Abstract
Systems biology represents a paradigm shift from the study of individual genes, proteins or other components to that of the analysis of entire pathways, cellular, developmental, or organismal processes. Large scale studies, primarily initiated in S. cerevisiae, have allowed the identification and characterization of components on an unprecedented level. Large scale interaction, transcription factor binding and phosphorylation data have enabled the elucidation of global regulatory networks. These studies have helped provide an understanding of cellular pathways and processes at a global and systems level.
Keywords: Systems biology, genomics, proteomics, transcriptome, phosphorylome, S.cerevisiae
Introduction
Systems Biology represents the study of entire pathways, processes or even organismal interactions usually at a molecular level. This larger analysis has allowed elucidation of basic principles that may not be apparent at the level of individual components. Such information is expected to be valuable to understanding how an entire system operates.
A Paradigm Shift
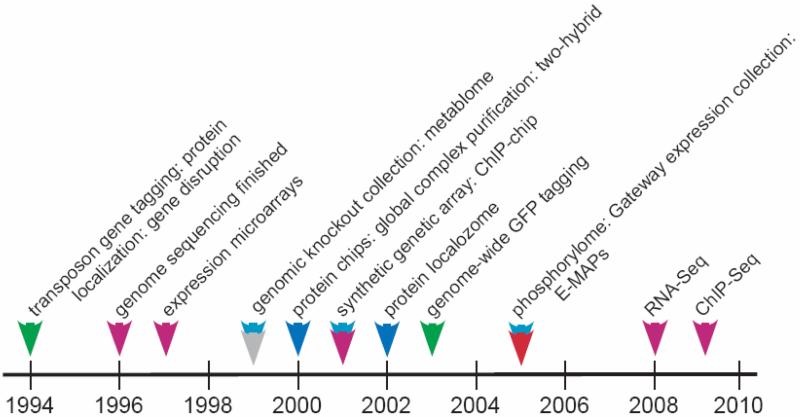
Until the early 1990's nearly all biological research was focused on the detailed study of individual components in which proteins or genes were studied one at a time, a very laborious and inefficient process. This approach shifted in the early and mid 1990's with several types of large-scale studies that allow the analysis of large numbers of components systematically and/ or simultaneously for the first time (Figure1 and Table 1). As the field matured, analysis moved from qualitative to quantitative experiments as a prerequisite to compare and draw meaningful conclusions from large datasets. The first study was our transposon tagging project which tagged a large number of yeast genes allowing for large-scale gene expression, protein localization, and disruption phenotype analyzes as well as identification of unannotated sequences [1]. The second was DNA microarrays, which allowed the monitoring of gene expression of large numbers of, and ultimately all, known and potential genes within an organism [2]. Subsequently, studies to systematically disrupt gene function and biochemically characterize proteins on large-scale emerged (see Table 1). Each of these projects usually required development of a new method (RNAi for multicellular organisms; protein expression collections for biochemical assays).
Figure 1.
Timeline of yeast projects
Table 1.
Large Scale Yeast Projects and Methods
| Project/Method | Goal | Reference |
|---|---|---|
| Transposon tagging | Gene expression; protein localization; disruption phenotypes | [1,28,55] |
| DNA Microarrays | Gene expression | [22] |
| Systematic Knockouts | Phenotypes | [56,57] |
| Protein Localization Using Directed Tagging | Subcellular protein localization | [1,28,55,58] |
| Biochemical protein characterization | Biochemical activities | [59,60] |
| Protein Microarrays | Biochemical activities; protein modifications; interactions | [21,45,60] |
| Mass Spectrometry | Protein profiling; Quantitative protein levels | [61-63] |
| Protein-Protein Interactions Two hybrid | Protein interaction maps; Protein function prediction | [14-16] |
| Protein-Protein Interactions Complex purification | Protein interaction maps; Protein function predication | [19,20,64] |
| Genetic Interactions | Global synthetic screens | [37,39,65,66] |
| Phosphorylation | Kinase substrate map; Large-scale phosophylation mapping | [41,42,46,67] |
| Other post-translational modifications | Mapping of glycosylation; SUMO, acetylation, ubiquitination, | [68-72] |
The vast majority of these projects were first pioneered in yeast because of a) its relatively small genome and numbers of genes (approximately 6000), b) the genome was one of the first sequenced (completed in 1996; [3]) and c) its facile genetics. Table 1 summarizes the different large-scale projects performed for yeast. Projects for characterizing gene expression, protein localization, gene disruption, transcription factor binding, biochemical studies and protein profiling have been carried out providing a wealth of information about each gene, and its corresponding RNA and protein in the cell. Perhaps most importantly, these studies have generated valuable collections of tagged strains and mutants that have proven very valuable to the scientific community. The culture of sharing reagents and information within the yeast community has greatly enhanced the entire field and community. Subsequent to the launch of the different yeast studies, parallel projects have been performed for C. elegans, Drosophila, Arabidopsis and vertebrates and enabled extensive gene characterization in those organisms [4-8]. Many of these have been performed as systematic efforts by consortia e.g. modENCODE to systematically analyze the genomic association of all transcription factors [9,10].
