Abstract
Hydroxylamine oxidation by hydroxylamine oxidoreductase (HAO) is a key step for energy-yielding in support of the growth of ammonia-oxidizing bacteria (AOB). Organohydrazines have been shown to inactivate HAO from Nitrosomonas europaea, and may serve as selective inhibitors to differentiate bacterial from archaeal ammonia oxidation due to the absence of bacterial HAO gene homolog in known ammonia-oxidizing archaea (AOA). In this study, the effects of three organohydrazines on activity, abundance, and composition of AOB and AOA were evaluated in soil microcosms. The results indicate that phenylhydrazine and methylhydrazine at the concentration of 100 μmol g−1 dry weight soil completely suppressed the activity of soil nitrification. Denaturing gradient gel electrophoresis fingerprinting and sequencing analysis of bacterial ammonia monooxygenase subunit A gene (amoA) clearly demonstrated that nitrification activity change is well paralleled with the growth of Nitrosomonas europaea-like AOB in soil microcosms. No significant correlation between AOA community structure and nitrification activity was observed among all treatments during the incubation period, although incomplete inhibition of nitrification activity occurred in 2-hydroxyethylhydrazine-amended soil microcosms. These findings show that the HAO-targeted organohydrazines can effectively inhibit bacterial nitrification in soil, and the mechanism of organohydrazine affecting AOA remains unclear.
Keywords: ammonia-oxidizing archaea, ammonia-oxidizing bacteria, hydroxylamine oxidoreductase, nitrification inhibitor, organohydrazine
Introduction
Nitrification, the consecutive oxidation of ammonia to nitrate via nitrite, is one of the central processes in the global nitrogen cycle (Gruber and Galloway, 2008). It has been long assumed that two groups of bacteria affiliated respectively with β- and γ-Proteobacteria contribute exclusively to the autotrophic ammonia oxidation, the first and rate-limiting step of nitrification. Until recently, however, several cultivation studies (Könneke et al., 2005; De La Torre et al., 2008; Hatzenpichler et al., 2008; Tourna et al., 2011) discovered that some members of archaea can oxidize ammonia autotrophically as well. The numerical dominance of ammonia-oxidizing archaea (AOA) over ammonia-oxidizing bacteria (AOB) appears to be a common feature in a wide variety of environments (Leininger et al., 2006; Agogué et al., 2008; Reed et al., 2010; Wu et al., 2010). The transcriptional activity and exceptionally high abundance of archaeal amoA gene in various habitats (Chen et al., 2008; Tourna et al., 2008; Church et al., 2010) have suggested AOA might play a key role in ammonia oxidation, although there are also some reports suggesting AOB dominate microbial ammonia oxidation in nitrogen-rich environment (Jia and Conrad, 2009; Xia et al., 2011). As such, the relative contributions of AOA and AOB to nitrification activity in natural environment have attracted considerable attentions in recent years (Prosser and Nicol, 2008).
The well-separated phylogeny indicates there are essential differences in physiology, biochemistry, and genetics between AOA and AOB, and results in the distinctly different responses to inhibitive agents (Schauss et al., 2009; Kleineidam et al., 2011). One of the key differences between AOB and AOA is the oxidization mechanism of hydroxylamine, the intermediate of ammonia oxidation. For AOB, ammonia is consecutively oxidized to hydroxylamine by ammonia monooxygenase and to nitrite by hydroxylamine oxidoreductase (HAO; Arp et al., 2007). HAO is the key enzyme for yielding energy to support the growth of AOB during active nitrification. Nevertheless, to our knowledge, there is no homolog of bacterial HAO gene (hao) in the genome of AOA isolates or enrichment cultures (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011). If this is the general feature of AOA, ideally, application of compounds that can inactivate HAO will inhibit the bacterial nitrification without disturbing AOA activity.
Alkyl- and aryl-hydrazine derivatives can irreversibly inactivate bacterial HAO by covalently modifying the active site of enzyme, which was initially observed in the cell-free enzyme extracts (Logan and Hooper, 1995). Activity of Nitrosomonas culture can be also inhibited in vivo by methylhydrazine (Kane and Williamson, 1983). However, the effects of organohydrazines on soil ammonia oxidizers remain unclear. In this study, we examined the effect of three kinds of organohydrazine (phenylhydrazine, PH; hydroxyethylhydrazine, HH; methylhydrazine, MH) on the activity, abundance, and community composition of AOA and AOB in soil microcosms. Our aims were to assess whether organohydrazines can inhibit ammonia oxidation in complex soil, and the potential effects of them on soil AOA and AOB populations.
