and polyubiquitylated PCNA and have a constitutively activated PRR. Our findings describe a crucial function for RNase H1, RNase H2, template switch, and translesion DNA synthesis in overcoming rNTPs misincorporated during DNA replication, and may be relevant for the pathogenesis of Aicardi-Goutières syndrome.
Graphical Abstract
Highlights
► Unprocessed rNMPs in the chromosomes sensitize cells to replication stress ► Postreplication repair pathways are essential to tolerating rNMP-containing templates ► DNA polymerase ζ efficiently bypasses rNMPs in DNA templates ► Postreplication repair is chronically activated in RNase H1/H2 defective cells
Introduction
The integrity of the eukaryotic cellular genome is preserved by surveillance mechanisms that coordinate DNA replication, repair, and recombination with cell-cycle progression (Muzi-Falconi et al., 2003; Lazzaro et al., 2009). The DNA nature of the chromosomes provides for an intrinsic stability as opposed to the fragility of RNA, which is due to the higher reactivity of ribose compared to deoxyribose. The incorporation of ribonucleotides (rNTPs) in place of deoxyribonucleotides (dNTPs) within genomic DNA is generally avoided by the high selectivity of DNA polymerases, largely due to a steric gate residue in the polymerase active site (Joyce, 1997). However, there are occasions when rNTPs can be linked to DNA chains, such as during the synthesis of Okazaki fragments or possibly during repair of double strand DNA breaks in G1 (Nick McElhinny and Ramsden, 2003; Zhu and Shuman, 2008). Recent work indicates that during normal DNA replication, DNA polymerases can also incorporate rNTPs in place of dNTPs (Nick McElhinny et al., 2010b). rNMPs embedded in DNA are expected to represent a problem for cycling cells, sensitizing the DNA backbone to spontaneous and/or enzymatic nicking. Indeed, the presence of rNMPs in the yeast genome elevates the rate of short deletions in repeated sequences through a mechanism depending on topoisomerase I (Nick McElhinny et al., 2010a; Clark et al., 2011; Kim et al., 2011). Furthermore, the presence of rNMPs alters DNA helix parameters. For example, structural studies (Egli et al., 1993; Jaishree et al., 1993; Ban et al., 1994a; Ban et al., 1994b; Wahl and Sundaralingam, 2000) indicate that rNMPs in dsDNA alter global conformation from B- to A-form, with most of the sugars adopting C3′-endo or closely related conformations. rNMPs must be removed prior to the next cell cycle or they will pose problems during subsequent rounds of replication; in fact, efficient and accurate synthesis by replicative DNA polymerases strongly depends on helix geometry, such that changes in sugar pucker could render a primer terminus more difficult to extend. Indeed, a recent study has shown that single rNMPs in DNA templates impede DNA synthesis by the yeast replicases (Watt et al., 2011). Altered helix geometry may be less problematic for polymerases specialized for translesion synthesis, e.g., DNA polymerase ζ, which can efficiently extend aberrant primer termini (Prakash et al., 2005). An important question is thus how cells cope with replicating chromosomes containing rNMPs that escape repair.
RNase H is a family of enzymes that cleave the RNA moiety in RNA:DNA hybrids, allowing the reconstruction of a dsDNA molecule. Eukaryotic cells possess RNase H1 and RNase H2 activities that have partially overlapping substrate specificity. While RNase H1 requires at least a tract of four rNMPs to cleave, RNase H2 can incise 5′ to a single rNMP incorporated within a DNA molecule (Cerritelli and Crouch, 2009). The in vivo roles of RNase H in eukaryotic cells are still not fully understood. In mammalian cells, RNase H1 is essential for mitochondrial DNA replication (Cerritelli et al., 2003); such function is not conserved in budding yeast cells. The role of the nuclear population of RNase H1 is still not clear. RNase H2 represents the major RNase H activity in eukaryotic cells and is involved in several cellular processes (Cerritelli and Crouch, 2009). Evidence indicates that these enzymes can process Okazaki fragments during replication although, at least in budding yeast, such activity is redundant and Okazaki fragment processing can be carried out by Rad27 and Dna2 (Rydberg and Game, 2002; Ayyagari et al., 2003). Furthermore, removal of R-loops, which accumulate when a transcription bubble collides with a replication fork, can be achieved by overexpressing RNase H (Huertas and Aguilera, 2003). Mutations in any of the three subunits of human RNase H2 are the molecular cause of a human genetic syndrome known as Aicardi-Goutières syndrome (AGS) (Crow et al., 2006a). The mechanism(s) involved in the pathogenesis of AGS is under intense investigation but still uncertain (Crow et al., 2006b; Yang et al., 2007; Stetson et al., 2008; Rice et al., 2009; Crow and Rehwinkel, 2009).
Another enzyme that processes rNMPs in DNA is topoisomerase I. It was recently reported that, in the absence of RNase H2, rNTPs incorporated in DNA are targeted by topoisomerase I, which cleaves but fails to rejoin the DNA backbone, generating a ssDNA break (Sekiguchi and Shuman, 1997; Kim et al., 2011). Interestingly, not all genomic rNMPs are topoisomerase I targets (Kim et al., 2011), and cells lacking RNase H2 do not exhibit growth defects, suggesting that cells must have other pathways allowing them to replicate rNMP-containing chromosomes.
In this work, we investigate the processes permitting yeast cells to survive in the presence of elevated rNTPs incorporated within genomic DNA. We show that both RNase H1 and RNase H2 play a critical role in repairing rNMPs incorporated by replicative polymerases, and in the absence of RNase H activity residual genomic rNMPs cause replication problems in the following cell cycle. When the replicative DNA polymerases encounter rNMPs in the template strand, endogenous replication stress is generated, which sensitizes cells to mild treatments with exogenous replication stress-inducing agents. In this situation, postreplication repair mechanisms are effectively responsible for the survival of RNase H defective cells. We provide genetic and biochemical evidence that rNMPs-containing chromosomes can be fully replicated through the action of template switch and DNA polymerase ζ, which efficiently bypasses rNMPs in a DNA template.
Our data show unexpected mechanisms that preserve genome integrity in normally replicating cells, extend the role of PRR, and particularly that of Pol ζ, to the replication of rNMPs in genomic DNA, and reveal a synthetic interaction between PRR, RNase H activities, and replication stress that may have relevant consequences for human disease, identifying a possible family of modifier genes that may influence the penetrance of a set of AGS mutations.
