Abstract
Ultrasound elastography is a technique used for clinical imaging of tissue stiffness with a conventional ultrasound machine. It was first proposed two decades ago, but active research continues in this area to the present day. Numerous clinical applications have been investigated, mostly related to cancer imaging, and though these have yet to prove conclusive, the technique has seen increasing commercial and clinical interest. This paper presents a review of the most widely adopted, non-quantitative, techniques focusing on technical innovations rather than clinical applications. The review is not intended to be exhaustive, concentrating instead on placing the various techniques in context according to the authors' perspective of the field.
Keywords: ultrasound, elastography, tissue stiffness, strain imaging
1. Introduction
1.1. Motivation
The term ‘elastography’, first suggested by Ophir et al. [1], refers to methods of imaging the mechanical properties of tissue, specifically those related to the elastic (Young's) modulus. The idea that mechanical properties are related to pathology is not new: clinicians have employed manual palpation to feel for stiff lumps as long ago as 400 BC [2]. However, recent improvements in imaging technology have allowed accurate, high resolution, in some cases quantitative visualization of the stiffness of relatively deep-lying structures.
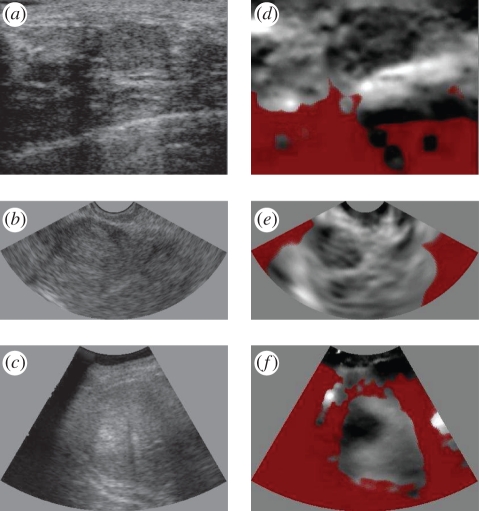
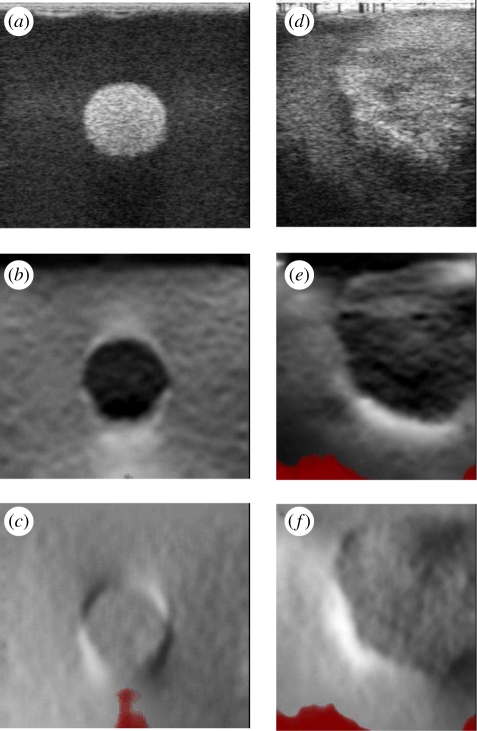
The rapid development of this technique has spawned clinical trials in many anatomical areas. These indicate that elastograms may differentiate between benign and malignant lesions, and distinguish between different types of malignancy [3–8], in some cases better than conventional B-mode ultrasound images [9]. Possible applications include discrimination without biopsy between complex cysts and malignant breast lesions [10], monitoring of atherosclerosis [11,12], detection and grading of deep vein thrombosis [13], assessment of skin pathologies [14] and evaluation of myocardial fitness [15]. Sample images are shown in figure 1 from some of our own clinical trials. However, according to a recent evidence-based review [16], only transient ultrasound elastography (a quantitative technique for making point stiffness measurements, not images) was considered a clinically proven technique for assessing liver fibrosis: in all other cases, the evidence was inconclusive. Nevertheless, most manufacturers now offer or are developing some form of elastography imaging on their flagship ultrasound systems.
Figure 1.
Clinical examples of ultrasound elastography. (a–c) Conventional ultrasound images and (d–f) elastograms from our own studies in three anatomical areas. (a,d) A fibroadenoma of the breast. (b,e) A fibroid in the endometrium. (c,f) A hepatocellular carcinoma of the liver. In these elastograms, black is stiff, white is soft and red represents unreliable data.
1.2. Elastographic techniques
Elastography involves the measurement of local tissue deformation in response to some sort of applied mechanical stress. The way in which the tissue deforms gives us information about the tissue's mechanical properties. The many elastographic (or related) techniques can be loosely categorized according to how the stress is applied and how the deformation is measured. For example, stress application can be external by the probe [1] or internal via ultrasonic radiation force [17], while deformation measurement can be achieved using magnetic resonance imaging (MRI) [18] or ultrasound [19]. In quasi-static ultrasound elastography, the deformation is induced by manually pressing on the anatomy with the transducer, and measured using ultrasound. Such techniques are known by various names commercially, including eSie Touch (Siemens Healthcare, http://www.medical.siemens.com), ElastoQ (Toshiba Medical Systems, http://www.medical.toshiba.com), Elastoscan (Medison, http://www.medison.com) and simply elastography (GE Healthcare, http://www.gehealthcare.com and Philips Healthcare, http://www.healthcare.philips.com). In contrast to more quantitative techniques based on shear wave speed [20], this is a qualitative technique, unsuitable for measuring absolute tissue stiffness, though it is possible to relate stiffness of a lesion to that of the background tissue. However, it has a high spatial resolution, is real time, and does not require any modifications to conventional ultrasound hardware.
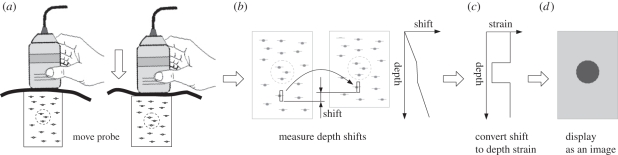
The essential components of quasi-static ultrasound elastography are summarized in figure 2. B-mode images are steadily acquired, with a slight varying pressure applied on the surface of the anatomy through the ultrasound probe. For scanning to 4 or 5 cm, physiological tremor is often enough to generate this pressure variation. It is from here that the term quasi-static derives, since the probe is in continuous motion, but with sufficiently low velocity and acceleration such that static mechanics can be assumed. The radio frequency (RF) ultrasound echo signals from each new image are compared with the previous image, and the tissue displacement is estimated at multiple locations along each signal (A-line). The axial strain is estimated by taking the gradient of this deformation data. An elastogram is displayed, which is a processed version of this strain data mapped to a colour scale (grey in this case). Stiff regions compress less, and hence exhibit lower strain values.
Figure 2.
Overview of the quasi-static elastography process. (a) The anatomy is scanned with a conventional ultrasound probe, which is moved very slightly up and down (much less than is shown here). In this example, the dashed circle is stiffer than the surrounding material. (b) Anatomical displacement in the axial direction is calculated everywhere in the ultrasound image. (c) Gradient estimation in the axial direction and filtering gives an estimate of axial strain. (d) A normalized version of the axial strain is converted to greyscale and displayed as an image, where black is stiff and white is soft.
1.3. Tissue mechanics
The mechanics of tissue are highly complex and many assumptions have to be made in order to relate measurable values to a mechanical material property. These assumptions have the effect of introducing artefacts into elastograms, which are often incorrectly interpreted as a direct visualization of the material stiffness. The linear viscoelastic model [21–23] relates stress to strain, describing anisotropic material properties with a dependency over time, which might be related to biochemical changes [24]. Owing to the quasi-static nature of the scanning process, we can ignore viscous effects, giving rise to the linear elastic model [21,22]; however, this still contains far more parameters than can be constrained by practical empirical measurement. Hence, tissue is generally assumed to be isotropic, in which case stress and strain can be modelled as a simple function of the Young modulus E and the Poisson ratio ν. It is usually assumed that ν is close to 0.5 in most tissue, leaving E alone to characterize an incompressible, isotropic material's stiffness. It has been suggested that this parameter accounts almost entirely for the useful information accessible by manual palpation [25,26].
The time dependency is sometimes re-introduced to this simplified model by considering the material to be suffused by an incompressible, near-inviscid fluid [27]. Resulting measurements of porosity might be useful for assessing oedematous tissue [28,29].
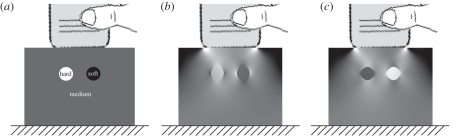
Even with the simple, single parameter model, estimating E is still problematic, since this relies on both stress and strain, and the stress field is not known. Figure 3 highlights this point with a finite-element simulation (Abaqus v. 6.7, Simulia, RI, USA). We can measure the strain in figure 3c, but we really need to divide this into the unknown stress in figure 3b in order to estimate the stiffness in figure 3a. Normalization and inversion techniques will be discussed in §2.4: their aim is to recover something similar to E from the strain measurements. However, even though the strain image does not directly relate to stiffness, it is apparent that the strain in the hard and soft lesions has good contrast and high spatial accuracy. Nearly all elastograms are actually versions of this strain image, rather than true stiffness images, and hence the term strain imaging or pseudo-strain imaging is sometimes used as a surrogate for elastography.
