Abstract
As the process of top-down mass spectrometry continues to mature, we benchmark the next installment of an improving methodology that incorporates a Tube-Gel Electrophoresis (TGE) device to separate intact proteins by molecular weight. Top-down proteomics is accomplished in a robust fashion to yield the identification of hundreds of unique proteins, many of which correspond to multiple protein forms. The TGE platform separates 0–50 kDa proteins extracted from the yeast proteome into 12 fractions prior to automated nanocapillary LC-MS/MS in technical triplicate. The process may be completed in less than 72 hours. From this study, 530 unique proteins and 1103 distinct protein species were identified and characterized, thus representing the highest coverage to date of the S. cerevisiae proteome using top-down proteomics. The work signifies a significant step in the maturation of proteomics based on direct measurement and fragmentation of intact proteins.
INTRODUCTION
Top-down mass spectrometry (MS), where intact proteins are fragmented directly in a mass spectrometer, has increased in popularity in recent years.1–3 Top-down proteomics is advantageous because different protein forms – including isoforms* and those species arising from post-translational modification (PTM) – can be detected in a general fashion and can produce a faithful representation of protein forms present after maturation and processing within cells. After formalization of the top-down strategy in 1999, studies were focused on the characterization of a single protein (or a small family of protein isoforms) and new developments in instrumentation or software.4–7 However, during the past few years, top-down platforms have been applied to increasingly complex protein mixtures from cells with a steady improvement in proteome coverage.8–11 Top-down mass spectrometry exists in its current state because of instrumentation development and advancements in data processing; however, front-end separations represent a significant bottleneck in development of proteomics based on intact proteins. Reversed-phase liquid chromatography (RPLC), weak-anion exchange, HILIC, and free-flow electrophoresis were previously applied as first-dimension separations prior to top-down MS of whole proteins.9, 12–14 Other front-end separations are also compatible with whole protein MS, including isoelectric focusing, ion-exchange chromatography, and blue-native gels.15–17 The platform presented here demonstrates that proteome fractionation, if well-developed with electrospray-MS of intact proteins in mind, can be an effective first dimension of separation prior to RPLC-MS/MS for protein identification.
Despite the many protein-level separation methods mentioned above, an integrated top-down MS platform capable of reasonable throughput was introduced only a few years ago and was based on Tube-Gel Electrophoresis (TGE).3 A commercial Gel Eluted Liquid Fraction Entrapment Electrophoresis (GELFREE) system prior to LC-MS/MS analysis builds on this approach. The GELFREE system is based on SDS-PAGE separations, but unlike conventional SDS-PAGE, the GELFREE method permits the collection of protein fractions in liquid form rather than having them embedded inside a polyacrylamide matrix at the end of the separation.20,21 Ironically, this system is not entirely free of polyacrylamide gels, but proteins do elute with high recovery from the gel tube into the solution phase for further processing.
This study builds on our previous work which typically achieved 10–20 identifications per LC-MS/MS injection.3 Elsewhere, SILAC has been utilized for top-down MS with 22 proteins identified.11 Another top-down platform utilizing anion exchange and RPLC has identified 174 proteins.10 RPLC-LC-MS/MS has also identified 154 proteins from Salmonella typhimurium.1 Other platforms for intact proteins report 700–1200 protein species detected; however, a principal drawback of intact protein detection without fragmentation is a lack of confident protein identification or characterization.22,23
With a relatively small genome and minimal splicing of RNA transcripts, Saccharomyces cerevisiae has long served as a prototypical organism for proteome platform assessment. Taking 12 fractions in the 5–50 kDa range, we assessed proteome coverage and reproducibility metrics for the TGE-capillary LC-MS/MS system. This platform may be implemented with many LC and MS configurations, which will enable future comparisons and benchmarking for accelerated development and application of top-down proteomics.
EXPERIMENTAL SECTION
Yeast Sample Preparation
Wild-type S. cerevisiae was prepared as previously described.24 Cells were grown to log phase (OD600 = 0.7) in yeast extract - peptone - dextrose (YPD) liquid medium. Cells were harvested by centrifugation at 4000 × g for 5 minutes followed by two water rinses and centrifugation. Alternatively, yeast cells were purchased (Cat. No. YSC2, Sigma-Aldrich, St. Louis, MO). Pelleted cells were treated with Cellytic Y (Sigma-Aldrich), in accordance to the manufacturer’s instructions, to harvest whole-cell lysate. The supernatant was collected and filtered through 8 µm, 0.8 µm, and then 0.2 µm pore filters. Proteins were quantified using the DC protein reagent assay (Bio-Rad, Hercules, CA) prior to analysis by TGE.
TGE Fractionation
For analysis using an 8% tris-tricine GELFREE cartridge (Protein Discovery, Knoxville, TN), 200 µg of extracted yeast protein from Sigma was loaded onto an individual separation channel (see Table 1). The system was run at 85 V. By stopping the separation process through voltage interruption, 12 fractions were collected over a 30 minute period starting at 18 minutes using the following timing: six fractions at 30 seconds each, three fractions at one minute each, and three fractions at two minutes each. For analysis using the Low Mass (12% tris-acetate) GELFREE cartridge (Protein Discovery), 500 µg of in-house-grown yeast protein was loaded onto each column. The system was run at a voltage of 50 V for 70.6 minutes then increased to 100 V for the duration of the experiment, 120.5 minutes total. Fractions were collected starting at 60 minutes and subsequently collected at 63, 66.6, 70.6, 72.9, 75.4, 78.2, 81.5, 85.5, 92.5, 102.5, and 120.5 minutes. The system was operated in accordance with the manufacturer’s instructions.