Materials and Methods
Soil microcosms
Soil (pH 8.0) was collected from the upper 10 cm from an agricultural field of State Key Experimental Station for Ecological Agriculture (35°00′N, 114°24′E) of the Chinese Academy of Sciences, Fengqiu County, Henan province of China. The field information has been detailed previously (Meng et al., 2005). The concentrations of ammonium and nitrate in this soil were and per gram dry weight soil, respectively. The soil sample was sieved (2 mm mesh size), well mixed and stored at 4°C in dark until use. The microcosm consisted of 5 g fresh soil and 50 ml of 1 mM phosphate buffer (pH 8.0) in sterile serum bottles. The incubation of soil microcosm was performed at 30°C in dark as the commonly used protocol for determination of potential nitrification activity (Hart et al., 1994), except that the incubation time was extended to 2 weeks and ammonium sulfate was replaced by ammonium bicarbonate as growth substrate. Soil slurry was sampled at day 0, 1, 7, and 14 immediately after a brief shaking.
Soil microcosms with 11 treatments (each in three replicates) were established including positive control with ammonium amendment (CK-N) and negative control without ammonia amendment (CK-0), as well as three organohydrazines as shown in Table 1. The final concentration of in slurry of ammonium-amended microcosms was soil. HAO-targeted inhibitors, i.e., phenylhydrazine hydrochloride (PH), methylhydrazine sulfate (MH), and 2-hydroxyethylhydrazine (HH; Tokyo Chemical Industry, Japan) were spiked into microcosms at low (1 μmol g−1 soil), medium (10 μmol g−1 soil), and high (100 μmol g−1 soil) concentration. Prior to the addition of ammonium substrate, soil microcosms were pre-conditioned at 30°C in dark for 3 days.
Table 1.
Experimental treatments.
| Treatment | Abbreviation | Treatment |
|
|---|---|---|---|
| addition* | Inhibitor (μmol g−1) | ||
| Positive control | CK-N | + | 0 |
| Negative control | CK-0 | − | 0 |
| PHENYLHYDRAZINE (PH) | |||
| Low | PH-L | + | 1 |
| Medium | PH-M | + | 10 |
| High | PH-H | + | 100 |
| METHYLHYDRAZINE (MH) | |||
| Low | MH-L | + | 1 |
| Medium | MH-M | + | 10 |
| High | MH-H | + | 100 |
| 2-HYDROXYETHYLHYDRAZINE (HH) | |||
| Low | HH-L | + | 1 |
| Medium | HH-M | + | 10 |
| High | HH-H | + | 100 |
* The final concentration of ammonium was 1.4 mg soil in -amended microcosms.
Soil inorganic nitrogen analysis
Soil slurry (5 ml) was centrifuged at 5000 × g for 10 min to separate soil pellet and supernatant. The soil pellets were kept under −20°C until DNA extraction within 1 month, and the supernatants were used for determination of nitrite and nitrate concentration by a SAN++ continuous flow analyzer (Skalar, Breda, The Netherlands). The nitrification activity was calculated on the basis of the production of nitrite and nitrate and expressed on a dry soil weight basis as well.
Soil DNA extraction
Soil DNA was extracted following a CTAB-based bead-beating protocol as described previously with minor modification (Griffiths et al., 2000). Briefly, about 0.5 g of soil pellets was mixed with 0.5 ml modified CTAB buffer containing equal volumes of 10% CTAB in 0.7 M NaCl and 240 mM potassium phosphate buffer (pH8.0), 0.5 g each of 0.5 mm and 0.1 mm silica beads, and 0.5 ml phenol:chloroform:isoamylalcohol (25:24:1). The mixture was then subjected to vigorous shaking on a FastPrep instrument (MP Biomedicals, Solon, OH, USA). After further purification with chloroform:isoamylalcohol (24:1), DNA was precipitated by PEG/NaCl solution and resuspended in 100 μl of TE buffer. The quantity and purity of DNA were determined using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies). No significant PCR inhibition was observed for 10× diluted extracts (data not shown).