Figure 3.
Finite-element simulations of quasi-static elastography. (a) A medium is simulated with inclusions both harder and softer than the background material. (b) The axial stress field which results from a 1% axial movement of the probe, on a scale from 0 to 2% of the Young modulus of the background material. (c) Corresponding axial strain, on a scale from 0 to 2% strain. The images are derived from data used in Gee et al. [30].
1.4. Review structure
The quasi-static elastography technique can be split into several distinct stages, which are all necessary for producing high quality, clinically useful elastograms. Section 2.1 reviews the methods for tracking the relative local deformation of ultrasound data between images, which lies at the heart of the technique. Section 2.2 discusses the measures used to calculate the quality of the displacement estimates and their use in limiting major tracking errors. Section 2.3 considers the process of creating strain data from displacement data, and methods of filtering these data. Section 2.4 looks at ways of creating images that are closer to stiffness than strain. Finally, §2.5 covers the process of creating dynamic, meaningful images from the raw image data. In §3, we also take a brief look at the related areas of three-dimensional imaging, and imaging of shear strain for detecting slip.
We summarize the various techniques that have been used for each stage, drawing examples from our own work in this area. The organization of the material in the following sections naturally follows our own understanding and emphasis in elastographic imaging. We neither claim to include everything which is good within the field, nor exclude everything which is not. Our aim is to give a broad overview of the field from which the technique can be explored.
2. Quasi-static elastography
2.1. Tracking tissue movement
A key part of an elastographic imaging system is the algorithm used to estimate tissue deformation between two frames of ultrasound data. The probe movement in quasi-static elastography is seldom purely in the axial direction, and in any case even axial movement will deform the tissue in all dimensions. However, the axial deformation contains most of the useful mechanical information, and it can be measured more accurately than motion in other directions [31]. Hence lateral or elevational tracking, if employed at all, is mainly of use to improve the estimation of the axial deformation.
There are many different types of displacement estimators [32,33], but they can be grouped into certain categories. Most estimators are concerned with the matching of blocks of RF ultrasound data by rigid relative movement of the blocks until some similarity metric is optimized. Correlation maximization was the first such metric to be proposed [1,19,34] and remains the most widely used. Others include sum of absolute differences [35,36], sum of squared differences [37], correlation phase [38–40] and weighted phase separation [41]. Displacement must be estimated to sub-sample precision, either by interpolation of the similarity metric before finding the peak [37,39,42–53] or by interpolation of the RF data during the matching process [41,54]. Phase-based similarity metrics are particularly efficient at providing sub-sample accuracy. Optimal alignment is indicated by zero correlation phase [38,55], or zero phase separation [41], but a non-zero result, coupled with knowledge of the transducer's centre frequency, allows rapid iteration towards the correct answer provided the initial alignment is within half a wavelength of the zero-phase shift [39]. An inaccurate estimate of the centre frequency does not affect the accuracy of the displacement estimate, just the speed of convergence [39].
Correlation maximization can be achieved by independent exhaustive searches at each window [19], or using uphill search from a guessed initial displacement, which must be within half a wavelength of the correct displacement, i.e. exactly the same constraint as in phase-based estimation. The initial guess can be achieved by imposing inter-window continuity, either by dynamic programming [56,57], or by minimizing a global cost function that penalizes discontinuous displacements [37,48,58], or by tracking the displacements from one window to the next [41,45,49,50,54,59,60]. The search at each window might be fully two-dimensional [44], or there may be separate, one-dimensional axial and lateral searches, perhaps with some iteration between the two [46,54]. There could be a single, high-resolution matching process [39,41,43,47,54], or a multi-level approach with a coarse matching stage initializing subsequent stages at progressively higher resolutions [36,37,48,61–63]. This matching process affects both the speed and robustness of displacement estimation. For example, exhaustive search is neither fast nor robust: it requires a large search range, and false-positive matches are common when the window size is small (as required for fine-scale displacement estimation) and the noise level significant [64].
A potential problem with all such approaches is that pre- and post-deformation windows can match poorly, because the data in the windows themselves may be deformed to different extents. Several techniques have been developed that involve warping the data to increase the correlation between windows. These are variously referred to as ‘temporal stretching’ [65], ‘adaptive stretching’ [42] or ‘companding’ [66], and involve searching over a stretch as well as a displacement parameter when matching each window. Companding increases the precision of deformation data, but at huge computational cost. However, it has been noted that the main advantage of this approach is not so much the increase in correlation that it affords, but rather the ability to match each window at all points simultaneously [67].
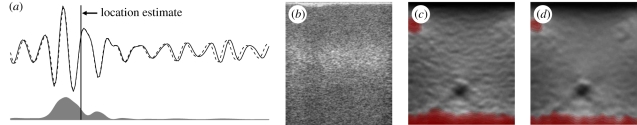
The precision of displacement estimates from rigidly displaced window matching can be increased by noting the position within the window at which the pre- and post-displacement data match best. For most similarity measures, particularly phase-based methods, the estimation of this location is a simple calculation based on the signal amplitude, as in figure 4a [67], and has hence been termed amplitude modulation correction (AMC). Taking note of the location at which each displacement estimate is most valid, rather than just assuming that the estimate applies to the centre of the fixed window from which it is derived, provides a considerable increase in precision, as shown in the phantom data of figure 4d (elastography phantom, CIRS Inc., Norfolk, VA, USA).
Figure 4.
Improving displacement accuracy using amplitude modulation correction (AMC). (a) Matched pre- and post-deformation RF signals are shown, together with the amplitude function. The point where the signals match best is at the centre of gravity of the amplitude, not the centre of the window. (b) B-scan of a phantom with a small hard inclusion. (c) Corresponding elastogram using a phase-based displacement estimator. (d) The result of adding AMC to the same displacement estimator. Dashed line, post-deformation RF; solid line, pre-deformation RF; grey region, weightings.
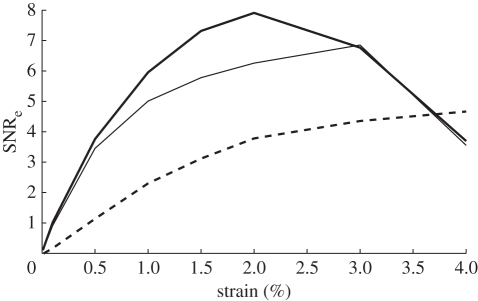
The performance of all displacement estimators varies with the amount of relative strain between the pre- and post-deformation data. Elastographic signal-to-noise ratio (SNRe), used as a measure of the performance of such algorithms, is shown in figure 5 for the best phase- and correlation-based algorithms with AMC applied, and for adaptive companding. This is based on simulated ultrasound data for which the ground truth deformation is known [67]. In practice, typical strains are less than 2 per cent, often much less than 1 per cent, and in this region the AMC methods are both quicker and potentially more precise.
Figure 5.
Comparison of SNRe for different displacement estimators. The best phase-based and correlation-based displacement estimators are shown together with a much slower adaptive estimate which does not use fixed windows. The window-based estimates outperform the adaptive estimate at typical strains of 1–2%, although adaptive estimation is increasingly worthwhile for larger strains. Derived from data used in Lindop et al. [67]. Bold line, phase-based using AMC; thin line, correlation-based using AMC; dashed line, adaptive.
2.2. Quality metrics
Since displacement estimation techniques are all based on optimizing similarity measures between pre- and post-deformation data, the value of this measure at the optimal displacement can also be used to give an indication of displacement estimation quality. In fact, the precision W (the reciprocal of the variance or the mean-squared error) of each displacement estimate is approximately given by [68]
| 2.1 |
where ρ is the normalized correlation coefficient between the RF data in the matched windows. An equivalent expression can be derived for phase-based methods, which makes use of residual phase error rather than normalized correlation [69].
Such quality metrics can be put to good use in many of the algorithms within quasi-static elastography. For example, all displacement tracking approaches that use previous estimates to initialize the following search can fail catastrophically if an incorrect displacement is found at any point and then propagated into other parts of the image. For simple forward tracking, such errors can be called drop-outs [60], since they generate a vertical line of errors in an elastogram. Typically, tracking algorithms assume that tissue exhibits continuous displacement in order to eliminate such errors. However, in in vivo scanning, apparent continuity is often violated either by slip planes or regions of complete ultrasound signal loss.
It is inevitable that such regions will result in localized errors in displacement estimation. However, propagation of these errors can be prevented if the tracking path is allowed to evolve dynamically according to the quality measure: so windows with high-quality matches are preferentially used to initialize the searches at their neighbours [41,54,59,60]. Displacement can even be tracked in disjoint regions separated by slip planes, provided multiple seeds are employed, at least one seed is planted in each region, and the tracking wavefronts evolve in a quality-guided manner [59,70].