Table 1.
Information on TGE-LC-MS/MS Runs
| TGE Cartridge |
Amount Loaded |
Data in Figure(s): |
Unique Identifications |
Distinct Forms |
|
|---|---|---|---|---|---|
| Run Ia | 8% T | 200 µg | 3, 5 | 330 | 656 |
| Run IIb | 12% T | 500 µg | 1 – 5 | 202 | 428 |
| Run IIIb | 12% T | 500 µg | 1, 3, 5 | 163 | 395 |
| Total | 530 | 1103 |
Yeast Source:
Sigma
In-house
SDS Page Analytical Slab Gels
A 10 µL aliquot of each 150 µL fraction from the electrophoresis was loaded onto a 4–20% gradient gel (Invitrogen, Carlsbad, CA). Gels were silver stained and scanned using a traditional flat-bed scanner.
LC-MS/MS
After TGE, separated fractions were cleaned up as described previously in order to remove surfactant and salts.25 Briefly, proteins were precipitated at the interface of methanol/water chloroform. The upper methanol/water layer (containing salts and SDS) was removed by pipette. Methanol was then added, and proteins precipitated at the bottom of the chloroform/methanol mixture. After centrifugation, the chloroform/methanol mixture was removed by pipette and precipitated proteins were dried by placing the open-capped tubes in a fume hood. Protein pellets were resuspended in 40 µL of LC buffer A (95% water, 4.8% acetonitrile, and 0.2% formic acid). Ten µL of each fraction were injected in triplicate onto a 2 cm × 150 µm i.d. PLRP-S (5 µm particle size) trapping column (New Objective, Inc., Woburn, MA). Samples were eluted onto a 10 cm × 75 µm i.d. PLRP-S analytical column (New Objective) by use of a vented trap column setup. Samples were separated by use of an Eksigent 1D Plus system (Eksigent Technologies, Dublin, CA). The gradient started at 5% buffer B (95% acetonitrile, 4.8% water, and 0.2% formic acid) and increased to 25% B in 10 minutes, 60% B in 45 minutes, 95% B in 5 minutes, and then decreased to 5% B in 5 minutes followed by 10 minutes of equilibration at 5% B.
Samples were analyzed using a 12 tesla (T) LTQ-FT Ultra mass spectrometer (Thermo Fisher Scientific, San Jose, CA). MS/MS acquisition proceeded by use of one of two fragmentation methods: collision-induced dissociation (CID) in the ion trap or source-induced dissociation (SID) fragmentation in the Q00 region of the LTQ. Generally, CID was used for fractions which contained proteins <15 kDa in molecular weight and SID was used for fractions which contained >15 kDa proteins. For CID, a top two data-dependent method was used with a 25 m/z isolation width and 6–8 microscans at a setting of 170,000 resolving power (at m/z 400) for the MS scan event. For the MS/MS scan events, 6–8 microscans at 85,000 resolving power (at m/z 400) were used. Mass-based dynamic exclusion was enabled and set to a repeat count of 2, an exclusion width of 10 Da, an exclusion duration of 600 s, and a repeat duration of 60 s. For SID, the method was not data-dependent and consisted of two scan events, the first with an SID activation voltage of 15 V and the second with an SID activation voltage of 75 V. The microscans and resolving power settings for MS and MS/MS were the same as the respective CID scan type.
Data Analysis and Software
Individual LC-MS/MS files were processed by use of ProSightPC 2.0 (Service Pack 1) (Thermo Fisher Scientific), with database searches performed on a 120-core computer cluster. Intact and fragment masses were first determined using the THRASH algorithm.26 Intact and fragment masses were compiled into a *.puf (ProSight Upload Format) file, which is a human-readable *.xml file. Each *.puf file was searched separately against forward and scrambled S. cerevisiae databases. The forward database was annotated to include four species for each protein, considering every combination of initial methionine on/off and N-terminal acetylation on/off, giving a total of 28,700 protein sequences. The search scheme used two search modes, “absolute mass” mode and “biomarker” mode.27 Here, “biomarker” is the previously established term for a protease independent search in ProSight that accounts for natural and artificial proteolysis; thus, “biomarker” mode attempts to match the observed intact mass and fragment ions with any theoretical protein subsequence in the database. In “absolute mass” mode, fragments are searched against all intact masses in the specified mass range (for example, a ±200 Da window), and entries with fragmentation profiles consistent with the data are returned as hits. “Absolute mass” mode also can be searched using the “Δm” option, where the mass difference (Δm) between the theoretical and observed intact protein masses determined by the THRASH algorithm was applied to all theoretical fragment ions to compensate for the unexpected/unknown shift in fragment ion masses (e.g., due to a PTM).
The following logic for an iterative “absolute mass” search tree was followed: a search with a ±2.2 Da intact mass tolerance (for example, an observed 18000.0 Da protein was searched against theoretical proteins from the database in a range of 17997.8 – 18002.2 Da), a search with a ±200 Da intact mass tolerance, a search with a ±2000 Da intact mass tolerance, a search with a ±80 kDa intact mass tolerance, and finally a search with a ±80 kDa intact mass tolerance in “Δm” mode. These separate mass ranges were used in searching because ProSight reported the best hit (highest probability that the hit is real) for each individual search. Extending the window after each subsequent search achieved a balance between error-tolerance in searching and the time required to search. So, for the “absolute mass” searches, the narrow search detected all exact mass hits rapidly, and the wide searches detected hits with PTMs that were not annotated in the database as well as those cases with large differences between theoretical and observed intact mass values. In a separate “biomarker” search, a ± 15 ppm mass tolerance was used. Thus for the “biomarker” searches, only exact mass hits were considered. The number of matching fragments for each species was used to calculate the P-score, based on a Poisson model.28 Each file (CID or SID) took approximately 10 minutes to search.
