Abstract
This article uses an integrative systems biological view of the relationship between genotypes and phenotypes to clarify some conceptual problems in biological debates about causality. The differential (gene-centric) view is incomplete in a sense analogous to using differentiation without integration in mathematics. Differences in genotype are frequently not reflected in significant differences in phenotype as they are buffered by networks of molecular interactions capable of substituting an alternative pathway to achieve a given phenotype characteristic when one pathway is removed. Those networks integrate the influences of many genes on each phenotype so that the effect of a modification in DNA depends on the context in which it occurs. Mathematical modelling of these interactions can help to understand the mechanisms of buffering and the contextual-dependence of phenotypic outcome, and so to represent correctly and quantitatively the relations between genomes and phenotypes. By incorporating all the causal factors in generating a phenotype, this approach also highlights the role of non-DNA forms of inheritance, and of the interactions at multiple levels.
Keywords: genotype, phenotype, computational systems biology
1. Introduction
Are organisms encoded as molecular descriptions in their genes? By analysing the genome, could we solve the forward problem of computing the behaviour of the system from this information, as was implied by the original idea of the ‘genetic programme’ [1] and the more modern representation of the genome as the ‘book of life’? In this article, I will argue that this is both impossible and incorrect. We therefore need to replace the gene-centric ‘differential’ view of the relation between genotype and phenotype with an integrative view.
2. Impossibility
Current estimates of the number of genes in the human genome range up to 25 000, though the number would be even larger if we included regions of the genome forming templates for non-protein coding RNAs and as yet unknown numbers of microRNAs [2]. With no further information to restrict them, the number of conceivable interactions between 25 000 components is approximately 1070 000 [3]. Many more proteins are formed than the number of genes, depending on the number of splice variants and post-transcriptional modifications. Proteins are the real workhorses of the organism so the calculation should really be based on this number, which may be in excess of 100 000, and further increased by a wide variety of post-translational modifications that influence their function.
Of course, such calculations are not realistic. In practice, the great majority of the conceivable interactions cannot occur. Compartmentalization ensures that some components never interact directly with each other, and proteins certainly do not interact with everything they encounter. Nevertheless, we cannot rely on specificity of interactions to reduce the number by as much as was once thought. Most proteins are not very specific [4,5]. Each has many interactions (with central hubs having dozens) with other elements in the organism [6], and many (around 30%) are unstructured in the sense that they lack a unique three-dimensional structure and so can change to react in variable ways in protein and metabolic networks [7].
In figure 1, I show the calculations for a more reasonable range of possible interactions by calculating the results for between 0 and 100 gene products for each biological function (phenotype characteristic) for genomes up to 30 000 in size. At 100 gene products per function, we calculate around 10300 possible interactions. Even when we reduce the number of genes involved in each function to 25 we still calculate a figure, 1080, which is as large as the estimated number of elementary particles in the universe. These are therefore literally ‘astronomic’ numbers. We do not yet have any way of exploring interaction spaces of this degree of multi-dimensionality without insight into how the interactions are restricted. Computational biology has serious difficulties with the problem of combinatorial explosion even when we deal with just 100 elements, let alone tens of thousands.
Figure 1.
Genetic combinatorial explosion. Solutions of the equation  , where n denotes number of genes in the genome, r is the number assumed to be involved in each function. Ordinate: number of possible combinations (potential biological functions). Abscissa: Number of genes required in each function. The curves show results for genomes of various sizes between 100 and 30 000 genes and for up to 100 genes involved in each function (adapted from Feytmans et al. [3]).
, where n denotes number of genes in the genome, r is the number assumed to be involved in each function. Ordinate: number of possible combinations (potential biological functions). Abscissa: Number of genes required in each function. The curves show results for genomes of various sizes between 100 and 30 000 genes and for up to 100 genes involved in each function (adapted from Feytmans et al. [3]).
Given these estimates of the scale of the forward problem, no-one should contemplate calculating the interactions in this massively ‘blind’ bottom-up fashion. That is the reason why the middle-out approach has been proposed [8]. This was originally a suggestion made by Brenner et al. [9]. The quotations from that Novartis Foundation discussion are interesting in the present context. Brenner wrote ‘I know one approach that will fail, which is to start with genes, make proteins from them and to try to build things bottom-up’ ([9], p. 51) and, then later, ‘Middle-out. The bottom-up approach has very grave difficulties to go all the way’ ([9], p. 154). My interpretation of the ‘middle-out’ approach is that you start calculating at the level at which you have the relevant data. In my work, this is at the level of cells, where we calculate the interactions between the protein and other components that generate cardiac rhythm, then we reach ‘out’ to go down towards the level of genes [10] and upwards towards the level of the whole organ [11,12].1 By starting, in our case, at the level of the cell, we focus on the data relevant to that level and to a particular function at that level in order to reduce the number of components we must take into account. Other computational biologists choose other levels as their middle.
In practice, therefore, even a dedicated bottom-up computational biologist would look for ways in which nature itself has restricted the interactions that are theoretically possible. Organisms evolve step by step, with each step changing the options subsequently possible. I will argue that much of this restriction is embodied in the structural detail of the cells, tissues and organs of the body, as well as in its DNA. To take this route is therefore already to abandon the idea that the reconstruction can be based on DNA sequences alone.
3. Incorrect
One possible answer to the argument so far could be that while we may not be able, in practice, to calculate all the possible interactions, nevertheless it may be true that the essence of all biological systems is that they are encoded as molecular descriptions in their genes. An argument from impossibility of computation is not, in itself, an argument against the truth of a hypothesis. In the pre-relativity and pre-quantum mechanical world of physics (a world of Laplacian billiard balls), many people considered determinate behaviour of the universe to be obviously correct even though they would readily have admitted the practical impossibility of doing the calculations.