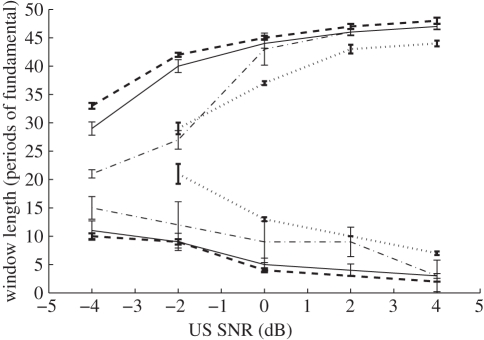
A good tracking algorithm can therefore be seen as a means to increase the robustness of the displacement estimation process, whereas it is the fundamental displacement estimation algorithm that determines the precision. Figure 6 provides an example of this increase in robustness. Here, varying levels of noise are added to simulated data, and displacement estimation attempted using various fixed window lengths [59]. At each noise level, there are clear upper and lower bounds on the window lengths that allow successful recovery of the displacement field. Since varying the window length changes the trade-off between the precision and the resolution of the elastogram, an increased range allows much greater flexibility in optimizing the system parameters. The quality-based tracking strategies, particularly the multiple-seed [59] and two-pass [60] methods, which allow for multiple initialization points in the image, are more robust than the simpler tracking strategies.
Figure 6.
The effect of different tracking strategies. A varying amount of noise is added to ultrasound data with a 1% relative strain, and displacement tracking attempted with varying window lengths. The lines show the region beyond which tracking fails, defined in this case as less than 90% of the displacement estimates being correct. Clearly a good tracking scheme significantly increases the range of possible window lengths and noise values over which good elastograms can be generated. Derived from data used in Chen et al. [59]. Dotted lines, simple axial tracking; dashed-dotted lines, quality-based, single-seed tracking; solid lines, quality-based, multiple-seed tracking; dashed lines, quality-based, two-pass axial tracking.
2.3. Gradient estimation and filtering
Most quasi-static elastography systems follow displacement estimation with axial gradient estimation in order to generate axial strain. Filtering of some kind is nearly universal at this stage, since differencing of consecutive samples [1] (i.e. the digital equivalent of differentiation) amplifies the high-frequency components of the measurement noise. For instance, moving-average filtering (MAF) can be applied either before [48] or after [38] differentiation. Staggered strain estimation (SSE) [71], where displacements are differenced over a wider interval, has also been applied in a wide range of practical strain imaging investigations [29,72–78]. MAF and SSE have been shown to be mathematically identical [79].
Another popular technique for gradient estimation is piecewise-linear least-squares regression (PLLSR) [80], where the gradient is calculated from fitting a linear model to a window of data. This too can be interpreted as simple differencing followed by a linear filter, and it has been shown that even simple Gaussian-based filters can achieve lower estimation noise than these methods at the same resolution [79]. Many other linear filters are possible, such as cubic polynomial fitting [81].
This linear filtering operation has a significant impact on the ultimate resolution and accuracy of strain images. For the case of possibly the most popular, PLLSR, the precision of the strain estimates is then [68]
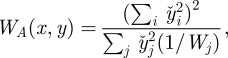 |
2.2 |
where the summations are over displacement windows in the regression kernel, 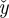 denotes axial distance from the kernel centre, and W is defined in equation (2.1). The precision of strain estimates calculated through simple differencing can be derived from this by taking
denotes axial distance from the kernel centre, and W is defined in equation (2.1). The precision of strain estimates calculated through simple differencing can be derived from this by taking 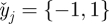 .
.
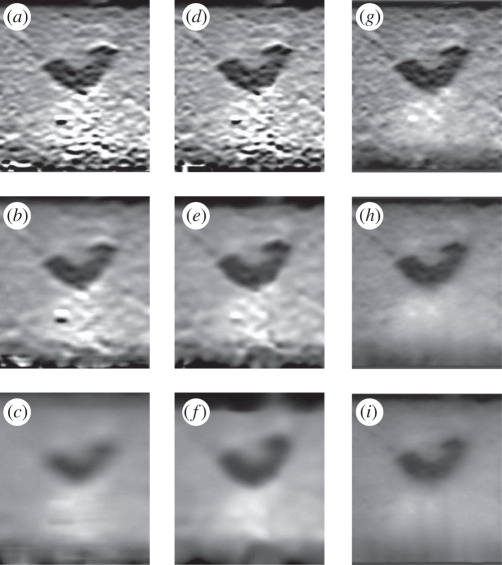
Since most displacement tracking and filtering techniques make use of windows with fixed size, subsequent strain images have fixed resolution but variable quality. It is also possible to use matrix inversion schemes that act on the entire image and produce results that are globally optimal in some sense. For instance, non-parametric regression (NPR), using the strain quality as a per-pixel weighting, can generate strain images in real time of fixed quality but variable resolution [69]. Figure 7 contains some sample filtered images for three different types of filter, and three different extents of filtering. Global quality-based algorithms compare well with PLLSR or even quality-weighted Gaussian filtering, with well-defined borders between different strain regions, good suppression of noise and better preservation of the size of stiff objects.
Figure 7.
Gradient estimation and filtering in elastograms. The data are from a phantom containing half an olive in gelatin mixed with flour. (a–c) Using PLLSR for gradient estimation and filtering. (d–f) Using simple differencing followed by a quality-weighted Gaussian filter. (g–i) Using simple differencing followed by NPR. Each row shows progressively more severe filtering. It is evident here that the global quality-based approach of NPR allows good images to be generated at a range of precisions. Derived from data used in Treece et al. [69].
2.4. Normalization and inversion of strain images
Thus far we have created a precise, filtered, version of the relative axial strain between two frames in an ultrasound scanning sequence, i.e. equivalent to the simulated data of figure 3c. However, ideally we want to display tissue stiffness, E, in figure 3a, rather than axial strain. To do this, we need to solve an inverse problem for which we also need either the stress, figure 3b, or the stiffness E everywhere on the boundary of the data [82]. Although stress can be measured to some extent on the tissue surface using load cells [83], some assumptions (e.g. of uniformity [84]) are necessary to satisfy these conditions, even slight violations of which can lead to incorrect estimates of E [82].
The more popular, less quantitatively motivated, though much faster and better constrained approach is to display an image based directly on the axial strain. Such images always need some form of normalization to convert the strain into a displayable range, and to reduce variation that is simply a result of varying applied stress [68]. Hence if sA(x,y) is the measured strain, the normalized value sB is
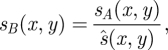 |
2.3 |
where 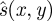 is a normalization value that may simply be a constant representing the maximum strain in each image. The quality of sB is then
is a normalization value that may simply be a constant representing the maximum strain in each image. The quality of sB is then
| 2.4 |
where WA was defined in equation (2.2). It is also possible to use more complex definitions of the normalizing function 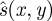 , for instance, to allow for uneven pressure across the probe face, or stress decay with depth in tissue [30,68]. Such functions can be fitted to the strain data by making broad assumptions about the typical stiffness distribution in clinical data.
, for instance, to allow for uneven pressure across the probe face, or stress decay with depth in tissue [30,68]. Such functions can be fitted to the strain data by making broad assumptions about the typical stiffness distribution in clinical data.
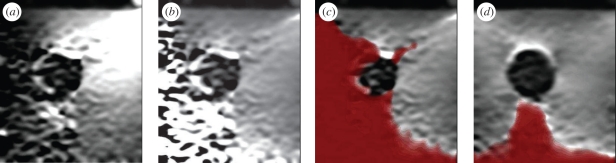
Figure 8a,b shows an example of axial strain-based display (breast biopsy phantom, CIRS Inc., Norfolk, VA, USA). Even the ‘raw’ strain image in figure 8a requires some form of normalization for display: this is really a normalized image with 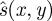 as a constant value. The stiff inclusion is visible even in this image. The normalized ‘pseudo-strain’ image in figure 8b has been created using the flexible scheme outlined in Lindop et al. [68]. The effect is to remove distracting variations in the background that did not represent variations in material stiffness. Normalization has also accentuated the noisy measurements at the bottom-left, which are the result of very little axial probe movement on that side owing to rotation of the probe about the elevational axis. In order to remove such noisy regions, we need to persist data from more than one pair of ultrasound frames, leading to the result in figure 8d: this is the subject of §2.5.
as a constant value. The stiff inclusion is visible even in this image. The normalized ‘pseudo-strain’ image in figure 8b has been created using the flexible scheme outlined in Lindop et al. [68]. The effect is to remove distracting variations in the background that did not represent variations in material stiffness. Normalization has also accentuated the noisy measurements at the bottom-left, which are the result of very little axial probe movement on that side owing to rotation of the probe about the elevational axis. In order to remove such noisy regions, we need to persist data from more than one pair of ultrasound frames, leading to the result in figure 8d: this is the subject of §2.5.