False Discovery Rates (FDRs) were estimated for multiple hypothesis testing using the method of Benjamini and Hochberg as applied by Storey.29,30 For each Poisson-based P-score associated with an experiment, its corresponding q-value, or instantaneous FDR, was calculated.28 A decoy search was performed in all cases against a database of scrambled protein sequences, and the distribution of decoy searches was taken as the null hypothesis, H0. Decoy searches were performed both for absolute mass searches as well as biomarker searches and each was fit to a gamma distribution (Supplementary Figure 1; “absolute mass” mode, shape (k): 9.94 ± 0.04 and rate (θ) of 3.29 ± 0.01; “biomarker” mode, shape (k): 5.07 ± 0.03 and rate (θ) of 1.91 ± 0.01). The scrambled hit distribution was an estimate of the distribution of scores under H0 that the match was due to chance. Thus, the area under the scrambled score distribution to the right of the observed forward score was the probability of getting as good a forward score, or better, by chance. From these posterior probabilities, all data were rank-ordered and q-values are calculated as in Storey.30 The final results were generated using a q-value cut off of 0.05, thus achieving a protein-level FDR of 5%. The visualization of proteins detected used the programs cRAWler and Proteome Display, described previously, to generate 2D maps of detected molecular weight as a function of LC retention time for individual LC-MS/MS injections.3,31
RESULTS AND DISCUSSION
TGE Separations and Metrics
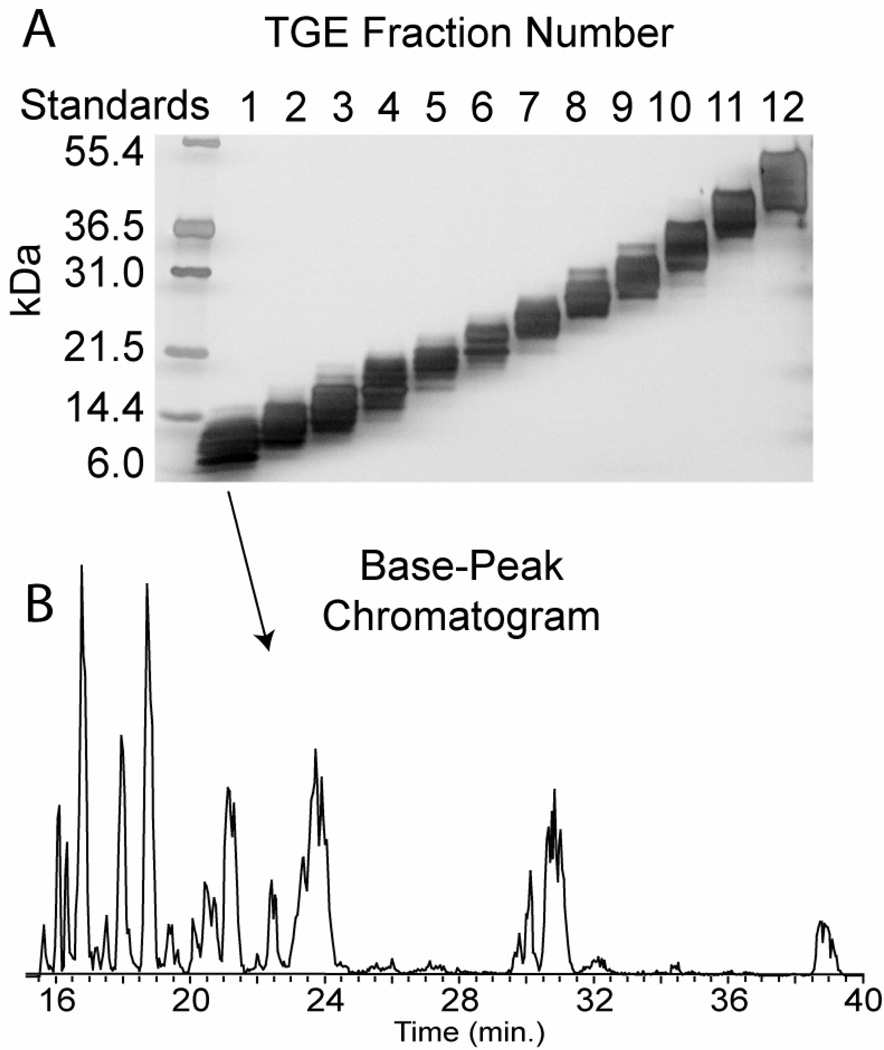
The TGE device separates a complex mixture of cell lysate containing intact proteins into 12 discrete liquid fractions (5–50 kDa) which can be used for downstream applications, including top-down MS. A small portion of each fraction is visualized with SDS-PAGE to assess separation quality and to determine which MS fragmentation options to use (based on molecular weight of the proteins in the fraction). A slab gel image of the 12% tris-acetate separation is shown in Figure 1. Fragmentation by data-dependent CID was found to be most-effective in the first three 12% tris-acetate fractions, but CID was effective in only the first and second fractions from a TGE run using an 8% tris-tricine gel. Also, the 12% tris-acetate cartridge gives a greater number of fractions for which intact proteins are able to be isotopically resolved on-line by a 12 T FT-ICR MS, approximately 30–35 kDa from these data.
Figure 1.
