Abstract
Protein phosphorylation continues to be regarded as one of the most important post-translational modifications found in eukaryotes and has been implicated in key roles in the development of a number of human diseases. In order to elucidate roles for the 518 human kinases, phosphorylation has routinely been studied using the budding yeast Saccharomyces cerevisiae as a model system. In recent years, a number of technologies have emerged to globally map phosphorylation in yeast. In this article, we review these technologies and discuss how these phosphorylation mapping efforts have shed light on our understanding of kinase signaling pathways and eukaryotic proteomic networks in general.
Keywords: dynamic networks, kinase, substrate relationships, phosphorylation mapping, proteomic networks, Saccharomyces cerevisiae, scale-free networks
Protein phosphorylation is one of the most widespread types of post-translational modification used in signal transduction. It is involved in the regulation of virtually every basic cellular process and can affect a protein’s activity, localization, stability, conformation, and/or interaction with other proteins. In fact, the reversible nature of protein phosphorylation is one of the many factors that enable a cell to have tunable control of its basic cellular processes. Recent decades have uncovered a wealth of evidence implicating important roles for phosphorylation in human disease as misregulated kinase activity is often associated with a wide variety of disease phenotypes. These disease phenotypes include various leukemias, the development of a number of different types of tumors, vascular diseases, diabetes mellitus and immune/inflammatory disorders [1]. Not surprisingly, recent years has witnessed kinases being avidly pursued as drug targets. Two examples of kinase inhibitor drugs currently on the market include imatinib (Gleevec®), which targets Bcr-abl and is used in the treatment of chronic myeloid leukemia [2], and gefitinib (Iressa®) which targets EGF receptors and is used in the treatment of non-small-cell lung carcinoma [3].
In an effort to better understand the roles of kinases in human disease, much attention has been placed on developing technology to study phosphorylation on a global scale. Given the considerably smaller proteome of yeast compared with that of humans, Saccharomyces cerevisiae has routinely been used as a model system with which to develop such technology. Over the last 5 years, there has been an explosion of proteomic technologies, which have contributed to the large-scale mapping of phosphorylation in the yeast proteome, in terms of identifying both which proteins are phosphorylated and which kinases are responsible for those phosphorylation events. With the ability to elucidate in detail the mechanisms underlying signaling pathways on a global scale, these technologies have led to a deeper understanding of how various signaling pathways are interconnected. In this article, we review these recent yeast technologies and discuss what these efforts to map protein phosphorylation have taught us about proteomic networks in eukaryotes.
In vitro-based technologies for phosphorylation mapping
Early technologies to globally map phosphorylation were aimed at identifying novel kinase–substrate relationships. These technologies took the strategy of increasing the throughput of in vitro kinase assays. Instead of incubating a kinase with a single purified candidate substrate, as was done with single gene studies, pools of thousands of potential substrates were systematically screened using protein microarrays, peptide libraries, or whole cell lysates.
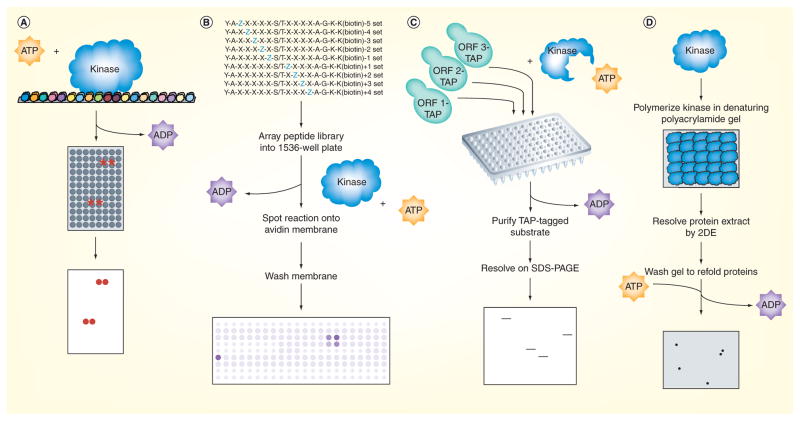
The use of protein microarrays to globally map phosphorylation involves spotting purified proteins at a high spatial density onto a glass slide (Figure 1A). In a study conducted by Ptacek et al., yeast protein microarrays consisting of approximately 4400 of the approximately 6000 proteins, spotted in duplicate, were used as substrates in radioactive kinase assays [4]. The kinase assays were performed by first incubating the protein microarray in kinase buffer in the presence of purified kinase and [γ-33P]-ATP. The protein microarray was then washed to remove the unincorporated radiolabel and exposed to autoradiography film. In vitro substrates of the kinase of interest were identified by quantifying the amount of radiolabel incorporated at each pair of spots relative to the corresponding pair on a control slide performed in parallel in the absence of kinase. Eighty-two unique yeast kinases were assayed for their in vitro targets, resulting in the identification of approximately 4200 phosphorylation events on 1325 different proteins. This study also showed kinases to exhibit a wide range of substrate specificities; 26 kinases were found to target only a single substrate, whereas one kinase was found to target more than 550 substrates. While this range in substrate specificities is likely to be partially due to artifacts arising from the kinase purification process, the range in substrate specificites of kinases may also be reflective of the fact that some kinases play key roles in coordinating multiple signaling pathways, whereas others play more focused roles in one particular signaling pathway.
Figure 1. Phosphorylation mapping on a global scale.
(A) Protein microarrays containing approximately 4400 of the approximately 6000 yeast proteins spotted in duplicate at high spatial density onto glass slides were incubated with kinase and radiolabeled ATP, washed and developed by autoradiography [4]. Positive substrates were determined by quantifying the amount of radiolabel incorporated at each pair of spots relative to the corresponding pair on a control slide assayed in parallel in the absence of kinase. (B) The peptide library used by Mok et al. was a positional-scanning solution-phase library made up of 198 distinct mixtures of biotinylated 16-mers, which each had a central serine or threonine residue as a phosphoacceptor site, and a different amino acid residue fixed at one of the nine positions surrounding the phosphoacceptor site [15]. Kinases were assayed against the peptides in 1536-well plates using radiolabeled ATP, and upon completion of the reaction, the peptides were spotted onto an avidin-impregnated membrane that was then washed and exposed to a phosphoimager. The extent of phosphorylation of each mixture was quantified to generate a motif representing the amino acid preferences targeted by the kinase. (C) Analog-sensitive kinase alleles were generated by mutating the ATP binding pocket; such binding is favored for an ATP analog that cannot be accommodated by wild-type kinases. Lysates prepared individually from strains containing candidate substrates epitope-tagged at their endogenous locus were mixed with the purified kinase and a radiolabeled form of the ATP analog. The candidate substrates were then purified and resolved by gel electrophoresis before being exposed by autoradiography. Positive substrates were determined as those bands appearing on the autoradiograph indicating incorporation of the radiolabeled phosphate. (D) In reverse in-gel kinase assay, the kinase of interest was first polymerized in a denaturing polyacrylamide gel that was subsequently used to resolve a protein extract containing the candidate substrates by 2DE. The gel was then washed thoroughly to remove the SDS detergent and refold the proteins into their native forms, incubated in kinase buffer containing radiolabeled ATP and exposed using autoradiography. Positive substrates were determined as those spots appearing on the radiograph having incorporated the radiolabeled phosphate. The identities of the novel substrates were determined by excising the corresponding spots from a silver-stained gel run in parallel without any kinase and analyzing them by mass spectrometry.
