ABSTRACT
Hydrothermal chimneys are a globally dispersed habitat on the seafloor associated with mid-ocean ridge (MOR) spreading centers. Active, hot, venting sulfide structures from MORs have been examined for microbial diversity and ecology since their discovery in the mid-1970s, and recent work has also begun to explore the microbiology of inactive sulfides—structures that persist for decades to millennia and form moderate to massive deposits at and below the seafloor. Here we used tag pyrosequencing of the V6 region of the 16S rRNA and full-length 16S rRNA sequencing on inactive hydrothermal sulfide chimney samples from 9°N on the East Pacific Rise to learn their bacterial composition, metabolic potential, and succession from venting to nonventing (inactive) regimes. Alpha-, beta-, delta-, and gammaproteobacteria and members of the phylum Bacteroidetes dominate all inactive sulfides. Greater than 26% of the V6 tags obtained are closely related to lineages involved in sulfur, nitrogen, iron, and methane cycling. Epsilonproteobacteria represent <4% of the V6 tags recovered from inactive sulfides and 15% of the full-length clones, despite their high abundance in active chimneys. Members of the phylum Aquificae, which are common in active vents, were absent from both the V6 tags and full-length 16S rRNA data sets. In both analyses, the proportions of alphaproteobacteria, betaproteobacteria, and members of the phylum Bacteroidetes were greater than those found on active hydrothermal sulfides. These shifts in bacterial population structure on inactive chimneys reveal ecological succession following cessation of venting and also imply a potential shift in microbial activity and metabolic guilds on hydrothermal sulfides, the dominant biome that results from seafloor venting.
IMPORTANCE
Hydrothermal chimneys are globally dispersed seafloor habitats associated with mid-ocean ridge spreading centers. Active, hot, venting chimneys have been examined for microbial ecology since their discovery in the late 1970s, but the microbiology of inactive chimneys, which may persist for thousands of years, has only recently been explored. We studied bacterial diversity on inactive hydrothermal sulfide chimney samples from 9°N on the East Pacific Rise to learn their bacterial community composition, potential ecological roles, and succession from active venting to inactive chimneys. Many bacteria on inactive sulfide chimneys are closely related to lineages involved in sulfur, nitrogen, iron, and methane cycling, and two common groups found on active chimneys are nearly absent from inactive vents, where they were replaced by groups likely involved in the elemental cycling mentioned above. Our findings reveal that ecological succession occurs on hydrothermal sulfides after active venting ceases and also imply a potential shift in microbial metabolic guilds.
Hydrothermal venting is recognized throughout the global mid-ocean ridge (MOR) system, a 60,000-km seam along the ocean floor at which new ocean crust is continuously created. Vents are commonly associated with metal- and sulfur-rich mineralized structures such as chimneys, which precipitate from hydrothermal fluids when the fluids are expelled into cold, oxidized deep seawater at hot springs (1). Reduced chemical species that are vented at deep-sea hydrothermal systems support diverse microbial populations that participate in important biogeochemical processes (2–9). However, venting in the deep sea is ephemeral and the fate and geochemical evolution of inactive mineralized deposits are of keen interest in economic geology and mineralogy. Therefore, some sites such as the massive Trans-Atlantic Geotraverse (TAG) hydrothermal system in the Atlantic (10), the longest-lived hydrothermal system in the ocean, 9 to 10°N on the East Pacific Rise (EPR) (11, 12), which globally emits the highest flux of hydrothermal fluids in the oceans, and the Galapagos Spreading Center (13–15) have been studied both during venting and postventing from geologic and geochemical perspectives. Dating of inactive sulfides from the EPR revealed that inactive sulfides can last at least 20,000 years on the seafloor (11), providing long-lived habitats for microbial settlement. However, the microbiology and microbial community succession that occur as sulfide deposits transition from active to inactive venting systems have received minimal attention, despite some hints, for example, that suggest that the microbial populations at inactive sulfides are distinct from those found within actively venting structures (16, 17) and that the extant microbial fauna of inactive structures shows a high potential for participating in important biogeochemical transformations (18), even after hydrothermal activity ceases. One recent study showed that active and inactive sulfide vents from the same hydrothermal vent field harbored statistically distinct microbial communities and specifically noted that members of Epsilonproteobacteria, a microbial clade well recognized at actively venting deep-sea sites, are less common within inactive chimneys (16).
Previous studies of microbial diversity on inactive sulfides used 16S rRNA clone libraries. The application of tag pyrosequencing, generating tens of thousands of sequences of small sections of the 16S rRNA gene per sample (19), has enabled a much deeper view of microbial diversity within samples. Recent studies that use tag pyrosequencing for a “deep” view of diversity in environmental samples have proposed that there exists a rare biosphere of microbes—that is, microbial taxa that are present at very low abundance in all samples—and have suggested that this rare biosphere exhibits biogeography (20) and can act as seed populations that become dominant when favorable conditions prevail (21, 22).
Here we used tag pyrosequencing of the V6 region of the 16S rRNA gene and full-length 16S rRNA sequencing to assess bacterial diversity associated with inactive sulfide chimneys using samples from 9°N on the EPR. Deep sequencing of the V6 hypervariable region of the 16S rRNA gene allowed us to describe in detail the bacterial communities present on inactive sulfides from 9°N on the EPR, including bacterial groups cosmopolitan to multiple chimney samples and those dominant on one or a few samples. Additional insights were gained from full-length 16S rRNA clones, allowing confirmation of V6 tag classification and comparisons with other studies. We observed a clear succession of bacteria from active chimneys to inactive chimneys. Given the observed changes in microbial community structure, we consider the possible changes in metabolic capacity as well as the implications for biogeochemical cycling at inactive sulfides.
RESULTS
Sample descriptions.
Samples were collected from 9°N on the EPR, a fast-spreading ridge. All five inactive chimneys represent Fe-Zn-rich sulfide chimney structures (12). Sulfides 3M23 and 3M33 were both sampled from the exterior of inactive chimneys near the K vent chimney, 2,564 m deep. They are rich in Fe and Zn, and their external walls are composed of mixed assemblages of marcasite, fine-grained sphalerite, and silica (12). 7M24 and 9M32 represent Fe-rich massive sulfides sampled at a depth of 2,504 m just north of the Bio9″ vent. The external sides of these massive sulfides are heavily encrusted Fe oxides and pyrite. Sulfide 9M4 was recovered from the top 1 m of a 9-m-tall inactive chimney 300 m off axis from the axial summit trough, 2,512 m deep. It was a Zn-rich inactive chimney with significant Fe enrichment. The interior (9M4I) is composed of euhedral sphalerite forming millimeter- to centimeter-wide layers, followed by fine-grained assemblages of sphalerite and pyrite (12). One sample was taken specifically from a sphalerite-rich section of the inner wall (9M4S). Botryoidal sphalerite and fine-grained pyrite form the external walls, which are coated with Fe oxyhydroxides and amorphous Si (9M4O).