Results
Unrepaired rNMPs Incorporated in Genomic DNA during Replication Sensitize Cells to Replication Stress-Inducing Agents
The preferential incorporation of dNTPs over that of rNTPs is at least partially provided by a steric gate that prevents replicative DNA polymerases from using rNTPs during the elongation step (Joyce, 1997). Nonetheless, budding yeast DNA polymerase ε has been demonstrated (Nick McElhinny et al., 2010b) to incorporate large numbers of rNTPs into DNA. This effect is exacerbated in a Pol ε variant, Pol2-M644G, where a methionine adjacent to the steric gate residue (Y645) has been changed to glycine (Nick McElhinny et al., 2010a). A pol2-M644G rnh201Δ strain, where the mutation in Pol ε is combined with inactivation of RNase H2, which has been implicated in processing of rNMPs incorporated during DNA synthesis, exhibits slower progression through S phase (Nick McElhinny et al., 2010a), coupled to phosphorylated Rad53 checkpoint kinase (Figure S1C), suggestive of increased replication stress.
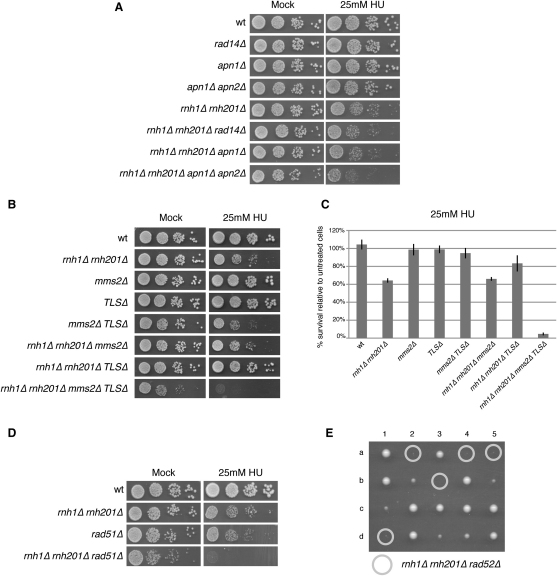
Low levels of replication stress-inducing agents (HU or MMS) are known to be toxic for cells with replication problems. To test whether the presence of rNMPs in the template strand affected DNA replication, we plated pol2-M644G rnh201Δ cells on medium containing low doses of HU or MMS, which in wild-type cells only mildly slow down cell-cycle progression. Figure 1A shows that a combination of the pol2-M644G and rnh201Δ mutations, leading to accumulation of elevated levels of rNMPs in genomic DNA, causes high sensitivity to low levels of HU and MMS (see also Figure S5 for quantitative survival data). Interestingly, loss of RNase H1 alone does not sensitize pol2-M644G cells to HU or MMS (Figure 1A). These phenotypes can be explained by the fact that, even though the substrate specificity of RNase H1 partially overlaps with that of RNase H2, and both enzymes cleave DNA containing four or more consecutive rNMPs, only RNase H2 cleaves at single rNMPs (Cerritelli and Crouch, 2009). These observations suggest that the presence of large amounts of single rNMPs within chromosomal DNA generates endogenous replication stress. When both RNase H1 and H2 enzymes are inactivated, virtually all single and multiple rNMPs incorporated during DNA synthesis will persist until the next round of replication. Strikingly, pol2-M644G rnh201Δ is synthetic lethal with the absence of RNase H1 (Figure 1B), indicating that RNase H1 plays an important role in repairing the rNTPs incorporated by Pol ε.
Figure 1.
Abundant Incorporation of rNTPs into DNA Sensitizes Cells to Replication Stress and Is Lethal in Cells Lacking RNase H
(A) To test sensitivity to sublethal doses of HU or MMS, 10-fold serial dilutions of the indicated mutant strains were plated on YPD, YPD + 25 mM HU and YPD + 0.04% MMS. Pictures were taken after 3 days of incubation.
(B) Tetrads derived from a cross between rnh1Δ rnh201Δ and rnh1Δ pol2-M644G were dissected on YPD plates. Seven tetrads (1–7) are shown. The circles on the figure indicate the position of the original rhn1Δ rnh201Δ pol2-M644G spores.
(C) Sensitivity to HU and MMS of the indicated strains was tested as described in (A). A checkpoint-defective mec1-1 strain was included as a positive control.
(D) Single cells were isolated on YPD plates and grown for 22 hr in the presence of 25 mM HU; colonies were visualized by microscopic analysis.
(E and F) wild-type and rnh1Δ rn201Δ cells were released in 25 mM HU after α factor arrest. After 180 min, cultures were analyzed by FACS, for DNA contents, and cell extracts were tested by western blotting with anti-Rad53 antibodies.
(G) Wild-type and rnh1Δ rnh201Δ cells were plated on YPD with or without 25 mM HU in the presence of Phloxine B, which stains in red colonies containing dead cells.
(H) Quantification of cell survival was obtained by plating G1 synchronized cells (100 cells per plate) on dishes containing 25 mM HU or mock. Colonies were counted after 3 days of incubation. The graph is representative of three independent experiments. Error bars describe standard deviation.