ORF: Open reading frame; TAP: Tandem affinity purification.
By contrast, the use of peptide libraries to globally map phosphorylation takes a more indirect approach to identifying novel kinase–substrate relationships and first involves identifying the consensus phosphorylation motif targeted by the kinase of interest, then systematically scanning the entire eukaryotic proteome for that motif to identify putative phosphorylation sites (Figure 1B). Kinases are known to exhibit preferences for specific amino acids at the positions neighboring the phosphoacceptor site in its targeted substrate. Thus, peptide libraries can facilitate the identification of these amino acid preferences, or the kinase’s consensus phosphorylation motif. To date, a number of approaches using peptide libraries have been described that involve screening either immobilized or solution-phase peptide libraries [5–14]. Mok et al. made use of a positional-scanning solution-phase peptide library to screen the S. cerevisiae kinases for their consensus phosphorylation motifs [15]. The peptide library they used was made up of 198 distinct mixtures of biotinylated 16-mers which each had a central serine or threonine residue as a phosphoacceptor site, and the mixtures were designed such that a different amino acid residue was fixed at each of the nine positions immediately surrounding the phosphoacceptor site. In vitro kinase assays were performed in 1536-well plates using [γ-33P]-ATP. Following the kinase reaction, the peptides were spotted onto an avidin-impregnated membrane, and the membrane was washed to remove the unincorporated radiolabel and exposed to a phosphorimager. The extent of phosphorylation of each peptide mixture was quantified to calculate a postion weight matrix representing the observed amino acid preferences. Motifs were generated for 61 yeast kinases that were then used to bioinformatically generate predictions for novel kinase–substrate relationships. In addition, this study revealed a substrate specificity code in which specific amino acids in the kinase domain were found to confer a specific amino acid preference in its targeted consensus phosphorylation motif.
Budovskaya et al. used a similar motif searching strategy to predict substrates for the yeast PKA, a highly conserved serine (S)–threonine (T) kinase with a consensus phosphorylation motif (RRx[S/T]φ, where φ is any hydrophobic residue) that has been thoroughly studied in both lower and higher eukaryotes [16]. However, their study additionally incorporated conservation analysis into their motif searches and systematically analyzed the evoluationary conservation of PKA phosphorylation sites in the S. cerevisiae proteome across a group of related budding yeasts (five Saccharomyces species and Candida albicans). Using the assumption that functional in vivo phosphorylation sites are more likely to be conserved, they were able to predict 44 candidate PKA phosphorylation sites, five of which fell within already known in vivo PKA substrates.
The use of whole cell lysates to identify novel kinase–substrate relationships has served the basis of two different technologies to globally map phosphorylation. The first involves performing solution kinase assays using analog-sensitive (AS) kinase alleles (Figure 1C). AS kinase alleles are alleles in which the kinase of interest is mutated at a conserved bulky residue in its ATP binding pocket to preferentially bind an ATP analog (N6-benzyl-ATP) that cannot be accommodated by the wild-type form of the kinase. Tagged candidate substrates from pooled extracts are first prepared and mixed with purified AS kinase in the presence of a radiolabeled form of the ATP analog. The candidate substrates are then purified, resolved by SDS-PAGE and exposed to a phosphoimager. Positive substrates are identified as those having incorporated the radiolabeled phosphate. AS kinase allele technology was successfully used by Ubersax et al. to identify 181 in vitro substrates of the Cdk1 from 385 candidate proteins that contained multiple Cdk1 consensus phosphorylation sites [17], and Loog and Morgan to reveal the specificity of approximately 40 of these 181 in vitro substrates for Clb5-Cdk1 over Clb2-Cdk1 [18]. AS kinase allele technology was also used by Dephoure et al. to identify substrates of kinases with no known target sequences by pooling multiple epitope-tagged strains and deconvoluting those pools that showed positive phosphorylation to identify specific substrates; they screened 4250 yeast proteins and identified 24 in vitro substrates of Pho85-Pcl1, which included the known Pho85 in vivo substrate Rvs167 [19].
The second technology based on using whole cell lysates to identify novel kinase–substrate relationships involves performing kinase assays within the context of a polyacrylamide gel, a technology commonly referred to as reverse in-gel kinase assay (RIKA) (Figure 1D). In RIKA, the kinase of interest is polymerized in a denaturing polyacrylamide gel that is subsequently used to resolve a tissue or cell protein extract [20]. Following 2DE, the gel is subjected to a series of washes to remove the SDS detergent and refold the kinase and candidate substrates into their native forms. An in situ kinase reaction is then performed by incubating the gel in kinase buffer containing radiolabeled ATP. Substrates phosphorylated by the kinase of interest are identified using autoradiography, and the identities of the novel substrates are determined by mass spectrometry analysis of the corresponding spots excised from a silver-stained gel run in parallel without kinase. Using this method, Li et al. identified ten novel in vitro substrates for casein kinase 2 and two novel in vitro substrates for PKA [21]. A reciprocal approach has also been developed in which the substrate, instead of the kinase, is immobilized by polymerization in a denaturing polyacrylamide gel that is subsequently used to resolve putative cognate kinases. Lo et al. used a modified version of this reciprocal approach, which utilized fluorography instead of radiography to visualize the phosphorylated substrates, to identify Snf1 as a relevant histone H3-phosphorylating kinase [22].