Bacterial diversity of inactive sulfides.
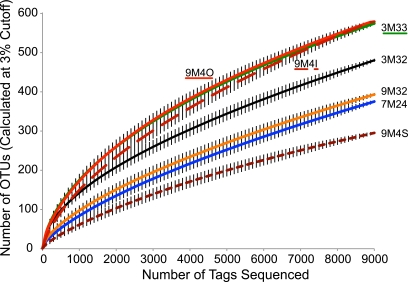
We obtained a total of 206,647 V6 sequence tags combined from the seven inactive hydrothermal sulfides. Rarefaction analysis indicates that 9M4O, 9M4I, and 3M33 host the most diverse bacterial communities (their rarefaction curves are statistically equal), while 9M4S hosts the least diverse community (Fig. 1). Abundance-based coverage estimator (ACE) and Chao1 estimates of the total number of operational taxonomic units (OTUs) per sample correspond well to rarefaction analysis, but these estimators generally diverge from Simpson’s reciprocal index, particularly for samples 9M4O, 9M4I, and 9M32 (Table 1; see Fig. S2 in the supplemental material).
FIG 1 .
Rarefaction curves for inactive sulfide samples. All samples were randomly resampled down to the smallest sample size, 9,149 tags (9M4O). The preclustering option was used in mothur for rarefaction calculation, and OTUs were calculated with a cutoff of 3%. Vertical bars represent 95% confidence intervals.
TABLE 1 .
Diversity statistics for several sample typesa
| Sample | Sample type | No. of OTUs |
ACE richness estimation |
Chao1 richness estimation |
Simpson’s reciprocal index |
Reference |
|---|---|---|---|---|---|---|
| 3M23 | Inactive sulfide | 485 | 1,789–2,213 | 1,074–1,726 | 21.2–22.8 | This study |
| 3M33 | Inactive sulfide | 578 | 1,338–1,611 | 901–1,191 | 8.6–9.4 | This study |
| 7M24 | Inactive sulfide | 379 | 1,840–2,339 | 842–1,362 | 4.0–4.3 | This study |
| 9M32 | Inactive sulfide | 397 | 1,896–2,372 | 824–1,297 | 2.7–2.9 | This study |
| 9M4O | Inactive sulfide | 578 | 1,273–1,534 | 993–1,383 | 21.9–23.7 | This study |
| 9M4I | Inactive sulfide | 579 | 1,738–2,123 | 1,147–1,651 | 4.3–4.7 | This study |
| 9M4S | Inactive sulfide | 298 | 872–1,177 | 550–866 | 1.6 | This study |
| LostCity1 | Active carbonate | 247 | 715–977 | 465–826 | 5.3–5.8 | 22 |
| LostCity2 | Active carbonate | 363 | 966–1,230 | 572–817 | 8.3–9.2 | 22 |
| LostCity3 | Active carbonate | 574 | 2,012–2,499 | 1,211–1,802 | 19.0–21.8 | 22 |
| LostCity4 | Inactive carbonate | 435 | 1,676–2,084 | 874–1,337 | 16.5–17.8 | 22 |
| FS312 | JdFR diffuse fluids | 1,386 | 7,386–8,434 | 3,843–5,084 | 23.1–25.5 | 28 |
| FS396 | JdFR diffuse fluids | 1,143 | 5,971–6,910 | 3,172–4,352 | 8.4–9.3 | 28 |
| FS431 | Mariana diffuse fluids | 1,505 | 4,225–4,758 | 2,915–3,589 | 13.8–15.6 | 29 |
| FS432 | Mariana diffuse fluids | 1,650 | 4,057–4,532 | 2,766–3,264 | 43.5–50.9 | 29 |
| FS445 | Mariana diffuse fluids | 582 | 713–820 | 759–964 | 20.9–23.1 | 29 |
| FS446 | Mariana diffuse fluids | 628 | 1,711–2,053 | 1,156–1,593 | 6.7–7.2 | 29 |
| FS447 | Mariana diffuse fluids | 1,181 | 2,661–3,041 | 1,870–2,241 | 24.3–27.4 | 29 |
| FS448 | Mariana diffuse fluids | 1,268 | 4,103–4,678 | 2,438–3,055 | 60.2–67.8 | 29 |
| FS449 | Mariana diffuse fluids | 848 | 1,525–1,760 | 1,186–1,428 | 21.2–23.3 | 29 |
| FS467 | Mariana diffuse fluids | 1,097 | 3,569–4,122 | 2,164–2,766 | 27.7–30.4 | 29 |
| FS468 | Mariana diffuse fluids | 1,550 | 7,412–8,416 | 4,032–5,195 | 44.3–50.1 | 29 |
| FS473 | Mariana diffuse fluids | 1,068 | 3,972–4,607 | 2,249–2,913 | 5.0–5.5 | 29 |
| FS475 | Mariana diffuse fluids | 643 | 1,968–2,368 | 1,150–1,542 | 7.0–7.5 | 29 |
| FS479 | Mariana diffuse fluids | 1,201 | 4,386–5,067 | 2,544–3,238 | 24.4–26.8 | 29 |
| FS480 | Mariana diffuse fluids | 617 | 1,256–1,495 | 951–1,244 | 14.2–15.3 | 29 |
| FS481 | Mariana diffuse fluids | 1,003 | 2,309–2,673 | 1,686–2,104 | 20.8–23.0 | 29 |
| 53R | Deep seawater | 1,325 | 4,241–4,843 | 2,740–3,455 | 19.2–21.3 | 19 |
| 55R | Deep seawater | 1,510 | 7,040–7,928 | 3,756–4,834 | 22.5–25.3 | 19 |
| 112R | Deep seawater | 1,705 | 9,435–10,586 | 4,447–5,636 | 52.4–59.0 | 19 |
| 115R | Deep seawater | 1,254 | 4,801–5,482 | 2,858–3,718 | 23.5–26.1 | 19 |
| 137 | Deep seawater | 1,163 | 3,008–3,441 | 2,333–3,036 | 24.8–28.0 | 19 |
| 138 | Deep seawater | 1,155 | 3,501–4,004 | 2,297–2,955 | 17.7–19.7 | 19 |
aAll data were calculated using the preclustering option and average neighbor distance in mothur and a cutoff of 3% for OTU determination. All samples were randomly resampled down to 9,149 tags, except for FS481 (only 8,923 tags in original sample), FS445 (only 8,398 tags in original sample), and the Lost City samples (all resampled to 5,567 tags and reanalyzed here). The ranges shown are 95% confidence intervals. A graphical version of this table is presented in Fig. S2 in the supplemental material.
