Abstract
The fields of medical image analysis and computer-aided interventions deal with reducing the large volume of digital images (X-ray, computed tomography, magnetic resonance imaging (MRI), positron emission tomography and ultrasound (US)) to more meaningful clinical information using software algorithms. US is a core imaging modality employed in these areas, both in its own right and used in conjunction with the other imaging modalities. It is receiving increased interest owing to the recent introduction of three-dimensional US, significant improvements in US image quality, and better understanding of how to design algorithms which exploit the unique strengths and properties of this real-time imaging modality. This article reviews the current state of art in US image analysis and its application in image-guided interventions. The article concludes by giving a perspective from clinical cardiology which is one of the most advanced areas of clinical application of US image analysis and describing some probable future trends in this important area of ultrasonic imaging research.
Keywords: ultrasound, quantitative ultrasound, image-guided interventions, image analysis, fusion echocardiography
1. Introduction
Advances in ultrasonic hardware have transformed the quality of information provided by a medical ultrasound (US) scan today relative to 15–20 years ago. Most hospitals in the Western World now have fully digital two-dimensional US systems in radiology, cardiology and obstetrics departments; many have US systems—particularly for clinical diagnosis in cardiovascular medicine and obstetrics; and portable systems are becoming more widely used, with this trend towards smaller systems likely to continue, particularly driven by healthcare needs and the requirement for low-cost imaging solutions in the developing world. As a result, Zetabytes (1021 bytes) of ultrasonic data are acquired every minute around the world and are used to make clinical decisions. US is the first modality of choice in, for instance, obstetrics, breast mass assessment and biopsy guidance, and used extensively in assessment of cardiovascular disease. However, and perhaps not always appreciated by researchers from outside the imaging field, clinicians use medical US to provide information for diagnosis, guidance of treatments or to assess the success of therapy, and not to generate pretty pictures of anatomy. US cannot compete with magnetic resonance imaging (MRI) and X-ray computed tomography (CT) in this respect. However, US is certainly not inferior to MRI and CT—it is just different. For instance, it is the only imaging modality capable of imaging soft tissue deformations quickly enough for interventional procedure guidance—information that fluoroscopy, CT or MRI cannot provide. Further, its portability and low cost relative to its expensive alternatives are significant positives that mean US has a very exciting future ahead.
This review focuses on the impact of US on medical image analysis and image-guided interventions. These fields deal with reducing the large volume of digital images (X-ray, CT, MRI, positron emission tomography and US) to more meaningful clinical information using software algorithms. Their role in clinical workflow and decision making is increasing. These research fields are also quite young—around 20 years old—with their origins in computer vision, video image processing and robotics. The fields recently accelerated their clinical relevance and acceptance due to increased computational power of desktop computers, and the maturity of certain classes of method that delineate anatomical boundaries (image segmentation), align images of the same or different imaging modalities (image registration), perform automated quantification and provide real-time processing solutions suitable for interventional use. This review article outlines the state of the art in US image analysis and US-based image interventions illustrated by some success stories and on-going work by the authors and others. It concludes by taking a forward look at possible future directions in this field.
2. Image analysis
Working with US imaging as an information source has positives and negatives.
On the positive side, US is a relative cheap imaging modality compared with MRI and CT, it is portable and it offers a real-time acquisition capability. Principal negatives are that acquisition is operator-dependent and (often significant) training is required to acquire good data. In addition, the presence of speckle—the coherent interference pattern that gives US images their characteristic granular-looking pattern—complicates analysis (but can be a strength, see below). Therefore, the challenge is to work with its strengths, and to try to overcome its limitations by embedding prior knowledge of image formation and the object of interest (geometry, expected image appearance, etc.), and to use carefully designed acquisition protocols wherever possible to minimize operator-dependency.