The advantage of these biochemical approaches is that the entire complement of potential substrates targeted by a specific kinase can be comprehensively and rapidly determined. Thus, these studies have proven to be powerful tools in increasing the number of kinase–substrate relationships known. Notably, each of these biochemical approaches also has unique strengths, and so the data generated from these studies is complementary. The strength of protein microarrays is its throughput with thousands of candidate substrates able to be screened systematically in a spatially addressable manner in single in vitro kinase reaction. On the other hand, the strength of peptide libraries is their ability to additionally provide information for mapping the actual phosphorylation site within the substrate’s amino acid sequence. AS kinase alleles have the strength of being able to have the kinase assay conditions more closely mimic physiological conditions; and RIKA has the strength of being relatively inexpensive in that multiple substrates are assayed against a single kinase at the cost of running a polyacrylamide gel.
The in vitro nature of these technologies however means that there are also limitations to these approaches in accurately predicting novel kinase–substrate relationships. Most purified recombinant protein kinases exhibit promiscuity in substrate specificity. In vivo, part of this promiscuity is regulated by cellular compartment partitioning. By contrast, in vitro, because of the lack of cellular compartment partitioning, kinases are able to phosphorylate proteins that they would never encounter in vivo. Consequently, such in vitro biochemical approaches routinely yield a high number of false positives and require in vivo follow-up experiments to identify those kinase–substrate relationships that are physiologically important. Furthermore, each approach also has other distinct limitations. For example, the use of protein microarrays is biased by candidate substrate proteins being represented in unequal quantities on the chip because of missing clones, variable expression of the clones, misfolded proteins, truncations and the loss of priming post-translational modifications during the purification process. Protein microarrays are further biased by their requirement of a kinase purification step, as many kinases rely on tertiary structure, which may not be preserved during purification, and/or the presence of scaffolding proteins to be efficiently recruited to its target substrate. Indeed, Ptacek et al. were only able to validate 9% of the substrates that were phosphorylated by the protein microarray assay by gel mobility shift assay and/or western blotting using phosphospecific antibodies [4]. Peptide libraries are similarly biased by their requirement for a kinase purification step. However, the use of peptide libraries also suffers from a dependence on motif-scanning algorithms, which are known to be prone to a high rate of false positives. This high rate of false positives is likely to be caused by the fact that removing peptide sequences from the context of the full-length protein often exposes residues normally masked in native conformation. The use of AS alleles is limited by the tolerance of the kinase of interest to mutations as many kinases lose catalytic activity or cellular function with any alterations to its ATP binding pocket [23], and RIKA is reliant upon being able to purify a substantial amount of substrate to near homogeneity and efficient renaturation of the kinase into its active form in the gel.
In vivo-based technologies for phosphorylation mapping
Other technologies used to globally map phosphorylation in yeast by identifying novel kinase–substrate relationships include in vivo-based genetic approaches. In contrast to biochemical approaches that assay for the actual molecular targeting of a substrate by a kinase, genetic approaches more generally assay for the dependence of one gene product on another in producing a specific phenotype. Genetic approaches in turn boast the advantage of providing functional information about the kinase of interest. In these approaches, cells carrying gain- or loss-of-function kinase mutations are screened for suppression or exacerbation phenotypes caused by the overexpression or mutation of another gene (Table 1). Thus, as both gain- and loss-of-function phenotypes are rare (~17 and ~15%, respectively), the main caveat for such a screen is having an assayable phenotype [24,25]. Such approaches nevertheless have successfully led to identification of Kin28 and Bur1 as substrates targeted by the Cdk-activating kinase Cak1; overexpression of CAK1 suppressed both a kin28 and bur1 temperature-sensitive mutant, and further analysis demonstrated that Kin28 and Bur1 are activated by Cak1-dependent phosphorylation [26–28]. Such approaches also led to the identification of 65 candidate substrates for the Cdk Pho85, including the calcineurin-responsive transcription factor Crz1; previous work had shown Pho85 to negatively regulate its targets, and Pho85 candidate substrates were identified by screening an overexpression genomic library for synthetic dosage lethality in a strain lacking Pho85 [29]. Crz1 was later validated to be a bona fide Pho85 substrate by mutational studies showing that Crz1 transcriptional activity and localization were dependent on Pho85-Pho80 kinase activity. Fiedler et al. extended these phosphorylation studies by using this same approach to analyze nearly every yeast protein kinase (121) and phosphatase (38), as well as their regulatory proteins (45 and 39, respectively) [30]. The pairwise genetic interactions of 483 alleles were evaluated, resulting in a total of approximately 100,000 pairwise genetic interaction measurements. Interestingly, in addition to identifying a number of novel positive functional relationships among kinases, phosphatases, and their substrates, the analysis by Fiedler et al. uncovered triple motifs in which three genes exhibited strong interactions, either positive or negative, with one another suggesting the existence of previously unknown pathway connections. One such triple motif was found between the cell cycle (Cak1), the Fus3 mitogen-activated protein kinase pathway, and the regulation of chromatin integrity during transcription by RNA polymerase II.
Table 1.
Mapping phosphorylation by synthetic dosage lethality.
| Kinase | Substrate | Effect |
|---|---|---|
| + | + | No effect |
| − | + | No effect |
| + | − | No effect |
| − | − | Lethality |
Strains containing gain- or loss-of-function kinase mutations were screened for the suppression or exacerbation of phenotypes resulting from the overexpression or mutation of the candidate substrates. Thus, strains containing the mutated kinase or substrate alone were viable, but strains containing both the mutated kinase and substrate were not.
Additional technologies used to globally map phosphorylation in yeast involve mass spectrometry. Mass spectrometry is a technique that enables the quantitative and simultaneous identification of thousands of proteins in a complex biological sample (e.g., whole cell extracts), and in recent years has been adapted to analyze phosphorylation signaling pathways. Early mass spectrometry studies contributed to the global mapping of phosphorylation in yeast by identifying thousands of in vivo phosphorylation sites, which currently continues to serve as an invaluable resource for both motif analysis and bioinformatic kinase–substrate predictions. More recent mass spectrometry studies continued to add to the number of identified in vivo phosphorylation sites, but more importantly, also provided information linking the cognate kinases to these newly identified phosphorylation sites.
Because phosphoproteins are generally of low abundance, a common feature of mass spectrometry phospho-mapping studies is a phosphopeptide enrichment step. Typically, phosphopeptide enrichment strategies involve the isolation of phosphopeptides by solid-phase methods, either through the direct binding of phosphate groups or chemically modified phosphate groups. Regardless of the phosphopeptide enrichment strategy used, whole cell extracts are prepared in the presence of protease and phosphatase inhibitors. Once the extract is prepared, it is treated with a protease, such as trypsin or chemotrypsin, to generate a mixture of small peptides for fragmentation by mass spectrometry. Phosphopeptides are then isolated from other peptides, introduced into the mass spectrometer via liquid chromatography coupled to electrospray ionization or by using MALDI, and identified by comparing the resulting fragmentation spectra obtained from the mass spectrometer to primary sequence databases.