Figure 8.
The effect of normalization and persistence on elastograms. (a) An axial strain image from a phantom containing a stiff inclusion in a uniform background, with careless non-axial probe motion. (b) Normalization of this data accounts for probe movement and leads to a more uniform background value. (c) Visualization of measurement quality obscures distracting noise in the image. (d) Quality-based persistence of multiple images from the same dataset rapidly builds up a good elastogram even from such low-quality raw data.
2.5. Persistence and visualization
The formation of an elastogram only requires two frames of ultrasound data: one before and one after an axial deformation of the tissue. However, there are several reasons for using many more frames of data to form a stiffness image. Firstly, if the probe is moved by hand, there will be some frame pairs that do not exhibit any relative deformation at all, or for which the deformation is inappropriate for measuring axial strain. Secondly, figure 5 shows that displacement estimators tend to perform best at fairly low strain, whereas it can be seen from equation (2.4) that higher strain increases the precision of the displayed, normalized value. The easiest way to satisfy both these constraints is to measure displacement between multiple low-strain frames, which accumulate to give higher strain deformations. Thirdly, ultrasound data are available at rates up to 200 frames per second, depending on the focusing technique and the imaging depth, generally much higher than the rates required for good visual responsiveness in the images.
One common approach to this problem is to create elastograms between all pairs of frames and then select the ‘best’ pair, either manually or with the use of a metric based on the quality discussed in §2.2 [85,86]. In fact, such a metric is often displayed dynamically as the scan progresses, to give users some feedback on the quality of their scanning technique. Such schemes do not make optimal use of the large amount of data available. To make use of all the data, sequences of images can be averaged in order to reduce noise [87], but this does not take into account the variations in quality within each elastogram.
It is possible to generate better sequences of elastograms by physically constraining the types of freehand probe motion [88]. Alternatively, the known quality WB(x,y) of the data can be used either as a per-image [89] or per-pixel weighting [68] in persisting the stream of images. An example of per-pixel quality-based persistence is shown in figure 8. The single elastogram in figure 8b contains some very good data, but also some areas of very poor data, either owing to inappropriate tissue deformation, or difficulty in tracking displacements owing to low or decorrelated ultrasound signals. The persisted elastogram in figure 8d has been created from a sequence containing the frame in figure 8b, and some other frames which contained good data in other regions. Per-pixel quality weighting has the effect of combining the best regions from each frame to build up the final displayed image. Poor-quality data in the scan sequence are effectively ignored, since in this case, the quality WB(x,y) is low [30].
There is currently little consensus on how elastograms should be displayed. Images in this paper have used greyscale for strain, with black indicating stiff and white indicating soft regions, but this scale is sometimes inverted. It is also popular to use a colourmap to display strain values, often based on a blue–yellow–red progression, though blue has been used to represent either stiff or soft regions. Such a colourmap can be overlaid on top of a conventional greyscale ultrasound image, with some transparency, so that, at least to some extent, both modalities are simultaneously visible. This display is familiar to clinicians in that it is similar to Doppler ultrasound, used for visualization of fluid flow. However, unlike Doppler, where the blue–red colour transition marks a change in flow direction, in quasi-static elastography, there are no clear pseudo-strain boundaries that could appropriately be aligned with the marked changes in colour. Hence it is potentially dangerous to associate regions which are ‘red’, ‘blue’ or any other colour with any particular pathology.
Quality values are generally also displayed alongside some form of pseudo-strain, at least as an independent visualization of the overall quality of the image. However, since the quality is available at a per-pixel level, much better use can be made of it either by fading out strain data according to quality, or displaying quality as an overlay on top of the strain data [30]. The images in this paper have used a red overlay to indicate poor quality, gradually fading to a solid colour as the quality degrades, generated by our own freely available Stradwin software (http://mi.eng.cam.ac.uk/rwp/stradwin) [90,91]. Quality display acts as an important indication of poor scanning performance. Per-pixel quality overlays can also be used to obscure low-quality strain data, which might otherwise appear to represent actual stiffness variations, and hence confuse the interpretation of an elastogram, as in figure 8b,c.
3. Related topics
3.1. Three-dimensional elastographic techniques
Many of the algorithms in the previous sections extend naturally to three dimensions. Indeed, since the stress and strain fields induced during deformation of tissue are inherently three-dimensional, it might therefore be assumed that much better stiffness data, particularly from inversion techniques, should be available by acquiring and processing in three dimensions. Many three-dimensional techniques are already well developed for conventional ultrasound imaging. However, the lateral and elevational components of displacement can generally be estimated with less precision [93], and hence much of the focus of three-dimensional elastography has been in increasing the precision of axial displacement, and hence axial strain.
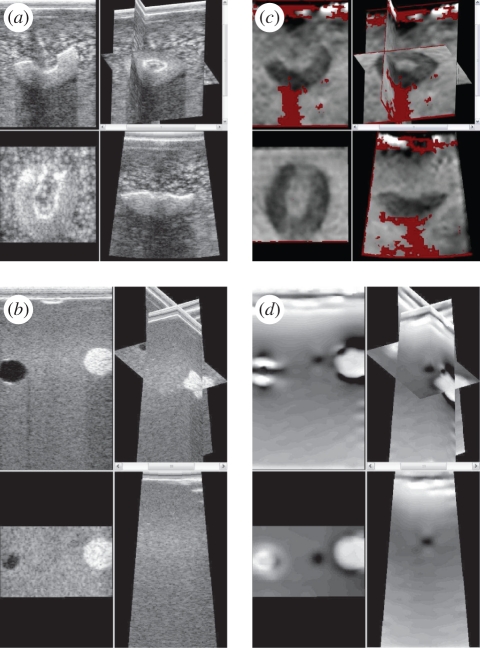
There are two main approaches for three-dimensional elastography. Firstly, a sequence of two-dimensional elastograms can be created as the probe is moved elevationally [94], for instance, for intra-vascular [95] or prostate [96] imaging. Alternatively, a mechanically swept two-dimensional probe or two-dimensional phased-array probe can be used to acquire a volume of data, then a controlled or freehand compression applied [54,76,93,97–99] before further volumes are acquired. Some mixed approaches have also been suggested, either using a three-dimensional pre-compression volume with a two-dimensional post-compression scan [100], or a mechanically swept two-dimensional probe but with two-dimensional elastogram processing [101]. Such approaches generate three-dimensional datasets of axial strain. Figure 9 shows an example with an in-house and commercial phantom using the system presented in Treece et al. [54]. These data may be useful in accurately quantifying the volume of stiff lesions.
Figure 9.
Three-dimensional elastography. (a,c) Three orthogonal views and one three-dimensional view from conventional ultrasound and elastography data of half an olive in gelatin mixed with flour. (b,d) Similar images from a phantom with various inclusions. Derived from data used in Treece et al. [54,92].
The difference between using probes that acquire fundamentally three-dimensional data, and sequences of data from a two-dimensional probe moved across the anatomy, is more than just in the techniques for processing the data. Mechanically swept two-dimensional probes and two-dimensional phased arrays have a much larger probe face than two-dimensional probes, and since it is the probe face that provides the pressure contact with the anatomy, this affects the nature of the generated stress and strain fields [1,102]. In theory, a larger probe acts more like an infinite compressor plate and should generate more uniform stresses. In practice, better images are sometimes generated by techniques which use two-dimensional probes [101], or one-dimensional probes with compression plates [88].
3.2. Slip and shear strain imaging
Elastograms based on axial strain show relative anatomical stiffness; however, it has been suggested that how lesions are attached to surrounding tissue may also correlate with malignancy [103]. The slip patterns generated by un-attached lesions are to some extent visible on elastograms, since slip results in discontinuity in the axial strain. A more direct way to visualize these patterns is by the axial-shear strain, i.e. the lateral gradient of axial displacement [78,104–107], or indeed some combination of axial- and lateral-shear strain [108,109].
Most of the discussion in this paper regarding axial strain can also be applied to the generation of an axial-shear strain image. Aside from the obvious difference in the direction in which the gradient is taken, there are two further differences. Both axial and axial-shear strain change sign dependent on whether they are induced by a compression or relaxation of the anatomy. In an elastogram, this sign change is removed at the normalization or inversion stage, but in axial-shear strain images, the sign change is useful and needs to be preserved. Secondly, whereas the average magnitude of axial strain is non-zero, typically axial-shear strain images do have a mean value close to zero.
These changes mean a new normalization process is required for imaging axial-shear strain [110], though all other processing steps can remain the same. Figure 10 contains some examples of phantom (breast biopsy phantom, CIRS Inc., Norfolk, VA, USA) and in vivo axial-shear strain images produced using this normalization technique. The alternating bright and dark quadrants surrounding the inclusions are typical of axial-shear strain images, the location of the bright quadrants indicating whether the inclusions are stiff or soft. Relative shear strain levels inside and outside the inclusions may indicate how well they are bonded to the surrounding material [111,112].
Figure 10.