US imaging does not share the high anatomical definition of CT and MRI. Image appearance is dependent on factors including orientation of the probe with respect to the object, signal attenuation and missing information due to signal dropouts and acoustic shadows. These offer unique challenges to US image processing and analysis relative to CT and MRI analysis. The large majority of the US image analysis literature today is based on B-mode images (also sometimes called display images, or log-compressed US images) in part because this is the most easily available clinical US data offered by commercial systems. Unlike the case of MRI research, which is heavily driven by advances in MR pulse sequences developed by the academic community on MRI research systems, there are few commercial US research machines which provide access to the radio-frequency (RF) or post-processed RF (in-phase quadrature) data. The number of articles on methods that analyse the RF signal is increasing, in part through the maturity of elastography (see separate article) but also recognition of the limitation of B-mode processing alone. Working with B-mode images is more challenging, because the image formation process is nonlinear and difficult to model mathematically. This means that you cannot invoke simple imaging physics models to aid B-mode image processing. Sometimes empirical models of B-model images have been assumed but only for application-specific cases (e.g. [1]).
2.1. Speckle-reduction techniques
Given the variability of image quality a large amount of research effort has gone into attempting to produce images of standard appearance and to reduce speckle. This is both to improve the appearance of images but importantly for image analysis, both intensity inhomogeniety and speckle can challenge intensity-based segmentation methods. Attenuation of US with depth means that a deeper region of the same object or tissue (e.g. a liver) can appear darker than a less deep region. In our own work, for instance, we have used an Expectation–Maximization approach to reduce the effects of attenuation on breast US images [2] and an image surface interpolation method to standardize two-dimensional echocardiographic images while not distorting speckle [3]. Speckle-reduction is a well-studied problem. Here speckle is treated as noise and something that needs to be removed or reduced. At the RF signal level, speckle is often modelled by a Rayleigh distribution but this is not suitable for log-compressed US images. Recent work has shown that a Gamma distribution [1] or a Fisher–Tippett distribution [4] are good approximations for US images. Other work, such as [5] has assumed a general multiplicative model. Adaptive filters [6–8], partial differential equation-based approaches [9,10] and wavelet-based approaches [11,12] to US speckle reduction all appear in the literature for speckle-reduction. Results from one recent approach that adapted the non-local-means filter to speckle noise [13] is shown in figure 1 to illustrate the general effect of a speckle-reduction method. Note, however, that it is not always desirable to remove speckle as it acts as a natural tissue ‘tag’ and moves as tissue is deformed (cf. in many respects to tagged MRI [14]). The presence of speckle is critical to the success of techniques such as strain imaging and speckle tracking [15], and also for many methods of US tissue characterization [16].
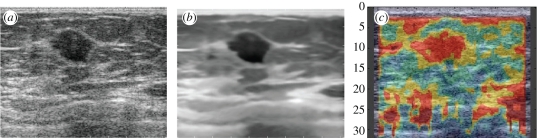
Figure 1.
Is speckle noise or a feature? (a) Original B-mode image of a breast mass. (b) Speckle reduction applied to the original image. Note that the texture has been removed from the image but major structures remain well-defined. (c) Elastography image of the same mass derived from RF data. Speckle tracking is an implicit or explicit part of elastography. (Results courtesy of Aymeric Larrue and Andy di Battista, U Oxford UK).
2.2. Image segmentation
Image segmentation is the technical term used to mean manually, semi-automatically or fully automatically delineating the boundaries of an object or tissue regions. Using computer algorithms to assist in US image segmentation is a well-studied problem and there are some good software solutions available, including within commercial packages, although primary use is still within the research community. Readers are referred to the articles in [17,18] for recent reviews of the general field of US segmentation and the case of carotid plaque image segmentation, respectively.