Thus far, three different phosphopeptide enrichment strategies have successfully been used to analyze the yeast proteome: immobilized affinity chromatography (IMAC), strong cation exchange (SCX) and, most recently, hydrophilic interaction chromatography (HILIC). IMAC relies on the affinity of phosphate groups for positively charged transition metals, namely Fe3+ and Ga3+, that are bound to tethered chelating reagents present on solid phase supports. The effectiveness of IMAC to map phosphorylation has been demonstrated by several of the early yeast mass spectrometry studies; IMAC was used by Ficarro et al. to identify 383 phosphorylation sites on 216 proteins [31], Li et al. to identify 2288 phosphorylation sites on 985 proteins [32] and Chi et al. to identify 1252 phosphorylation sites on 629 proteins [33]. By contrast, SCX separates phosphorylated from non-phosphorylated peptides based on the charge difference associated with the negatively charged phosphate group. At low pH, singly phosphorylated peptides display a net charge of +1, which is different to non-phosphorylated peptides, which have a net charge of +2. One issue with SCX is that only singly phosphorylated peptides are enriched; all singly phosphorylated peptides with basic residues or multiply phosphorylated peptides have a net charge other than +1 and therefore will be missed in the analysis. Thus, Gruhler et al. strategically used SCX, in combination with IMAC, to analyze α-factor-arrested yeast cells and identified 729 phosphorylation sites on 503 proteins [34]. HILIC separation depends on the interaction with hydrophilic and charged amino acid residues in the phosphopeptides and has most recently been used as part of a multidimensional chromatography method in a study by Albuquerque et al. [35]. Albuquerque et al. used HILIC, in combination with IMAC, to identify 8764 distinct phosphopeptides from 2278 proteins, 756 of which were doubly phosphorylated. Taken together, these studies have been paramount in the identification of thousands of biologically relevant phosphorylation events, and are an invaluable resource for motif analysis and bioinformatic predictions of novel kinase–substrate relationships.
The more recent mass spectrometry studies have incorporated advances in quantitative mass spectrometry to study the yeast proteome and thus have enabled these in vivo phosphorylations to be linked to their cognate kinases. These studies use stable isotope labeling to compare the phosphoprotein complement between wild-type and kinase-deficient cells or organisms and identify those phosphopeptides whose abundance is dependent upon the kinase activity. One of these more recent mass spectrometry studies was performed by Smolka et al. in which they analyzed the yeast DNA damage checkpoint kinases Mec1, Tel1 and Rad53 using IMAC and SCX [36]. They identified 2653 phosphorylation sites in wild-type and various kinase null alleles and observed 62 phosphorylation sites from 55 proteins that were regulated by the DNA damage checkpoint. Chen et al. extended these studies by conducting similar analyses that instead made use of HILIC and identified 53 additional, along with 34 previously known, targets of Mec1, Tel1, Rad53 and Dun1 [37]. Quantitative mass spectrometry was also used by Bodenmiller et al. to conduct a more comprehensive study of yeast phosphorylation, which analyzed 97 kinase and 27 phosphatase mutants and resulted in the identification of 8814 regulated phosphorylation events that mapped to 1026 proteins [38]. These studies impressively demonstrate the ability of quantitative mass spectrometry to identify not only which proteins are phosphorylated in vivo, but also which kinases are responsible for these phosphorylation events, and thus underscore the emergence of mass spectrometry as the current dominant technology for studying phosphorylation signaling. It is important to point out, however, that some of the regulated phosphorylation events identified in these studies are likely to have been caused by indirect relationships – that is, the abundance of a particular phosphopeptide may change, despite not being a direct molecular target of the perturbed kinase. Thus, orthogonal genetic and/or biochemical analyses are needed to verify these novel kinase–substrate relationships.
Network topology
With thousands of newly identified kinase–substrate relationships, these efforts to globally map phosphorylation in yeast have laid the essential groundwork for assembling a first-generation yeast phosphorylation network, in which each phosphorylation event is represented as a directed arc starting from the kinase and ending at its corresponding substrate. In-depth analysis of this phosphorylation network has taught us much about the complexity with which various signaling pathways are interconnected. However, analyses of network dynamics and evolution has also taught us that this first-generation yeast phosphorylation network is an oversimplified representation of the actual yeast phosphorylation network, which largely ignores the time- and condition-specific nature of phosphorylation events, and thus, much still remains to be learned about the mechanisms underlying basic cellular processes, and in turn human disease in general.
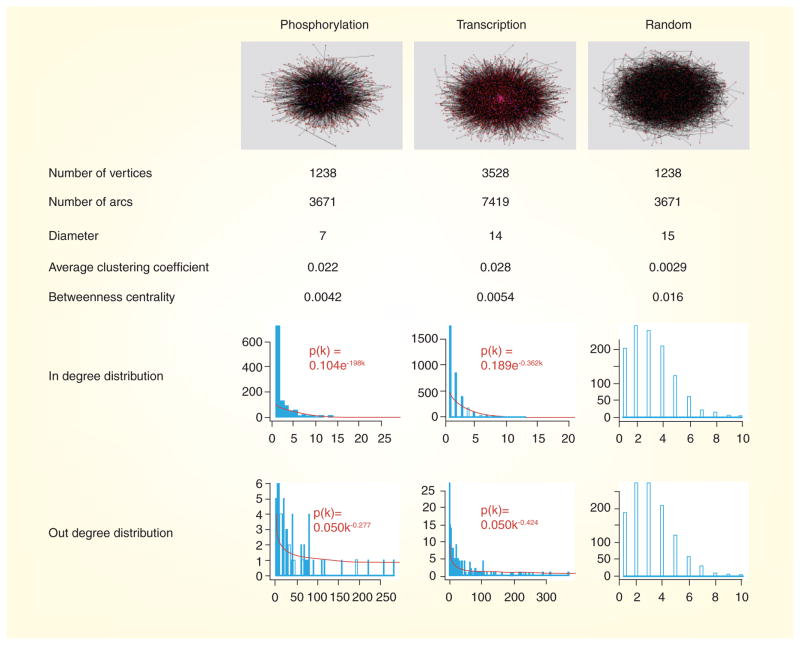
A wide range of statistical methods have been developed for the structural and functional analysis of biological networks, from measuring global topological parameters to characterizing smaller sub-networks, and comparative analysis between various types of networks. To capture the architecture of the phosphorylation network, initial efforts to characterize the yeast phosphorylation network have been aimed at analyzing topological parameters. These topological parameters include basic features, such as degree, distance, diameter, clustering coefficients and betweenness centrality, as well as more complicated features, such as degree distributions and enriched network motifs [39]. Degree characterizes the number of links connected to one node, or how well one node is connected in the network. In directed networks such as the phosphorylation network, the number of arcs that end at one node is termed as ‘indegree’, and the number of arcs that start from one node is termed as ‘out-degree’. Distributions of degrees, or how fast the fractions of nodes that have k connections decay with k, vary in real-world networks and therefore were applied to make classifications among different networks. Distance is defined as the shortest path length between two nodes, and the maximum distance between any two nodes is defined as graph diameter. Diameter characterizes how far apart every node is separated from other nodes, and the clustering coefficient topological parameter measures how likely the nodes in one graph are to group together. Real-world networks, including the yeast phosphorylation network, tend to have higher clustering coefficients than random networks [40]. Finally, betweenness centrality measures the fraction of the shortest paths between all pairs of vertices that pass through one vertex or link and gives an estimation of the traffic load through one node or links assuming the information flows over a network by primarily following the shortest available paths.