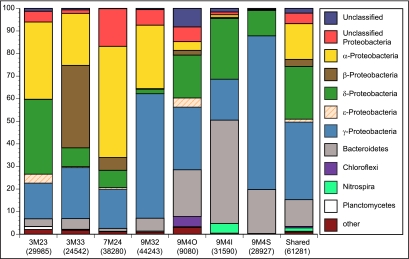
Alphaproteobacterial V6 tags are present on all samples from the outside of chimneys but are rare on 9M4I and absent from 9M4S (Fig. 2). Betaproteobacterial V6 tags are present on four of the seven samples, and Betaproteobacteria is the dominant bacterial class on sample 3M33. Both Deltaproteobacteria and Gammaproteobacteria are present in all of the samples analyzed, and Gammaproteobacteria comprise 15 to 60% of the bacterial communities on inactive chimneys assessed via V6 tag sequencing. Epsilonproteobacteria are present on only four the seven samples and are never >4% of the total V6 tags for any sample. All three samples from inactive sulfide 9M4 contained few, if any, Alphaproteobacteria and no Betaproteobacteria but had a much higher proportion of members of the phylum Bacteroidetes than any of the other samples analyzed in this study. 9M4I had the highest proportion of members of the phylum Bacteroidetes and is the only sample also with high numbers of Nitrospira V6 tags. All three samples from rock 9M4, especially 9M4O, contained many unclassified members of the domain Bacteria, indicating a high proportion of unknown diversity on that chimney.
FIG 2 .
Bacterial distribution among the different samples. Only groups that represent >1% of the tags in at least one of the samples are included. The category “other” refers to all groups that represent <1% of the tags in all of the samples. These are shown in detail in Fig. S3 in the supplemental material. The total number of tags obtained for each sample is given in parentheses. The rightmost column (Shared) represents tags shared by at least two samples in this study.
The designation “other” in Fig. 2 refers to 27 classes of bacteria that comprise <1% of the tags in all of the samples. We designate these classes as comprising the rare biosphere for the inactive sulfide ecosystem, following the convention established in prior studies (23). Several classes of bacteria are ubiquitous in the inactive sulfide rare biosphere, including Verrucomicrobia, Firmicutes, Actinobacteria, and Acidobacteria (see Fig. S3 in the supplemental material). Chlorobi tags are present in five samples. Thermophilic rare biosphere lineages detected among inactive sulfides include Deinococcus-Thermus, Thermomicrobia, Thermotogae, and Thermodesulfobacteria. The two interior sulfide samples, 9M4I (sulfide) and 9M4S (sphalerite), display the lowest proportions of rare biosphere bacteria. The rare biosphere on 9M4I is predominately Actinobacteria, Acidobacteria, and Chlorobi, while the rare biosphere tags in sample 9M4S are dominated by Firmicutes. Both interior 9M4 samples were the only samples to lack Verrucomicrobia among the sample set examined.
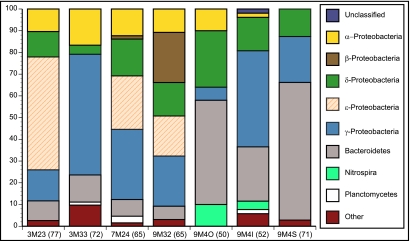
Analysis of these same samples with full-length 16S rRNA sequencing revealed small differences from the V6 tags, but in general, the two data sets were broadly similar (Fig. 2 and 3). The recovery of Epsilonproteobacteria from 3M23, 7M24, and 9M32 was higher in the full-length clone libraries. Interestingly, epsilonproteobacterial V6 tags were recovered from sample 3M23 but 7M24 had very few and 9M32 had none. In one case, sample 9M4O, Epsilonproteobacteria were recovered by V6 tag sequencing but not with the full-length libraries. Alphaproteobacteria were underrepresented in the full-length 16S rRNA libraries, compared to the V6 tag analysis in most samples, but were absent from sample 9M4S in both analyses. Betaproteobacteria were not recovered from sample 3M33 full-length libraries but were recovered using V6 tag sequencing. Proportions of Gammaproteobacteria were different between the two sequencing methods as well, but Gammaproteobacteria were present in full-length 16S rRNA libraries from all samples, as they were with the V6 tags. Members of the class Nitrospira were detected from 9M4I using both sequencing approaches.
FIG 3 .
Bacterial distribution among the different samples as determined via full-length Sanger sequencing. The total number of clones is shown in parentheses.
Cosmopolitan and abundant OTUs on inactive sulfides.
We searched our entire data set for V6 tag OTUs (defined by 97% similarity, which allows for one or two substitutions) shared by two or more samples to determine the most cosmopolitan bacteria. In our data set, there were 11,404 V6 OTUs, of which 1,199 (10.5% of the OTUs and 176,451 total tags) were shared by two or more samples (Fig. 2). Three V6 tag OTUs were found on all 7 samples (see Table S1 in the supplemental material), and two of these OTUs were identical to sequences recovered from other hydrothermal sulfides (including inactive sulfides) via BLASTn searches (24) of the NCBI database. Thirteen V6 tag OTUs were recovered from six of the seven samples. Five of these OTUs were most similar to clones recovered from sediments or hydrothermal vent environments. Many of the most abundant (overall) V6 tag OTUs were highly similar (96 to 100% similarity) to clones recovered from hydrothermal vent environments and sediments.
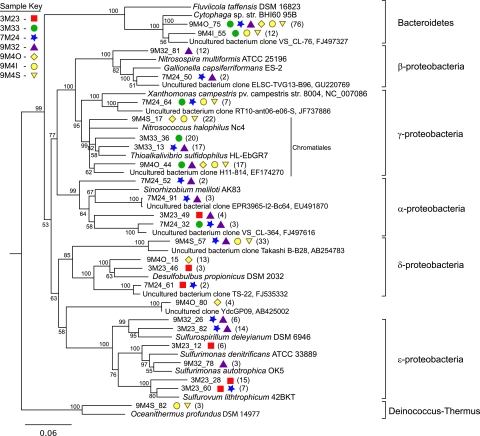
We grouped full-length clones into OTUs with a cutoff of 97% similarity and then compared sequences that were abundant (more than three clones belonging to a single OTU) and/or cosmopolitan (clones from different samples in the same OTU). The most common and/or most cosmopolitan full-length clones represent 68% of the clones recovered in this study, and all fall within the phyla Proteobacteria and Bacteroidetes, with the exception of one OTU, which is within the Deinococcus-Thermus phylum (Fig. 4). OTU 9M4O_80, represented by three clones from 9M4O, is identical to a clone recovered from an inactive sulfide in the Southern Mariana Arc (16). The closest related sequence in the NCBI database following the Southern Mariana Arc clone is the cultivated bacterium Hippea maritima, a deltaproteobacterium, but OTU 9M4O_80 and H. maritima are only 85% similar. However, OTU 9M4O_80 groups closest to Epsilonproteobacteria in our analysis and appears to represent a potentially widespread organism resident on inactive sulfides.
FIG 4 .