These studies enabled assignment of functions to genes at a rapid pace. For example, when the yeast genome sequenced was completed in 1996 the function of two thirds of the encoded genes was not known [3]. Now 13 years later this figure has been reduced to 10% [11]. Through the integration of different types of large-scale data [12,13], some level of function can be inferred about most yeast genes, although this information is by no means comprehensive, and new function of proteins continue to emerge. As a consequence of these various global studies, we also have a much deeper understanding of entire biological processes and pathways.
Analysis of Networks and Regulatory Circuits
In conjunction with the advent of large-scale characterization of genes and proteins, came the large-scale analysis of their interactions and regulation. To date a number of interaction networks have been generated including protein-protein interaction and transcription factor binding to DNA. These global interaction studies have help elucidate entire pathways as well as regulatory networks controlling biological processes.
Protein-protein interactions
The first of the interaction studies were protein-protein interaction projects. Initial research focused on high throughput yeast two-hybrid studies; these studies were initially incomplete although a more recent study and related protein complementation method generated much larger datasets [14-18]. These studies generally identify direct interactions among protein. Subsequent to the initial two-hybrid studies large-scale studies using affinity purification of tagged proteins and identification of associated proteins by mass spectrometry were performed [19,20]. This approach tends to identify members of a complex, usually at physiological levels in vivo and was a dramatic shift away from assigning protein function to pathways from protein-protein associations rather than classical mutant characterization. Smaller scale in vitro interaction studies using protein microarrays have also been performed [21] and allowed computational detection of noncanonical small molecule binding motifs. In general, the overlap between the approaches is rather modest. This is partly because these approaches themselves are incomplete and are rarely performed to saturation. Moreover, each method has its biases and limitations and this likely also contributes to the observed incomplete overlap.
Transcription Factors
A second major area for the analysis of regulatory networks is the monitoring of gene expression using DNA microarrays. Expression profiling has now been performed for a large number of organisms and now thousands of microarray experiments have been performed for yeast, C. elegans, Drosophila, Arabidopsis, mice and humans [2,22-24]. More recent studies avoid DNA microarrays and involve direct sequencing of RNA (RNA-Seq; [25,26]), which is much more sensitive and accurate due to lack of cross hybridization [27]. These different studies have allowed the profiling of all annotated genes under diverse conditions, different tissues and/or different developmental stages. By correlating gene patterns, it has been possible to determine which genes work together and often identify common DNA sequence motifs in promoter regions.
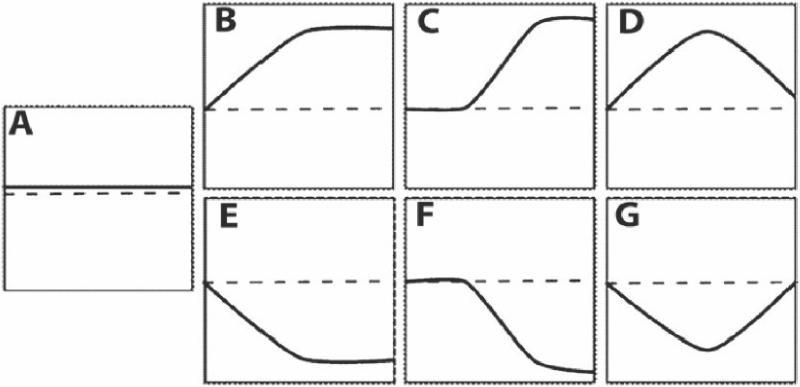
Other large-scale studies have been performed to characterize transcription factor binding sites. We have found that 27% of epitope-tagged proteins localize to the nucleus and the majority of these exhibit punctate patterns of staining of chromosomes using meiotic chromosome spreads [28]. Together with Dr. Patrick Brown's laboratory, we invented the ChIP-chip method for large-scale identification of binding sites throughout the yeast genome [29,30]. This method involves immunoprecipitation of a transcription factor along with its associated DNA, followed by probing of DNA microarrays containing genomic sequences. This method led to several large-scale studies to map DNA binding sites of many factors during either vegetative growth or under different conditions [30-32]. A modification of the ChIP method is ChIP-Seq, which uses high-throughput DNA sequencing as its readout [33,34]. A version for yeast uses bar coding of samples so that large numbers of samples can be analyzed at once [35]. ChIP-chip and ChIP-Seq, when combined with gene expression studies, are a particularly powerful method for dissecting regulatory circuits. To date most of the global DNA binding studies have been performed at a single time point; however recent studies have demonstrated that transcription factor binding sites can have vary temporally presumably reflected the combinatorial effects of multiple binding partners [32]. After an initial challenge to yeast, changes in the transcription factor binding can be graphed over time (Figure 2). Possible outcomes are no change, a rapid increase/ decrease, a lag in change transcription factor binding or a transient binding dissociation of the transcription factor. Adding temporal component to mapping transcription factor binding allows a dynamic picture of cellular response to environmental changes.