Real-time quantitative PCR analysis of amoA gene abundance
Real-time PCR with three replicates for each sample was performed to quantify the copy number of bacterial and archaeal amoA genes using primer sets amoA-1F/amoA-2R-GG (Rotthauwe et al., 1997) and Arch-amoAF/Arch-amoAR (Francis et al., 2005) with a CFX96 Optical Real-Time Detection System (Bio-Rad Laboratories, Inc. Hercules, CA, USA), respectively. Real-time PCR standards were generated using plasmid DNA from representative clones containing bacterial or archaeal amoA gene, and a dilution series of standard template over five orders of magnitude per assay was used to optimize real-time PCR conditions. Blank was always run with water as template instead of soil DNA extract. The 25 μl reaction mixture contained 12.5 μl of SYBR Premix Ex Taq (TaKaRa Biotech, Dalian, China), 0.25 μM of each primer, and 1.0 μl 10× diluted template. Thermal profiles were the same as those described previously (Jia and Conrad, 2009). For bacterial amoA gene, PCR amplification efficiencies of 105.3–118.8% were obtained with R2 values of 0.981–0.996. For archaeal amoA gene quantification, amplification efficiencies of 86.2–90.2% were obtained with R2 values of 0.992–0.997. Specificity of PCR products was always checked by both melting curve analysis and gel electrophoresis.
Denaturing gradient gel electrophoresis analysis of amoA genes
For denaturing gradient gel electrophoresis (DGGE) analysis, PCR amplification of bacterial and archaeal amoA genes was carried out using the same primers as above, except that the forward primer of bacterial amoA was attached with a 27 bp GC-clamp (Nicolaisen and Ramsing, 2002). The PCR reaction was performed in a 25 μl mixture containing 2.5 μl 10× PCR buffer, 0.25 μM of each primer, 200 μM (each) deoxyribonucleoside triphosphate, 1.5 U of Taq DNA polymerase, and 0.5 μl of soil DNA. The PCR was run in a Thermal Cycler Dice (Takara Bio, Shiga, Japan) following the thermal profiles described previously (Nicolaisen and Ramsing, 2002; Francis et al., 2005). PCR products were run on 1.5% agarose gel to check their specificity, and the concentrations were spectrophotometrically measured.
About 150 ng PCR products from each sample was subjected to DGGE analysis as described previously (Wu et al., 2010). Dominant bands in the DGGE fingerprints were excised and re-amplified. For AOA, the purified PCR products were directly sequenced (Invitrogen, Shanghai, China), while sequences of some AOB DGGE bands were obtained by standard cloning technique, as previously described (Jia and Conrad, 2009). The sequences of DGGE bands, as well as their closest relatives obtained by BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi), were aligned using CLUSTAL X 1.83 (Thompson et al., 1997). Phylogenetic trees were constructed using the neighbor-joining method based on the Jukes–Cantor correction with MEGA (Molecular Evolutionary Genetics Analysis) version 4 (Tamura et al., 2007). Bootstrap support was calculated (999 replications). The sequences generated in this study were deposited in the GenBank database under the accession numbers JN408200–JN408210.
Results
Nitrification activity
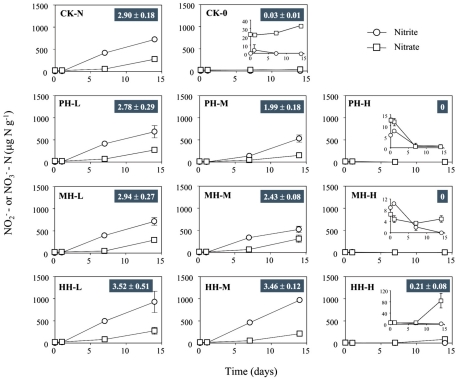
The concentrations of nitrite and nitrate in soil microcosms were determined at day 0, 1, 7, and 14 to assess the kinetics changes of nitrification activity (Figure 1). In the absence of organohydrazines, near-linear accumulation of indicated that active ammonia oxidation occurred in soil microcosms upon ammonium fertilization (CK-N), whereas in soil microcosms without ammonium amendment (CK-0), there was only slight increase in after incubation for 14 days. Therefore, the seemingly high concentration of nitrite and nitrate in CK-N could have resulted from the consumption of ammonium added to soil microcosms rather than released from soil mineralization.
Figure 1.
Concentration of and in soil microcosms incubated for 14 days at various concentrations of phenylhydrazine hydrochloride (PH), methylhydrazine sulfate (MH), and 2-hydroxyethylhydrazine (HH). The designations of PH-L, PH-M, and PH-H represent PH in the microcosms at low (1 μmol g−1 soil), medium (10 μmol g−1 soil), and high (100 μmol g−1 soil) concentration, respectively. Each point represents the mean of triplicate microcosms with some error bars smaller than the symbol size. The inserts show the same data on a smaller scale. The values in gray box refer to nitrification rate during the 14 day incubation (mean ± SE, μ).