Analyses of these topological parameters indicate that the yeast phosphorylation network is not randomly organized. Instead, the yeast phosphorylation network bears a similar structure to the yeast transcription factor-binding network, which was assembled by linking the transcription factors to their binding targets, suggesting that such key regulators probably use similar mechanisms in organizing their targets (Figure 2). First, unlike the well-studied protein–protein interaction network, interactions in both the phosphorylation and transcriptional factor-binding networks are directed from regulators to the regulated targets. Second, most topological parameters are comparable between the two networks with the one exception being that the transcription factor-binding network is larger and sparser, and therefore has a bigger network diameter. By contrast, a control network generated by random permutation of the phosphorylation network while keeping the same number of vertices and arcs has a bigger diameter, smaller average cluster coefficient, and larger average betweenness, suggesting that the nodes in the random network are on average less clustered and have higher traffic load. Third, unlike the bell-shaped degree distributions in the random network, both the phosphorylation and transcription factor-binding networks have an exponential in-degree distribution and a power law out-degree distribution. A network with a power law degree distribution is commonly recognized as a ‘scale-free’ network, which is more likely to contain ‘hub’ nodes with a large number of connections [41]. Previous studies have shown that the structure of scale-free networks is more robust against the loss of random nodes than scaled networks, which may explain why they are preferred in many biological networks [42–46].
Figure 2. Network comparisons.
The yeast phosphorylation network shares similar topological features with the transcription factor-binding network, but differs significantly from a random network permutated from the phosphorylation network. Regulators (kinases or transcription factors) are colored magenta, targets red and regulation interactions black. Both the phosphorylation and transcription factor-binding networks have power law in-degree distributions and exponential out-degree distributions, with many other comparable topological parameters between the two networks.
This figure was generated using the program Pajek [62].
Network motifs
In addition to topological parameters, initial efforts to characterize the yeast phosphorylation network have also been aimed at analyzing the yeast phosphorylation network for smaller common patterns, or motifs, that are present in much higher frequencies relative to random networks. Common motifs characterized thus far include feed forward loop (FFL), dense overlapping regulons (DOR) and single input module (SIM), of which FFL was the only motif not found to be enriched in the current yeast phosphorylation network [39,47]. FFL was originally characterized in the Escherichia coli transcription factor-binding network as an efficient model to carry out stable and precise transcriptional regulations even with transient fluctuations at the regulator levels [48]. FFLs are probably not significantly enriched in the current yeast phosphorylation network because the approach used to assemble the network underestimates the number of phosphorylation events between kinases [4]. However, it is also possible that the lack of FFL in phosphorylation networks reflects the biology of these networks, since phosphorylation does not always directly affect protein expression and instead may drive transient signals that lead to extremely rapid responses in the order of a few minutes.
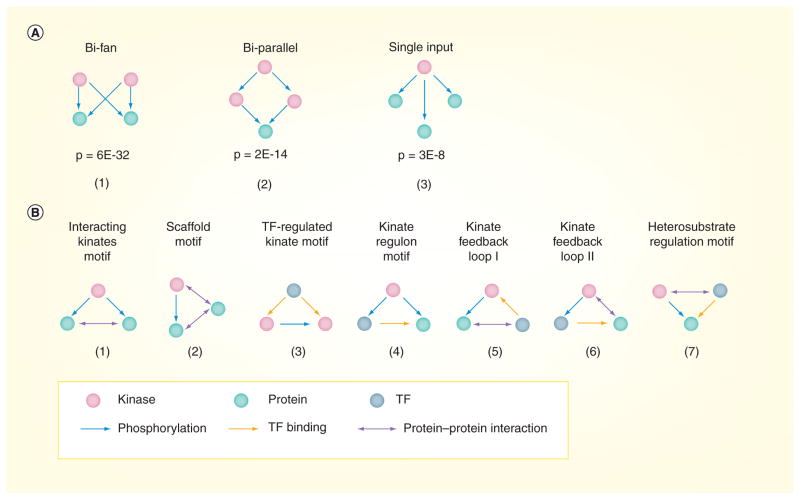
Three network motifs, two DOR motifs and one SIM motif, were enriched in the yeast phosphorylation network relative to random networks (Figure 3A). The first motif that was enriched is the ‘bi-fan motif ’, a simple version of the DOR motif in which two kinases phosphorylate two targets. This bi-fan motif may suggest a way in which a limited number of regulators can be used to precisely control a large number of targets under several different conditions. The second motif that was enriched is the ‘bi-parallel motif ’, a four-element motif in which a regulator controls two other regulators that in turn both regulate one target gene. This motif may indicate potential redundancy. Finally, a SIM motif was also significantly enriched in the yeast phosphorylation network, reflecting the preference of kinases for specific substrates. However, it should be noted that the number of SIMs may be overestimated because of the incompleteness of the phosphorylation dataset and will likely decline as more phosphorylation interactions are identified.
Figure 3. Network motifs in the yeast phosphorylation network.