To the problem of computability therefore we must add that it is clearly incorrect to suppose that all biological systems are encoded in DNA alone. An organism inherits not just its DNA. It also inherits the complete fertilized egg cell and any non-DNA components that come via sperm. With the DNA alone, the development process cannot even get started, as DNA itself is inert until triggered by transcription factors (various proteins and RNAs). These initially come from the mother [13] and from the father, possibly through RNAs carried in the sperm [14–16]. It is only through an interaction between DNA and its environment, mediated by these triggering molecules, that development begins. The centriole also is inherited via sperm [17], while maternal transfer of antibodies and other factors has also been identified as a major source of transgenerational phenotype plasticity [18–20].
4. Comparing the different forms of inheritance
How does non-DNA inheritance compare with that through DNA? The eukaryotic cell is an unbelievably complex structure. It is not simply a bag formed by a cell membrane enclosing a protein soup. Even prokaryotes, formerly thought to fit that description, are structured [21] and some are also compartmentalized [22]. But the eukaryotic cell is divided up into many more compartments formed by the membranous organelles and other structures. The nucleus is also highly structured. It is not simply a container for naked DNA, which is why nuclear transfer experiments are not strict tests for excluding non-DNA inheritance.
If we wished to represent these structures as digital information to enable computation, we would need to convert the three-dimensional images of the cell at a level of resolution that would capture the way in which these structures restrict the molecular interactions. This would require a resolution of around 10 nm to give at least 10 image points across an organelle of around 100 nm diameter. To represent the three-dimensional structure of a cell around 100 µm across would require a grid of 10 000 image points across. Each gridpoint (or group of points forming a compartment) would need data on the proteins and other molecules that could be present and at what level. Assuming the cell has a similar size in all directions (i.e. is approximately a cube), we would require 1012 gridpoints, i.e. 1000 billion points. Even a cell as small as 10 µm across would require a billion grid points. Recall that the genome is about three billion base pairs. It is therefore easy to represent the three-dimensional image structure of a cell as containing as much information as the genome, or even more since there are only four possible nucleotides at each position in the genome sequence, whereas each grid point of the cellular structure representation is associated with digital or analogue information on a large number of features that are present or absent locally.
There are many qualifications to be put on these calculations and comparisons. Many of the cell structures are repetitive. This is what enables cell modellers to lump together compartments like mitochondria, endoplasmic reticulum, ribosomes, filaments, and other organelles and structures, though we are also beginning to understand that, sometimes, this is an oversimplification. A good example is the calcium signalling system in muscles, where the tiny spaces in which calcium signalling occurs, that couples excitation to contraction have to be represented at ever finer detail to capture what the experimental information tells us. Current estimates of the number of calcium ions in a single dyad (the space across which calcium signalling occurs) is only between 10 and 100 [23], too small for the laws of mass action to be valid.
Nevertheless, there is extensive repetition. One mitochondrion is basically similar to another, as are ribosomes and all the other organelles. But then, extensive repetition is also characteristic of the genome. A large fraction of the three billion base pairs forms repetitive sequences. Protein template regions of the human genome are estimated to be less than 1.5 per cent. Even if 99 per cent of the structural information from a cell image were to be redundant because of repetition, we would still arrive at figures comparable to the effective information content of the genome. And, for the arguments in this paper to be valid, it does not really matter whether the information is strictly comparable, nor whether one is greater than the other. Significance of information matters as much as its quantity. All I need to establish at this point is that, in a bottom-up reconstruction—or indeed in any other kind of reconstruction—it would be courting failure to ignore the structural detail. That is precisely what restricts the combinations of interactions (a protein in one compartment cannot interact directly with one in another, and proteins floating in lipid bilayer membranes have their parts exposed to different sets of molecules) and may therefore make the computations possible. Successful systems biology has to combine reduction and integration [24,25]. There is no alternative. Electrophysiological cell modellers are familiar with this necessity since the electrochemical potential gradients across membranes are central to function. The influence of these gradients on the gating of ion channel proteins is a fundamental feature of models of the Hodgkin–Huxley type. Only by integrating the equations for the kinetics of these channels with the electrochemical properties of the whole cell can the analysis be successful. As such models have been extended from nerve to cardiac and other kinds of muscle the incorporation of ever finer detail of cell structure has become increasingly important.
5. The differential view of genetics
These points are so obvious, and have been so ever since electron microscopes first revealed the fine details of those intricate sub-cellular structures around 50 years ago, that one has to ask how mainstream genetics came to ignore the problem. The answer lies in what I will call the differential view of genetics.
At this point, a little history of genetics is relevant. The original concept of a gene was whatever is the inheritable cause of a particular characteristic in the phenotype, such as eye colour, number of limbs/digits, and so on. For each identifiable phenotype characteristic, there would be a gene (actually an allele—a particular variant of a gene) responsible for that characteristic. A gene could be defined therefore as something whose presence or absence makes a difference to the phenotype. When genetics was combined with natural selection to produce the modern synthesis [26], which is usually called neo-Darwinism, the idea took hold that only those differences were relevant to evolutionary success and all that mattered in relating genetics to phenotypes was to identify the genetic causes of those differences. Since each phenotype must have such a cause (on this view at least) then selection of phenotypes amounts, in effect, to selection of individual genes. It does not really matter which way one looks at it. They are effectively equivalent [27]. The gene's-eye view then relegates the organism itself to the role of disposable carrier of its genes [28]. To this view we can add the idea that, in any case, only differences of genetic make-up can be observed. The procedure is simply to alter the genes, by mutation, deletion, addition and observe the effect on the phenotype.