Nitrification rates varied greatly among soil microcosms in the presence or absence of organohydrazines (Figure 1). When compared to CK-N treatment, organohydrazines at low concentration of 1 μmol g−1 soil showed no inhibition on nitrification activity, although it seemed that 31 and 16% of nitrification activity was eliminated under elevated concentration of 10 μmol organohydrazines g−1 soil for PH-M and MH-M microcosms, respectively. Complete inhibition was observed at only high concentration of 100 μmol PH or MH g−1 soil, whereas a 7.2% of nitrification activity remained in HH-H microcosm. In microcosms with 1 and 10 μmol HH g−1 soil nitrification was not significantly higher (21.3 and 19.2%, respectively, P > 0.05 with Student’s t-test) than that in CK-N treatment.
AOA and AOB abundance in soil microcosms
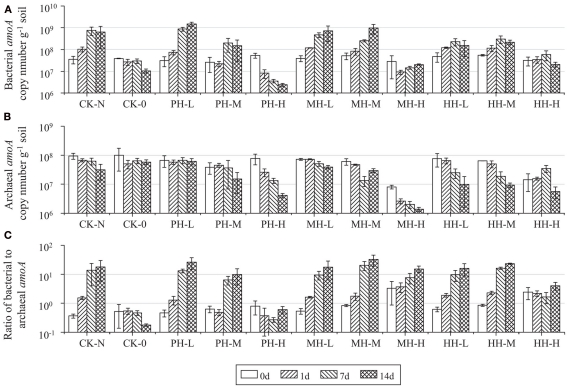
Abundance of soil AOB and AOA was assessed by enumerating the copy number of bacterial and archaeal amoA genes in soil microcosms (Figure 2). For CK-N treatment, the bacterial amoA gene copy number increased from 3.51 × 107 to 6.32 × 108 g−1, while the archaeal amoA gene copy number decreased from 9.41 × 107 to 3.13 × 107 g−1 after incubation for 14 days. Consequently, the ratio of bacterial to archaeal amoA gene copy number increased from 0.3 at zero time to 17.9 after 14 day incubation. Similar results were obtained from soil microcosms in the presence of HAO-targeted inhibitors at low (1 μmol g−1 soil) or medium (10 μmol g−1 soil) concentrations. However, high concentration of PH and MH at 100 μmol g−1 soil appeared to suppress the population sizes of both AOB and AOA. For example, the copy number of bacterial amoA gene in PH-H microcosm was 22 times lower after 14 day incubation than at zero time, while a 5.9-fold decline was observed for archaeal amoA gene abundance in MH-H microcosms. In contrast, the population sizes of AOB and AOA remained relatively stable in soil microcosms with 100 μmol HH g−1 soil.
Figure 2.
Changes of bacterial amoA copy number (A), archaeal amoA copy number (B), and of bacterial to archaeal amoA ratio (C) in the soil microcosm incubated for 14 days. All designations are the same as those in Figure 1.
Changes in the composition of AOB and AOA in soil microcosms
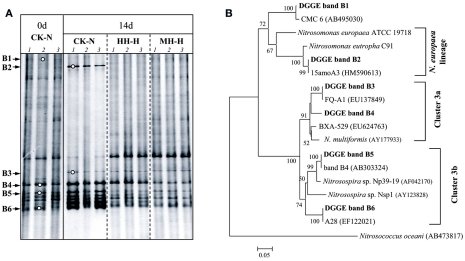
The variation of AOB and AOA community was assessed by DGGE finger printing of respective amoA gene in soil microcosms without organohydrazine addition (CK-N) and with high concentration of organohydrazines treatments (Figures 3 and 4). Dominant DGGE bands were excised and subjected to sequencing analysis to infer the phylogeny of AOB and AOA. Compared to zero time samples, AOB community in HH-H and MH-H microcosms remained largely unchanged, while bacterial amoA gene in PH-H microcosms was not analyzed due to the extremely low PCR amplification efficiency (Figure 3A). By contrast, DGGE fingerprints in CK-N microcosms after incubation for 14 days were dominated by bands B4–B6 within the Nitrosospira cluster 3 and the newly emerged band B2 within the Nitrosomonas europaea lineage (Figure 3B). The DGGE band B2 indeed appeared after incubation for 7 days (data not shown), suggesting that the rapid growth of this N. europaea-like AOB might be responsible for the strong nitrification activity in CK-N microcosms (Figure 1). This was further supported by the absence of DGGE band B2 in MH-H and HH-H microcosms in which nitrification activity was deeply inhibited.