(A) Three common motifs were enriched in the phosphorylation network: (1) a bi-fan motif in which two regulators bind common targets; (2) a bi-parallel motif in which one regulator controls two other regulators that in turn regulate one target gene; and (3) a single input module motif in which one regulator binds to multiple targets. (B) Seven three-element kinase-centered composite motifs are listed: (1) an interacting kinate motif in which one kinase phosphorylates two interacting substrates; (2) a scaffold motif in which one protein interacts with both a kinase and its substrate; (3) a TF-regulated kinate motif in which one TF regulates the expression of both a kinase and its substrate; (4) a kinate regulon motif in which one kinase phosphorylates both a TF and the target regulated by the TF; (5) a kinate feedback loop I motif in which a kinase phosphorylates a protein that interacts with a TF that in turn regulates the expression of that kinase; (6) a kinate feedback loop II motif in which a kinase phosphorylates a TF whose target physically interacts with the kinase; and (7) a heterosubstrate regulation motif in which an interacting kinase and TF regulate one target together. Motifs (1) – (5) were enriched in the yeast integrated network.
TF: Transcription factor.
The enrichment p-values in (A) are generated by mfinder [63].
Network integration
Efforts to characterize the phosphorylation network have also been aimed at integrating the yeast phosphorylation network with other types of interaction data. These efforts have led to the discovery of composite motifs that contain multiple types of interactions and elements as basic units [4]. Integration of transcription factor-binding, protein–protein interactions and phosphorylation data from yeast has generated a mega-network of over 60,000 interactions. Investigations of this mega-network revealed seven three-element kinase-centered composite motifs (Figure 3B), of which five (motifs 1–5) were over-represented. These composite motifs involve at least one kinase–substrate interaction pair (referred to as ‘kinates’) and one other type of interaction (protein–protein interaction or transcription factor-binding). There recently have been two studies analyzing the crosstalk between the phosphorylation network and protein–protein interaction network. In one study, Breitkreutz et al. used immunoprecipitation and mass spectrometry [49] to identify 1844 protein–protein interactions for 130 protein kinases, 24 lipid and metabolic kinases, 47 kinase regulatory subunits, 38 protein phosphatases, 32 phosphatase regulatory subunits and four metabolic phosphatases [49]. In the second study, Fasolo et al. used protein microarrays to identify 1023 protein–protein interactions for 85 protein kinases, the majority of which were distinct from those previously identified by mass spectrometry [50]. Comparison with various high-throughput phosphorylation mapping studies showed limited overlap between the kinase phosphorylation interactions and kinase protein–protein interactions, suggesting that the two types of interactions are independent. In addition, further investigation of Kss1 showed cellular pathways to be highly intertwined on multiple levels, giving credence that the network integration can both assist in uncovering proteins that are important in multiple types of interactions and provide a more comprehensive view of their cellular functions.
Network dynamics
Efforts have also been made to analyze the network dynamics of the yeast phosphorylation network. All biological networks exhibit complex dynamic behavior, and it is this complex dynamic behavior that enables cells to react to various conditions or cell states. Because phosphorylation events are the result of transient associations between kinases and their targets, phosphorylation networks are far from being static. Thus the components, connections and network architecture in the yeast phosphorylation network all change dramatically in different cellular environments. For example, the activity of the Cdk Cdc28 depends on its association with various types of cyclins, and this quality of Cdc28 is what enables specifically timed phosphorylation events to be executed in a precise order, ensuring a robust mitotic cell cycle [51]. In addition, multiple phosphorylation sites within the same protein may also be regulated differently. The phosphorylation of initiation factor Sld2 by S-phase Cdks (S-Cdks) triggers the protein–protein interactions between Sld2 and Dbp11 [52]. The multiple phosphorylation sites on Sld2 by S-Cdks are regulated differently, with the key site on the threonine 84 residue phosphorylated only when Sld2 is hyperphosphorylated by S-Cdks on other loci, causing a conformation change of Sld2 to reveal the Thr84 site [53]. Unfortunately, the current yeast phoshorylation network neglects these sorts of diversities among the different phosphorylation sites and simplifies all the sites from one protein as one node.
Evolution of yeast phosphorylation modifications
Efforts aimed at analyzing the conservation of network components and connections of the yeast phosphorylation network have also been performed. Such conservation analysis has proven to be useful in mapping novel interactions in other organisms [54]. Core components of a network tend to be conserved, whereas elements at the periphery or false interactions are not. Thus, as alluded to earlier with the Budovskaya et al. study [16], if the kinase and the phosphorylation sites are highly conserved, well-characterized links in model organisms can be mapped in other species to predict novel kinase–substrate interactions.
However, although conservation of network components and connections may have value for mapping conserved interactions and common features among organisms, many regulatory interactions are not conserved. In fact, comparative genomic studies among several yeast species suggest phosphorylation sites to be more prone to mutations than unphosphorylated control sites, probably because of the enrichment of phosphorylation sites in fast evolving disordered regions [55,56]. Furthermore, different types of phosphorylation sites in reality evolve at different rates. For example, it has been suggested that proline-directed phosphorylation sites in MAPK substrates are not conserved at the exact positions, rather the overall charge in the targeted region in the substrate is conserved by having several phosphorylation events occur within a short sequence [57]. By contrast, phosphorylation sites with a local basic amino acid (+3R) are highly conserved, probably because the phosphorylation site will already have a significant impact on the local charge.
Despite the fast evolution of phosphorylation sites, the functions of kinase–substrate phosphorylations may still be well conserved as othologous kinases may still regulate the same targets on different, but related, sites. Interestingly, further studies indicate that kinase–substrate interactions evolve slower than transcription factor–target interactions, but much faster than other types of biological interactions, confirming that the important role of rapid regulatory network rewiring in evolutionary innovations and adaptations [58,59].
Expert commentary
The explosion of new technologies that have enabled phosphorylation mapping on a global scale has furthered our understanding of the role phosphorylation in eukaryotic signaling networks over the last 5 years. We have witnessed the mapping of thousands of novel in vitro phosphorylation sites, the generation of consensus phosphorylation motifs for half of the S. cerevisiae kinases enabling the prediction of thousands of additional kinase–substrate relationships, and identification of thousands of additionally mapped in vivo phosphorylation sites that await being linked to their cognate kinases. Most importantly these technologies have identified 54 novel biologically relevant in vivo kinase–substrate relationships to date.
It should nonetheless be noted that since the activity of the various signaling pathways is dependent on the cellular state of the yeast, kinases exhibit different activities under different conditions. An example already mentioned is the Cdk Cdc28, which when coupled to different cyclins during different points of the cell cycle, targets different sets of substrates. Thus, since current mass spectrometry analyses have been focused on actively growing cells in rich media, phosphorylation events that are triggered only under special conditions, such as heat, nitrogen, salt and low-glucose stress, still need to be mapped.
