Abstract
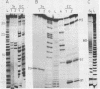
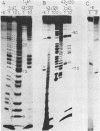
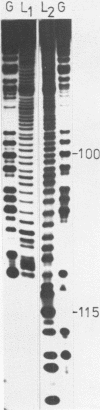
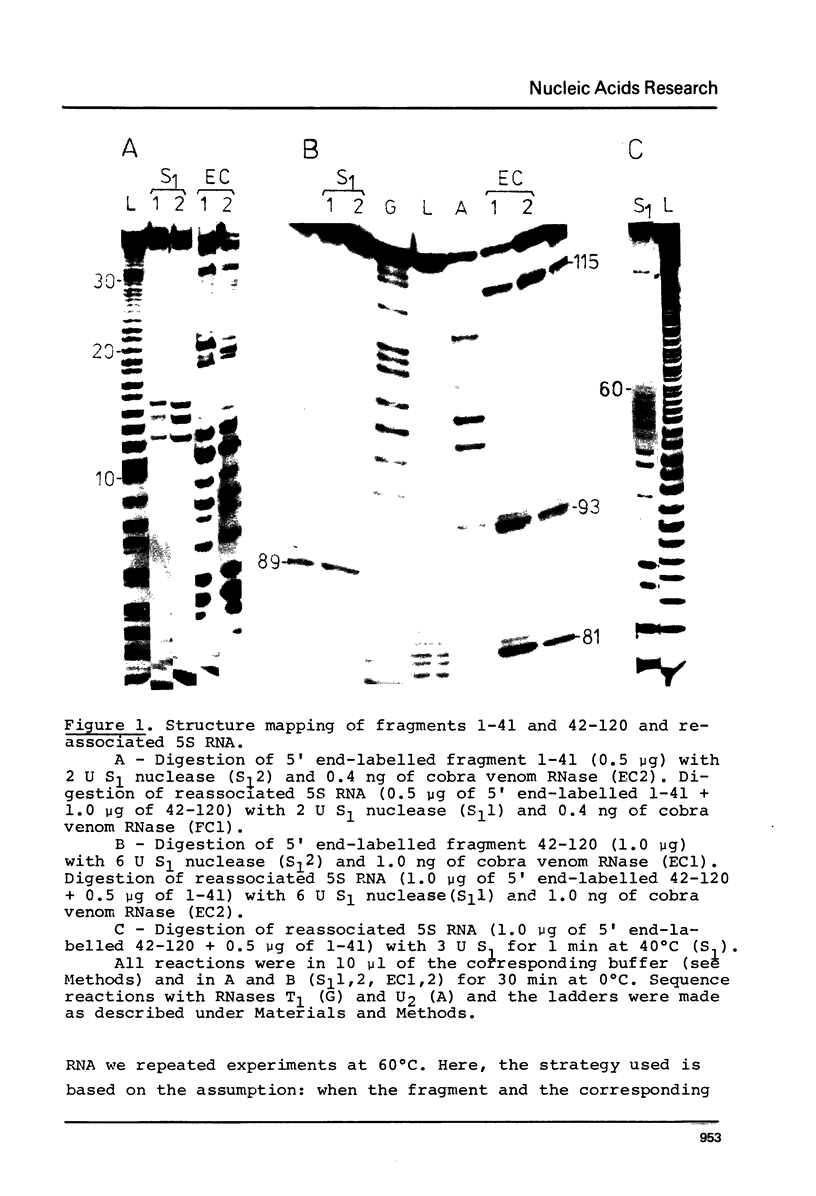
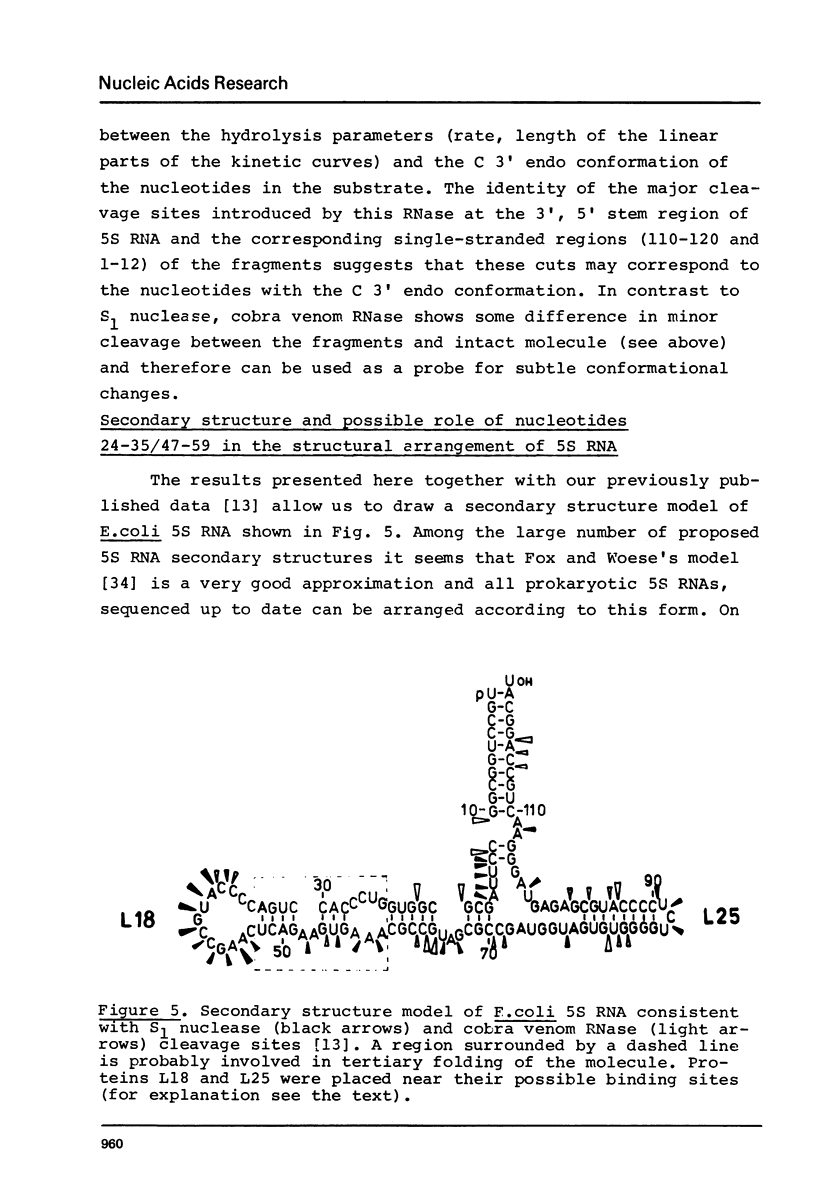
The structure of Escherichia coli 5S RNA fragments 1-41 and 42-120 has been studied by the read-off gel sequencing technique using S1 nuclease and cobra venom RNase as probes. Comparison of the digestion patterns with those of reassociated and intact 5S RNA suggests that the structure of both fragments is very similar to that of the corresponding regions in the intact molecule. Six different fragments obtained by partial digestion with T1 RNase and S1 nuclease have been used for reconstitution of 5S RNA, its certain structural regions and complexes with ribosomal proteins L18 and L25 recognizes the double-helix consisting of nucleotides 79-97 (i.e. prokaryotic stem), whereas a loop-region around position 40 (possible positions 39-47) is involved in the interaction with protein L18.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Bellemare G., Monier R. Selective reaction of glyoxal with guanine residues in native and denatured Escherichia coli 5S RNA. Biochimie. 1973;55(2):135–142. doi: 10.1016/s0300-9084(73)80385-2. [DOI] [PubMed] [Google Scholar]
- Boyle J., Robillard G. T., Kim S. H. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5' end. J Mol Biol. 1980 Jun 5;139(4):601–625. doi: 10.1016/0022-2836(80)90051-0. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Gallinaro H., Lazar E., Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981 Feb 25;9(4):841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Changchien L. M., Craven G. R. Analysis of protein--protein relationships in 30S ribosome assembly intermediates using protection from proteolytic digestion. Biochemistry. 1979 Apr 3;18(7):1275–1281. doi: 10.1021/bi00574a024. [DOI] [PubMed] [Google Scholar]