Imaging of axial-shear strain. (a) B-mode image, (b) normalized axial strain and (c) normalized shear strain of a hard inclusion in a phantom. (d–f) Corresponding images acquired during neurosurgery. The level of axial shear within the inclusion, and the variation in axial shear surrounding the inclusion, provides an indication of how well it is bonded to the surrounding tissue. Derived from data used in Chen et al. [110].
4. Conclusions
Quasi-static ultrasound elastography is an apparently simple idea, which has required surprisingly complex techniques to enable its development to date. Many of these techniques have been briefly outlined and contrasted in this paper. It is sufficiently mature to allow successful commercialization, even though the clinical case has yet to be conclusively made. However, it is clear that many assumptions are required in the formation of an elastogram, particularly in the mechanics of the tissue deformation. Relaxing some of these assumptions continues to provide a fruitful area of ongoing research.
Acknowledgements
We thank Ellen Neale for acquiring the clinical data in figure 1. Full ethical approval was obtained from the Cambridgeshire 3 Research Ethics Committee (reference no. 07/H0306/90). We also thank Chris Uff for acquiring the clinical data in figure 10. Full ethical approval was obtained from the National Hospital for Neurology and Neurosurgery and the Institute of Neurology Joint Regional Ethics Committee (reference 08/H0716/92).
References
- 1.Ophir J., Céspedes I., Ponnekanti H., Yazdi Y., Li X. 1991. Elastography: a quantitative method for imaging the elasticity of biological tissues. Ultrason. Imaging 13, 111–134 10.1016/0161-7346(91)90079-W (doi:10.1016/0161-7346(91)90079-W) [DOI] [PubMed] [Google Scholar]
- 2.Adams F. (ed.) 1849. The genuine works of Hippocrates. London, UK: Adlard [Google Scholar]
- 3.Garra B. S., Céspedes E. I., Ophir J., Spratt S. R., Zuurbier R. A., Magnant C. M., Pennanen M. F. 1997. Elastography of breast lesions: initial clinical results. Radiology 202, 79–86 [DOI] [PubMed] [Google Scholar]
- 4.Hall T. J., Zhu Y., Spalding C. S. 2003. In vivo real-time freehand palpation imaging. Ultrasound Med. Biol. 29, 427–435 10.1016/S0301-5629(02)00733-0 (doi:10.1016/S0301-5629(02)00733-0) [DOI] [PubMed] [Google Scholar]
- 5.Itoh A., Ueno E., Tohno E., Kamma H., Takahashi H., Shiina T., Yamakawa M., Matsumura T. 2006. Breast disease: clinical application of US elastography for diagnosis. Radiology 239, 341–350 10.1148/radiol.2391041676 (doi:10.1148/radiol.2391041676) [DOI] [PubMed] [Google Scholar]
- 6.Regner D. M., et al. 2006. Breast lesions: evaluation with US strain imaging—clinical experience of multiple observers. Radiology 238, 425–437 10.1148/radiol.2381041336 (doi:10.1148/radiol.2381041336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensson W. E., Amiras D. 2006. Ultrasound elasticity imaging. Breast Cancer Online 9, 1–7 10.1017/S1470903106002835 (doi:10.1017/S1470903106002835) [DOI] [Google Scholar]
- 8.Burnside E. S., Hall T. J., Sommer A. M., Hesley G. K., Sisney G. A., Svensson W. E., Fine J. P., Jiang J., Hangiandreou N. J. 2007. Differentiating benign from malignant solid breast masses with US strain imaging. Radiology 245, 401–410 10.1148/radiol.2452061805 (doi:10.1148/radiol.2452061805) [DOI] [PubMed] [Google Scholar]
- 9.Miyanaga N., Akaza H., Yamakawa M., Oikawa T., Sekido N., Hinotsu S., Kawai K., Shimazui T., Shiina T. 2006. Tissue elasticity imaging for diagnosis of prostate cancer: a preliminary report. Int. J. Urol. 13, 1514–1518 10.1111/j.1442-2042.2006.01612.x (doi:10.1111/j.1442-2042.2006.01612.x) [DOI] [PubMed] [Google Scholar]
- 10.Barr R. G. 2006. Clinical applications of a real time elastography technique in breast imaging. In Proc. 5th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Snowbird, UT, 8–11 October 2006, p. 112 Austin, TX: ITEC [Google Scholar]
- 11.de Korte C. L., Céspedes E. I., van der Steen A. F. W., Pasterkamp G., Bom N. 1998. Intravascular ultrasound elastography: assessment and imaging of elastic properties of diseased arteries and vulnerable plaque. Eur. J. Ultrasound 7, 219–224 10.1016/S0929-8266(98)00043-3 (doi:10.1016/S0929-8266(98)00043-3) [DOI] [PubMed] [Google Scholar]
- 12.de Korte C. L., Pasterkamp G., van der Steen A. F. W., Woutman H. A., Bom N. 2000. Characterization of plaque components with intravascular ultrasound elastography in human femoral and coronary arteries in vitro. Circulation 102, 617–623 [DOI] [PubMed] [Google Scholar]
- 13.Emelianov S. Y., Chen X., Knipp B., Myers D. 2002. Triplex ultrasound: elasticity imaging to age deep venous thrombosis. Ultrasound Med. Biol. 28, 757–767 10.1016/S0301-5629(02)00516-1 (doi:10.1016/S0301-5629(02)00516-1) [DOI] [PubMed] [Google Scholar]
- 14.Vogt M., Ermert H. 2005. Development and evaluation of a high-frequency ultrasound-based system for in vivo strain imaging of the skin. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 52, 375–385 10.1109/TUFFC.2005.1417260 (doi:10.1109/TUFFC.2005.1417260) [DOI] [PubMed] [Google Scholar]
- 15.Kaluzynski K., Chen X., Emelianov S. Y., Skovoroda S. R., O'Donnell M. 2001. Strain rate imaging using two-dimensional speckle tracking. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 48, 1111–1123 10.1109/58.935730 (doi:10.1109/58.935730) [DOI] [PubMed] [Google Scholar]
- 16.Cole J. A. J., et al. 2009. Evidence review, ultrasound elastography. Technical report CEP08052, NHS Purchasing and Supply Agency, Centre for Evidence-based Purchasing [Google Scholar]
- 17.Sugimoto T., Ueha S., Itoh K. 1990. Tissue hardness measurement using the radiation force of focused ultrasound. In Proc. IEEE Ultrasonics Symp., Honolulu, HI, 4–7 December 1990, vol. 3, pp. 1377–1380 [Google Scholar]
- 18.Muthupillai R., Lomas D. J., Rossman P., Greenleaf J. F., Manduca A., Ehman R. L. 1995. Magnetic resonance elastography by direct visualization of propagating acoustic strain waves. Science 269, 1854–1857 10.1126/science.7569924 (doi:10.1126/science.7569924) [DOI] [PubMed] [Google Scholar]
- 19.Dickinson R. J., Hill C. R. 1982. Measurement of soft tissue motion using correlation between A-scans. Ultrasound Med. Biol. 8, 263–271 10.1016/0301-5629(82)90032-1 (doi:10.1016/0301-5629(82)90032-1) [DOI] [PubMed] [Google Scholar]
- 20.Bercoff J., Tanter M., Fink M. 2004. Supersonic shear imaging: a new technique for soft tissue elasticity mapping. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 51, 396–409 10.1109/TUFFC.2004.1295425 (doi:10.1109/TUFFC.2004.1295425) [DOI] [PubMed] [Google Scholar]
- 21.Fung Y. C. 1993. Biomechanics: mechanical properties of living tissues. Berlin, Germany: Springer [Google Scholar]
- 22.Landau E. D., Lifshitz E. M. 1986. Theory of elasticity, 3rd edn Oxford, UK: Pergamon Press [Google Scholar]
- 23.Flügge W. 1967. Viscoelasticity. Waltham, MA: Blaisdell [Google Scholar]
- 24.Insana M. F., Pellot-Barakat C., Sridhar M., Lindfors K. K. 2004. Viscoelastic imaging of breast tumor microenvironment with ultrasound. J. Mammary Gland Biol. 9, 393–404 10.1007/s10911-004-1409-5 (doi:10.1007/s10911-004-1409-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarvazyan A. P., Skovoroda A. R., Emelianov S. Y., Fowlkes J. B., Pipe J. G., Adler R. S., Buxton R. B., Carson P. L. 1995. Biophysical bases of elasticity imaging. Acoust. Imaging 21, 223–240 [Google Scholar]
- 26.Greenleaf J. F., Fatemi M., Insana M. 2003. Selected methods for imaging elastic properties of biological tissues. Annu. Rev. Biomed. Eng. 5, 57–78 10.1146/annurev.bioeng.5.040202.121623 (doi:10.1146/annurev.bioeng.5.040202.121623) [DOI] [PubMed] [Google Scholar]