Phylogenetic tree of OTUs represented by more than three clones from a single sample and/or clones from multiple samples. OTUs are defined by a 97% similarity cutoff. OTUs were aligned using MAFFT (48) with the G-INS-I algorithm and 200PAM scoring matrix in Geneious 5.4 (49), and then the tree was constructed using the neighbor-joining algorithm with 1,000 bootstraps. Symbols next to the OTUs from this study represent the samples from which they were recovered, as indicated by the sample key at the top left. The number of clones belonging to each OTU is in parentheses. Bootstrap values of >50% are shown at nodes. Aquifex pyrophilus and Hydrogenobacter subterraneus were used as outgroups.
DISCUSSION
V6 tag sequencing versus full-length 16S rRNA clones.
Prior work has shown a good correlation between full-length rRNA clone libraries and hypervariable region tag sequencing through the generation of extensive full-length data sets for comparison to tag sequencing (25, 26). Our full-length data set was smaller per sample but comparable to those of other environmental diversity studies. We also found agreement between the two sequencing methods. Differences were more apparent with individual samples (Fig. 2 and 3) than when the entire data sets from V6 tags and full-length clones were compared (Fig. 5). The largest noted differences between the two methods were higher percentages of Epsilonproteobacteria and members of the phylum Bacteroidetes recovered by the full-length clones than by the V6 tags. Data sets from V6 tag sequencing appear to be comparable to those from full-length sequencing. However, both of the PCR methods used here introduce inherent biases and future studies could use a quantitative method such as fluorescence in situ hybridization to further constrain the patterns observed here.
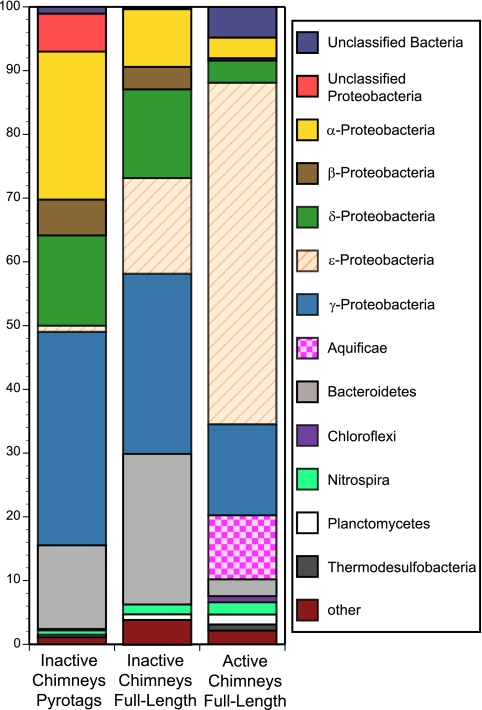
FIG 5 .
Bacterial distribution in composite inactive chimneys and a composite active black smoker chimney. “Inactive Chimneys Pyrotags” (n = 206,647) is the sum of all of the tags in this study. “Inactive Chimneys Full-Length” (n = 452) is the sum of all of the full-length clones in this study. The composite active chimney (“Active Chimneys Full-Length,” n = 834) was generated by compiling data from previously published studies of full-length 16S rRNA clones on active black smoker chimneys and represents the sum of all of the clones in these studies (2–9, 36, 37). Only studies where clone frequency was reported were used. The color code is same as that in Fig. 1, and the category “other” is shown in detail in Fig. S4 in the supplemental material.
Inactive sulfide chimneys represent a biogeochemically active microbial ecosystem.
This is the first deep sequencing investigation into the bacterial diversity of inactive chimneys, permitting the evaluation of both major and minor taxa within inactive sulfide ecosystems. Following the cessation of active venting, hydrothermal sulfides are transformed from an ecosystem that is supported through energy from hot, reduced hydrothermal fluids to one that is supported through the chemical energy that can be derived from oxidative weathering of the sulfide structures. Gases in hydrothermal fluids such as ammonium, methane, and hydrogen are no longer available to support chemolithoautotrophic production but are replaced by the chemical energy present in reduced minerals within the sulfide structure. Here we discuss the likely biogeochemical roles on inactive sulfides based on the sequences recovered.
More than one-quarter of the V6 tags and more than half of the full-length clones recovered represent bacterial taxa for which defined ecological roles can be hypothesized, based on high sequence similarity to cultivated representatives (Table 2; see Table S2 in the supplemental material). Many of these taxa are closely related to known autotrophs, suggesting that they might represent a base of the food web to the inactive sulfide ecosystem. The exceptions to this observation are among the sulfate-reducing bacterial tags detected; most are related to bacteria that cannot fix carbon or display a variable ability to fix carbon (e.g., Desulfobulbaceae).
TABLE 2 .
Potential ecological roles of tags for which obvious metabolisms can be inferreda
| Potential ecological role |
Taxa | No. of tags |
% of tags in data set |
|---|---|---|---|
| S oxidation | Gammaproteobacteria: Chromatiales | 157 | 0.0760 |
| Gammaproteobacteria: Chromatiales: Ectothiorhodospiraceae | 20,121 | 9.7369 | |
| Gammaproteobacteria: Chromatiales: Chromatiaceae | 39 | 0.0189 | |
| Gammaproteobacteria: Thiotrichales: Piscirickettsiaceae: Thiomicrospira | 1,546 | 0.7481 | |
| Gammaproteobacteria: Thiotrichales: Francisellaceae: Francisella | 217 | 0.1050 | |
| Epsilonproteobacteria | 2,015 | 0.9751 | |
| SO42− reduction | Deltaproteobacteria: Desulfobacterales: Desulfobacteraceae | 20,709 | 10.0214 |
| Deltaproteobacteria: Desulfovibrionales: Desulfovibrionaceae: Desulfovibrio | 1,074 | 0.5198 | |
| Deltaproteobacteria: Desulfuromonadales: Desulfuromonadaceae | 621 | 0.3005 | |
| Deltaproteobacteria: Syntrophobacterales | 49 | 0.0237 | |
| Thermodesulfobacteria | 23 | 0.0111 | |
| Sum of S oxidation and SO42− reduction | 22.5365 | ||
| Nitrite oxidation | Nitrospira | 1,392 | 0.6736 |
| Deltaproteobacteria: Desulfobacterales: Nitrospinaceae: Nitrospina | 766 | 0.3707 | |
| Nitrification | Betaproteobacteria: Nitrosomonadales: Nitrosomonadaceae | 51 | 0.0247 |
| N fixation | Alphaproteobacteria: Rhizobiales | 2,404 | 1.1633 |
| Sum of nitrite oxidation, nitrification, and N fixation | 2.5798 | ||
| Fe oxidation | Betaproteobacteria: Nitrosomonadales: Gallionellaceae: Gallionella | 2,087 | 1.0099 |
| H oxidation | Gammaproteobacteria: Thiotrichales: Piscirickettsiaceae: Hydrogenovibrio | 58 | 0.0281 |
| CH4 oxidation | Gammaproteobacteria: Methylococcales: Methylococcaceae | 1,602 | 0.7752 |
| Autotrophy | Chlorobi | 72 | 0.0348 |
| Total sum | 26.6168 |
aData are pooled from all samples; therefore, multiple tags are represented per lineage listed. The percentage of tags in the data set is for the entire data set (the sum of all tags sequenced on all samples). Taxa are designated by class (phylum for Nitrospira and Chlorobi), order, family, and genus. The highest level at which an ecological role can be assigned per group of tags is shown.