RNase H1 Cooperates with RNase H2 in the Removal of rNMPs from the Chromosomes Preserving Genome Integrity
The critical role of both RNase H enzymes is supported by the fact that double mutant rnh1Δ rnh201Δ, rnh1Δ rnh202Δ, and rnh1Δ rnh203Δ cells (RNH202 and RNH203 encode the two noncatalytic subunits of RNase H2) are sensitive to low levels of replication stress even in the presence of normal replicases (Figure 1C). Microscopic observation revealed that rnh1Δ rnh201Δ cells form small and irregular microcolonies on plates containing 25 mM HU while wild-type cells generate a regular colony (Figure 1D). FACS analysis of synchronous cultures incubated with low levels of HU or MMS shows that cells lacking RNase H arrest in G2-M after the bulk of genome replication has been completed (Figures 1E and S1A), and western blot analysis of Rad53 kinase revealed that mutant cells accumulate hyperphosphorylated Rad53 (Figures 1F and S1B). It is worth noting that cycling cells of mutants that accumulate elevated rNMP levels in the genome exhibit a constitutively phosphorylated Rad53, indicative of chronic replication stress (Figure S1C). These findings indicate that low doses of HU lead rnh1Δ rnh201Δ cells to block at the mitotic checkpoint and cause massive cell lethality, as suggested by the rugged shape of the microcolonies (Figure 1D) and further demonstrated by the fact that the small colonies eventually growing on 25 mM HU contain a large proportion of dead cells, which are stained by Phloxine B (Figure 1G). To estimate the extent of such lethality, we plated wild-type and rnh1Δ rnh201Δ cells in the absence or presence of 25 mM HU and calculated the percent survival on HU. Three independent experiments confirmed 40% lethality in cells lacking RNase H and exposed to low doses of HU (Figure 1H). Quantitative survival data for all the strains used throughout this study are shown in Figure S5. To test whether Rad53 phosphorylation and loss of cell viability derive from enzymatic processing of rNMP-containing DNA followed by chromosome breakage, we monitored phosphorylation of histone H2A on S129, a marker of DNA damage. Figure S1D shows that exposure of rnh1Δ rnh201Δ cultures to 25 mM HU does not induce H2A phosphorylation, suggesting that these cells do not accumulate double strand breaks, even when challenged with HU.
The sensitivity to HU observed upon loss of RNase H is unlikely to be due to the role of RNase H in Okazaki fragment processing or to a possible involvement in R-loop metabolism. Indeed, rad27 mutated cells, which accumulate unprocessed Okazaki fragments (Ayyagari et al., 2003), are not sensitive to replication stress (Figure S2A). Moreover, combining rnh1Δ rnh201Δ with a mutation in HPR1 gene, which leads to the accumulation of R-loops (Huertas and Aguilera, 2003), does not increase sensitivity to 25 mM HU and actually seems to mildly suppress the rnh1Δ rnh201Δ phenotype at this low dose, even though the mechanism is not known (Figure S2B). These findings strongly support the notion that RNase H activity is important to keeping genomic DNA free from rNMPs incorporated by DNA polymerases during replication and that sensitivity to replication stress-inducing drugs is a valid assay to track this process.
Survival of Cells with rNMPs-Containing Chromosomes Requires Translesion DNA Synthesis and Template Switch PRR Pathways
The survival of cells lacking RNase H activities indicates that yeast must have additional mechanisms to cope with the incorporation of rNTPs into the genome.
We investigated whether nucleotide excision repair (NER) or base excision repair (BER) play a role in the removal of rNMPs from the chromosomes. Abolishing NER (rad14Δ) or deleting APN1, which is responsible for ≥97% of AP endonuclease and 3′-diesterase activities required for BER (Popoff et al., 1990), does not sensitize rnh1Δ rhn201Δ cells to replication stress-inducing agents (Figure 2A). This result is consistent with data showing that rNMPs-containing DNA cannot be processed by NER and BER nucleases (Rydberg and Game, 2002). The observation that deletion of APN2 in a rnh1Δ rnh201Δ apn1Δ causes an increase in sensitivity to 25 mM HU can be explained by the fact that simultaneous deletion of APN1 and APN2 causes an accumulation of elevated levels of endogenous lesions, increasing cellular stress (Leroy et al., 2001). We cannot exclude, though, that a secondary BER pathway may be able to process a minority of rNMPs.
Figure 2.
Postreplication Repair Is Specifically Required to Tolerate rNMPs-Containing Chromosomes
Sensitivity to sublethal doses of HU was assayed as described in Figure 1. Pictures were taken after 3 days of incubation. The contribution of NER (A), BER (A), the two branches of PRR (B), and RAD51 (D) was tested. In (C), Quantification of cell survival was obtained as described in Figure 1H. The graph is representative of three independent experiments. Error bars describe standard deviation. It is worth noting that mms2Δ TLSΔ cells, despite being sensitive to HU in the spot tests, do not exhibit increased cell lethality, suggesting that the HU sensitivity derives from a very slow cell-cycle progression. In (E), tetrads derived from a cross between rnh1Δ rnh201Δ and rad52Δ were dissected on YPD plates. Five tetrads (1–5) are shown.The circles on the figure indicate the position of the original rhn1Δ rnh201Δ rad52Δ spores. Cells derived from such microcolonies do not grow when restreaked, revealing that a rad52Δ mutation is synthetic lethal with deletion of the RNH1 and RNH201 genes. TLSΔ comprises rev1Δ rev3Δ rev7Δ rad30Δ.
Given that rNMPs in DNA templates impede DNA synthesis by the yeast replicases Pols ε and δ (Watt et al., 2011), lethality may result from failure to complete DNA replication. We thus investigated whether postreplication repair (PRR) mechanisms may allow full genome replication in rnh1Δ rnh201Δ cells. When DNA polymerases encounter replication-blocking lesions, PCNA is monoubiquitylated by Rad6-Rad18 triggering translesion DNA synthesis (TLS), while polyubiquitylation, carried out by Mms2-Ubc13-Rad5, promotes template switch (Ulrich, 2011).
We checked by spot assay whether deleting either branch of PRR would affect DNA replication in cells that do not remove rNMPs from genomic DNA, and cell lethality was quantitated as in Figure 1. Loss of only the template switch pathway (mms2Δ) or only translesion DNA synthesis (TLSΔ: corresponding to deletions of REV1, REV3, REV7, and RAD30 genes encoding all TLS polymerases in budding yeast) does not sensitize cells lacking RNase H to HU. On the other hand, concomitant elimination of TLS and template switch results in almost no growth of rnh1Δ rhn201Δ cells in 25 mM HU, due to cell lethality (Figures 2B and 2C). These findings show that when rnh1Δ rnh201Δ cells are subjected to a low level of replication stress, survival depends almost entirely on either PRR pathway. This effect, although striking in the presence of HU, can also be detected in unperturbed conditions (bottom line, Figure 2B; see also Figures S3A and S3B). We conclude that cells devoid of RNase H1 and H2 can use TLS and template switch pathways to completely replicate their rNMPs-containing genome. Consistently, deletion of RAD51, which is required for a recombination-dependent PRR pathway (Gangavarapu et al., 2007), increases the sensitivity to HU of rnh1Δ rnh201Δ cells, while loss of RAD52 is lethal in this genetic background (Figures 2D and 2E). These phenotypes may be influenced by defects in the additional processes that involve homologous recombination.