Display of separations prior to MS analysis. In panel A, a slab gel image of TGE fractionation is shown. Fractions are collected in the solution phase and loaded onto a traditional SDS-PAGE slab gel. Slab gel separation is shown after silver-staining of a 12% tris-acetate resolving gel. The imaging of fractions allows assessment of the separation and determination of MS methods for downstream analyses. In panel B, an example LC base-peak chromatogram from TGE fraction 1 is shown. Typical chromatographic peak widths have a full width at half max (FWHM) of 15–45 seconds.
Another important criterion for TGE performance is the number of intact protein identifications that are unique to specific fractions. Table 2 illustrates the number of counts for unique protein identifications, and the number of TGE fractions in which they appear. A majority of identifications (>59% from each run) were found in only one fraction. Ideally, a perfect separation would yield a set of proteins in each fraction, and no proteins from one set would be found in any other fraction. This does not occur for two main reasons: overlap between fractions and proteolysis occurring naturally or during sample preparation (although steps are taken to prevent the latter). Further, silver-staining does not occur in a linear fashion with respect to protein concentration in a gel band, providing a misleading visual assessment of protein amount present.32
Table 2.
Count of fractions in which unique protein identifications (expressed as %) were found
| Count | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run I | 77.5 | 11.4 | 3.6 | 1.8 | 1.5 | 0.6 | 0.9 | 0.3 | 0.6 | 0.3 | 0.6 | 0.9 |
| Run II | 62.7 | 19.8 | 8.5 | 3.5 | 2.5 | 0.5 | 0.5 | 0 | 1 | 0.5 | 0.5 | 0 |
| Run III | 59.5 | 15.7 | 12.8 | 3.3 | 3.3 | 0.7 | 2 | 0 | 0.7 | 0 | 2 | 0 |
LC-MS/MS and Protein Identifications
Data acquisition consists of two types of fragmentation (CID and SID) and database searching consists of two search modes (“absolute mass” and “biomarker”). An average of 60% of CID and 50% of SID spectra resulted in an identification event in these LC-MS/MS analyses. The proportion of identifications for each fragmentation method and search mode is shown Table 3. A ProSight search is created for each detected mass and the associated fragment masses. The result may be repeat identifications which correspond to one unique accession number. Although there are many redundant identification events, each event corresponds to individual MS/MS spectra. The FDR is determined by the q-value cutoff of 0.05 (5% FDR) from the plot in Supplementary Figure 1. For “absolute mass” mode the P-score cutoff that corresponded to a 5% instantaneous FDR was 2 × 10−6, and for “biomarker” mode the P-score cutoff was 9 × 10−8.
Table 3.
Proportion of total identifications from each search mode for fragmentation types
| Absolute mass narrow |
Biomarker narrow |
Absolute mass wide |
Absolute mass wide + delta mass |
|
|---|---|---|---|---|
| CID | 18% | 13% | 68% | 1% |
| NS | 4% | 5% | 91% | 1% |
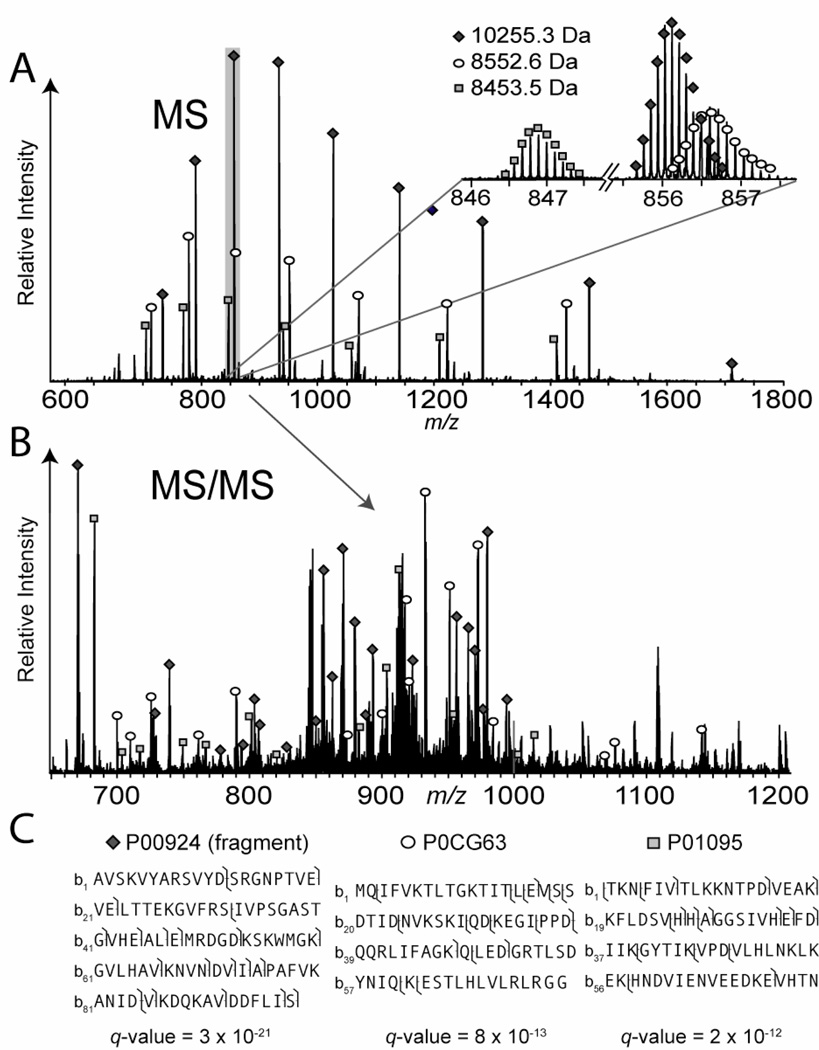
Representative results of an LC separation from TGE fraction 1 are illustrated in a base-peak chromatogram in Figure 1B. Here, typical chromatographic peak widths have a full width at half maximum (FWHM) of 15–45 seconds. While ~30 second peak widths are typical for proteins in the 10–15 kDa mass range, as protein molecular weight approaches 50 kDa, peak widths stretch to ~1–2 minutes or wider (not shown). Wide LC peaks at higher mass contribute to identifications becoming less numerous (see Table 4). In Figure 2, intact proteins, fragments, and protein identifications are shown. Sample complexity is reduced by TGE prior to LC-MS, but two or three proteins can still co-elute (Figure 2A). Three separate proteins were detected within a ~12 m/z window: ubiquitin (P0CG63), enolase 1 (P00924), and protease inhibitors 2 and 1 (P01095). All three proteins were attributed to different gene products with completely dissimilar sequences. MS/MS fragments from the three proteins were detected in the same scan (Figure 2B) and identified with confidence by virtue of the multiplexing search feature in ProSight. (Figure 2C). The relatively wide isolation/activation window of 25 m/z is beneficial here since it allows a greater chance for multiple proteins to be fragmented. Also, the wide window can incorporate multiple protein isoforms/species from one unique protein into the same fragmentation event, thus saving instrument time.