Tags affiliated with taxa known to be capable of sulfur oxidation or sulfate reduction each comprised greater than 10% each of the total V6 tags from all of the samples and 30% (S oxidation) and 13% (sulfate reduction) of the full-length clones. Many of these lineages are shared between the V6 tags and full-length sequences, including Chromatiales, Epsilonproteobacteria, and Deltaproteobacteria. Close to 5% of the V6 tags and full-length clones were associated with N redox cycling capabilities (N fixation, nitrification, and nitrite oxidation). Other ecological functions that are suggested based on the sequences recovered include Fe, H2, and methane oxidation. The bacterial communities on these seven samples support our hypothesis that active biogeochemical cycling of S, N, and Fe is supported within and on inactive hydrothermal sulfide minerals. Previous work has shown that total organic carbon on the exterior of inactive sulfides is up to five times higher than on the exterior of actively venting sulfides (16), indicating that the members of the community observed here are contributing to substantial production of organic matter on sulfides once venting ceases.
The ecological role of the cosmopolitan V6 tags (see Table S1 in the supplemental material) is rarely possible to define, but many of the V6 tags that are most abundant can be linked to N, Fe, and S redox cycling. For example, the second most abundant V6 tag in the entire data set is most closely related to members of the family Ectothiorhodospiraceae, chemolithoautotrophic S-oxidizing gammaproteobacteria. This tag occurs on all samples from chimney 9M4 and accounts for 7.2% of the V6 tags sequenced in this study. Tags related to known nitrite oxidizers in the order Nitrospirales and the family Nitrospinaceae are common on chimney 9M4 as well. Tags with sequence similarity to sulfate reducers in the bacterial family Desulfobulbaceae are also among the most abundant detected in this study. A Fe-oxidizing member of the family Gallionellaceae was one of the most common tags on sample 7M24, a massive sulfide deposit with extensive alteration and Fe oxyhydroxide deposition (12).
Similar to the analysis with V6 tags, many of the most abundant clones can be attributed to an ecological function based on their lineage. Many clones belong to the Chromatiales order within the class Gammaproteobacteria, which are all capable of sulfide oxidation (27). Recovered clones related to Sulfurospirillum deleyianum may be active in sulfur reduction, while the other epsilonproteobacterial clones in this tree are more closely related to organisms active in sulfur oxidation. Other cosmopolitan clones represented in the full-length libraries that can be assigned potential ecological roles are related to genera of known sulfur reducers (Desulfobulbus) and iron oxidizers (Gallionella).
The bacterial communities on the inactive sulfides sampled here harbor communities with an estimated 872 to 2,372 OTUs, as measured with ACE (Table 1). This is similar to the number of OTUs measured on the carbonate chimneys at the Lost City hydrothermal field (715 to 2,499 OTUs [22]) but much lower than that measured in samples from diffuse-flow hydrothermal fluids at Axial Volcano on the Juan de Fuca Ridge (5,971 to 8,434 OTUs [28]), along the Mariana Trench (713 to 8,416 OTUs [29]), and in deep water from the North Atlantic (3,008 to 10,586 OTUs [19]). The Chao1 estimator indicates trends in diversity between these habitats similar to those indicated by ACE, but here, as discussed earlier, there are samples with values of Simpson’s reciprocal index that differ from the trends observed with ACE and the Chao1 estimator. These small differences result because ACE and the Chao1 estimator are estimates of the total species richness in a sample and both account for singletons (30), whereas Simpson’s reciprocal index is an overall measure of diversity and is not as heavily influenced by singletons. Although rarefaction curves did not plateau for any samples, indicating that the full diversity of these samples has not yet been revealed, these analyses suggest that inactive sulfides represent low-diversity environments in comparison to other deep-sea sites examined to date (19, 28, 29).
While diversity indices indicate that these inactive sulfides are not as diverse as some other deep ocean habitats, the sequences recovered here by both methods reveal that a diverse set of geochemical transformations are likely on these structures. In particular, there was co-occurrence of tags and clones associated with both oxidation and reduction of S and N compounds on the same samples (Table 2; see Table S2 in the supplemental material). Inactive sulfide chimneys appear to represent an active ecosystem with elevated metal concentrations that is slowly transitioning from a reduced environment, created by formerly present hydrothermal fluids and precipitates, to an oxidized environment. It is notable that the alphaproteobacterial family Rhodobacteraceae comprised ~14% of the tags recovered. While a definitive ecological function cannot be assigned to this family, a recent isolate from the TAG hydrothermal field on the Mid-Atlantic Ridge is a chemolithoautotrophic S and H2 oxidizer (31), indicating that at least some of the Rhodobacteraceae V6 tags in our samples may represent organisms that can oxidize S and/or H2. The ubiquitous Roseobacter clade of marine bacteria falls within the Rhodobacteraceae family, and 72% of the sequenced genomes from this group contain the sox cluster of genes responsible for S oxidation (32), lending further support to potential participation in S cycling among the members of the family Rhodobacteraceae detected here.
Variation in community composition is evident among our inactive sulfide chimney samples (Fig. 2 and 3). It has been shown previously that variation among taxa identified at some active hydrothermal vents are related to variation in subseafloor fluid chemistry (33). For microbial communities inhabiting chimneys that no longer vent fluids, we hypothesize that geochemical diversity of the substrate exerts an influence on microbial ecology. Overall, we observe with both V6 tags and full-length clones that Alpha-, Delta- and Gammaproteobacteria and members of the phylum Bacteroidetes dominate inactive sulfide chimneys.
With the exception of 3M33, Betaproteobacteria were either absent or not more than a few percent of the total V6 tags on the inactive chimneys. Two extremely abundant V6 tags accounted for the high proportion of Betaproteobacteria in sulfide 3M33. While neither could be classified beyond the class level by the Global Alignment for Sequence Taxonomy (GAST) method (26), one of these tags (1,855 copies) is identical to the betaproteobacterial species “Sideroxydans paludicola,” a chemolithoautotrophic Fe oxidizer (34), Zoogloea ramigera, an Mn oxidizer, and “Candidatus Nitrotoga arctica,” a nitrite oxidizer (35), while the other tag (6,251 copies) is 96% similar to the same betaproteobacterial species. These data suggest the likely presence and activity of metal-oxidizing bacteria associated with this sample. Full-length clones also revealed low incidences of Betaproteobacteria, and those that were observed potentially play a role in N (9M32_81) and Fe (7M24_50) transformations.
A previous study found a high percentage of clones related to “Candidatus Magnetobacterium” in inactive sulfides from both the western Pacific and Indian Oceans (17). We recovered 125 V6 tags classified as “Candidatus Magnetobacterium” from 9M4I and suggest that it may be a common resident of inactive sulfides. The clones represented by OTU 9M4O_80 are identical to a clone in the NCBI database recovered from another inactive sulfide structure, but these clones are not closely related (>85% similarity) to any known organisms. These may also be common on inactive sulfide structures.