Figure 2.
Types of temporal patterns of transcription factors binding to DNA A) Constitutive binding (no change). B) Rapid binding (immediate response and no attenuation of signal). C) Delayed binding (lag in binding and then no change in binding). D) Transient binding (immediate response then return to steady-state levels). E) Rapid dissociation (immediate response and no reassociation). F) Delayed dissociation (lag before complete dissociation). G) Transient dissociation (initial dissociation before reassociation). (Reproduced from [32])
Genetic Interactions
Genetic screens uncover interactions that are not necessarily physical, but rather indicate functional relationships between proteins at the pathway, cellular, or even organismal level. The most common of these are synthetic lethal screen in which strains containing combinations of gene mutations in have a much more severe phenotype (typically death) than that of mutations in individual genes [36]. The first screens systematically examined the effects of combining different null mutations for the “knockout collections” [37]. More recent studies using a collection of DAMP alleles, which contain hypomorphs of essential proteins [38], have revealed synthetic interactions of essential genes. These types of studies have been extended to not only examine negative interactions (synthetic lethal interactions), but also positive interactions (suppression) between mutated genes. The analysis of multiple global screens can also reveal parallel pathways by clustering interactions by the similarities in of genetic interactions (E-MAPs) that would otherwise be missed by analysis of only physical interactions [39,40].
Phosphorylome
A fourth area of global analysis is protein phosphorylation. Initial estimates have suggested that 30% of proteins in yeast and humans are phosphoproteins; although, this estimate is likely to be a significant underestimate. However, from the limited number of recent studies, over 12,000 high confidence phosphorylation sites on ~50% of yeast proteins have been identified in yeast using mass spectrometry [41-44] and the number of phosphorylated sites is expected to grow even higher as additional sites continue to be mapped. A significant challenge has been linking kinases to their substrates: yeast have 122 putative kinases whereas humans have 518 [45]. Thus far several methods have been employed for this. To begin to systematically identify 95 kinase substrates, we used protein microarray containing 4400 yeast proteins to find in vitro targets for 95 yeast kinases [46,47]. From the first 87 kinases, 4200 substrates were identified, an average of 47 targets per kinases. Among the many interesting findings from this study was the observation that closely related kinases such as the three protein kinase A homologs, Tpk1, Tpk2 and Tpk3, each have different targets. Likewise, the cyclin-dependent kinase, Pho85, phosphorylated different targets when associated with different cyclins. A related approach has been to incubate purified kinases with GST fusion proteins or to use modified kinases that only use a modified ATP [48-50]. Another strategy has been to identify differences in phosphorylation patterns in the presence and absence of a protein kinase [51]. Phosphoproteins are purified from cells lacking a protein kinase and identified using mass spectrometry. Phosphorylation is a dynamic post-translational modification crucial for rapid response in cell signaling pathways and mapping these sites trace provide connections between pathways.
Integration of Datasets
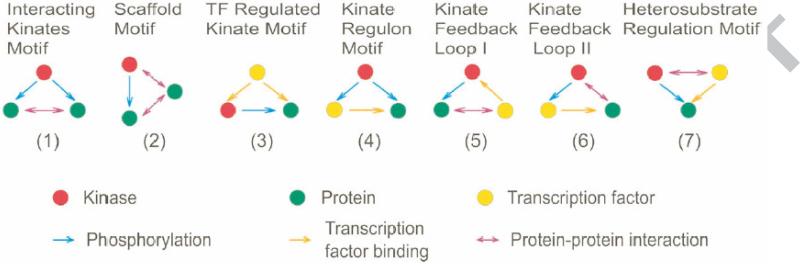
Individually, these studies have provided a wealth of information for specific types of regulation. However, considerable emphasis is now being placed on integration of different data types. Initial studies integrated gene expression and transcription factor binding data to determine the roles of transcription factors (e.g. positive or negative regulation). Moreover, studies have been performed to integrate additional data types such as protein-protein interaction data, phosphorylation data [46], and even metabolite data [52] into large “meta network” or “ridiculograms,” which are visually stunning networks containing extensive interactions. The combined data in these meta networks can be searched for motifs or modules that are overrepresented in the networks. Examples of overrepresented modules are shown in Figure 3, including the scaffold module, in which protein kinase and their substrates each interacting with a third proteins and the “ménage à trois” module in which kinases interact with two interaction partners [46,53].