Table 4.
Average unique identifications found per fraction
| Fraction Range (kDa) |
1 6–15 |
2 12–17 |
3 15–18 |
4 17–20 |
5 20–25 |
6 18–23 |
7 20–26 |
8 25–32 |
9 27–32 |
10 29–35 |
11 30–40 |
12 35–55 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Run I | 27 | 29 | 23 | 23 | 35 | 16 | 17 | 24 | 21 | 20 | 16 | 20 |
| Run II | 36 | 32 | 30 | 30 | 25 | 15 | 18 | 15 | 15 | 14 | 8 | 7 |
| Run III | 30 | 27 | 22 | 18 | 19 | 20 | 18 | 10 | 13 | 24 | 11 | 14 |
Figure 2.
Data from an LC-MS/MS injection with an example of triplexed protein identification. In panel A, an example of an MS scan featuring multiple proteins is shown. In A, three proteins are detected which fall in the same m/z window for fragmentation. Panel B features the corresponding MS/MS scan, with markers over different fragment ions which correspond to the three precursor ion species. In C, the identifications for the three proteins are shown along with the fragmentation maps and respective q-values.
For top-down proteomics, increased data acquisition speed for instrumentation and improvements in software have allowed for a greater number of sample analyses (i.e., biological and technical replicates) over a shorter period of time. While a single injection of each sample or TGE fraction is often sufficient to identify >25 unique proteins or even to observe simple biological differences, multiple sample analyses aid in understanding platform reproducibility. Results are presented from three separate TGE-LC-MS/MS runs, each consisting of about 36 LC-MS/MS injections (triplicate analysis of each fraction) with typical completion times of 54 hours. One TGE-LC-MS/MS run is from an 8% tris-tricine cartridge, and two are from 12% tris-acetate cartridges. Table 1 presents the results from one 8% tris-tricine and two 12% tris-acetate analyses (12 fractions each). Integrating results from the three complete runs, 530 unique proteins and 1103 distinct species were identified at 5% FDR. For a 1% FDR, there were 295 unique proteins and 766 distinct species identified. A file containing information for all species is available in the supplementary material. The numbers of unique identifications are fewer than bottom-up proteomics can provide, but 100% sequence coverage is obtained for each protein form identified. This is a feat achieved for very few proteins identified by bottom-up experiments using only one type of enzymatic digestion. To our knowledge, this is the highest number of top-down protein identifications from S. cerevisiae to date.
Identification of Protein Isoforms/Species
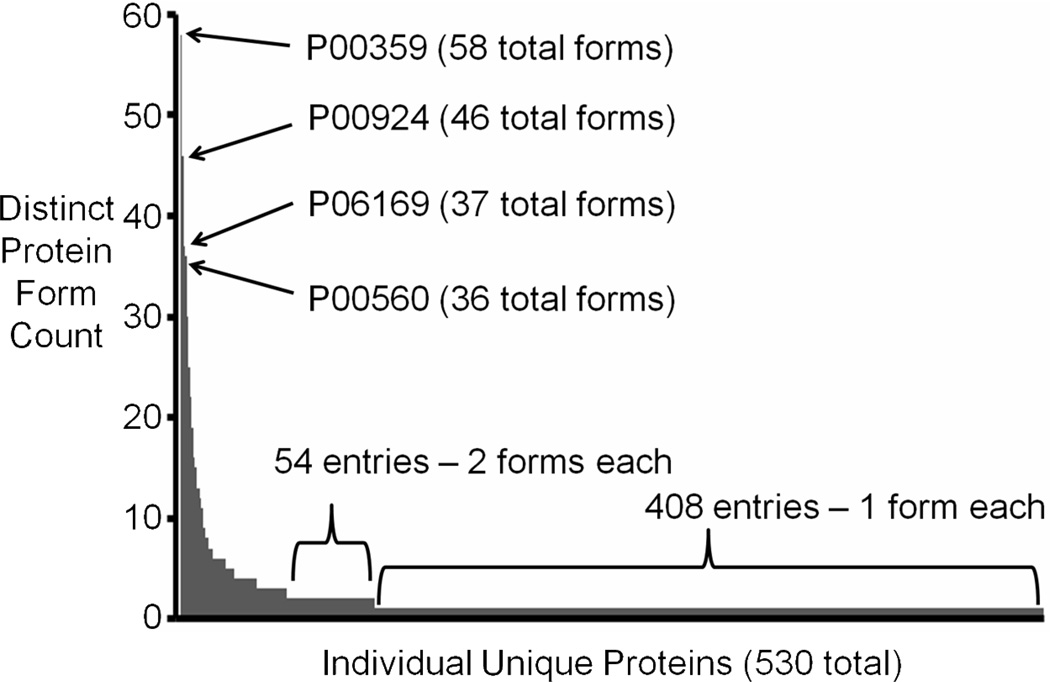
In many cases, one unique protein identification expands into multiple protein species. Protein forms identified from this study are either annotated in the S. cerevisiae database (found via error tolerant “absolute mass” mode searching) or are protein subsequences (found via “biomarker” searching). No artificial adductions (oxidations, salt adducts, etc.) were counted as distinct protein species. For each class of protein form searched in the database, the following percentages were identified relative to the all identification events: unmodified, 55%; N-terminal acetylation, 31%; start methionine on, 4%; N-terminal acetylation with Met on, 4%; and proteolytic fragment, 6%. In Figure 3, a histogram of the number of identified protein forms per unique protein is shown. Fourteen unique proteins exhibit greater than 10 proteolytic species. Another 54 unique proteins have multiple forms, and the remaining 408 proteins have just one form identified.