I will call this gene-centric approach the ‘differential view’ of genetics to distinguish it from the ‘integral view’ I will propose later. To the differential view, we must add an implicit assumption. Since, on this view, no differences in the phenotype that are not caused by a genetic difference can be inherited, the fertilized egg cell (or just the cell itself in the case of unicellular organisms) does not evolve other than by mutations and other forms of evolution of its genes. The inherited information in the rest of the egg cell is ignored because (i) it is thought to be equivalent in different species (the prediction being that a cross-species clone will always show the phenotype of whichever species provides the genes), and (ii) it does not evolve or, if it does through the acquisition of new characteristics, these differences are not passed on to subsequent generations, which amounts to the same thing. Evolution requires inheritance. A temporary change does not matter.
At this stage in the argument, I will divide the holders of the differential view into two categories. The ‘strong’ version is that, while it is correct to say that the intricate structure of the egg cell is inherited as well as the genes, in principle that structure can be deduced from the genome information. On this view, a complete bottom-up reconstruction might still be possible even without the non-genetic information. This is a version of an old idea, that the complete organism is somehow represented in the genetic information. It just needs to be unfolded during development, like a building emerging from its blueprint.
The ‘weak’ version is one that does not make this assumption but still supposes that the genetic information carries all the differences that make one species different from another.
The weak version is easier to deal with, so I will start with that. In fact, it is remarkably easy to deal with. Only by restricting ourselves to the differential view of genetics it is possible to ignore the non-genetic structural information. But Nature does not play just with differences when it develops an organism. The organism develops only because the non-genetic structural information is also inherited and is used to develop the organism. When we try to solve the forward problem, we will be compelled to take that structural information into account even if it were to be identical in different species. To use a computer analogy, we need not only the ‘programme’ of life, we also need the ‘computer’ of life, the interpreter of the genome, i.e. the highly complex egg cell. In other words, we have to take the context of the cell into account, not only its genome. There is a question remaining, which is whether the weak version is correct in assuming the identity of egg cell information between species. I will deal with that question later. The important point at this stage is that, even with that assumption, the forward problem cannot be solved on the basis of genetic information alone. Recall that genes need to be activated to do anything at all.
Proponents of the strong version would probably also take this route in solving the forward problem, but only as a temporary measure. They would argue that, when we have gained sufficient experience in solving this problem, we will come to see how the structural information is somehow also encoded in the genetic information.
This is an article of faith, not a proven hypothesis. As I have argued elsewhere [29,30], the DNA sequences do not form a ‘programme’ that could be described as complete in the sense that it can be parsed and analysed to reveal its logic. What we have found in the genome is better described as a database of templates [31] to enable a cell to make proteins and RNA. Unless that complete ‘programme’ can be found (which I would now regard as highly implausible given what we already know of the structure of the genome), I do not think the strong version is worth considering further. It is also implausible from an evolutionary viewpoint. Cells must have evolved before genomes. Why on earth would nature bother to ‘code’ for detail which is inherited anyway in the complete cell? This would be as unnecessary as attempting to ‘code for’ the properties of water or of lipids. Those properties are essential for life (they are what allow cells to form), but they do not require genes. Mother Nature would have learnt fairly quickly how to be parsimonious in creating genetic information: do not code for what happens naturally in the physico-chemical universe. Many wonderful things can be constructed on the basis of relatively little transmitted information, relying simply on physico-chemical processes, and these include what seem at first sight to be highly complex structures like that of a flower (see, for example, [32]; figures 2 and 3).
Figure 2.

Solutions of a generalized Schrödinger equation for diffusive spheric growth from a centre (adapted from Nottale & Auffray [32]).
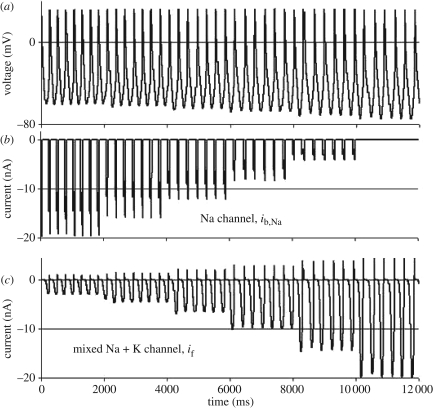
Figure 3.
Example of the use of computational systems biology to model a genetic buffering mechanism. (a) Membrane potential variations in a model of the sinus node pacemaker of the heart. (b) The background sodium channel, ib,Na, is progressively reduced until it is eventually ‘knocked out’. (c) The mixed (sodium and potassium) cation current channel, if, progressively takes over the function, and so ensures that the change in frequency is minimized (adapted from Noble et al. [61]), recomputed using COR: http://cor.physiol.ox.ac.uk/. Coordinates: membrane potential in millivolt, current in nanoampere, time (abscissa) in milliseconds.
The point here is not that a flower can be made without genes (clearly, the image in figure 2 is not a flower—it does not have the biochemistry of a flower, for example), but rather that genes do not need to code for everything. Nature can, as it were, get ‘free rides’ from the physics of structure: the attractors towards which systems move naturally. Such physical structures do not require detailed templates in the DNA sequences, they appear as the natural expression of the underlying physics. The structures can then act as templates for the self-organization of the protein networks, thus making self-organization a process depending both on the genome and the inherited structure.