- Cramer F., Erdmann V. A. Amount of adenine and uracil base pairs in E. coli 23S, 16S and 5S ribosomal RNA. Nature. 1968 Apr 6;218(5136):92–93. doi: 10.1038/218092a0. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Garrett R. A., Wagner R., Feunteun J. A ribonuclease-resistant region of 5S RNA and its relation to the RNA binding sites of proteins L18 and L25. Nucleic Acids Res. 1979 Jun 11;6(7):2453–2470. doi: 10.1093/nar/6.7.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G., Traut R. R. Topography of the C. coli 5S RNA-protein complex as determined by crosslinking with dimethyl suberimidate and dimethyl-3,3'-dithiobispropionimidate. Nucleic Acids Res. 1981 Feb 25;9(4):993–1004. doi: 10.1093/nar/9.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. A slow tritium exchange study of the solution structure of Escherichia coli 5 S ribosomal RNA. J Mol Biol. 1981 Feb 25;146(2):223–239. doi: 10.1016/0022-2836(81)90433-2. [DOI] [PubMed] [Google Scholar]
- Favorova O. O., Fasiolo F., Keith G., Vassilenko S. K., Ebel J. P. Partial digestion of tRNA--aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry. 1981 Feb 17;20(4):1006–1011. doi: 10.1021/bi00507a055. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Fox J. W., Wong K. P. The hydrodynamic shape, conformation, and molecular model of Escherichia coli ribosomal 5 S RNA. J Biol Chem. 1979 Oct 25;254(20):10139–10144. [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Gallinaro H., Lazar E., Jacob M., Branlant C. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 1981 Feb 25;9(4):769–787. doi: 10.1093/nar/9.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machatt M. A., Ebel J. P., Branlant C. The 3'-terminal region of bacterial 23S ribosomal RNA: structure and homology with the 3'-terminal region of eukaryotic 28S rRNA and with chloroplast 4.5s rRNA. Nucleic Acids Res. 1981 Apr 10;9(7):1533–1549. doi: 10.1093/nar/9.7.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarill S. A., Vasilenko S. K., Rait V. K. Osobennosti gidroliza sinteticheskikh poliribonukleotidov éndoribonukleazoi iz iada kobry Naja oxiana. Biokhimiia. 1981 Mar;46(3):408–413. [PubMed] [Google Scholar]