Figure 3.
Analysis of distinct protein forms. The counts of total protein forms (species arising primarily from proteolysis) per unique protein is shown. Proteolytic forms from abundant primary metabolism proteins comprise highest counts in the plot. A majority of entries have only one form identified.
Reproducibility
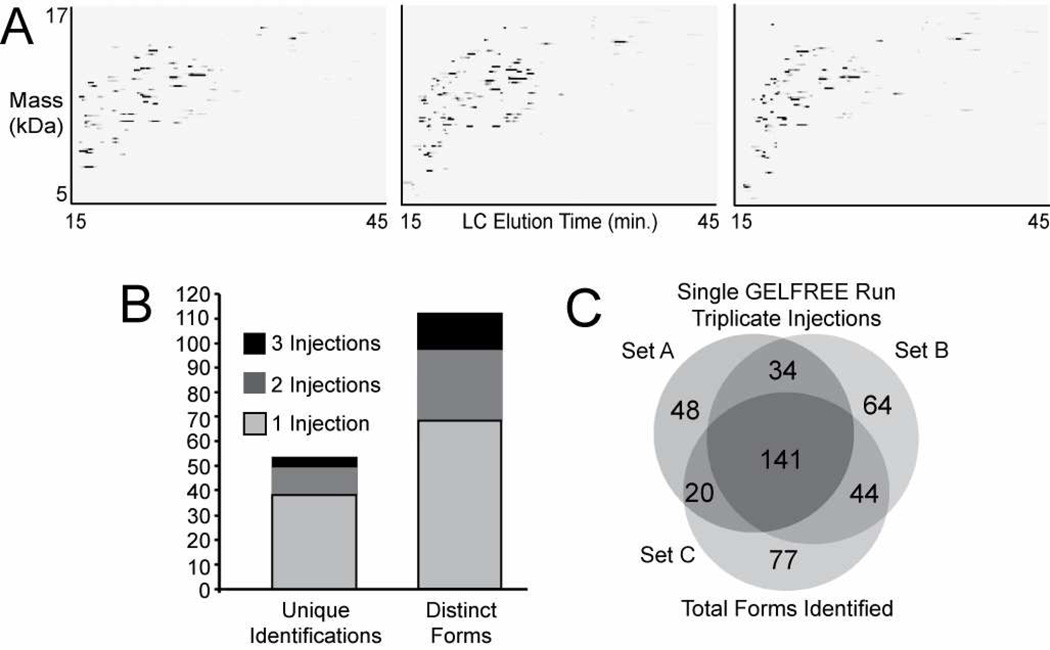
Data on the reproducibility of TGE separations has been published previously.21 As long as identical protein amounts are loaded across each channel, identical molecular weight ranges elute in each fraction, and visible bands are present in the same fraction across all TGE channels. Also, TGE-LC-MS injections have been shown to be highly reproducible.3 Detected proteins are observed with retention times ±15 seconds and peak widths within 10% of each other are observed in technical triplicate injections. In this study we report not only LC-MS reproducibility, but also protein detection and identification reproducibility. LC-MS maps, as shown in Figure 4A, are not only a visual metric of reproducibility, but also illustrate comparison between two or more samples. Similar to 2D gels, such LC-MS maps can be used to compare biological samples, and careful consideration must be made with regard to LC conditions, signal processing, and map generation (e.g., intensities, background, etc.). Examining LC-MS/MS reproducibility, the unique identification and distinct protein isoform counts (Figure 4B) show that additional injections result in additional identifications. In the case of top-down proteomics as implemented here, the second and third injections account for about 25% and 10% of the total fraction identifications, respectively. The overlapping protein forms from replicate injections of TGE fractions 1–12 (Figure 4C) represent the most abundant proteins (about one-third of the distinct protein species are found in all three sets of injections); however, protein species identified in only one set of injections were still confidently identified and may represent proteins of lower abundance.
Figure 4.
Metrics of replicate injections in analysis of the yeast proteome under 50 kDa. In panel A, three images are shown from triplicate LC-MS/MS injections of the same TGE fraction 2. The images show detected intact protein molecular weight as a function of LC retention time. The injections are similar and have common proteins, but each differs slightly. The unique identifications and distinct protein forms from these injections are tallied in B showing the increase in identifications from each subsequent injection. The total distinct protein forms from a single TGE-LC-MS/MS run (triplicate injections for fractions 1–12) are shown in the Venn diagram in C. Each run contributes additional protein forms, but more than 30% of distinct protein forms were identified in all three sets of injections.