- 27.Berry G. P., Bamber J. C., Armstrong C. G., Miller N. R., Barbone P. E. 2006. Towards an acoustic model-based poroelastic imaging method. I. Theoretical foundation. Ultrasound Med. Biol. 32, 547–567 10.1016/j.ultrasmedbio.2006.01.003 (doi:10.1016/j.ultrasmedbio.2006.01.003) [DOI] [PubMed] [Google Scholar]
- 28.Berry G. P., Bamber J. C., Miller N., Miller N. R., Barbone P. E., Bush N. L., Armstrong C. G. 2006. Towards an acoustic model-based poroelastic imaging method: II. Experimental investigation. Ultrasound Med. Biol. 32, 1869–1885 10.1016/j.ultrasmedbio.2006.07.013 (doi:10.1016/j.ultrasmedbio.2006.07.013) [DOI] [PubMed] [Google Scholar]
- 29.Righetti R., Ophir J., Srinivasan S., Krouskop T. A. 2004. The feasibility of using elastography for imaging the Poisson's ratio in porous media. Ultrasound Med. Biol. 30, 215–228 10.1016/j.ultrasmedbio.2003.10.022 (doi:10.1016/j.ultrasmedbio.2003.10.022) [DOI] [PubMed] [Google Scholar]
- 30.Gee A., Lindop J., Treece G., Prager R., Freeman S. 2008. Stable, intelligible ultrasonic strain imaging. Ultrasound 16, 187–192 10.1179/174313408X320932 (doi:10.1179/174313408X320932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Housden R. J., Gee A. H., Treece G. M., Prager R. W. 2006. Sub-sample interpolation strategies for sensorless freehand 3D ultrasound. Ultrasound Med. Biol. 32, 1897–1904 10.1016/j.ultrasmedbio.2006.06.027 (doi:10.1016/j.ultrasmedbio.2006.06.027) [DOI] [PubMed] [Google Scholar]
- 32.Langeland S., d'Hooge J., Torp H., Bijnens B., Suetens P. 2003. Comparison of time-domain displacement estimators for two-dimensional RF tracking. Ultrasound Med. Biol. 29, 1177–1186 10.1016/S0301-5629(03)00972-4 (doi:10.1016/S0301-5629(03)00972-4) [DOI] [PubMed] [Google Scholar]
- 33.Viola F., Walker W. F. 2003. A comparison of the performance of time-delay estimators in medical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 50, 392–401 10.1109/TUFFC.2003.1197962 (doi:10.1109/TUFFC.2003.1197962) [DOI] [PubMed] [Google Scholar]
- 34.Bilgen M., Insana M. F. 1996. Deformation models and correlation analysis in elastography. J. Acoust. Soc. Am. 99, 3212–3224 10.1121/1.414865 (doi:10.1121/1.414865) [DOI] [PubMed] [Google Scholar]
- 35.Bohs L. N., Trahey G. E. 1991. A novel method for angle independent ultrasonic imaging of blood flow and tissue motion. IEEE Trans. Biomed. Eng. 38, 280–286 10.1109/10.133210 (doi:10.1109/10.133210) [DOI] [PubMed] [Google Scholar]
- 36.Chaturvedi P., Insana M. F., Hall T. J. 1998. Testing the limitations of 2-D companding for strain imaging using phantoms. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 45, 1022–1031 10.1109/58.710585 (doi:10.1109/58.710585) [DOI] [PubMed] [Google Scholar]
- 37.Yeung F., Levinson S. F., Parker K. J. 1998. Multilevel and motion model-based ultrasonic speckle tracking algorithms. Ultrasound Med. Biol. 24, 427–441 10.1016/S0301-5629(97)00281-0 (doi:10.1016/S0301-5629(97)00281-0) [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell M., Skovoroda A. R., Shapo B. M., Emelianov S. Y. 1994. Internal displacement and strain imaging using ultrasonic speckle tracking. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 41, 314–325 10.1109/58.285465 (doi:10.1109/58.285465) [DOI] [Google Scholar]
- 39.Pesavento A., Perrey C., Krueger M., Ermert H. 1999. A time efficient and accurate strain estimation concept for ultrasonic elastography using iterative phase zero estimation. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 46, 1057–1067 10.1109/58.796111 (doi:10.1109/58.796111) [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Zohdy M. J., Emelianov S. Y., O'Donnell M. 2004. Lateral speckle tracking using synthetic lateral phase. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 51, 540–550 10.1109/TUFFC.2004.1320827 (doi:10.1109/TUFFC.2004.1320827) [DOI] [PubMed] [Google Scholar]
- 41.Lindop J. E., Treece G. M., Gee A. H., Prager R. W. 2008. Phase-based ultrasonic deformation estimation. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 55, 94–111 10.1109/TUFFC.2008.620 (doi:10.1109/TUFFC.2008.620) [DOI] [PubMed] [Google Scholar]
- 42.Alam S. K., Ophir J., Konofagou E. E. 1998. An adaptive strain estimator for elastography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 45, 461–472 10.1006/uimg.1995.1007 (doi:10.1006/uimg.1995.1007) [DOI] [PubMed] [Google Scholar]
- 43.Céspedes I., Huang Y., Ophir J., Spratt S. 1995. Methods for estimation of subsample time delays of digitized echo signals. Ultrason. Imaging 17, 142–171 10.1006/uimg.1995.1007 (doi:10.1006/uimg.1995.1007) [DOI] [PubMed] [Google Scholar]
- 44.Ebbini E. S. 2006. Phase-coupled two-dimensional speckle tracking algorithm. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 53, 972–990 10.1109/TUFFC.2006.1632687 (doi:10.1109/TUFFC.2006.1632687) [DOI] [PubMed] [Google Scholar]
- 45.Jiang J., Hall T. J. 2007. A parallelizable real-time motion tracking algorithm with applications to ultrasonic strain imaging. Phys. Med. Biol. 52, 3773–3790 10.1088/0031-9155/52/13/008 (doi:10.1088/0031-9155/52/13/008) [DOI] [PubMed] [Google Scholar]
- 46.Konofagou E. E., Ophir J. 1998. A new elastographic method for estimation and imaging of lateral displacements, lateral strains, corrected axial strains and Poisson's ratios in tissues. Ultrasound Med. Biol. 24, 1183–1199 10.1016/S0301-5629(98)00109-4 (doi:10.1016/S0301-5629(98)00109-4) [DOI] [PubMed] [Google Scholar]
- 47.Lubinski M. A., Emelianov S. Y., O'Donnell M. 1999. Speckle tracking methods for ultrasonic elasticity imaging using short-time correlation. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 46, 82–96 10.1109/58.741427 (doi:10.1109/58.741427) [DOI] [PubMed] [Google Scholar]
- 48.Pellot-Barakat C., Frouin F., Insana M. F., Herment A. 2004. Ultrasound elastography based on multiscale estimations of regularized displacement fields. IEEE Trans. Med. Imaging 23, 153–163 10.1109/TMI.2003.822825 (doi:10.1109/TMI.2003.822825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zahiri-Azar R., Salcudean S. E. 2006. Motion estimation in ultrasound images using time domain cross correlation with prior estimates. IEEE Trans. Biomed. Eng. 53, 1990–2000 10.1109/TBME.2006.881780 (doi:10.1109/TBME.2006.881780) [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y., Hall T. J. 2002. A modified block matching method for real-time freehand strain imaging. Ultrason. Imaging 24, 161–176 [DOI] [PubMed] [Google Scholar]
- 51.Viola F., Walker W. F. 2005. A spline-based algorithm for continuous time-delay estimation using sampled data. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 52, 80–93 10.1109/TUFFC.2005.1397352 (doi:10.1109/TUFFC.2005.1397352) [DOI] [PubMed] [Google Scholar]
- 52.Viola F., Walker W. F. 2006. A comparison between spline-based and phase-domain time-delay estimators. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 53, 515–517 10.1109/TUFFC.2006.1610558 (doi:10.1109/TUFFC.2006.1610558) [DOI] [PubMed] [Google Scholar]
- 53.Pinton G. F., Trahey G. E. 2006. Continuous delay estimation with polynomial splines. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 53, 2026–2035 10.1109/TUFFC.2006.143 (doi:10.1109/TUFFC.2006.143) [DOI] [PubMed] [Google Scholar]
- 54.Treece G. M., Lindop J. E., Gee A. H., Prager R. W. 2008. Freehand ultrasound elastography with a 3D probe. Ultrasound Med. Biol. 34, 463–474 10.1016/j.ultrasmedbio.2007.08.014 (doi:10.1016/j.ultrasmedbio.2007.08.014) [DOI] [PubMed] [Google Scholar]
- 55.O'Donnell M., Skovoroda A. R., Shapo B. M. 1991. Measurement of arterial wall motion using Fourier based speckle tracking algorithms. In Proc. IEEE Ultrasonics Symp., Orlando, FL, 8–11 December 1991, pp. 1101–1104 [Google Scholar]