DNA Polymerase ζ Is the TLS Polymerase Replicating rNMPs-Containing DNA
To identify which translesion DNA polymerase allows the bypass of rNMPs, we combined mutations in genes coding each of the three yeast TLS polymerases, REV1, REV3/REV7 (the catalytic and noncatalytic subunits of Pol ζ, respectively), and RAD30 (Pol η) in rnh1Δ rnh201Δ cells. The experiment was performed in the absence of the MMS2-dependent template switch pathway, so that rnh1Δ rnh201Δ cells rely only on TLS to complete replication. The spot tests shown in Figure 3A reveal that rnh1Δ rnh201Δ mms2Δ cells carrying a deletion of REV1 or direct inactivation of DNA polymerase ζ (rev3Δ rev7Δ) do not survive HU treatment and are less viable even in untreated conditions, recapitulating the total absence of TLS activities. Deletion of RAD30 does not increase cell lethality under these conditions; on the contrary, we reproducibly observed that loss of Pol η confers an unexpected growth advantage when genomic DNA contains unrepaired rNMPs (Figure 3A), consistently with the phenotype observed in rnh1Δ rnh201Δ TLSΔ (Figures 2B and 2C). This unpredicted observation may be justified envisioning a competition between noneffective pol η and other PRR pathways.
Figure 3.
Pol ζ Allows Cells to Cope with Unrepaired rNMPs
The sensitivity to HU was measured as described in Figure 1: the specific contribution of each TLS polymerase (A) and the requirement of the catalytic activity of Rev1 (B) were tested.
Rev1 plays a noncatalytic role in supporting Pol ζ function (Lawrence and Hinkle, 1996; Lawrence, 2002) and also has a deoxycytidyl transferase activity (Nelson et al., 1996) that could insert a dCTP opposite a rNMP, allowing Pol ζ to extend. Figure 3B shows that, contrary to what was observed with rev1Δ, inactivating the polymerase activity of Rev1 does not significantly affect the HU sensitivity of rnh1Δ rnh201Δ mms2Δ. Altogether, these data indicate that cells can use Pol ζ to replicate rNMPs-containing templates in vivo and that Rev1 most likely plays a noncatalytic role to promote Pol ζ activity.
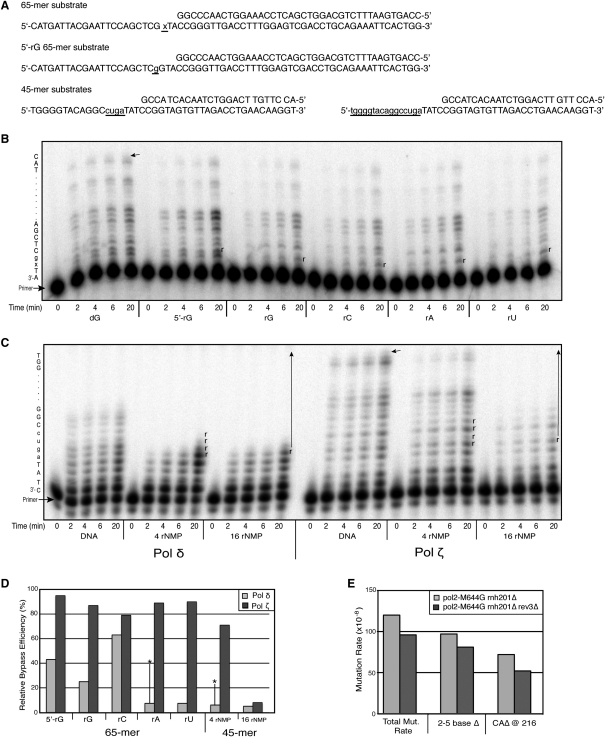
To confirm biochemically that DNA polymerase ζ is capable of bypassing rNMPs in DNA templates, we measured the rNMP bypass efficiency of purified yeast Pol ζ in vitro. Labeled substrates containing one, four, or sixteen consecutive rNMPs (Figure 4A) were incubated with purified DNA polymerase ζ or δ, and bypass efficiency was calculated after quantifying the primer extension products resulting from a single cycle of processive elongation (Figures 4B and 4D), as previously described (Nick McElhinny et al., 2010b). Consistently with the genetic observations (Figure 3A), the data indicate that Pol ζ bypasses ribonucleotides incorporated in DNA, efficiently copying DNA templates containing one (Figures 4B and 4D) or four rNMPs (Figures 4C, right, and 4D). This is in contrast with Pol δ which is somewhat less efficient in copying templates containing rC and much less efficient at copying templates containing rG, rA, rU, or four consecutive rNMPs (Watt et al., 2011) (Figures 4C and 4D). Pol δ bypass of rA or four consecutive rNMPs was stimulated several fold by adding PCNA to the reactions (see asterisks in Figure 4D), but in neither case was bypass as efficient as for Pol ζ.
Figure 4.
DNA Polymerase ζ Efficiently Bypasses rNMPs in the Template Strand
(A) Primer-template sequences. In the 65-mer substrate, “x” is the position of the single rNMP (rG, rC, rA, or rU) and “g” is the position of the 5′-rG in the DNA template. In the 45-mer substrate, the underlined lowercase nucleotides indicate the position and sequence of the rNMPs in 4- and 16-rNMP substrates, respectively.
(B and C) Phosphorimages of products generated during bypass of a single rNMP (B) and tracts of rNMPs by Pol δ and ζ (C). The template sequence is shown on the left, the arrow depicts the location of full-length product, and “r” represents the position of the rNMPs in the template. No enzyme was added to the unextended primer reaction (0 min).
(D) Relative bypass efficiencies for Pols δ and ζ. Images of reaction products shown in (B) and (C) were quantified, and relative bypass efficiencies were calculated as described (Stone et al., 2009). The values for Pol δ with the 65-mer substrates in the absence of PCNA have been reported previously (Watt et al., 2011) and are shown here for comparison. The asterisks indicate the relative bypass values for Pol δ for reaction mixtures containing 200 nM PCNA.