Figure 3.
Denaturing gradient gel electrophoresis fingerprints (A) and phylogenetic tree (B) of bacterial amoA gene in soil microcosms.
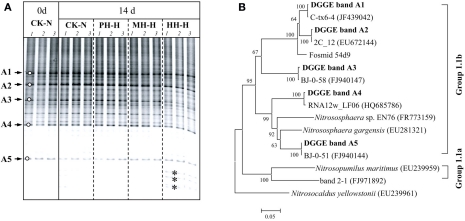
Figure 4.
Denaturing gradient gel electrophoresis fingerprints (A) and phylogenetic tree (B) of archaeal amoA gene in soil microcosms.
DGGE fingerprints of archaeal amoA gene remained largely unchanged in soil microcosms after incubation for 14 days regardless of organohydrazine treatment (Figure 4A). Phylogenetic analysis indicated that the amoA gene sequences of dominant DGGE bands exclusively clustered with the soil Group 1.1b (Figure 4B). It is noteworthy that the intensities of several bands were apparently higher at the bottom of DGGE gel for HH-H microcosms (denoted by asterisks in Figure 4A), in which nitrification activity was not completely inhibited (Figure 1). However, these sequences could not be retrieved despite the huge efforts for PCR amplification of the excised DGGE bands.
Discussion
HAO purified from N. europaea could be inactivated by organohydrazines (Logan and Hooper, 1995). Our results demonstrate that organohydrazines could inhibit bacterial ammonia oxidation in complex soil environment as well. When applied at 100 μmol g−1 soil, the three organohydrazines tested in this study hindered the growth of a N. europaea-like species (band B2 in Figure 3) as well as the rapid accumulation of in respective soil microcosms (Figure 1), suggesting the inhibition by organohydrazines of bacterial ammonia oxidation in soil microcosms.
The response of AOA to organohydrazines is especially interesting because AOA presumably lack the bacterial hao homolog (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011). Organohydrazine inhibition of microbial ammonia oxidation may thus provide clues to the physiological features of this recently discovered archaeal ammonia oxidizer. The AOA population remained highly stable in terms of size and composition (Figures 2 and 4) in the presence of 1 or 10 μmol g−1 soil organohydrazines. Upon the addition of high concentration of PH and MH, however, drastic decline in archaeal amoA abundance rather than minor compositional changes was observed, suggesting the general inhibition of both organohydrazines on AOA. The AOA species presented in this soil mostly fall within Group 1.1b (Figure 4), of which the cultured Candidatus Nitrososphaera viennensis and Ca. N. gargensis have been reported to lack the hao gene (personal communication with Professor Christa Schleper at University of Vienna), similar to those representative cultures in Group 1.1a. As such, assuming that there is no homolog of hao in genome of AOA, we speculate that unknown mechanism might have been involved in the inhibition of AOA by PH and MH. For example, some organohydrazines have shown the toxic effect on bacteria that lack hao (Reinhartz et al., 1987), and their mechanism of action on AOA merits further study.
A minor nitrification activity was observed in HH-H microcosms, accounting for 7.2% of the observed activity in CK-N microcosms without HH (Figure 1). The abundance and composition of both AOB and AOA provided no evidence about the role of these two guilds. However, in HH-H microcosms, the cell-specific nitrification rates (CNR) of AOB (Table A1 in Appendix), which are inferred from nitrification activity and the copy number of amoA gene, fall well within the ranges reported for pure cultures (Belser, 1979; Ward et al., 1989) and environmental samples (; Okano et al., 2004). In contrast, the cell-specific rate of AOA during 7–14 day (; Table A1 in Appendix) was 4.2-fold higher than that of Nitrosopumilus viennensis ( see Appendix) obtained under optimal conditions (Tourna et al., 2011). This implies that AOA alone can not account for the nitrification presented in HH-H microcosms. It is therefore plausible that AOB were not completely inhibited and likely contributed to the nitrification activity, potentially along with AOA in HH-H microcosms.