Figure 3.
Simple motifs/ modules found in complex regulatory networks. Seven different motifs of interactions described between kinates: kinases, their substrates and transcription factors. (Reproduced from [53])
Interdisciplinary Research
System Biology by its very nature operates at the interface of Biology and Computational Biology. Biologists are typically responsible for collecting large data sets often involving many data points and computational biologist are typically responsible for both initial scoring of results and more general analyses. The production of these dataset also provides materials to propose mathematical models for pathways that can identify and or predict key regulators. Furthermore, engineers are having a profound impact on reducing sample size therefore saving reagents and increasing throughput. In some areas of systems biology, chemists and biologist are designing small molecular probes or inhibitors to specific proteins or a class of proteins to probe function.
Regardless of the project, systems biology by necessity is becoming more quantitative, both in data collecting and modeling pathways and biological processes. These models make predictions which in turn can be tested through additional experimentation with respect to data collection as well as with regard to the modeling of biological pathways and processes.
Conclusion
Science today is different from that of 20 years ago. Whole genomes have been sequenced and the opportunity for comprehensive analysis of biological pathways has arrived. Not only is it possible to uncover the biological components and events that occur, but the ability to obtain a quantitative description is now possible. This information will be extremely valuable in medicine for understanding how pathways go awry in human disease. Often common diseases arise from complex interactions between genetics (polymorphisms in coding and non-coding regions) influencing transcription and translation the proteome and the environment. Global analysis of molecular components and states during human diseases is expected to be valuable for diagnostics and monitoring treatments [54].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Goffeau A, et al. Life with 6000 genes. Science. 1996;274:546, 563–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 4.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–30. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 5.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–6. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 6.Reboul J, et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat Genet. 2001;27:332–6. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- 7.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–6. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 8.Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 9.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie KR, Hong EL, Cherry JM. Functional annotations for the Saccharomyces cerevisiae genome: the knowns and the known unknowns. Trends Microbiol. 2009;17:286–94. doi: 10.1016/j.tim.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 13.Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–53. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 14.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, et al. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–7. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–10. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 18.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–5. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 22.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 23.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–92. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 24.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–84. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 25.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross-Macdonald P, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–8. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 29.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–8. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 30.Horak CE, Luscombe NM, Qian J, Bertone P, Piccirrillo S, Gerstein M, Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–33. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, Gerstein MB, Snyder M. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 2009;23:1351–63. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 34.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 35.Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 38.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–8. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 40.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 41.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–7. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 44.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–6. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu H, et al. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–9. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 46.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–84. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 47.Devgan G.a.S., M. Functional Microarrays in Drug Discovery. CRC; 2007. Kinase substrate identification using yeast protein microarrays. pp. 351–360. [Google Scholar]
- 48.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 49.Holt K. Identifying adaptive differences could provide insight. Nature. 2009;458:145. doi: 10.1038/458145b. [DOI] [PubMed] [Google Scholar]
- 50.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 51.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–43. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, G.T., Yip K, Gerstein M, Snyder M. Extensive Metabolite-Protein Interactions Revealed by Large-Scale Systematic Analysesed. 2009. [DOI] [PMC free article] [PubMed]
- 53.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–24. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 54.Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Mol Syst Biol. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar A, et al. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon. Genome Res. 2004;14:1975–86. doi: 10.1101/gr.2875304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 57.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 58.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 59.Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–5. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 60.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–26. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 62.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–4. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 63.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 65.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–81. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 66.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 67.Gnad F, de Godoy LM, Cox J, Neuhauser N, Ren S, Olsen JV, Mann M. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 2009;9:4642–4652. doi: 10.1002/pmic.200900144. [DOI] [PubMed] [Google Scholar]
- 68.Kung LA, Tao SC, Qian J, Smith MG, Snyder M, Zhu H. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol Syst Biol. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–8. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 70.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 71.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 72.Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4:246–54. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]