6. Is the differential view correct?
Both the strong and weak versions exclude the possibility of inheritance of changes in the non-DNA structural information. Indications that this may not be entirely correct have existed for many years. Over 50 years ago, McLaren & Michie [33] showed that the skeletal morphology (number of tail vertebrae) of different strains of mice depended on that of the mother into which the fertilized egg cell was implanted, and cannot therefore be entirely determined by the genome. Many other maternal effects have since been found in mammals [13,34]. We can now begin to understand how these effects may occur. The genome is marked epigenetically in various ways that modify gene-expression patterns. These markings can also be transmitted from one generation to another, either via the germline or via behavioural marking of the relevant genes [14,35,36].
Transmission of changes in structural information also occurs in unicellular animals. Again, this has been known for many years. Surgical modification of the direction of cilia patterns in paramecium, produced by cutting a pole of the animal and reinserting it the wrong way round, are robustly inherited by the daughter cells down many generations [37,38].
Interest in this kind of phenomenon has returned, perhaps in the wake of discoveries in epigenetics that make the phenomena explicable. A good example is the work of Sun et al. [39] on cross-species cloning of fish from different genera. They enucleated fertilized goldfish eggs and then inserted a carp nucleus. The overall body structure of the resulting adult fish is intermediate. Some features are clearly inherited from the goldfish egg. Intriguingly, in the light of McLaren and Michie's work, this included the number of vertebrae. The goldfish has fewer than the carp. So does the cross-species clone.2
Sun et al.'s [39] work is remarkable for another reason also. Success in creating adult cross-species clones is very rare. Virtually all other attempts at cross-species cloning failed to develop to the adult [40]. An obvious possible explanation is that the egg cell information is too specific [41] as it has also evolved to become usually incompatible between different species. Strathmann [42] also refers to the influence of the egg cytoplasm on gene expression during early development as one of the impediments to hybridization in an evolutionary context. There is no good reason why cells themselves should have ceased to evolve once genomes arose. But if we need a specific (special purpose) ‘computer’ for each ‘programme’, the programme concept loses much of its attraction. The programming of living systems is distributed. Organisms are systems in continuous interaction with their environment. They are not Turing machines.
Contrary to the differential view, therefore, inheritance involves much more than nuclear DNA (see also [43]). It is simply incorrect to assume that all inherited differences are attributable to DNA [44,45].
7. The integral view of genetics
The alternative to the differential view is the integral approach. It is best defined as the complement to the differential approach. We study the contributions of a gene to all the functions in which its products take part. This is the approach of integrative biology, and here I am using ‘integral’ and ‘integrative’ in much the same sense. Integrative biology does not always or necessarily use mathematics of course, but even when it does not, the analogy with mathematical integration is still appropriate, precisely because it is not limited to investigating differences, and the additional information taken into account is analogous to the initial (= initial states of the networks of interactions) and boundary (= structural) conditions of mathematics. Indeed, they are exactly analogous when the mathematical modelling uses differential equations (as in figure 3 above). The middle-out approach is necessarily integrative. It must address the complexities arising from taking these conditions into account. The argument for the integrative approach is not that it is somehow easier or eliminates the complexity. On the contrary, the complexity is a major challenge. So, we need strong arguments for adopting this approach.
One such argument is that, most often, the differential approach does not work in revealing gene functions. Many interventions, such as knockouts, at the level of the genome are effectively buffered by the organism. In yeast, for example, 80 per cent of knockouts are normally ‘silent’ [46]. While there must be underlying effects in the protein networks, these are clearly hidden by the buffering at the higher levels. In fact, the failure of knockouts to systematically and reliably reveal gene functions is one of the great (and expensive) disappointments of recent biology. Note however that the disappointment exists only in the differential genetic view. By contrast, it is an exciting challenge from the integrative systems perspective. This very effective ‘buffering’ of genetic change is itself an important integrative property of cells and organisms. It is part of the robustness of organisms.
Moreover, even when a difference in the phenotype is manifest, it may not reveal the function(s) of the gene. In fact, it cannot do so, since all the functions shared between the original and the mutated gene are necessarily hidden from view. This is clearly evident when we talk of oncogenes [47]. What we mean is that a particular change in DNA sequence predisposes to cancer. But this does not tell us the function(s) of the un-mutated gene, which would be better characterized as a cell cycle gene, an apoptosis gene, etc. Only a full physiological analysis of the roles of the proteins, for which the DNA sequence forms templates, in higher level functions can reveal that. That will include identifying the real biological regulators as systems properties. Knockout experiments by themselves do not identify regulators [48]. Moreover, those gene changes that do yield a simple phenotype change are the few that happen to reflect the final output of the networks of interactions.
So, the view that we can only observe differences in phenotype correlated with differences in genotype leads both to incorrect labelling of gene functions, and it falls into the fallacy of confusing the tip with the whole iceberg. We want to know what the relevant gene products do in the organism as a physiological whole, not simply by observing differences. Most genes and their products, RNA and proteins, have multiple functions.
My point here is not that we should abandon knockouts and other interventions at the genome level. It is rather that this approach needs to be complemented by an integrative one. In contrast to the days when genes were hypothetical entities—postulated as hidden causes (postulated alleles—gene variants) of particular phenotypes—we now identify genes as particular sequences of DNA. These are far from being hypothetical hidden entities. It now makes sense to ask: what are all the phenotypic functions in which they (or rather their products, the RNAs and proteins) are involved.