Furthermore, while we do have a better handle on the scope and complexity of phosphorylation in yeast, we still lack a complete understanding of how the various phosphorylation signaling pathways are interconnected. Crucial to achieving this complete understanding is being able to definitively link a specific phosphorylation event to a particular kinase in vivo. The most significant efforts aimed in this direction thus far include using quantitative mass spectrometry to perform parallel analysis of wild-type and kinase deficient strains, as in the Smolka et al. [36] and Chen et al. [37] studies that looked at kinases involved in the DNA damage checkpoint, as well as the more comprehensive Bodenmiller et al. [38] study that surveyed 97 kinases and 27 phosphatases during log phase growth in synthetic defined media. However, additional studies looking at the remaining kinases during log phase growth in synthetic defined media as well as all the kinases under other growth conditions are undoubtedly necessary. Other efforts aimed at being able to definitively link a specific phosphorylation event to a particular kinase in vivo include using bioinformatics to integrate the yeast proteomic datasets. Sopko et al. [29] demonstrated the power of data integration by filtering the genetic data generated from their Pho85 synthetic dosage lethality screen using the biochemical data generated from Ptacek et al.’s [4] Pho85 protein microarray kinase assay to identify Crz1 as a bona fide Pho85 substrate. Here, one phosphoproteomic dataset was integrated with a complementary phosphoprotoemic dataset; however studies that integrate multiple orthologous datasets, both phosphoproteomic and non-phosphoproteomic, should be able to shed even more light on these phosphorylation signaling pathway relationships.
Five-year view
In the immediate future, we should look to continue efforts aimed at definitively linking a specific phosphorylation event to a particular kinase by mapping more time- and condition-specific phosphorylation events. In vivo technologies, such as quantitative mass spectrometry analysis and genetic screens, will play significant roles here, as will detailed biochemical assays characterizing enzyme kinetics. With dynamic phosphorylation data, we will then be primed to assemble a next-generation yeast phosphorylation network that evolves in real time, with weighted links from kinases to the targeted sites reflecting the specific enzyme kinetics. Furthermore, this next-generation yeast phosphorylation network can be integrated into a more comprehensive multilayered model of the eukaryotic cell, encompassing various types of biological information, such as protein–protein interactions, transcriptional activity and metabolic regulations [60,61].
Future efforts should also be targeted at mapping phosphorylation in both related yeast species and in higher eukaryotes to understand how phosphorylation events have evolved across time. Such information will not only help to explain the phenotypic variations among different yeast species, but also reveal the mechanisms underlying misregulated conserved pathways associated with human disease phenotypes.
Key issues.
Saccharomyces cerevisiae has successfully served as a model system with which to develop new technology aimed at globally mapping phosphorylation in eukaryotes.
A number of technologies have adopted the strategy of increasing the throughput of in vitro kinase assays. These include the use of protein microarrays, peptide libraries, analog-sensitive kinase alleles and polyacrylamide gels in which the kinase or substrate of interest has been polymerized. While these technologies enable the prediction of thousands of candidate substrates for a kinase, a high number of false positives result owing to their in vitro nature.
Genetic-based technologies have also been used, and involve screening cells carrying gain- or loss-of-function kinase mutations for suppression or exacerbation phenotypes caused by the overexpression or mutation of another gene. Such genetic screens have the advantage of only identifying those novel kinase–substrate relationships that are of functional significance.
Other technologies include the use of mass spectrometry. These technologies have the advantage of only providing information about phosphorylation events that occur in vivo, and adaptations to mass spectrometry that allow for quantitative analysis of phosphorylation have proven to greatly aid in uncovering regulated phosphorylation events, in which both an in vivo phosphorylation site and its putative cognate kinase are identified.
The yeast phosphorylation network is significantly different from random networks, but shares similarities with the transcription factor-binding network.
Network motifs, defined as small circuits connecting network nodes, reflect distinct architecture features in various biological networks. A bi-fan motif, a bi-parallel motif and a single-input module motif, were found to be enriched in the yeast phosphorylation network.
The yeast phosphorylation network is highly dynamic; interactions are constantly being rewired in different cell stages and evolve at a rapid rate in the tree of life. Thus, future phosphorylation studies should be targeted at not only definitively linking a phosphorylation event to its cognate kinase in vivo, but also mapping more time- and condition-specific phosphorylation events.
Footnotes
Financial & competing interests disclosure
M Snyder is a consultant/scientific advisor for DNANexus, Personalis and Genapsys. M Snyder is supported by the NIH (Protein Chip grant #48722). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact reprints@expert-reviews.com
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1••.Cohen P. Protein kinases – the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1(4):309–315. doi: 10.1038/nrd773. Details the discovery of phosphorylation and gives an overview of key discoveries that have shaped the phosphorylation field over the years. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Treatment for chronic myelogenous leukemia: the long road to imatinib. J Clin Invest. 2007;117(8):2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gridelli C, De Marinis F, Di Maio M, Cortinovis D, Cappuzzo F, Mok T. Gefitinib as first-line treatment for patients with advanced non-small-cell lung cancer with activating epidermal growth factor receptor mutation: implications for clinical practice and open issues. Lung Cancer. 2011;72(1):3–8. doi: 10.1016/j.lungcan.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 4•.Ptacek J, Devgan G, Michaud G, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438(7068):679–684. doi: 10.1038/nature04187. This global study assayed more than two-thirds of the Saccharomyces cerevisiae kinases against the yeast proteome for their in vitro substrates using protein microarray technology. The dataset generated included approximately 4200 phosphorylation events on 1325 different proteins and has repeatedly served as a valuable resource for the general yeast community. [DOI] [PubMed] [Google Scholar]
- 5.Tegge WJ, Frank R. Analysis of protein kinase substrate specificity by the use of peptide libraries on cellulose paper (SPOT-method) Methods Mol Biol. 1998;87:99–106. doi: 10.1385/0-89603-392-9:99. [DOI] [PubMed] [Google Scholar]
- 6.Wu JJ, Phan H, Lam KS. Comparison of the intrinsic kinase activity and substrate specificity of c-Abl and Bcr–Abl. Bioorg Med Chem Lett. 1998;8(17):2279–2284. doi: 10.1016/s0960-894x(98)00413-2. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez M, Li SS, Harper JW, Songyang Z. An oriented peptide array library (OPAL) strategy to study protein–protein interactions. J Biol Chem. 2004;279(10):8802–8807. doi: 10.1074/jbc.M311886200. [DOI] [PubMed] [Google Scholar]
- 8.Rychlewski L, Kschischo M, Dong L, Schutkowski M, Reimer U. Target specificity analysis of the Abl kinase using peptide microarray data. J Mol Biol. 2004;336(2):307–311. doi: 10.1016/j.jmb.2003.12.052. [DOI] [PubMed] [Google Scholar]