- Maimets T. O., Ustav M. B., Villems R. L., Saarma M. Iu, Lind A. Ia. Sviazyvanie belkov 50S subchastitsy ribosomy Escherichia coli s dvumia krupnymi fragmentami 5S RNK. Mol Biol (Mosk) 1981 May-Jun;15(3):569–574. [PubMed] [Google Scholar]
- Olins P. O., Jones D. S. Nucleotide sequence of Scenedesmus obliquus cytoplasmic initiator tRNA. Nucleic Acids Res. 1980 Feb 25;8(4):715–729. [PMC free article] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shinagawa M., Padmanabhan R. Inhibition of a nuclease contaminant in the commercial preparations of Escherichia coli alkaline phosphatase. Anal Biochem. 1979 Jun;95(2):458–464. doi: 10.1016/0003-2697(79)90756-5. [DOI] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Spierer P., Bogdanov A. A., Zimmermann R. A. Parameters for the interaction of ribosomal proteins L5, L18, and L25 with 5S RNA from Escherichia coli. Biochemistry. 1978 Dec 12;17(25):5394–5398. doi: 10.1021/bi00618a012. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Dyer T. A., Brownlee G. G. An improved direct RNA sequence method; its application to Vicia faba 5.8S ribosomal RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1259–1272. doi: 10.1093/nar/8.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Winter G., Brownlee G. G. 3'End labelling of RNA with 32P suitable for rapid gel sequencing. Nucleic Acids Res. 1978 Sep;5(9):3129–3139. doi: 10.1093/nar/5.9.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Rich A. Stability of the unique anticodon loop conformation of E.coli tRNAfMet. Nucleic Acids Res. 1979 Nov 24;7(6):1457–1467. doi: 10.1093/nar/7.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Woo N. H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Zimmermann J., Erdmann V. A. Identification of Escherichia coli and Bacillus stearothermophilus ribosomal protein binding sites on Escherichia coli 5S RNA. Mol Gen Genet. 1978 Apr 17;160(3):247–257. doi: 10.1007/BF00332968. [DOI] [PubMed] [Google Scholar]