Nitrification activity is correlated well with AOB rather than AOA community structure in this study, suggesting that AOB dominate nitrification process in soil microcosms tested. This finding is consistent with previous observations on a German agricultural soil microcosm (Jia and Conrad, 2009) and a Chinese paddy soil (Wu et al., 2011). In this study, ammonium was supplied at This relatively high concentration significantly stimulated nitrification activity (Figure 1) and growth of AOB (Figure 2). In the meantime, the abundance and composition of AOA stayed largely unchanged (Figures 2 and 4). The increasing lines of evidence suggest that AOB are favored in high ammonium environment, while AOA may play an important role in ammonium-poor environment (Gubry-Rangin et al., 2010; Zhang et al., 2010). However, there is also evidence that AOA pure culture may grow at the concentration up to 15 mM ammonium (Tourna et al., 2011). Thus we can not exclude the possibility that AOA actively catalyze ammonia oxidation in this study. In combination with inhibition techniques, stable isotope probing (SIP) would provide more robust evidence for relative contributions of AOA and AOB to nitrification activity in complex environments. For example, recent study has demonstrated active transcription of archaeal amoA gene in a German agricultural soil by mRNA–SIP (Pratscher et al., 2011), while the chemolithoautotrophic growth of AOA was shown by DNA–SIP in combination with pyrosequencing (Xia et al., 2011). In addition, the recently cultured Ca. Nitrososphaera viennensis might provide an ideal model for understanding the inhibition mechanism of archaeal nitrifiers by organohydrazine (Tourna et al., 2011).
Taken together, the results of this study demonstrate that bacterial ammonia oxidation in soil can be effectively inhibited by organohydrazines as previously shown in cell extracts. Organohydrazines seem to affect the population sizes of AOA, suggesting their inhibition on archaeal nitrification. Further study is needed to elucidate the HAO-targeted inhibition mechanism of soil nitrification, and to develop new assays for a better understanding of nitrification kinetics in complex soil.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is financially supported by the Knowledge Innovation Programs of the Chinese Academy of Sciences (kzcx2-yw-BR-06) and by the National Natural Science Foundation of China (40901052 and 41090281). We thank Weiwei Xia, Chen Yan, Jing Xu, and Caixia Zhang for technical assistance.
Appendix
Table A1.
Estimated cell-specific nitrification rates of AOA and AOB in soil.
| Group | Nitrification rate | Cell-specific nitrification rate | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AOA* | AOB* | ||||||||
| 0–1 day | 1–7 days | 7–14 days | 0–1 day | 1–7 days | 7–14 days | 0–1 day | 1–7days | 7–14 days | |
| CK-N | 0.22 | 3.10 | 3.13 | 0.19 | 3.42 | 4.76 | 0.56 | 1.3 | 0.81 |
| CK-0 | 0.12 | NA | 0.05 | 0.11 | NA | 0.06 | 0.63 | NA | 0.48 |
| PH-L | 0.18 | 3.16 | 2.83 | 0.21 | 3.61 | 3.16 | 0.61 | 1.2 | 0.43 |
| PH-M | NA | 1.16 | 3.00 | NA | 2.03 | 8.30 | NA | 1.88 | 3.06 |
| PH-H | 0.03 | NA | 0.00 | 0.05 | NA | 0.00 | 0.2 | NA | 0.01 |
| MH-L | 0.60 | 2.83 | 3.37 | 0.59 | 3.27 | 5.33 | 1.36 | 1.73 | 1.02 |
| MH-M | 0.02 | 2.68 | 2.55 | 0.03 | 6.33 | 8.45 | 0.05 | 2.86 | 0.75 |
| MH-H | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| HH-L | 0.88 | 3.71 | 3.74 | 0.88 | 5.89 | 15.31 | 1.86 | 3.77 | 3.49 |
| HH-M | 0.36 | 3.41 | 3.95 | 0.46 | 7.04 | 19.98 | 0.77 | 2.92 | 2.72 |
| HH-H | NA | NA | 0.46 | NA | NA | 1.63 | NA | NA | 2.08 |
* The cell-specific nitrification rate of AOA and AOB was calculated by dividing the production of nitrite and nitrate by the mean population size between two sampling points assuming each AOA and AOB cell contains 1 and 2.5 amoA gene copy, respectively. Cell-specific rate of AOB has a wide range for pure cultures (; Belser, 1979; Ward et al., 1989) and for environmental samples (; Okano et al., 2004). AOA cell-specific rate of Nitrosopumilus viennensis was estimated to be (Tourna et al., 2011).
NA means cell-specific nitrification rate not applicable due to the absence of nitrification activity.