- 9.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4(11):973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 10.Songyang Z, Lu KP, Kwon YT, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16(11):6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem. 1997;272(2):952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 12.Obata T, Yaffe MB, Leparc GG, et al. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275(46):36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 13.Fujii K, Zhu G, Liu Y, et al. Kinase peptide specificity: improved determination and relevance to protein phosphorylation. Proc Natl Acad Sci USA. 2004;101(38):13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutti JE, Jarrell ET, Chang JD, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1(1):27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 15•.Mok J, Kim PM, Lam HY, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3(109):ra12. doi: 10.1126/scisignal.2000482. Details a new peptide array technology in which kinases can be assayed for their consensus phosphorylation motifs without any prior known substrates. Motifs for half of the S. cerevisiae were generated, and will serve as a resource in both the mapping of actual phosphorylation sites in known targets and the integration of multiple datasets aimed at identifying biologically relevant in vivo kinase–substrate relationships. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102(39):13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubersax JA, Woodbury EL, Quang PN, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425(6960):859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 18.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434(7029):104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 19.Dephoure N, Howson RW, Blethrow JD, Shokat KM, O’Shea EK. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci USA. 2005;102(50):17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wooten MW. In-gel kinase assay as a method to identify kinase substrates. Sci STKE. 2002;2002(153):pl15. doi: 10.1126/stke.2002.153.pl15. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Guan B, Srivastava MK, Padmanabhan A, Hampton BS, Bieberich CJ. The reverse in-gel kinase assay to profile physiological kinase substrates. Nat Methods. 2007;4(11):957–962. doi: 10.1038/nmeth1106. [DOI] [PubMed] [Google Scholar]
- 22.Lo WS, Duggan L, Emre NC, et al. Snf1 – a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293(5532):1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Kenski DM, Paulson JL, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2(6):435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 24.Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 25.Sopko R, Huang D, Preston N, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21(3):319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Thuret JY, Valay JG, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86(4):565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 27.Yao S, Prelich G. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol Cell Biol. 2002;22(19):6750–6758. doi: 10.1128/MCB.22.19.6750-6758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gileadi O, Morgan DO. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18(11):6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sopko R, Papp B, Oliver SG, Andrews BJ. Phenotypic activation to discover biological pathways and kinase substrates. Cell Cycle. 2006;5(13):1397–1402. doi: 10.4161/cc.5.13.2922. [DOI] [PubMed] [Google Scholar]
- 30.Fiedler D, Braberg H, Mehta M, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136(5):952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ficarro SB, McCleland ML, Stukenberg PT, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Gerber SA, Rudner AD, et al. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6(3):1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 33.Chi A, Huttenhower C, Geer LY, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104(7):2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruhler A, Olsen JV, Mohammed S, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4(3):310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7(7):1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104(25):10364–10369. doi: 10.1073/pnas.0701622104. Maps in vivo phosphorylation events regulated to DNA damage checkpoint kinases using comparative mass spectrometry analysis of wild-type and null alleles. Such a strategy boasts the advantage of being able to only map those phosphorylation events that are specifically regulated by the kinase of interest and actually occur in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase–substrate network in the DNA damage response. J Biol Chem. 2010;285(17):12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodenmiller B, Wanka S, Kraft C, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3(153):rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21(9):1010–1024. doi: 10.1101/gad.1528707. Summarizes the properties of five biological networks, including the protein–protein interaction, transcriptional, phosphorylation, metabolic and genetic networks. These biological networks are all significantly different from random networks and often exhibit similarities in terms of their structure and organization. [DOI] [PubMed] [Google Scholar]
- 40.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 41.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 42.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 43.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 44.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 45••.Guelzim N, Bottani S, Bourgine P, Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31(1):60–63. doi: 10.1038/ng873. Maps over 1000 protein–protein interactions for 85 yeast protein kinases using protein microarrays. Together with the interactions mapped by mass spectrometry, this work provides an essential dataset that was integrated with the yeast phosphorylation network for a more comprehensive view of the Kss1 filamentous pathway, in addition to many other cellular processes. [DOI] [PubMed] [Google Scholar]
- 46.Tong AH, Lesage G, Bader GD, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 47.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 48.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31(1):64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 49.Breitkreutz A, Choi H, Sharom JR, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fasolo J, Sboner A, Sun MG, et al. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 2011;25(7):767–778. doi: 10.1101/gad.1998811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barik D, Baumann WT, Paul MR, Novak B, Tyson JJ. A model of yeast cell-cycle regulation based on multisite phosphorylation. Mol Syst Biol. 2010;6:405. doi: 10.1038/msb.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006;25(9):1987–1996. doi: 10.1038/sj.emboj.7601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445(7125):328–332. doi: 10.1038/nature05465. This study identified 547 phosphorylation sites for Cdk1. Comparative genomic analysis of these sites suggested that while some sites are well conserved with precise locations in evolution, a larger number of sites shift their positions constantly. [DOI] [PubMed] [Google Scholar]
- 54.Yu H, Luscombe NM, Lu HX, et al. Annotation transfer between genomes: protein–protein interologs and protein– DNA regulogs. Genome Res. 2004;14(6):1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gnad F, Ren S, Cox J, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8(11):R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25(5):193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325(5948):1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beltrao P, Trinidad JC, Fiedler D, et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7(6):e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shou C, Bhardwaj N, Lam HY, et al. Measuring the evolutionary rewiring of biological networks. PLoS Comput Biol. 2011;7(1):e1001050. doi: 10.1371/journal.pcbi.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the Virtual Cell. Trends Cell Biol. 2003;13(11):570–576. doi: 10.1016/j.tcb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Baker ME, Wiley MJ. Multiscale control of flooding and riparian-forest composition in Lower Michigan, USA. Ecology. 2009;90(1):145–159. doi: 10.1890/07-1242.1. [DOI] [PubMed] [Google Scholar]
- 62.Nooy Wd, Mrvar A, Batagelj V. Exploratory Social Network Analysis with Pajek. Cambridge University Press; Cambridge, UK: 2005. [Google Scholar]
- 63.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298(5594):824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]