Restricting ourselves to the differential view of genetics is rather like working only at the level of differential equations in mathematics, as though the integral sign had never been invented. This is a good analogy since the constants of integration, the initial and boundary conditions, restrain the solutions possible in a way comparable to that by which the cell and tissue structures restrain whatever molecular interactions are possible. Modelling of biological functions should follow the lead of modellers in the engineering sciences. Engineering models are constructed to represent the integrative activity of all the components in the system. Good models of this kind in biology can even succeed in explaining the buffering process and why particular knockouts and other interventions at the DNA level do not reveal the function (figure 3 and [8], pp. 106–108).
An example of this approach is shown in figure 3. A computational model of rhythmic activity in the sino-atrial node of the heart was used to investigate the effect of progressive reduction in one of the ion channel proteins contributing current, ib,Na, that determines the pacemaker frequency. In normal circumstances, 80 per cent of the depolarizing current is carried by this channel. One might therefore expect a very large influence on frequency as the channel activity is reduced and finally knocked-out. In fact, the computed change in frequency is surprisingly small. The model reveals the mechanism of this very powerful buffering. As ib,Na is reduced, there is a small shift of the waveform in a negative direction: the amplitude of the negative phase of the voltage wave increases. This small voltage change is sufficient to increase the activation of a different ion channel current, if, to replace ib,Na, so maintaining the frequency. The rest of the heart receives the signal corresponding to the frequency, but the change in amplitude is not transmitted. It is ‘hidden’. This is how effective buffering systems work. Moreover, via the modelling we can achieve quantitative estimates of the absolute contribution of each protein channel to the rhythm, whereas simply recording the overall effect of the ‘knockout’ would hide those contributions; we would conclude that the contribution is very small. The integral approach succeeds, by estimating 80 per cent as the normal contribution of the sodium channel protein, where the differential approach fails by estimating only 10 per cent.
Finally, the integral view helps to resolve two related problems in heredity and evolutionary theory. The first is the question of the concept of a gene [49,50]. The existence of multiple splice variants of many genes, and the possibility even of splicing exons from different gene sequences, has led some biologists to propose that we should redefine the ‘gene’, for example as the completed mRNA [51]. An obvious difficulty with this approach is why should we stop at the mRNA stage? Why not go further and redefine the gene in terms of the proteins for which DNA sequences act as the templates, or even higher (see commentary by Noble [52])? The distinction between genotype and phenotype would then be less clear-cut and could even disappear. Something therefore seems wrong in this approach, at least if we wish to maintain the difference, and surely it does make sense to distinguish between what is inherited and what is produced as a consequence of that inheritance.
But perhaps we do not need to redefine genes at all. Why not just let the concept of individual genes be recognized as a partial truth, with reference to the genome as a whole, and specifically its organization, providing the more complete view? There could be different ways in which we can divide the genome up, only some of which would correspond to the current concept of a gene. Viewing the genome as an ‘organ of the cell’ [53] fits more naturally with the idea that the genome is a read-write memory [54], which is formatted in various ways to suit the organism, not to suit our need to categorize it. We certainly should not restrict our understanding of the way in which genomes can evolve by our imperfect definitions of a gene.
The second problem that this view helps to resolve is the vexed question of inheritance of acquired characteristics and how to fit it into modern evolutionary theory. Such inheritance is a problem for the neo-Darwinian synthesis precisely because it was formulated to exclude it. Too many exceptions now exist for that to be any longer tenable ([45]; see also the examples discussed previously).
In fact, the need to extend the synthesis has been evident for a long time. Consider, for example, the experiments of Waddington [55], who introduced the original idea of epigenetics. His definition was the rearrangement of gene alleles in response to environmental stress. His experiments on Drosophila showed that stress conditions could favour unusual forms of development, and that, after selection for these forms over a certain number of generations, the stress condition was no longer required (see discussion in Bard [56]). The new form had become permanently inheritable. We might argue over whether this should be called Lamarckism (see [57] for historical reasons why this term may be incorrect), but it is clearly an inherited acquired characteristic. Yet no mutations need occur to make this possible. All the gene alleles required for the new phenotype were already in the population but not in the right combinations in most, or even any, individuals to produce the new phenotype without the environmental stress. Those that did produce the new phenotype on being stressed had combinations that were at least partly correct. Selection among these could then improve the chances of individuals occurring for which the combinations were entirely correct so that the new phenotype could now be inherited even without the environmental stress. Waddington called this process an ‘assimilation’ of the acquired characteristic. There is nothing mysterious in the process of assimilation. Artificial selection has been used countless times to create new strains of animals and plants, and it has been used recently in biological research to create different colonies of high- and low-performing rats for studying disease states [58]. The main genetic loci involved can now be identified by whole genome studies (see, for example, [59]). The essential difference is that Waddington used an environmental stress that altered gene expression and revealed cryptic genetic variation and selected for this stress-induced response, rather than just selecting for the response from within an unstressed population The implication is obvious: in an environment in which the new phenotype was an advantage, natural selection could itself produce the assimilation. Natural selection is not incompatible with inheritance of acquired characteristics. As Darwin himself realized (for details, see Mayr [60]), the processes are complementary.
Neo-Darwinists dismissed Waddington's work largely because it did not involve the environment actually changing individual DNA gene sequences. But this is to restrict acquisition of evolutionarily significant change to individual DNA sequences (the gene's-eye view). On an integrative view, a new combination of alleles is just as significant from an evolutionary point of view. Speciation (defined, e.g., as failure of interbreeding) could occur just as readily from this process—and, as we now know, many other processes, such as gene transfer, genome duplication, symbiogenesis—as it might through the accumulation of mutations. What is the difference, from the organism's point of view, between a mutation in a particular DNA sequence that enables a particular phenotype to be displayed and a new combination of alleles that achieves the same result? There is an inherited change at the global genome level, even if no mutations in individual genes were involved. Sequences change, even if they do not occur within what we characterize as genes. Taking the integrative view naturally leads to a more inclusive view of the mechanisms of evolutionary change. Focusing on individual genes obscures this view.