Calculation of the Cell-Specific Nitrification Rate for Nitrosopumilus viennensis Under Optimal Growth Conditions (Tourna et al., 2011)
Tourna et al. (2011) reports that the fastest growth of Nitrosopumilus viennensis through day 6–10 coupled well with significant accumulation of nitrite and depletion of ammonia, suggesting the exponential growth phase of Nitrosopumilus viennensis under optimal conditions. Thus Nitrosopumilus viennensis may show the highest CNR in this period.
Approximately, the net accumulation of nitrate during day 6–10 was:
Δ Nitrate = 890 − 330 = 560 μM
Approximately, the mean cell abundance of Nitrosopumilus viennensis was:
Abundance = (3.4 − 0.4) × 107/2 = 1.5 × 107 cells ml−1
The cell-specific nitrification rate in this period was estimated by:
CNRNitrosopumilus viennensis = Δ nitrate/abundance/time =
References
- Agogué H., Brink M., Dinasquet J., Herndl G. J. (2008). Major gradients in putatively nitrifying and non-nitrifying archaea in the deep North Atlantic. Nature 456, 788–791 10.1038/nature07535 [DOI] [PubMed] [Google Scholar]
- Arp D. J., Chain P. S. G., Klotz M. G. (2007). The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol. 61, 503–528 10.1146/annurev.micro.61.080706.093449 [DOI] [PubMed] [Google Scholar]
- Belser L. W. (1979). Population ecology of nitrifying bacteria. Annu. Rev. Microbiol. 33, 309–333 10.1146/annurev.mi.33.100179.001521 [DOI] [PubMed] [Google Scholar]
- Blainey P. C., Mosier A. C., Potanina A., Francis C. A., Quake S. R. (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE 6, e16626 10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-P., Zhu Y.-G., Xia Y., Shen J.-P., He J.-Z. (2008). Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10, 1978–1987 10.1111/j.1462-2920.2008.01613.x [DOI] [PubMed] [Google Scholar]
- Church M. J., Wai B., Karl D. M., Delong E. F. (2010). Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ. Microbiol. 12, 679–688 10.1111/j.1462-2920.2009.02108.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre J. R., Walker C. B., Ingalls A. E., Könneke M., Stahl D. A. (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10, 810–818 10.1111/j.1462-2920.2007.01506.x [DOI] [PubMed] [Google Scholar]
- Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. I., Whiteley A. S., O’donnell A. G., Bailey M. J. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 10.1128/AEM.66.8.3432-3437.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber N., Galloway J. N. (2008). An Earth-system perspective of the global nitrogen cycle. Nature 451, 293–296 10.1038/nature06592 [DOI] [PubMed] [Google Scholar]
- Gubry-Rangin C., Nicol G. W., Prosser J. I. (2010). Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 74, 566–574 10.1111/j.1574-6941.2010.00971.x [DOI] [PubMed] [Google Scholar]
- Hallam S. J., Konstantinidis K. T., Putnam N., Schleper C., Watanabe Y., Sugahara J., Preston C., De La Torre J., Richardson P. M., Delong E. F. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U.S.A. 103, 18296–18301 10.1073/pnas.0608549103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart S. C., Stark J. M., Davidson E. A., Firestone M. K. (1994). “Nitrogen mineralization, immobilization, and nitrification,” in Methods of Soil Analysis, Part 2, Microbiological and Biochemical Properties, eds Weaver R. W., Angle J. S., Bottemley P. S. (Madison, WI: Soil Science Society of America; ), 985–1018 [Google Scholar]
- Hatzenpichler R., Lebedeva E. V., Spieck E., Stoecker K., Richter A., Daims H., Wagner M. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U.S.A. 105, 2134–2139 10.1073/pnas.0708857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Conrad R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671 10.1111/j.1462-2920.2009.01891.x [DOI] [PubMed] [Google Scholar]
- Kane D. A., Williamson K. J. (1983). Bacterial toxicity and metabolism of hydrazine fuels. Arch. Environ. Contam. Toxicol. 12, 447–453 [Google Scholar]
- Kleineidam K., Kosmrlj K., Kublik S., Palmer I., Pfab H., Ruser R., Fiedler S., Schloter M. (2011). Influence of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) on ammonia-oxidizing bacteria and archaea in rhizosphere and bulk soil. Chemosphere 84, 182–186 10.1016/j.chemosphere.2011.02.086 [DOI] [PubMed] [Google Scholar]