(E) Mutation rates for the pol2-M644G rnh201Δ and pol2-M644G rnh201Δ rev3Δ strains. The total mutation rates for resistance to 5-FOA were determined as described in Experimental Procedures. The 95% confidence intervals for the pol2-M644G rnh201Δ and pol2-M644G rnh201Δ rev3Δ strains were 110 to 200 and 57 to 140, respectively. For the pol2-M644G rnh201Δ strain, the rates for total 2–5 base pair deletions and for CA deletions at position 216–219 in URA3 are from Clark et al. (2011). For the pol2-M644G rnh201Δ rev3Δ strain, rates for short deletions were calculated after sequencing the ura3 gene in 163 independent 5-FOA resistant clones. Of these, 136 harbored 2–5 base pair deletions, 88 of which were CA deletions at the CACA hotspot at position 216–219 in URA3 (see spectrum in Figure S6).
We previously showed that, compared to RNase H2-proficient cells, pol2-M644G rnh201Δ strains (Nick McElhinny et al., 2010a) and rnh201Δ strains (Clark et al., 2011) have elevated rates of 2–5 base pair deletions in repetitive sequences and, recently, these deletions were shown to depend on topoisomerase 1 (Kim et al., 2011). This led to a model wherein topoisomerase 1 incises unrepaired rNMPs to create nicks in DNA with 3′-P and 5′-O ends that must be processed to allow ligation, and this processing may provide the opportunity for strand misalignments in repetitive sequences that yield the observed deletions. To determine if Pol ζ, which is relatively inaccurate (Zhong et al., 2006), might also contribute to this deletion mutagenesis, we measured the effect of deleting REV3 on mutagenesis in the pol2-M644G rnh201Δ strain. Mutagenesis rates were estimated by measuring frequencies of formation of 5-FOA resistant clones, indicative of mutations leading to uracil auxotrophy. The results (Figure 4E) reveal that deleting REV3 does not significantly (at p = 0.05; see figure legend) affect the overall rate of mutations to 5-FOA resistance, or the rates for total 2–5 base pair deletions or deletions of CA from a previously observed CACA hotspot sequence in the URA3 gene. The lack of an effect of rev3Δ on mutagenesis suggests that Pol ζ does not contribute to topoisomerase 1-dependent mutagenesis resulting from unrepaired ribonucleotides incorporated during replication by Pol2-M644G. When the rate of base substitutions that might be explained by misincorporation of dCTP by Rev1p was calculated, no significant difference was observed between the pol2-M644G (from Pursell et al., 2007, and unpublished data), pol2-644G rnh201Δ (from Nick McElhinny et al., 2010a) and pol2-M644G rnh201Δ rev3Δ strains (from Figures 3 and S6). This supports the notion that the requirement for REV1 in rNMPs bypass is structural rather dependent on its deoxycytidyltransferase activity.
RNase H-Defective Cells Exhibit Chronically Activated PRR Pathways
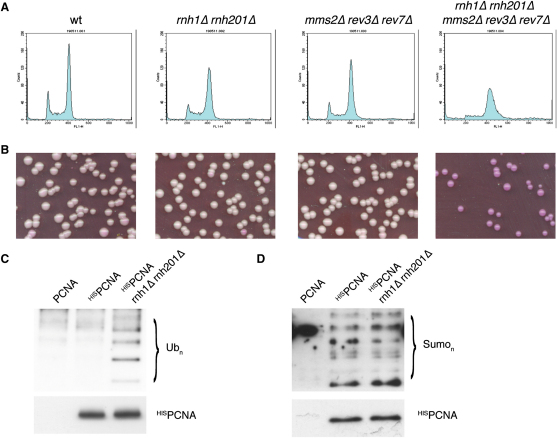
The relevance of PRR in coping with rNMPs in chromosomal DNA is evident by analyzing unperturbed mutant cells, which lack RNase H activities. FACS analysis of cycling cells suggests that rnh1Δ rnh201Δ cultures contain a higher fraction of S phase cells, and further inactivation of PRR pathways leads to a very sick phenotype (Figures 5A, S3A, and S3B). Indeed, these cells exhibit G2-M arrest coupled to cell lethality, as seen by Phloxine B staining of mutant colonies (Figure 5B).
Figure 5.
In the Absence of RNase H, the PRR Pathway Is Constitutively Activated and Promotes Cell Survival in an Unperturbed Cell Cycle
The role of PRR was assessed in unperturbed rnh1Δ rnh201Δ cultures. Exponentially growing cells lacking RNase H and defective in PRR were analyzed by FACS (A), to monitor cell cycle distribution, and by Phloxine B staining (B), to evaluate cell lethality. PCNA was affinity purified from exponentially growing unperturbed wild-type cells or from cells lacking RNase H activity. PCNA levels were estimated by western blotting with anti-HIS Ab. PCNA ubiquitylation was monitored by western blotting with anti-ubiquitin Ab (C), and PCNA sumoylation was monitored by western blotting with anti-SUMO Ab (D).
Affinity-purified HIS-tagged PCNA from exponentially growing rnh1Δ rnh201Δ cells revealed a striking increase in PCNA ubiquitylation, compared to wild-type cells. Figure 5C shows that both mono- and polyubiquitylated forms of PCNA are abundant in cells that cannot remove rNMPs from genomic DNA. Conversely, no significant effect is observed in PCNA sumoylation (Figure 5D). Accordingly, deletion of RAD18, coding for the ubiquitin ligase responsible for conjugating ubiquitin to PCNA, has a synthetic effect when combined with the loss of RNase H: rnh1Δ rnh201Δ rad18Δ cells are exquisitely sensitive to 25 mM HU and exhibit cell lethality even in untreated conditions (Figure S3C).
All these results indicate that cells lacking RNase H have constitutively active PRR, which is crucial to tolerating the presence of rNMP-containing genomic DNA.
Discussion
Yeast Cells Can Insert rNTPs into Genomic DNA
In eukaryotic cells the size of cellular dNTP pools is tightly controlled, and altered dNTP levels are responsible for increased mutagenesis and genome instability (Chabes and Stillman, 2007). Because the pools of rNTPs are much higher, DNA polymerases must be selective to correctly polymerize dNTPs during genome replication. Recent evidence has shown that during normal DNA replication in yeast, DNA polymerases incorporate rNTPs into genomic DNA. The pol2-M644G mutation affecting the steric gate of Pol ε increases rNTPs incorporation 10-fold (Nick McElhinny et al., 2010b). Genomic DNA isolated from rnh201Δ cells has a high number of alkali-sensitive sites, indicating that RNase H2 is involved in removing rNMPs from DNA (Nick McElhinny et al., 2010a).