In this article, I have been strongly critical of the gene-centred differential view. Let me end on a more positive note. The integral view does not exclude the differential view any more than integration excludes differentiation in mathematics. They complement each other. Genome sequencing, epigenomics, metabolomics, proteomics, transcriptomics are all contributing basic information that is of great value. We have only to think of how much genome sequencing of different species has contributed to evolutionary theory to recognize that the huge investment involved was well worth the effort. As integrative computational biology advances, it will be using this massive data collection, and it will be doing so in a meaningful way. The ‘meaning’ of a biological function lies at the level at which it is integrated, often enough at the level of a whole cell (a point frequently emphasized by Sydney Brenner), but in principle, the integration can be at any level in the organism. It is through identifying that level and the meaning to the whole organism of the function concerned that we acquire the spectacles required to interpret the data at other levels.
Acknowledgements
Work in the author's laboratory is funded by the EU (the Biosim network of excellence under Framework 6 and the PreDiCT project under Framework 7) and the British Heart Foundation. I would like to thank the participants of the seminars on Conceptual Foundations of Systems Biology at Balliol College, particularly Jonathan Bard, Tom Melham and Eric Werner, and Peter Kohl for the context of discussions in which some of the ideas for this article were developed. I thank Charles Auffray and the journal referees for many valuable suggestions on the manuscript.
Footnotes
Note that the terms ‘bottom’, ‘up’, ‘middle’ and ‘out’ are conveying the sense of a hierarchy between levels of organization in biological systems that tends to ignore interactions that take place between levels in all directions. So very much as ‘bottom-up’ and ‘top-down’ approaches are arguably complementary, we should consider ‘out-in’ as well as ‘middle-out’ approaches in our attempts to integrate upward and downward causation chains.
Note also that cross-species clones are not a full test of the differential view, since what is transferred between the species is not just DNA. The whole nucleus is transferred. All epigenetic marking that is determined by nuclear material would go with it. Cytoplasmic factors from the egg would have to compete with the nuclear factors to exert their effects.
One contribution of 16 to a Theme Issue ‘Advancing systems medicine and therapeutics through biosimulation’.
References
- 1.Jacob F., Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 10.1016/S0022-2836(61)80072-7 (doi:10.1016/S0022-2836(61)80072-7) [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. 2002. DNA events. An RNA microcosm. Science 297, 2002–2003 10.1126/science.1077906 (doi:10.1126/science.1077906) [DOI] [PubMed] [Google Scholar]
- 3.Feytmans E., Noble D., Peitsch M. 2005. Genome size and numbers of biological functions. Trans. Comput. Syst. Biol. 1, 44–49 10.1007/978-3-540-32126-2_4 (doi:10.1007/978-3-540-32126-2_4) [DOI] [Google Scholar]
- 4.Bray D. 2009. Wetware. A computer in every cell. New Haven, CT: Yale University Press [Google Scholar]
- 5.Kupiec J. 2009. The origin of individuals: a Darwinian approach to developmental biology. London, UK: World Scientific Publishing Company [Google Scholar]
- 6.Bork P., Jensen L. J., von Mering C., Ramani A. K., Lee I.-S., Marcotte E. M. 2004. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 14, 292–299 10.1016/j.sbi.2004.05.003 (doi:10.1016/j.sbi.2004.05.003) [DOI] [PubMed] [Google Scholar]
- 7.Gsponer J., Babu M. M. 2009. The rules of disorder or why disorder rules. Progr. Biophys. Mol. Biol. 99, 94–103 10.1016/j.pbiomolbio.2009.03.001 (doi:10.1016/j.pbiomolbio.2009.03.001) [DOI] [PubMed] [Google Scholar]
- 8.Noble D. 2006. The music of life. Oxford, UK: Oxford University Press [Google Scholar]
- 9.Brenner S., Noble D., Sejnowski T., Fields R. D., Laughlin S., Berridge M., Segel L., Prank K., Dolmetsch R. E. 2001. Understanding complex systems: top-down, bottom-up or middle-out? In Novartis Foundation Symposium: Complexity in biological information processing, vol. 239, pp. 150–159 Chichester, UK: John Wiley [Google Scholar]
- 10.Clancy C. E., Rudy Y. 1999. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature 400, 566–569 10.1038/23034 (doi:10.1038/23034) [DOI] [PubMed] [Google Scholar]
- 11.Bassingthwaighte J. B., Hunter P. J., Noble D. 2009. The Cardiac Physiome: perspectives for the future. Exp. Physiol. 94, 597–605 10.1113/expphysiol.2008.044099 (doi:10.1113/expphysiol.2008.044099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noble D. 2007. From the Hodgkin–Huxley axon to the virtual heart. J. Physiol. 580, 15–22 10.1113/jphysiol.2006.119370 (doi:10.1113/jphysiol.2006.119370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gluckman P., Hanson M. 2004. The fetal matrix. Evolution, development and disease. Cambridge, UK: Cambridge University Press [Google Scholar]