- Könneke M., Bernhard A. E., De La Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G. W., Prosser J. I., Schuster S. C., Schleper C. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- Logan M. S. P., Hooper A. B. (1995). Suicide inactivation of hydroxylamine oxidoreductase of Nitrosomonas europaea by Organohydrazines. Biochemistry 34, 9257–9264 10.1021/bi00028a039 [DOI] [PubMed] [Google Scholar]
- Meng L., Ding W., Cai Z. (2005). Long-term application of organic manure and nitrogen fertilizer on N2O emissions, soil quality and crop production in a sandy loam soil. Soil Biol. Biochem. 37, 2037–2045 10.1016/j.soilbio.2005.03.007 [DOI] [Google Scholar]
- Nicolaisen M. H., Ramsing N. B. (2002). Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50, 189–203 10.1016/S0167-7012(02)00026-X [DOI] [PubMed] [Google Scholar]
- Okano Y., Hristova K. R., Leutenegger C. M., Jackson L. E., Denison R. F., Gebreyesus B., Lebauer D., Scow K. M. (2004). Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70, 1008–1016 10.1128/AEM.70.2.1008-1016.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratscher J., Dumont M. G., Conrad R. (2011). Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U.S.A. 108, 4170–4175 10.1073/pnas.1010981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J. I., Nicol G. W. (2008). Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10, 2931–2941 10.1111/j.1462-2920.2008.01775.x [DOI] [PubMed] [Google Scholar]
- Reed D. W., Smith J. M., Francis C. A., Fujita Y. (2010). Response of ammonia-oxidizing bacterial and archaeal populations to organic nitrogen amendments in low-nutrient groundwater. Appl. Environ. Microbiol. 76, 2517–2523 10.1128/AEM.02436-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhartz A., Lampert I., Herzberg M., Fish F. (1987). A new, short term, sensitive, bacterial assay kit for the detection of toxicants. Environ. Toxicol. Water Qual. 2, 193–206 10.1002/tox.2540020207 [DOI] [Google Scholar]
- Rotthauwe J., Witzel K., Liesack W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss K., Focks A., Leininger S., Kotzerke A., Heuer H., Thiele-Bruhn S., Sharma S., Wilke B.-M., Matthies M., Smalla K., Munch J. C., Amelung W., Kaupenjohann M., Schloter M., Schleper C. (2009). Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ. Microbiol. 11, 446–456 10.1111/j.1462-2920.2008.01783.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M., Freitag T. E., Nicol G. W., Prosser J. I. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10, 1357–1364 10.1111/j.1462-2920.2007.01563.x [DOI] [PubMed] [Google Scholar]
- Tourna M., Stieglmeier M., Spang A., Könneke M., Schintlmeister A., Urich T., Engel M., Schloter M., Wagner M., Richter A., Schleper C. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U.S.A. 108, 8420–8425 10.1073/pnas.1013488108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. B., De La Torre J. R., Klotz M. G., Urakawa H., Pinel N., Arp D. J., Brochier-Armanet C., Chain P. S. G., Chan P. P., Gollabgir A., Hemp J., Hügler M., Karr E. A., Könneke M., Shin M., Lawton T. J., Lowe T., Martens-Habbena W., Sayavedra-Soto L. A., Lang D., Sievert S. M., Rosenzweig A. C., Manning G., Stahl D. A. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U.S.A. 107, 8818–8823 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. B., Glover H. E., Lipschultz F. (1989). Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep Sea Res. A 36, 1031–1051 10.1016/0198-0149(89)90076-9 [DOI] [Google Scholar]
- Wu Y., Lu L., Wang B., Lin X., Zhu J., Cai Z., Yan X., Jia Z. (2011). Long-term field fertilization significantly alters the community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci. Soc. Am. J. 75, 1431–1439 10.2136/sssaj2010.0434 [DOI] [Google Scholar]
- Wu Y., Xiang Y., Wang J., Zhong J., He J., Wu Q. L. (2010). Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ. Microbiol. Rep. 2, 569–576 10.1111/j.1758-2229.2009.00095.x [DOI] [PubMed] [Google Scholar]
- Xia W., Zhang C., Zeng X., Feng Y., Weng J., Lin X., Zhu J., Xiong Z., Xu J., Cai Z., Jia Z. (2011). Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5, 1226–1236 10.1038/ismej.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.-M., Offre P. R., He J.-Z., Verhamme D. T., Nicol G. W., Prosser J. I. (2010). Autotrophic ammonia oxidation by soil thaumarchaea. Proc. Natl. Acad. Sci. U.S.A. 107, 17240–17245 10.1073/pnas.1002506107 [DOI] [PMC free article] [PubMed] [Google Scholar]