Bacterial succession on hydrothermal chimneys.
Initial studies of bacterial diversity on inactive sulfides have suggested that communities on inactive sulfides are different from those on active chimneys (16, 17), most notably the lower proportion of Epsilonproteobacteria, which dominate bacterial communities on active sulfides (2–9, 36, 37). An active sulfide chimney from 9°N on the EPR investigated previously was found to be dominated almost exclusively by Epsilonproteobacteria of the genera Sulfurimonas and Sulfurospirillum and epsilonproteobacterial group F (3), which includes the genus Sulfurovum (38). These are the genera within which our full-length clones belonging to the Epsilonproteobacteria belong. In contrast, a prior study of bacterial populations on inactive chimneys in the Okinawa Trough and the Central Indian Ridge found these structures to be dominated by sequences from Alpha- and Gammaproteobacteria and also sequences related to Delta- and Epsilonproteobacteria, Actinobacteria, Nitrospira, Bacteroidetes, Planctomycetes, and Verrucomicrobia (17)—results that are consistent with the data presented here in comparison to prior active-chimney studies. Another study of the Juan de Fuca Ridge detected a predominance of Gammaproteobacteria Marinobacter, S-oxidizing Thiomicrospira, mesophilic Fe and S oxidizers, and an uncultured epsilonproteobacterium most closely related to Caminibacter mediatlanticus, which is capable of both nitrate ammonification and sulfur reduction (18). In our study, V6 tags classified as Epsilonproteobacteria are nearly all within the thermophilic genera Sulfurimonas or Sulfurovum, indicating that these tags may be relict populations, residual from when the sulfides were active and warm.
We constructed two composite inactive chimneys using all of the sequences recovered from our samples for both V6 tag sequencing and full-length clone libraries and a composite active chimney based on full-length sequences from previously published papers (Fig. 5). Conceptually, by binning these samples, the resulting composites “average” taxa from these two end-member sulfide biomes, minimizing sample-to-sample variation that may arise from fine-scale variation in geochemistry discussed above, in order to reveal variation that may be more specifically related to major differences between active and inactive sulfides. We chose only basalt-hosted hydrothermal sulfide studies (i.e., excluding ultramafic and sediment-hosted systems) that used universal bacterial primers for PCR and reported clone frequencies (2–9, 36, 37). The difference between the bacterial populations on active chimneys and those on inactive chimneys is apparent for both of the sequencing methods used here. The most obvious difference is the general lack of Epsilonproteobacteria in the inactive chimneys analyzed here versus an overwhelming predominance on active chimneys. Even for the full-length clones, which recovered proportionately more Epsilonproteobacteria than V6 tag sequencing did, there are much fewer Epsilonproteobacteria than on active chimneys. Similarly, members of the phylum Aquificae are absent from inactive sulfides, while they are common on active chimneys. We infer that Epsilonproteobacteria and members of the phylum Aquificae, which can comprise up to 60% of the active-chimney communities, are succeeded in active sulfides by members of the Alpha-, Beta-, Delta- and Gammaproteobacteria and members of the phylum Bacteroidetes on inactive sulfide chimneys. All of these phyla are more prevalent on the inactive chimneys than on the active chimneys.
Succession on sulfide chimneys was recently observed in the Southern Mariana Trough, where active and inactive sulfides were shown to host statistically significantly different microbial communities (16). Thermophilic Archaea and Epsilonproteobacteria were found to dominate active chimneys, while both groups were nearly absent from inactive sulfides. A recent study using V6 tag pyrosequencing also observed microbial succession on the carbonate chimneys at the Lost City hydrothermal field, an ultramafic system on the Mid-Atlantic Ridge flank (22). Young, active chimneys at Lost City are dominated by Archaea from a clade known as the Lost City Methanosarcinales, while an inactive-chimney sample, dated at 1,245 years, is dominated by a group of ANME-1, anaerobic methanotrophic archaea (22). Interestingly, the ANME-1 tag that dominates the older sample is detected in very low numbers on the younger samples via tag sequencing, supporting the hypothesis that members of the rare biosphere can become dominant under favorable conditions. This observation at Lost City, in addition to the bacterial succession observed here, prompts additional examination of sulfide systems to examine the role and dispersal of rarely occurring tags in acting as seed populations in hydrothermal systems. Future studies should endeavor to examine archaeal succession as well. Certainly, there should be a succession from thermophilic Archaea to mesophiles or psychrophiles, but given the presence of organisms derived from the water column in some of the samples studied here, it may be likely to see an increase in marine group I Archaea on inactive sulfides or even the association of methanogens with the bacterial sulfate reducers seen here (39).
This study described the bacterial community that is present on inactive hydrothermal sulfides. Compared to the bacterial community present on active sulfides, there is a clear shift from communities dominated by Epsilonproteobacteria and members of the phylum Aquificae to bacterial communities dominated by Alpha-, Beta-, Delta-, and Gammaproteobacteria and members of the phylum Bacteroidetes. Many of these are likely to participate in redox transformations of S, N, and Fe, and despite the loss of chemical energy derived from hydrothermal fluids, the mineralogy of inactive sulfides appears to support a community with chemolithoautotrophs that provide the organic carbon needed to maintain a microbial ecosystem, likely without a need for carbon input from the surface ocean. It is likely that once the heat from hydrothermal fluids disappears, microbes are able to colonize the internal microniches of inactive sulfides and thrive on both reduced and oxidized minerals present in the structures.
MATERIALS AND METHODS
Rock collection.
Sampling of all sulfides was conducted at 9°N on the EPR using deep submergence vehicle Alvin during research vessel Atlantis cruise AT11-20 in November 2004 using sealable bioboxes, which prevent mixing with seawater as the submersible moves throughout the water column. Samples were processed upon arrival on deck using sterile chisels and aluminum rock boxes specifically designed for the sampling of seafloor rocks and then placed at −80°C until processing back in the lab.
DNA extraction and processing.
Environmental DNA from seafloor sulfides was extracted using protocols and methods described previously using the Ultraclean Soil DNA extraction kit (MoBio Laboratories) following manufacturer protocols (40) or by a phenol-chloroform freeze-thaw extraction method for sample 9M4S (41, 42). PCR of the V6 hypervariable region of the 16S rRNA gene, followed by 454 pyrosequencing of the amplicons, was carried out as described previously (19). Phylogenetic affiliations of the tag sequences were identified using the GAST method (26). Diversity statistics were calculated in mothur (43) on samples trimmed down to an equal number of tags through random resampling using Daisy-Chopper (available from http://www.genomics.ceh.ac.uk/GeneSwytch). All statistics were calculated using the preclustering option in mothur, which preclusters at a 2% difference level (1-bp difference for the V6 tags used here) using modified single linkage (44), and the average neighbor clustering method. Sequences from the tag pyrosequencing runs can be accessed through the Visualization and Analysis of Microbial Population Structures website (http://vamps.mbl.edu).