- 14.Anway M. D., Memon M. A., Uzumcu M., Skinner M. K. 2006. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J. Androl. 27, 868–879 10.2164/jandrol.106.000349 (doi:10.2164/jandrol.106.000349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barroso G., Valdespin C., Vega E., Kershenovich R., Avila R., Avendano C., Oehninger S. 2009. Developmental sperm contributions: fertilization and beyond. Fertil. Steril. 92, 835–848 10.1016/j.fertnstert.2009.06.030 (doi:10.1016/j.fertnstert.2009.06.030) [DOI] [PubMed] [Google Scholar]
- 16.Pembrey M. E., Bygren L. O., Kaati G., Edvinsson S., Northstone K., Sjostrom M., Golding J. & ALSPAC study team 2006. Sex-specific, male-line transgenerational responses in humans. Eur. J. Hum. Genet. 14, 159–166 10.1038/sj.ejhg.5201538 (doi:10.1038/sj.ejhg.5201538) [DOI] [PubMed] [Google Scholar]
- 17.Sathananthan A. H. 2009. Editorial. Human centriole: origin, and how it impacts fertilization, embryogenesis, infertility and cloning. Ind. J. Med. Res. 129, 348–350 [PubMed] [Google Scholar]
- 18.Agrawal A. A., Laforsch C., Tollrian R. 1999. Transgenerational induction of defences in animals and plants. Nature 401, 60–63 10.1038/43425 (doi:10.1038/43425) [DOI] [Google Scholar]
- 19.Boulinier T., Staszewski V. 2008. Maternal transfer of antibodies: raising immuno-ecology issues. Trends Ecol. Evol. 23, 282–288 10.1016/j.tree.2007.12.006 (doi:10.1016/j.tree.2007.12.006) [DOI] [PubMed] [Google Scholar]
- 20.Hasselquist D., Nilsson J. A. 2009. Maternal transfer of antibodies in vertebrates: trans-generational effects on offspring immunity. Phil. Trans. R. Soc. B 364, 51–60 10.1098/rstb.2008.0137 (doi:10.1098/rstb.2008.0137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michie K. A., Lowe J. 2006. Dynamic filaments of the bacterial cytoskeleton. Ann. Rev. Biochem. 75, 467–492 10.1146/annurev.biochem.75.103004.142452 (doi:10.1146/annurev.biochem.75.103004.142452) [DOI] [PubMed] [Google Scholar]
- 22.Fuerst J. 2005. Intracellular compartmentation in planctomycetes. Ann. Rev. Microbiol. 59, 299–328 10.1146/annurev.micro.59.030804.121258 (doi:10.1146/annurev.micro.59.030804.121258) [DOI] [PubMed] [Google Scholar]
- 23.Tanskanen A. J., Greenstein J. L., Chen A., Sun S. X., Winslow R. L. 2007. Protein geometry and placement in the cardiac dyad influence macroscopic properties of calcium-induced calcium release. Biophys. J. 92, 3379–3396 10.1529/biophysj.106.089425 (doi:10.1529/biophysj.106.089425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohl P., Crampin E., Quinn T. A., Noble D. 2010. Systems biology: an approach. Clin. Pharmacol. Ther. 88, 25–33 10.1038/clpt.2010.92 (doi:10.1038/clpt.2010.92) [DOI] [PubMed] [Google Scholar]
- 25.Kohl P., Noble D. 2009. Systems biology and the Virtual Physiological Human. Mol. Syst. Biol. 5, 292, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huxley J. S. 1942. Evolution: the modern synthesis. London, UK: Allen & Unwin [Google Scholar]
- 27.Dawkins R. 1982. The extended phenotype. London, UK: Freeman [Google Scholar]
- 28.Dawkins R. 1976. The selfish gene. Oxford, UK: OUP [Google Scholar]
- 29.Noble D. 2008. Genes and causation. Phil. Trans. R. Soc. A 366, 3001–3015 10.1098/rsta.2008.0086 (doi:10.1098/rsta.2008.0086) [DOI] [PubMed] [Google Scholar]
- 30.Noble D. 2010. Biophysics and systems biology. Phil. Trans. R. Soc. A 368, 1125–1139 10.1098/rsta.2009.0245 (doi:10.1098/rsta.2009.0245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atlan H., Koppel M. 1990. The cellular computer DNA: program or data? Bull. Math. Biol. 52, 335–348 [DOI] [PubMed] [Google Scholar]
- 32.Nottale L., Auffray C. 2008. Scale relativity and integrative systems biology 2. Macroscopic quantum-type mechanics. Progr. Biophys. Mol. Biol. 97, 115–157 10.1016/j.pbiomolbio.2007.09.001 (doi:10.1016/j.pbiomolbio.2007.09.001) [DOI] [PubMed] [Google Scholar]
- 33.McLaren A., Michie D. 1958. An effect of uterine environment upon skeletal morphology of the mouse. Nature 181, 1147–1148 10.1038/1811147a0 (doi:10.1038/1811147a0) [DOI] [PubMed] [Google Scholar]
- 34.Mousseau T. A., Fox C. W. 1998. Maternal effects as adaptations. Oxford, UK: Oxford University Press [Google Scholar]
- 35.Weaver I. C. G. 2009. Life at the interface between a dynamic environment and a fixed genome. In Mammalian brain development (ed. Janigro D.), pp. 17–40 New York, NY: Humana Press, Springer [Google Scholar]
- 36.Weaver I. C. G., Cervoni N., Champagne F. A., D'Alessio A. C., Sharma S., Sekl J. R., Dymov S., Szyf M., Meaney M. J. 2004. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 10.1038/nn1276 (doi:10.1038/nn1276) [DOI] [PubMed] [Google Scholar]