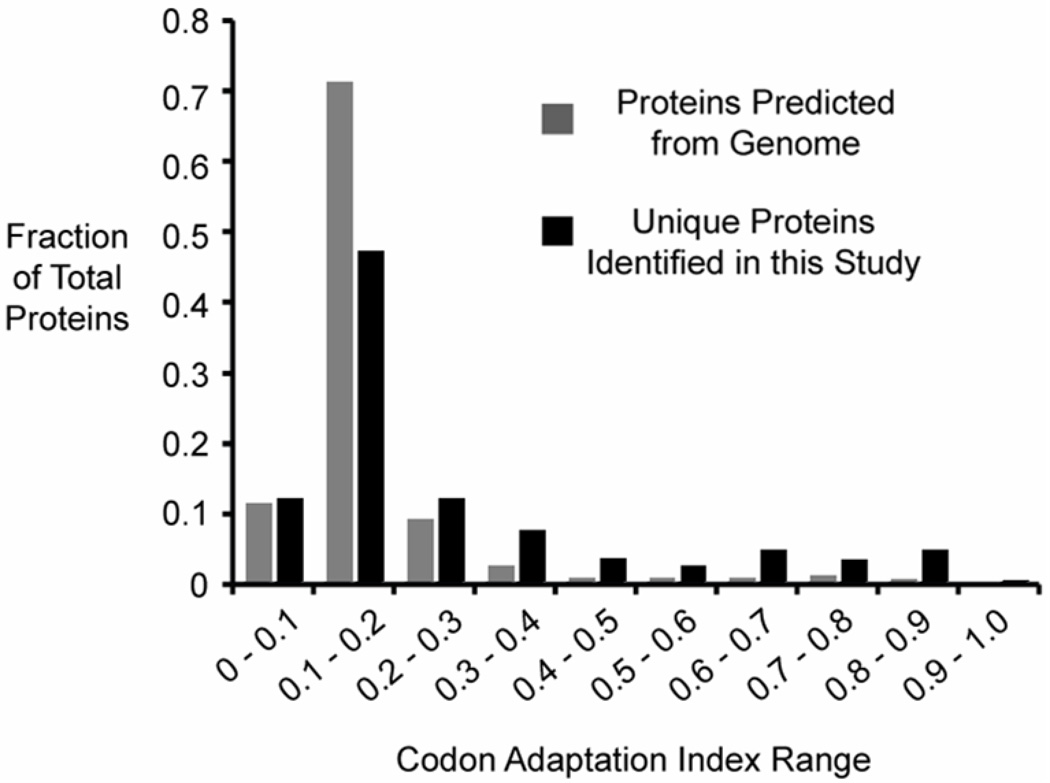
Estimation of Dynamic Range
The Codon Adaption Index (CAI) was devised in 1987 to measure codon bias – the frequency of occurrence for different codons.33 Codon bias can be used to predict the expression level of genes containing a given sequence of codons. The CAI has been used in other large-scale yeast studies to compare observed proteins’ CAI to theoretical CAI distribution.34, 35 The theoretical yeast CAI plot (Figure 5) shows most genes fall in the CAI range of 0.1– 0.2. Figure 5 also shows the CAI values for the 530 identified proteins from this study, and most of the genes fall in the 0.1–0.2 range, similar to previous bottom-up studies.34, 35 When compared to the theoretical CAI distribution, the current study favors higher CAI values (i.e., proteins more likely to be highly expressed). This highlights a classic obstacle in all of proteomics and benchmarks the current state of top-down development: the proteins with the highest expression ratios are most easily detected and identified. In the future, top-down proteomics must utilize sub-cellular fractionation, additional front-end separations, and increased MS sensitivity/speed to enhance proteome coverage. Given the extensive proteolysis in yeast, a lower level in mammalian systems of artifactual creation of abundant species which distribute over a large molecular weight range may ease future top-down proteomic studies.
Figure 5.
Top-down Codon Adaptation Index (CAI) distributions for the theoretical yeast proteome (grey bar) and unique proteins identified from this study (black bar). While both plots show that the highest number of proteins fall in the 0.1–0.2 range, this study disproportionately identified proteins with higher CAI values.
CONCLUSIONS
A top-down proteomic platform capable of hundreds of protein identifications has been presented and benchmarked. In this case, whole yeast cells can be lysed, separated via TGE, and prepared for LC-MS in 12 hours. Triplicate LC-MS/MS injections of 12 TGE fractions can be completed in less than 54 hours, and the data can be searched in as little as 6 hours using a 120-core computer cluster. For top-down proteomics, this platform is unprecedented in terms of speed and scale, with a total of 530 unique proteins and 1103 distinct protein species identified. Further improvement in separations and instrumentation will continue to refine the landscape for whole-protein mass spectrometry.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Kelleher Research Group, in particular former group members Matt Rich for culturing yeast cells, Leonid Zamdborg and Richard LeDuc for helpful discussions, and current group member Nathan Bohn for help with data processing. The authors also thank the staff at Protein Discovery who assisted with the GELFREE separations. The Kelleher Lab is supported by the National Institutes of Health (GM 067193-09). The authors also gratefully acknowledge the generous financial support of the Chicago Biomedical Consortium which is supported by the Searle Funds at The Chicago Community Trust.