For full-length clone library generation, the 16S rRNA gene was amplified by PCR using primers 27F (5′ AGAGTTTGATCCTGGCTCAG 3′) and 1492R (5′ GGTTACCTTGTTACGACTT 3′) (45). The PCR conditions were as follows: 1 cycle of 95°C for 5 min; 25 cycles of 95°C for 1.5 min, 47°C for 1.5 min, and 72°C for 3 min; 1 cycle of 72°C for 10 min; and holding at 4°C. Amplification products were purified using the QIAquick nucleotide removal kit (Qiagen), clone libraries were constructed using a TOPO cloning kit (Invitrogen), and plasmid extractions (alkaline lysis) and sequencing (ABI v3.1 BigDye terminator; Applied Biosystems) were performed at the Josephine Bay Paul Center, Marine Biological Laboratory, Woods Hole, MA. Clones were classified using the Greengenes server and classification scheme (46). OTUs were determined using mothur with a 97% similarity cutoff.
It must be stated that is impossible to rule out the possibility that biases in PCRs and sequencing methodology contributed to some of the differences between active and inactive sulfides observed here. However, prior studies of active sulfides using full-length clones and tag pyrosequencing (28, 29, 47) recovered the phyla (Epsilonproteobacteria and members of the phylum Aquificae) that were found to be missing here with similar effectiveness. Therefore, we conclude that the differences observed in the present study are real.
Nucleotide sequence accession numbers.
Sequences from full-length clone libraries were submitted to the GenBank database with accession numbers JQ286978 to JQ287493.
SUPPLEMENTAL MATERIAL
Photos of samples collected for this study. Seafloor photos of 3M23 (A), 7M24 (B), 9M32 (C), and 9M4, a 9-m chimney from which a section was collected from the top 1 m (D). Shipboard photos of 3M23 (E), 3M33 (F), 7M24 (G), and 9M4 (H), from which outside (9M4O), inside (9M4I), and sphalerite-rich strata (9M4S) were sampled. Download Figure S1, PDF file, 4.5 MB.
Graphical representation of data presented in Table 1. Error bars represent 95% confidence intervals. All data were calculated using the preclustering option and average neighbor clustering in mothur and a cutoff of 3% for OTU determination. All samples were randomly resampled down to 9,149 tags, except for FS481 (only 8,923 tags in original sample), FS445 (only 8,398 tags in original sample), and the Lost City samples (all resampled to 5,567 tags). Download Figure S2, PDF file, 0.1 MB.
Bacterial distributions within the “other” section of Fig. 2. The y axis is the percentage of tags within the total sample. Download Figure S3, PDF file, 0.1 MB.
Bacterial distributions within the “other” section of Fig. 5. The y axis is the percentage of bacteria within the total data set (all extinct chimneys or all active chimneys). Download Figure S4, PDF file, 0.1 MB.
Sixteen most cosmopolitan tags and 16 most abundant tags for extinct sulfides in this study. GAST classification and the closest BLASTn hits in the NCBI database are given, followed by the samples from which the OTU was recovered and the potential ecological role of the tags where all members of that bacterial group perform a defined role.
Potential ecological roles of full-length clones for which obvious metabolisms can be inferred. Data are pooled from all samples; therefore, multiple clones are represented per lineage listed. The percentage of clones in a data set is for the entire data set (n = 452). Taxa are designated by class (phylum for Nitrospira and Chlorobi), order, family, and genus. The highest level at which an ecological role can be assigned per group of tags is given.
ACKNOWLEDGMENTS
We thank Mitch Sogin and Susan Huse for sequencing the samples described here; Bill Nelson, Julie Huber, and Pete Girguis for helpful discussions of the data; and Julie Huber, Pete Girguis, Alban Ramette, and John Baross for critical comments on the manuscript. We thank Bill Nelson for his helpful assistance with PERL scripts and Wolfgang Bach for field work collaboration and guidance (co-principal investigator on grant funding and sample collection).
This work was supported by grants from the Keck Foundation, the Gordon and Betty Moore Foundation (no. 1609 to K.J.E.), the National Research Council associate and NASA postdoctoral fellowship programs (B.M.T.), and NSF OCE-0241791 (RIDGE program) to K.J.E. (and W. Bach) for sample collection.
This is Center for Dark Energy Biosphere Investigations contribution 116.
Footnotes
Citation Sylvan JB, Toner BM, Edwards KJ. 2012. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 3(1):e00279-11. doi:10.1128/mBio.00279-11.
REFERENCES
- 1. Hannington MD, Jonasson IR, Herzig PM, Petersen S. 1995. Physical and chemical processes of seafloor mineralization at mid-ocean ridges, p. 115–157 In Humphris SE, Zierenberg RA, Mullineaux LS, Thomson RE, Seafloor hydrothermal systems: physical, chemical, biological and geological interactions. American Geophysical Union, Washington, DC [Google Scholar]
- 2. Hoek J, Banta A, Hubler F, Reysenbach AL. 2003. Microbial diversity of a sulphide spire located in the Edmond deep-sea hydrothermal vent field on the central Indian ridge. Geobiology 1:119–127 [Google Scholar]
- 3. Kormas KA, Tivey MK, Von Damm K, Teske A. 2006. Bacterial and archaeal phylotypes associated with distinct mineralogical layers of a white smoker spire from a deep-sea hydrothermal vent site (nine degrees N, East Pacific rise). Environ. Microbiol. 8:909–920 [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa S, et al. 2005. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54:141–155 [DOI] [PubMed] [Google Scholar]
- 5. Nakagawa S, et al. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619–1632 [DOI] [PubMed] [Google Scholar]
- 6. Reysenbach AL, Longnecker K, Kirshtein J. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takai K, et al. 2009. Variability in microbial communities in black smoker chimneys at the NW caldera vent field, brothers volcano, Kermadec arc. Geomicrobiol. J. 26:552–569 [Google Scholar]
- 8. Takai K, et al. 2008. Variability in the microbial communities and hydrothermal fluid chemistry at the newly discovered mariner hydrothermal field, southern Lau basin. J. Geophys. Res. Biogeosci. 113:G02031 [Google Scholar]
- 9. Voordeckers JW, et al. 2008. Culture dependent and independent analyses of 16S rRNA and ATP citrate lyase genes: a comparison of microbial communities from different black smoker chimneys on the mid-Atlantic ridge. Extremophiles 12:627–640 [DOI] [PubMed] [Google Scholar]
- 10. Rona PA, et al. 1993. Active and relict sea-floor hydrothermal mineralization at the TAG hydrothermal field, Mid-Atlantic Ridge. Econ. Geol. 88:1989–2017 [Google Scholar]
- 11. Lalou C, Brichet E, Hekinian R. 1985. Age dating of sulfide deposits from axial and off-axial structures on the East Pacific Rise near 12°50′ N. Earth Planet. Sci. Lett. 75:59–71 [Google Scholar]
- 12. Rouxel O, Shanks WC, Bach W, Edwards KJ. 2008. Integrated Fe- and S-isotope study of seafloor hydrothermal vents at East Pacific Rise 9–10 degrees N. Chem. Geol. 252:214–227 [Google Scholar]