Unrepaired rNMPs in genomic DNA will impact on cell-cycle progression since, at the next round of DNA replication, replicative polymerases must duplicate a RNA-containing DNA template. It has been shown that replicative polymerases cannot effectively replicate a template containing rNMPs (Nick McElhinny et al., 2010a; Watt et al., 2011), and this situation generates replication stress, detectable as a higher fraction of cells in S phase (Nick McElhinny et al., 2010a and Figure S1C). Combining a deletion in RNH201, coding for the catalytic subunit of RNase H2, with a pol2-M644G mutation, we found that cells became sensitive to low doses of replication stress inducing agents (i.e., HU or MMS). These data suggest that the short RNA tracts, which cannot be processed in the absence of RNase H2, cause replication stress when the cell tries to replicate them. Low levels of HU and MMS increase such stress leading to cell lethality.
Since loss of RNase H2 activity from pol2-M644G mutated cells does not cause cell lethality per se, additional pathways repairing rNMPs-containing chromosomes must exist. In this work we describe different mechanisms that are involved in allowing cells to cope with the presence of rNMPs in their genome.
RNase H1 Participates in the Removal from the Genome of rNMPs Introduced during DNA Replication
RNase H1 has some overlapping substrates with RNase H2 and is the preferential enzyme for processing RNA:DNA hybrids where more than four rNMPs are present. Double mutant cells, combining rnh1Δ with the deletion of any of the RNase H2 subunits, exhibit hypersensitivity to low levels of HU or MMS, a cell-cycle delay in G2-M, and activation of the Rad53 checkpoint kinase.
Strikingly, deletion of RNH1, the gene coding for RNase H1, is synthetic lethal when combined with the pol2-M644G mutation and RNase H2 inactivation (rnh201Δ), demonstrating that RNase H1 also plays a crucial role in the repair of rNMPs incorporated by replicative DNA polymerases. Our genetic analysis excludes a contribution of NER in correcting rNMPs, while a minor involvement of BER in repairing rNMP-containing chromosomes cannot be completely ruled out.
The observation that cells lacking RNase H activities are sensitive to low doses of replication stress-inducing agents may have consequences for cancer chemotherapy. In fact, many cancer cells are characterized by elevated levels of endogenous replication stress (Negrini et al., 2010) and may be thus sensitized to inhibitors of RNase H activity, which could selectively kill cells experiencing replication stress.
Recently, topoisomerase 1 has been reported to be able to process rNMPs-containing DNA and generate ssDNA breaks, which can be easily converted to chromosome breaks. We believe it unlikely that rnh1Δ rnh201Δ lethality in HU is due to such chromosome fragmentation, since in our experiments the rnh1Δ rnh201Δ double mutant and the wild-type strains exhibit a similar level of phosphorylated histone H2A (Figure S1D), suggesting the absence of double-strand breaks. Altogether, these findings indicate that high levels of unrepaired rNMPs in the chromosome hinder DNA synthesis blocking replication forks, leading to replication stress.
Either One of the PRR Pathways Is Sufficient for Tolerating rNTPs Incorporated by Replicative DNA Polymerases, and DNA Polymerase ζ Is the Enzyme Replicating rNMPs-Containing DNA
When replication-blocking lesions are present in the DNA template, replication forks stall at the site of damage and cannot proceed. Completion of replication is facilitated by PRR mechanisms, namely error-prone translesion DNA synthesis (TLS) and error-free template switch, a recombination-like pathway, requiring, respectively, mono- and polyubiquitylation of PCNA.
Strikingly, exponentially growing rnh1Δ rnh201Δ cells exhibit high levels of constitutive mono- and polyubiquitylated PCNA, indicative of chronic PRR activation. Either TLS or template switch can be used to complete replication over rNMPs. Indeed, a strong synthetic effect is observed when both PRR pathways are inactivated or when PCNA ubiquitylation is prevented in cells lacking RNase H activities: these cells are exquisitely sensitive to mild replication stress and are also extremely sick in untreated conditions, indicating a novel role for PRR in tolerating RNA-containing DNA templates (Figure 6).
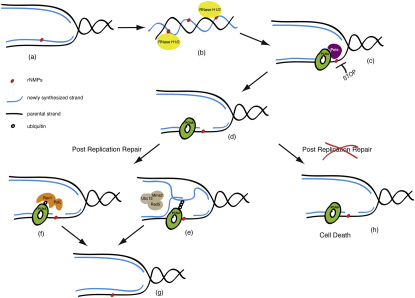
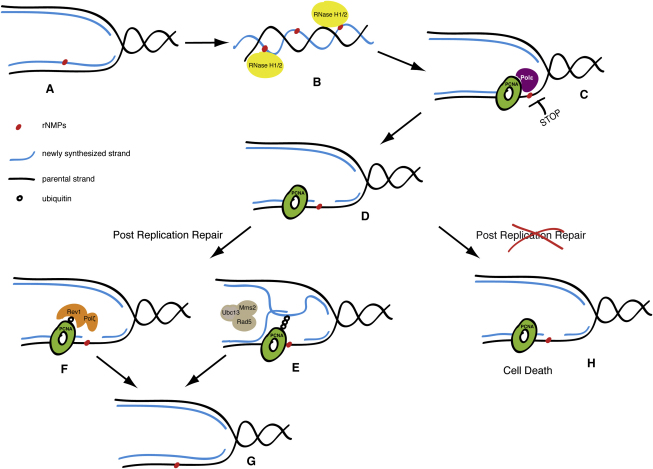
Figure 6.