- 56.Jiang J., Hall T. J. 2006. A regularized real-time motion tracking algorithm using dynamic programming for ultrasonic strain imaging. In Proc. IEEE Ultrasonics Symp., Vancouver, Canada, 2–6 October 2006, pp. 606–609 [Google Scholar]
- 57.Rivaz H., Boctor E., Foroughi P., Zellars R., Fichtinger G., Hager G. 2008. Ultrasound elastography: a dynamic programming approach. IEEE Trans. Med. Imaging 27, 1373–1377 10.1109/TMI.2008.917243 (doi:10.1109/TMI.2008.917243) [DOI] [PubMed] [Google Scholar]
- 58.Maurice R. L., Bertrand M. 1999. Lagrangian speckle model and tissue-motion estimation—theory. IEEE Trans. Med. Imaging 18, 593–603 10.1109/42.790459 (doi:10.1109/42.790459) [DOI] [PubMed] [Google Scholar]
- 59.Chen L., Treece G. M., Lindop J. E., Gee A. H., Prager R. W. 2009. A quality-guided displacement tracking algorithm for ultrasonic elasticity imaging. Med. Image Anal. 13, 286–296 10.1016/j.media.2008.10.007 (doi:10.1016/j.media.2008.10.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treece G. M., Lindop J. E., Gee A. H., Prager R. W. 2006. Efficient elimination of dropouts in displacement tracking. In Proc. 5th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Snowbird, UT, 8–11 October 2006, p. 68 Austin: TX: ITEC [Google Scholar]
- 61.Bai J., Ding C., Fan Y. 1999. A multi-scale algorithm for ultrasonic strain reconstruction under moderate compression. Ultrasonics 37, 511–519 10.1016/S0041-624X(99)00026-8 (doi:10.1016/S0041-624X(99)00026-8) [DOI] [Google Scholar]
- 62.Chen H., Shi H., Varghese T. 2007. Improvement of elastographic displacement estimation using a two-step cross-correlation method. Ultrasound Med. Biol. 33, 48–56 10.1016/j.ultrasmedbio.2006.07.022 (doi:10.1016/j.ultrasmedbio.2006.07.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi H., Varghese T. 2007. Two-dimensional multi-level strain estimation for discontinuous tissue. Phys. Med. Biol. 52, 389–401 10.1088/0031-9155/52/2/006 (doi:10.1088/0031-9155/52/2/006) [DOI] [PubMed] [Google Scholar]
- 64.Walker W. F., Trahey G. E. 1995. A fundamental limit on delay estimation using partially correlated speckle signals. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 42, 301–308 10.1109/58.365243 (doi:10.1109/58.365243) [DOI] [Google Scholar]
- 65.Alam S. K., Ophir J. 1997. Reduction of signal decorrelation from mechanical compression of tissues by temporal stretching: applications to elastography. Ultrasound Med. Biol. 23, 95–105 10.1016/S0301-5629(96)00164-0 (doi:10.1016/S0301-5629(96)00164-0) [DOI] [PubMed] [Google Scholar]
- 66.Chaturvedi P., Insana M. F., Hall T. J. 1998. 2-D companding for noise reduction in strain imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 45, 179–191 10.1109/58.646923 (doi:10.1109/58.646923) [DOI] [PubMed] [Google Scholar]
- 67.Lindop J. E., Treece G. M., Gee A. H., Prager R. W. 2007. Estimation of displacement location for enhanced strain imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 54, 1751–1771 10.1109/TUFFC.2007.460 (doi:10.1109/TUFFC.2007.460) [DOI] [PubMed] [Google Scholar]
- 68.Lindop J. E., Treece G. M., Gee A. H., Prager R. W. 2008. An intelligent interface for freehand strain imaging. Ultrasound Med. Biol. 34, 1117–1128 10.1016/j.ultrasmedbio.2007.12.012 (doi:10.1016/j.ultrasmedbio.2007.12.012) [DOI] [PubMed] [Google Scholar]
- 69.Treece G. M., Lindop J. E., Gee A. H., Prager R. W. 2009. Uniform precision ultrasound strain imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 56, 2420–2436 10.1109/TUFFc.2009.1330 (doi:10.1109/TUFFc.2009.1330) [DOI] [PubMed] [Google Scholar]
- 70.Chen L., Housden R. J., Treece G. M., Gee A. H., Prager R. W. 2010. A hybrid displacement estimation method for ultrasonic elasticity imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 57, 866–882 10.1109/TUFFC.2010.1491 (doi:10.1109/TUFFC.2010.1491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Srinivasan S., Ophir J., Alam S. K. 2002. Elastographic imaging using staggered strain estimates. Ultrason. Imaging 24, 229–245 [DOI] [PubMed] [Google Scholar]
- 72.Righetti R., Ophir J., Krouskop T. A. 2005. A method for generating permeability elastograms and Poisson's ratio time-constant elastograms. Ultrasound Med. Biol. 31, 803–816 10.1016/j.ultrasmedbio.2005.02.004 (doi:10.1016/j.ultrasmedbio.2005.02.004) [DOI] [PubMed] [Google Scholar]
- 73.Righetti R., Ophir J., Garra B. S., Chandrasekhar R. M., Krouskop T. A. 2005. A new method for generating poroelastograms in noisy environments. Ultrason. Imaging 27, 201–220 [DOI] [PubMed] [Google Scholar]
- 74.Souchon R., Bouchoux G., Maciejko E., Lafon C., Cathignol D., Bertrand M., Chapelon J. 2005. Monitoring the formation of thermal lesions with heat-induced echo-strain imaging: a feasibility study. Ultrasound Med. Biol. 31, 251–259 10.1016/j.ultrasmedbio.2004.11.004 (doi:10.1016/j.ultrasmedbio.2004.11.004) [DOI] [PubMed] [Google Scholar]
- 75.Mofid Y., Ossant F., Imberdis C., Josse G., Patat F. 2006. In-vivo imaging of skin under stress: potential of high-frequency (20 MHz) static 2-D elastography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 53, 925–935 10.1109/TUFFC.2006.1632683 (doi:10.1109/TUFFC.2006.1632683) [DOI] [PubMed] [Google Scholar]
- 76.Patil A. V., Garson C. D., Hossack J. A. 2007. 3D prostate elastography: algorithms, simulations and experiments. Phys. Med. Biol. 52, 3643–3663 10.1088/0031-9155/52/12/019 (doi:10.1088/0031-9155/52/12/019) [DOI] [PubMed] [Google Scholar]
- 77.Righetti R., Righetti M., Ophir J., Krouskop T. A. 2007. The feasibility of estimating and imaging the mechanical behaviour of poroelastic materials using axial strain elastography. Phys. Med. Biol. 52, 3241–3259 10.1088/0031-9155/52/11/020 (doi:10.1088/0031-9155/52/11/020) [DOI] [PubMed] [Google Scholar]
- 78.Thitaikumar A., Krouskop T. A., Garra B. S., Ophir J. 2007. Visualization of bonding at an inclusion boundary using axial-shear strain elastography: a feasibility study. Phys. Med. Biol. 52, 2615–2633 10.1088/0031-9155/52/9/019 (doi:10.1088/0031-9155/52/9/019) [DOI] [PubMed] [Google Scholar]
- 79.Lindop J. E., Treece G. M., Gee A. H., Prager R. W. 2008. The general properties including accuracy and resolution of linear-filtering methods for strain estimation. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 55, 2362–2368 10.1109/TUFFC.943 (doi:10.1109/TUFFC.943) [DOI] [PubMed] [Google Scholar]
- 80.Kallel F., Ophir J. 1997. A least-squares strain estimator for elastography. Ultrason. Imaging 19, 195–208 [DOI] [PubMed] [Google Scholar]
- 81.Luo J., Bai J., He P., Ying K. 2004. Axial strain calculation using a low-pass digital differentiator in ultrasound elastography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 51, 1119–1127 10.1109/TUFFC.2004.1334844 (doi:10.1109/TUFFC.2004.1334844) [DOI] [PubMed] [Google Scholar]
- 82.Barbone P. E., Bamber J. C. 2002. Quantitative elasticity imaging: what can and cannot be inferred from strain images. Phys. Med. Biol. 47, 2147–2164 10.1088/0031-9155/47/12/310 (doi:10.1088/0031-9155/47/12/310) [DOI] [PubMed] [Google Scholar]
- 83.Li J., Cui Y., Kadour M., Noble J. A. 2008. Elasticity reconstruction from displacement and confidence measures of a multi-compressed ultrasound RF sequence. IEEE Trans. Ultrason. Ferroelectr. Control 55, 319–326 10.1109/TUFFC.2008.651 (doi:10.1109/TUFFC.2008.651) [DOI] [PubMed] [Google Scholar]
- 84.Skovoroda A. R., Klishko A. N., Gusakyan D. A., Mayevshki Y. I., Yermilova V. D., Oranskaya G. A., Sarvazyan A. P. 1995. Quantitative analysis of the mechanical characteristics of pathologically changed soft biological tissues. Biophysics 40, 1359–1364 [PubMed] [Google Scholar]