- 37.Beisson J., Sonneborn T. M. 1965. Cytoplasmic inheritance of the organization of the cell cortex in paramecium Aurelia. Proc. Natl Acad. Sci. USA 53, 275–282 10.1073/pnas.53.2.275 (doi:10.1073/pnas.53.2.275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonneborn T. M. 1970. Gene action on development. Proc. R. Soc. Lond. B 176, 347–366 10.1098/rspb.1970.0054 (doi:10.1098/rspb.1970.0054) [DOI] [PubMed] [Google Scholar]
- 39.Sun Y. H., Chen S. P., Wang Y. P., Hu W., Zhu Z. Y. 2005. Cytoplasmic impact on cross-genus cloned fish derived from transgenic common carp (Cyprinus carpio) nuclei and goldfish (Carassius auratus) enucleated eggs. Biol. Reprod. 72, 510–515 10.1095/biolreprod.104.031302 (doi:10.1095/biolreprod.104.031302) [DOI] [PubMed] [Google Scholar]
- 40.Chung Y., et al. 2009. Reprogramming of human somatic cells using human and animal oocytes. Cloning Stem Cells 11, 1–11 10.1089/clo.2009.0004 (doi:10.1089/clo.2009.0004) [DOI] [PubMed] [Google Scholar]
- 41.Chen T., Zhang Y.-L., Jiang Y., Liu J.-H., Schatten H., Chen D.-Y., Sun Y. 2006. Interspecies nuclear transfer reveals that demethylation of specific repetitive sequences is determined by recipient ooplasm but not by donor intrinsic property in cloned embryos. Mol. Reprod. Dev. 73, 313–317 10.1002/mrd.20421 (doi:10.1002/mrd.20421) [DOI] [PubMed] [Google Scholar]
- 42.Strathmann R. R. 1993. Larvae and evolution: towards a new zoology (book review). Q. Rev. Biol. 68, 280–282 10.1086/418103 (doi:10.1086/418103) [DOI] [Google Scholar]
- 43.Maurel M.-C., Kanellopoulos-Langevin C. 2008. Heredity: venturing beyond genetics. Biol. Reprod. 79, 2–8 10.1095/biolreprod.107.065607 (doi:10.1095/biolreprod.107.065607) [DOI] [PubMed] [Google Scholar]
- 44.Jablonka E., Lamb M. 1995. Epigenetic inheritance and evolution. The Lamarckian dimension. Oxford, UK: Oxford University Press [Google Scholar]
- 45.Jablonka E., Lamb M. 2005. Evolution in four dimensions. Boston, MA: MIT Press [Google Scholar]
- 46.Hillenmeyer M. E., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365 10.1126/science.1150021 (doi:10.1126/science.1150021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg R. A. 1996. How cancer arises. Scient. Am. 275, 62–70 10.1038/scientificamerican0996-62 (doi:10.1038/scientificamerican0996-62) [DOI] [PubMed] [Google Scholar]
- 48.Davies J. 2009. Regulation, necessity, and the misinterpretation of knockouts. Bioessays 31, 826–830 10.1002/bies.200900044 (doi:10.1002/bies.200900044) [DOI] [PubMed] [Google Scholar]
- 49.Pearson H. 2006. What is a gene? Nature 441, 399–401 10.1038/441398a (doi:10.1038/441398a) [DOI] [PubMed] [Google Scholar]
- 50.Pennisi E. 2007. DNA study forces rethink of what it means to be a gene. Science 316, 1556–1557 10.1126/science.316.5831.1556 (doi:10.1126/science.316.5831.1556) [DOI] [PubMed] [Google Scholar]
- 51.Scherrer K., Jost J. 2007. Gene and genon concept. Coding versus regulation. Theory Biosci. 126, 65–113 10.1007/s12064-007-0012-x (doi:10.1007/s12064-007-0012-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noble D. 2009. Commentary on Scherrer & Jost (2007) Gene and genon concept: coding versus regulation. Theory Biosci. 128, 153. 10.1007/s12064-009-0073-0 (doi:10.1007/s12064-009-0073-0) [DOI] [PubMed] [Google Scholar]
- 53.McClintock B. 1984. The significance of responses of the genome to challenge. Science 226, 792–801 10.1126/science.15739260 (doi:10.1126/science.15739260) [DOI] [PubMed] [Google Scholar]
- 54.Shapiro J. A. 2009. Letting E. coli teach me about genome engineering. Genetics 183, 1205–1214 10.1534/genetics.109.110007 (doi:10.1534/genetics.109.110007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waddington C. H. 1959. Canalization of development and genetic assimilation of acquired characteristics. Nature 183, 1654–1655 10.1038/1831654a0 (doi:10.1038/1831654a0) [DOI] [PubMed] [Google Scholar]
- 56.Bard J. B. L. 2008. Waddington's legacy to developmental and theoretical biology. Biol. Theory 3, 188–197 10.1162/biot.2008.3.3.188 (doi:10.1162/biot.2008.3.3.188) [DOI] [Google Scholar]
- 57.Noble D. 2010. Letter from Lamarck. Physiol. News 78, 31 [Google Scholar]
- 58.Koch L. G., Britton S. L. 2001. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol. Genom. 5, 45–52 [DOI] [PubMed] [Google Scholar]
- 59.Rubin C.-J., et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464, 587–591 10.1038/nature08832 (doi:10.1038/nature08832) [DOI] [PubMed] [Google Scholar]
- 60.Mayr E. 1964. Introduction. In The origin of species. Cambridge, MA: Harvard [Google Scholar]
- 61.Noble D., Denyer J. C., Brown H. F., DiFrancesco D. 1992. Reciprocal role of the inward currents ib,Na and if in controlling and stabilizing pacemaker frequency of rabbit sino-atrial node cells. Proc. R. Soc. B 250, 199–207 10.1098/rspb.1992.0150 (doi:10.1098/rspb.1992.0150) [DOI] [PubMed] [Google Scholar]