RNase H and Postreplication Repair Protect Cells from rNMPs Incorporated in Chromosomal DNA
During DNA synthesis, replicative polymerases can incorporate rNTPs (red dot) in place of dNTPs (A). RNase H1 and RNase H2 are required to remove rNMPs from newly replicated DNA (blue line) (B). If rNMPs persist until the following cell cycle, they will create problems at during the DNA synthesis step (C), since replicative polymerases cannot efficiently elongate the nascent strand opposite rNMPs in the template strand (black line). Replication fork restart downstream of the lesions leaves incomplete replication products for postreplication repair (D). PCNA is ubiquitylated. Either MMS2-dependent template switch mechanisms (E) or Pol ζ-dependent translesion synthesis (F) allow bypass of rNMPs and completion of replication (G). Under these conditions, inactivation of PRR causes cell lethality (H).
How cells can replicate a chromosome containing rNMPs is not known. Yeast cells contain three TLS polymerases, Pol η, Pol ζ, and Rev1 (Friedberg et al., 1995). Our data show that, in the absence of a functional template switch pathway, rNMPs-containing DNA can only be replicated by the concerted action of Rev1 and Pol ζ. The fact that a catalytic rev1 mutant is capable of rescuing the phenotype imparted by a rev1Δ mutation indicates that the role of Rev1 is likely to help Pol ζ to replicate rNMPs containing templates. Indeed, untreated rnh1Δ rnh201Δ cells lacking the template switch pathway and missing Pol ζ form fewer and smaller colonies. A similar synthetic phenotype is observed in pol2-M644G mutant cells lacking PRR pathways (Figure S4). In conclusion, template switch and Pol ζ are crucial to allow replication of endogenous, unrepaired rNMPs (Figure 6), and mutations increasing the rNMPs load, such as pol2-M644G, may saturate PRR pathways so that cell survival relies on RNase H1 and RNase H2. The crucial role of Pol ζ for replicating rNMP-containing chromosomes may be performed either by adding a dNTP opposite the rNMP, or by elongating, downstream of the damaged site, a chain created by a replicative polymerase. In vitro data confirm the genetic findings and demonstrate that Pol ζ can efficiently insert a nucleotide opposite the lesion, bypassing 1 or 4 rNMPs in the DNA template, revealing a new cognate, endogenous substrate for this essential, specialized TLS polymerase.
Recombination-dependent PRR mechanisms are less understood. Rad51 and Rad52 are involved in PRR (Gangavarapu et al., 2007) and in other recombination events and, in general, rad52Δ strains are more recombination defective than rad51Δ cells. We found that deletion of RAD52 is synthetic lethal with loss of RNase H1 and RNase H2, and rad51Δ has a synthetic effect on HU sensitivity, supporting a role for recombination-dependent PRR in tolerating chromosomal rNMPs. However, defects in other recombination-dependent processes can contribute to these effects: for example, restart of blocked replication forks can proceed through recombination mechanisms (Heller and Marians, 2006; Petermann et al., 2010). Furthermore, RNase H enzymes may have diverse cellular targets in addition to rNTPs incorporated in genomic DNA. Among these are R-loops and Okazaki fragments; the accumulation of both these structures can have lethal outcomes and is prevented by recombination processes (Huertas and Aguilera, 2003; Ii and Brill, 2005).
Given the involvement of RNase H2 in the pathogenesis of the Aicardi-Goutières syndrome, the data reported in this work may help to understand the mechanisms underlying the disease. The reported synthetic effects between RNase H mutations, inducers of replication stress, and postreplication repair alterations, may facilitate the identification of modifier genes, whose alterations may be responsible for the phenotypic variability observed in different AGS patients carrying identical RNase H2 mutations.
Experimental Procedures
Yeast Strains
Strains are derivatives of W303, unless otherwise indicated in Table S1, and were generated by standard genetic procedures (Adams et al., 1998). YFL1449 and YFL1376 were obtained by crossing and backcrossing five times pol2-M644G (Nick McElhinny et al., 2010a) or HISPCNA (Ulrich and Davies, 2009) with SY2080.
FACS Analysis
Cells were fixed in 70% ethanol and treated with RNase A and proteinase K. DNA was stained with Sytox Green and cell-cycle distribution was estimated by cytofluorimetric analysis with a FACScan.
SDS-PAGE and Western Blotting
To monitor protein levels and phosphorylation, TCA protein extracts were prepared and analyzed by SDS-PAGE (Sabbioneda et al., 2007); western blots were performed with anti-Rad53, anti-H2A, and anti-γH2AX antibodies.
To study PCNA posttranslational modifications, HIS-tagged PCNA was pulled down from denaturing extracts as described (Ulrich and Davies, 2009), separated on 10% SDS-Urea-PAGE, and transferred to nitrocellulose membranes. PCNA ubiquitylation and sumoylation were analyzed by western blotting with anti-ubiquitin and anti-SUMO antibodies.
Sensitivity Assay
To assess cell survival in HU and MMS, overnight yeast cultures were diluted to 1 × 106 cfu/ml, and 10-fold serial dilutions were spotted on plates containing HU or MMS at the indicated concentrations. Images were captured after 3 days' incubation at 28°C. To obtain quantitative data, exponentially growing cells were arrested in G1 with 6 μg/ml α-factor. Cultures were diluted and 100 cfu were distributed on each of three independent YPD plates (mock or 25 mM HU). After 3 days' incubation colonies were counted. The experiments were repeated three times. The graphs show the percentage of surviving cells with respect to the mock sample. Standard deviations were used to obtain error bars.
Cell Lethality Assay
Overnight yeast cultures were diluted as above, and ∼100 cfu were plated on YPD containing 20 mg/l Phloxine B, with or without 25 mM HU. Images were captured after 3 days' incubation at 28°C.
Microcolony Assays
Yeast cells were grown to a concentration of 5 × 106 cells/ml and arrested in G1 with α-factor (6 μg/ml). Diluted samples were spread on YPD plates containing 25 mM HU, and single cells were separated using a micromanipulator. Plates were then incubated at 28°C and photographs were taken after 22 hr; thirty individual cells were followed for each experiment. The experiment was repeated four times, and a representative example is shown.