Footnotes
Note that IUPAC has recently recommended that the term isoform be used only for related protein forms that arise from gene family members with high sequence identity or other sources of genetic variation such as polymorphism. Events such as alternative splicing or post-translational modification are suggested by IUPAC to be called “protein species”.18,19
REFERENCES
- 1.Tsai YS, Scherl A, Shaw JL, MacKay CL, Shaffer SA, Langridge-Smith PRR, Goodlett DR. J. Am. Soc. Mass. Spectrom. 2009;20:2154–2166. doi: 10.1016/j.jasms.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Ouvry-Patat SA, Torres MP, Gelfand CA, Quek HH, Easterling M, Speir JP, Borchers CH. Methods Mol. Biol. 2009;492:215–231. doi: 10.1007/978-1-59745-493-3_12. [DOI] [PubMed] [Google Scholar]
- 3.Lee JE, Kellie JF, Tran JC, Tipton JD, Catherman AD, Thomas HM, Ahlf DR, Durbin KR, Vellaichamy A, Ntai I, Marshall AG, Kelleher NL. J. Am. Soc. Mass. Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 5.Pesavento JJ, Mizzen CA, Kelleher NL. Biophys. J. 2004;86:421A–421A. [Google Scholar]
- 6.Amunugama R, Hogan JM, Newton KA, McLuckey SA. Anal. Chem. 2004;76:720–727. doi: 10.1021/ac034900k. [DOI] [PubMed] [Google Scholar]
- 7.Macek B, Waanders LF, Olsen JV, Mann M. Mol. Cell. Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Kellie JF, Tran JC, Lee JE, Ahlf DR, Thomas HM, Ntai I, Catherman AD, Durbin KR, Zamdborg L, Vellaichamy A, Thomas PM, Kelleher NL. Mol. Biosyst. 2010;6:1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth MJ, Parks BA, Ferguson JT, Boyne MT, Kelleher NL. Anal. Chem. 2008;80:2857–2866. doi: 10.1021/ac800141g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, Stephenson JL. Anal. Chem. 2008;80:1459–1467. doi: 10.1021/ac7018409. [DOI] [PubMed] [Google Scholar]
- 11.Collier TS, Hawkridge AM, Georgianna DR, Payne GA, Muddiman DC. Anal. Chem. 2008;80:4994–5001. doi: 10.1021/ac800254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waanders LF, Hanke S, Mann M. J. Am. Soc. Mass. Spectrom. 2007;18:2058–2064. doi: 10.1016/j.jasms.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Ouvry-Patat SA, Torres MP, Quek HH, Gelfand CA, O'Mullan P, Nissum M, Schroeder GK, Han J, Elliott M, Dryhurst D, Ausio J, Wolfenden R, Borchers CH. Proteomics. 2008;8:2798–2808. doi: 10.1002/pmic.200800079. [DOI] [PubMed] [Google Scholar]
- 14.Tian ZX, Zhao R, Tolic N, Moore RJ, Stenoien DL, Robinson EW, Smith RD, Pasa-Tolic L. Proteomics. 2010;10:3610–3620. doi: 10.1002/pmic.201000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran JC, Wall MJ, Doucette AA. J. Chromat. B, Analyt. Technol. Biomed. Life Sci. 2009;877:807–813. doi: 10.1016/j.jchromb.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Sokolova L, Wittig I, Barth HD, Schagger H, Brutschy B, Brandt U. Proteomics. 2010;10:1401–1407. doi: 10.1002/pmic.200900756. [DOI] [PubMed] [Google Scholar]
- 17.Tran JC, Doucette AA. J. Proteome Res. 2008;7:1761–1766. doi: 10.1021/pr700677u. [DOI] [PubMed] [Google Scholar]
- 18.Jungblut P, Holzhutter H, Apweiler R, Schluter H. Chem. Cent. J. 2008;2:16. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schluter H, Apweiler R, Holzhutter H-G, Jungblut P. Chem. Cent. J. 2009;3:11. doi: 10.1186/1752-153X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran JC, Doucette AA. Anal. Chem. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 21.Tran JC, Doucette AA. Anal. Chem. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 22.Sharma S, Simpson DC, Tolic N, Jaitly N, Mayampurath AM, Smith RD, Pasa-Tolic L. J. Proteome Res. 2007;6:602–610. doi: 10.1021/pr060354a. [DOI] [PubMed] [Google Scholar]
- 23.Zhou F, Hanson TE, Johnston MV. Anal. Chem. 2007;79:7145–7153. doi: 10.1021/ac071147c. [DOI] [PubMed] [Google Scholar]
- 24.de Godoy LMF, Olsen JV, de Souza GA, Li GQ, Mortensen P, Mann M. Genome Biol. 2006;7:R50–R50.15. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessel D, Flugge UI. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 26.Horn DM, Zubarev RA, McLafferty FW. J. Am. Soc. Mass. Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 27.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL. Nucleic Acids Res. 2007;35:W701–W706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Nat. Biotechnol. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 30.Storey JD. Annals Stat. 2003;31:2013–2035. [Google Scholar]
- 31.Durbin KR, Tran JC, Zamdborg L, Sweet SM, Catherman AD, Lee JE, Li M, Kellie JF, Kelleher NL. Proteomics. 2010;10:3589–3597. doi: 10.1002/pmic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White IR, Pickford R, Wood J, Skehel JM, Gangadharan B, Cutler P. Electrophoresis. 2004;25:3048–3054. doi: 10.1002/elps.200405947. [DOI] [PubMed] [Google Scholar]
- 33.Sharp PM, Li WH. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn MP, Wolters D, Yates JR. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 35.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. Mol. Cell. Bio. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.