- 85.Jiang J., Hall T. J., Sommer A. M. 2006. A novel performance descriptor for ultrasonic strain imaging: a preliminary study. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 53, 1088–1102 10.1109/TUFFC.2006.1642508 (doi:10.1109/TUFFC.2006.1642508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang J., Hall T. J., Sommer A. M. 2007. A novel image formation method for ultrasonic strain imaging. Ultrasound Med. Biol. 33, 643–652 10.1016/j.ultrasmedbio.2006.11.005 (doi:10.1016/j.ultrasmedbio.2006.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varghese T., Ophir J. 1996. Performance optimization in elastography: multicompression with temporal stretching. Ultrason. Imaging 18, 193–214 10.1006/uimg.1996.0011 (doi:10.1006/uimg.1996.0011) [DOI] [PubMed] [Google Scholar]
- 88.Kadour M., Noble J. A. 2009. Assisted-freehand ultrasound elasticity imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 56, 36–43 10.1109/TUFFC.2009.1003 (doi:10.1109/TUFFC.2009.1003) [DOI] [PubMed] [Google Scholar]
- 89.Hiltawsky K. M., Krüger M., Starke C., Heuser L., Ermert H., Jensen A. 2001. Freehand ultrasound elastography of breast lesions: clinical results. Ultrasound Med. Biol. 27, 1461–1469 10.1016/S0301-5629(01)00434-3 (doi:10.1016/S0301-5629(01)00434-3) [DOI] [PubMed] [Google Scholar]
- 90.Gee A. H., Prager R. W., Treece G. M., Cash C. C. J., Berman L. 2004. Processing and visualising three-dimensional ultrasound data. Br. J. Radiol. 77, S186–S193 10.1259/bjr/80676194 (doi:10.1259/bjr/80676194) [DOI] [PubMed] [Google Scholar]
- 91.Gee A. H., Chen L., Freeman S., Treece G. M., Prager R. W., Berman L. H. 2008. A clinical database for evaluating freehand quasistatic strain imaging systems. In Proc. 7th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Austin, TX, 27–30 October 2008, p. 25 Austin, TX: ITEC [Google Scholar]
- 92.Treece G. M., Lindop J. E., Gee A. H., Prager R. W. 2008. Fast, variable smoothing of strain data using nonparametric regression. In Proc. 7th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Austin, TX, 27–30 October 2008, p. 57 Austin, TX: ITEC [Google Scholar]
- 93.Lorenz A., Pesavento A., Pesavento M., Ermert H. 1999. Three-dimensional strain imaging and related strain artifacts using an ultrasonic 3D abdominal probe. In Proc. IEEE Ultrasonics Symp., Caesars Tahoe, NV, 17–20 October 1999, vol. 2, pp. 1657–1660 [Google Scholar]
- 94.Lindop J. E., Treece G. M., Gee A. H., Prager R. W. 2006. 3D elastography using freehand ultrasound. Ultrasound Med. Biol. 37, 529–545 10.1016/j.ultrasmedbio.2005.11.018 (doi:10.1016/j.ultrasmedbio.2005.11.018) [DOI] [PubMed] [Google Scholar]
- 95.Schaar J. A., de Korte C. L., Mastik F., van Damme L. C. A., Krams R., Serruys P. W., van der Steen A. F. W. 2005. Three-dimensional palpography of human coronary arteries. Herz 30, 125–133 10.1007/s00059-005-2642-4 (doi:10.1007/s00059-005-2642-4) [DOI] [PubMed] [Google Scholar]
- 96.Li Y., Patil A., Hossack J. A. 2006. High resolution three-dimensional prostate ultrasound imaging. Proc. SPIE 6147, 32–40 10.1117/12.661504 (doi:10.1117/12.661504) [DOI] [Google Scholar]
- 97.Bharat S., Fisher T. G., Varghese T., Hall T. J., Jiang J., Madsen E. L., Zagzebski J. A., Lee F. T., Jr 2008. Three-dimensional electrode displacement elastography using the Siemens C7F2 fourSight four-dimensional ultrasound transducer. Ultrasound Med. Biol. 34, 1307–1316 10.1016/j.ultrasmedbio.2008.01.007 (doi:10.1016/j.ultrasmedbio.2008.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krueger M., Pesavento A., Ermert H., Hiltawsky K. M., Heuser L., Rosenthal H., Jensen A. 1998. Ultrasonic strain imaging of the female breast using phase root seeking and three-dimensional ‘optical flow’. In Proc. IEEE Ultrasonics Symp., Sendai, Japan, 5–8 October 1998, vol. 2, pp. 1757–1760 [Google Scholar]
- 99.Fisher T. G., Hall T. J., Panda S., Jiang J., Resnick J., Barnes S., Madsen E. L. 2006. Volume elasticity imaging with a 2-D capacitive micro-machined ultrasound transducer (CMUT) array. In Proc. 5th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Snowbird, UT, 8–11 October 2006, p. 125 Austin, TX: ITEC [Google Scholar]
- 100.Insana M. F., Chaturvedi P., Hall T. J., Bilgen M. 1997. 3-D companding using linear arrays for improved strain imaging. In IEEE Ultrasonics Symp., Toronto, Canada, 5–8 October 1997, pp. 1435–1438 [Google Scholar]
- 101.Housden R. J., Gee A. H., Treece G. M., Prager R. W. 2010. 3-D ultrasonic strain imaging using freehand scanning and a mechanically-swept probe. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 57, 501–506 10.1109/TUFFC.2010.1431 (doi:10.1109/TUFFC.2010.1431) [DOI] [PubMed] [Google Scholar]
- 102.Ponnekanti H., Ophir J., Cespedes I. 1994. Ultrasonic imaging of the stress distribution in elastic media due to an external compressor. Ultrasound Med. Biol. 20, 27–33 10.1016/0301-5629(94)90014-0 (doi:10.1016/0301-5629(94)90014-0) [DOI] [PubMed] [Google Scholar]
- 103.Konofagou E. E., Varghese T., Ophir J. 2000. Theoretical bounds on the estimation of transverse displacement, transverse strain and Poisson's ratio in elastography. Ultrason. Imaging 22, 153–177 [DOI] [PubMed] [Google Scholar]
- 104.Thitaikumar A., Ophir J., Krouskop T. A. 2005. Noise performance and signal-to-noise ratio of shear strain elastograms. Ultrason. Imaging 27, 145–165 [DOI] [PubMed] [Google Scholar]
- 105.Thitaikumar A., Krouskop T. A., Righetti R., Ophir J. 2006. Resolution of axial shear strain elastography. Phys. Med. Biol. 51, 5245–5257 10.1088/0031-9155/51/20/011 (doi:10.1088/0031-9155/51/20/011) [DOI] [PubMed] [Google Scholar]
- 106.Thitaikumar A., Krouskop T. A., Ophir J. 2007. Signal-to-noise ratio, contrast-to-noise ratio and their trade-offs with resolution in axial-shear strain elastography. Phys. Med. Biol. 52, 13–28 10.1088/0031-9155/52/1/002 (doi:10.1088/0031-9155/52/1/002) [DOI] [PubMed] [Google Scholar]
- 107.Thitaikumar A., Mobbs L. M., Kraemer-Chant C. M., Garra B. S., Ophir J. 2008. Breast tumor classification using axial shear strain elastography: a feasibility study. Phys. Med. Biol. 53, 4809–4823 10.1088/0031-9155/53/17/022 (doi:10.1088/0031-9155/53/17/022) [DOI] [PubMed] [Google Scholar]
- 108.Rao M., Varghese T., Madsen E. L. 2008. Shear strain imaging using shear deformations. Med. Phys. 35, 412–423 10.1118/1.2825621 (doi:10.1118/1.2825621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen H., Varghese T. 2009. Principal component analysis of shear strain effects. Ultrasonics 49, 472–483 10.1016/j.ultras.2008.12.003 (doi:10.1016/j.ultras.2008.12.003) [DOI] [PubMed] [Google Scholar]
- 110.Chen L., Housden R. J., Treece G. M., Gee A. H., Prager R. W. 2010. A normalization method for axial-shear strain elastography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 57, 2833–2838 10.1109/TUFFC.2010.1757 (doi:10.1109/TUFFC.2010.1757) [DOI] [PubMed] [Google Scholar]
- 111.Galaz B., Thitaikumar A., Ophir J. 2009. Axial-shear strain distributions in an elliptical inclusion model. I. A simulation study. In Proc. 8th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Vlissingen, The Netherlands, 14–17 September 2009, p. 99 Austin, TX: ITEC [Google Scholar]
- 112.Thitaikumar A., Galaz B., Ophir J. 2009. Axial-shear strain distributions in an elliptical inclusion model. II. Experimental validation and in vivo examples with implications to breast tumor classification. In Proc. 8th Int. Conf. on Ultrasonic Measurement and Imaging of Tissue Elasticity, Vlissingen, The Netherlands, 14–17 September 2009, p. 100 Austin, TX: ITEC [Google Scholar]