- 13. Embley RW, et al. 1988. Submersible investigation of an extinct hydrothermal system on the Galapagos Ridge—sulfide mounds, stockwork zone, and differentiated lavas. Can. Mineral. 26:517–539 [Google Scholar]
- 14. Haymon RM, et al. 2008. High-resolution surveys along the hot spot-affected Galapagos Spreading Center: 3. black smoker discoveries and the implications for geological controls on hydrothermal activity. Geochem. Geophys. Geosystems 9:Q12006 [Google Scholar]
- 15. Herzig PM, Becker KP, Stoffers P, Backer H, Blum N. 1988. Hydrothermal silica chimney fields in the Galapagos Spreading Center at 86° W. Earth Planet. Sci. Lett. 89:261–272 [Google Scholar]
- 16. Kato S, et al. 2010. Biogeography and biodiversity in sulfide structures of active and inactive vents at deep-sea hydrothermal fields of the southern Mariana trough. Appl. Environ. Microbiol. 76:2968–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki Y, Inagaki F, Takai K, Nealson KH, Horikoshi K. 2004. Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb. Ecol. 47:186–196 [DOI] [PubMed] [Google Scholar]
- 18. Rogers DR, Santelli CM, Edwards KJ. 2003. Geomicrobiology of deep-sea deposits: estimating community diversity from low-temperature seafloor rocks and minerals. Geobiology 1:109–117 [Google Scholar]
- 19. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. 2009. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. U. S. A. 106:22427–22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Antonopoulos DA, et al. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brazelton WJ, et al. 2010. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc. Natl. Acad. Sci. U. S. A. 107:1612–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199 [DOI] [PubMed] [Google Scholar]
- 24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 25. Edgcomb V, et al. 2011. Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J. 5:1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huse SM, et al. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brenner DJ, et al. 2005. The Proteobacteria, part B, the Gammaproteobacteria. In Garrity GM, Bergey’s manual of systematic bacteriology, 2nd ed Springer Verlag, East Lansing, MI [Google Scholar]
- 28. Huber JA, et al. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100 [DOI] [PubMed] [Google Scholar]
- 29. Huber JA, et al. 2010. Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana arc seamounts. FEMS Microbiol. Ecol. 73:538–549 [DOI] [PubMed] [Google Scholar]
- 30. Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takai K, et al. 2009. Isolation and physiological characterization of two novel, piezophilic, thermophilic chemolithoautotrophs from a deep-sea hydrothermal vent chimney. Environ. Microbiol. 11:1983–1997 [DOI] [PubMed] [Google Scholar]
- 32. Newton RJ, et al. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4:784–798 [DOI] [PubMed] [Google Scholar]
- 33. Opatkiewicz AD, Butterfield DA, Baross JA. 2009. Individual hydrothermal vents at axial seamount harbor distinct subseafloor microbial communities. FEMS Microbiol. Ecol. 70:413–424 [DOI] [PubMed] [Google Scholar]
- 34. Weiss JV, et al. 2007. Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov. and Sideroxydans paludicola sp. nov. Geomicrobiol. J. 24:559–570 [Google Scholar]
- 35. Alawi M, Lipski A, Sanders T, Eva Maria P, Spieck E. 2007. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 1:256–264 [DOI] [PubMed] [Google Scholar]
- 36. Schrenk MO, Kelley DS, Delaney JR, Baross JA. 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69:3580–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou HY, et al. 2009. Microbial diversity of a sulfide black smoker in main endeavour hydrothermal vent field, Juan de Fuca ridge. J. Microbiol. 47:235–247 [DOI] [PubMed] [Google Scholar]
- 38. Inagaki F, Takai K, Nealson KH, Horikoshi K. 2004. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54:1477–1482 [DOI] [PubMed] [Google Scholar]
- 39. Alperin M, Hoehler T. 2010. The ongoing mystery of sea-floor methane. Science 329:288–289 [DOI] [PubMed] [Google Scholar]
- 40. Santelli CM, et al. 2008. Abundance and diversity of microbial life in ocean crust. Nature 453:653–657 [DOI] [PubMed] [Google Scholar]
- 41. Barns SM, Fundyga RE, Jeffries MW, Pace NR. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. U. S. A. 91:1609–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edwards KJ, Bond PL, Gihring TM, Banfield JF. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796–1799 [DOI] [PubMed] [Google Scholar]
- 43. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lane DJ. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E, Goodfellow M, Nucleic acid technology in bacterial systematics. John Wiley and Sons, New York, NY. [Google Scholar]
- 46. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huber JA, Johnson HP, Butterfield DA, Baross JA. 2006. Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8:88–99 [DOI] [PubMed] [Google Scholar]
- 48. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drummond AJ, et al. 2011. Geneious, v 5.4 http://www.geneious.com [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photos of samples collected for this study. Seafloor photos of 3M23 (A), 7M24 (B), 9M32 (C), and 9M4, a 9-m chimney from which a section was collected from the top 1 m (D). Shipboard photos of 3M23 (E), 3M33 (F), 7M24 (G), and 9M4 (H), from which outside (9M4O), inside (9M4I), and sphalerite-rich strata (9M4S) were sampled. Download Figure S1, PDF file, 4.5 MB.
Graphical representation of data presented in Table 1. Error bars represent 95% confidence intervals. All data were calculated using the preclustering option and average neighbor clustering in mothur and a cutoff of 3% for OTU determination. All samples were randomly resampled down to 9,149 tags, except for FS481 (only 8,923 tags in original sample), FS445 (only 8,398 tags in original sample), and the Lost City samples (all resampled to 5,567 tags). Download Figure S2, PDF file, 0.1 MB.
Bacterial distributions within the “other” section of Fig. 2. The y axis is the percentage of tags within the total sample. Download Figure S3, PDF file, 0.1 MB.
Bacterial distributions within the “other” section of Fig. 5. The y axis is the percentage of bacteria within the total data set (all extinct chimneys or all active chimneys). Download Figure S4, PDF file, 0.1 MB.
Sixteen most cosmopolitan tags and 16 most abundant tags for extinct sulfides in this study. GAST classification and the closest BLASTn hits in the NCBI database are given, followed by the samples from which the OTU was recovered and the potential ecological role of the tags where all members of that bacterial group perform a defined role.
Potential ecological roles of full-length clones for which obvious metabolisms can be inferred. Data are pooled from all samples; therefore, multiple clones are represented per lineage listed. The percentage of clones in a data set is for the entire data set (n = 452). Taxa are designated by class (phylum for Nitrospira and Chlorobi), order, family, and genus. The highest level at which an ecological role can be assigned per group of tags is given.