Cell-Cycle Analysis
Yeast cultures were grown to a concentration of 5 × 106 cells/ml and arrested in G1 with α-factor (6 μg/ml). Cells were released in YPD supplemented with 25 mM HU or 0.04% MMS or mock. Ninety minutes after, the release α-factor (6 μg/ml) was added back to the culture to avoid re-entry in S phase, allowing the analysis of a complete single cell cycle. Samples were collected for SDS-PAGE and FACS analysis and processed as described above. Growth curves were obtained by measuring cell concentration microscopically and normalizing each read to the initial concentration. Generation time was calculated by interpolating the growth curves.
rNMP Bypass, Mutation Rates, and Spectra
S.cerevisiae two-subunit wild-type Pol ζ (Rev3 –Rev7) and three-subunit Pol δ were expressed in yeast and purified as previously described (Burgers and Gerik, 1998; Garg et al., 2005). Oligonucleotide primer templates were prepared as described (Nick McElhinny et al., 2010b). The polymerase was added to initiate the reaction and aliquots were removed at 2, 4, 6, and 20 min. The DNA products were separated by electrophoresis through an 8% denaturing polyacrylamide gel containing 25% formamide for the 65-mer and 12% denaturing polyacrylamide gel for the 45-mer substrates. A PhosphorImager was used to visualize and quantify the DNA products. The efficiency of insertion opposite individual template positions and the bypass probability were calculated as previously described (Kokoska et al., 2003; Stone et al., 2009; Watt et al., 2011). Mutation rates and spectra were determined as described (Nick McElhinny et al., 2010a); in the relevant strains, the URA3 reporter was inserted at the AGP1 locus in orientation 2 as previously described (Nick McElhinny et al., 2010a).
Acknowledgments
We thank H. Ulrich, A. Aguilera, and C. Santocanale for strains and reagents; J. Williams and A. Clark for comments on the manuscript; S. Sabbioneda, S. Carnevali, F. Spadaro, and the members of our laboratories for discussions. This work was supported by grants from AIRC, MIUR, and Fondazione Cariplo to P.P. and M.M.-F. and MIUR (FIRB RBFR10S3UQ) to F.L. The financial support of Telethon-Italy (grant number GGP11003) is gratefully acknowledged. F.L. was partially supported by a fellowship from Fondazione Buzzati-Traverso. Part of this work was supported by Project Z01ES065070 to T.A.K. from the Division of Intramural Research of the NIH and grant NIHGM32431 to P.M.B. Cancer Research UK, an ERC start-up grant (206281), the Lister Institute of Preventive Medicine, and the EMBO Young Investigator Program supported V.C.
Contributor Information
Paolo Plevani, Email: paolo.plevani@unimi.it.
Marco Muzi-Falconi, Email: marco.muzifalconi@unimi.it.
Supplemental Information
References
- Adams A., Gottschling D.E., Stearns T., Kaiser C.A. Cold Spring Harbor Laboratory Press; Plainview, NY: 1998. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. [Google Scholar]
- Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- Ban C., Ramakrishnan B., Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular G-C base-paired quadruplet. J. Mol. Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- Ban C., Ramakrishnan B., Sundaralingam M. Crystal structure of the highly distorted chimeric decamer r(C)d(CGGCGCCG)r(G).spermine complex—spermine binding to phosphate only and minor groove tertiary base-pairing. Nucleic Acids Res. 1994;22:5466–5476. doi: 10.1093/nar/22.24.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli S.M., Frolova E.G., Feng C., Grinberg A., Love P.E., Crouch R.J. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Chabes A., Stillman B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:1183–1188. doi: 10.1073/pnas.0610585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.B., Lujan S.A., Kissling G.E., Kunkel T.A. Mismatch repair-independent tandem repeat sequence instability resulting ribonucleotide incorporation by DNA polymerase ε. DNA Repair (Amst.) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 2009;18(R2):R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Egli M., Usman N., Rich A. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- Friedberg E.C., Walker G.C., Siede W. ASM Press; Washington, DC: 1995. DNA repair and mutagenesis. [Google Scholar]
- Gangavarapu V., Prakash S., Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P., Stith C.M., Majka J., Burgers P.M. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase ζ. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- Heller R.C., Marians K.J. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Ii M., Brill S.J. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr. Genet. 2005;48:213–225. doi: 10.1007/s00294-005-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaishree T.N., van der Marel G.A., van Boom J.H., Wang A.H. Structural influence of RNA incorporation in DNA: quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32:4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- Joyce C.M. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Huang S.N., Williams J.S., Li Y.C., Clark A.B., Cho J.E., Kunkel T.A., Pommier Y., Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska R.J., McCulloch S.D., Kunkel T.A. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst.) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W., Hinkle D.C. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- Lazzaro F., Giannattasio M., Puddu F., Granata M., Pellicioli A., Plevani P., Muzi-Falconi M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst.) 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Leroy C., Mann C., Marsolier M.C. Silent repair accounts for cell cycle specificity in the signaling of oxidative DNA lesions. EMBO J. 2001;20:2896–2906. doi: 10.1093/emboj/20.11.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi-Falconi M., Liberi G., Lucca C., Foiani M. Mechanisms controlling the integrity of replicating chromosomes in budding yeast. Cell Cycle. 2003;2:564–567. [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V.G., Halazonetis T.D. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., Hinkle D.C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Ramsden D.A. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundström E.B., Johansson E., Chabes A., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff S.C., Spira A.I., Johnson A.W., Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Pursell Z.F., Isoz I., Lundström E.B., Johansson E., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M., Fuller J.C., Jackson R.M., Lamb T., Briggs T.A. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B., Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda S., Bortolomai I., Giannattasio M., Plevani P., Muzi-Falconi M. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst.) 2007;6:121–127. doi: 10.1016/j.dnarep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.E., Kissling G.E., Lujan S.A., Rogozin I.B., Stith C.M., Burgers P.M., Kunkel T.A. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 2009;37:3774–3787. doi: 10.1093/nar/gkp238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Ulrich H.D., Davies A.A. In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol. Biol. 2009;497:81–103. doi: 10.1007/978-1-59745-566-4_6. [DOI] [PubMed] [Google Scholar]
- Wahl M.C., Sundaralingam M. B-form to A-form conversion by a 3′-terminal ribose: crystal structure of the chimera d(CCACTAGTG)r(G) Nucleic Acids Res. 2000;28:4356–4363. doi: 10.1093/nar/28.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt D.L., Johansson E., Burgers P.M., Kunkel T.A. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst.) 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., Barnes D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zhong X., Garg P., Stith C.M., Nick McElhinny S.A., Kissling G.E., Burgers P.M.J., Kunkel T.A. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J. Biol. Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.