Given that US images do not necessarily define complete object boundaries, the most successful automated image segmentation methods have been those that use prior knowledge of object shape, motion and image appearance. A range of methodologies have thus been developed to embed these constraints, a non-exhaustive list of alternatives including Bayesian approaches [19,20], active contours and active appearance models [21–24], and level-sets [25,26]. An excellent example of this is the work on applying active appearance models applied to echocardiography by Leung et al. [27]. In this approach, image training datasets are used to define the expected shape variation of a typical left ventricle. The shape model is then used to constrain the segmentation solution as the model is fit to a new image example. Figure 2 illustrates this approach.
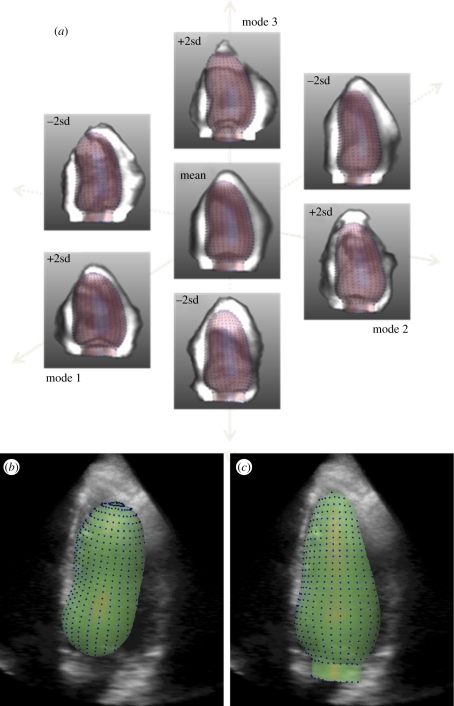
Figure 2.
Three-dimensional active appearance model segmentation. (a) Average left ventricular object shape and image appearance over a large training set of patients (centre) and the first three most important modes of variation of shape and appearance. (b) Initialization of the model in a three-dimensional dataset; (c) final segmentation result. (Results courtesy of Hans Bosch and Esther Leung, Erasmus MC, Rotterdam NL).
Another way to embed prior knowledge is to take a data-driven or learning-based approach in which image intensity patterns are learned using a classifier to distinguish an object of interest from its surroundings where the classifier has been trained on a large database of example images. This can be viewed as object detection rather than the classic image segmentation problem of object boundary delineation, and used, for instance, to quickly find an image region in which to subsequently manually or automatically take an image measurement. Advanced machine-learning techniques such as AdaBoost, Probabilistic Boosting Trees and Random Forests, and their application to assist US image segmentation are being actively researched [28–30], and we are likely to see more applications of this class of method in the future.
In the case of four-dimensional US imaging (three spatial dimensions and the fourth being time), the problem is that of detection, segmentation and tracking. For instance, to perform fast and accurate tracking of the left ventricle from four-dimensional transthoracic echocardiography data, the algorithm in [31] learns a cardiac motion model using ISOMAP and K-means clustering. This is extended in [32] to automatically quantify the three-dimensional myocardial motion from high frame rate instantaneous US images using learning-based information fusion.
In the case of US segmentation problems that are very difficult to automate, or when a fast segmentation is required, an alternative strategy is to use a semi-automated approach where user-initialization or interaction is used to guide the process of segmentation. Indeed, many of the methods described above in practice are initialized manually (mathematically meaning that the optimization method underpinning the algorithm is sensitive to initialization conditions). Particularly for intensity-based methods, it is a common practice to ‘seed’ an algorithm with manually selected regions-of-interest defining ‘object’ and ‘background’ intensity distributions. This is illustrated in figure 3, which shows semi-automated segmentation of a placenta from a three-dimensional US image using a variant of the graph-based method known as the random walker algorithm [33]. Figure 3a shows a slice from the original three-dimensional US scan. Figure 3b shows the seeding step, where the green areas indicate background voxels, red areas indicate foreground voxels. Figure 3c shows both the boundaries found by the random walker algorithm, and the manually defined boundary independently defined by a clinical expert. Note that in the case shown here, only one object was seeded, but the random walker can be seeded by multiple seeds in general. It can also be automated but was chosen here to illustrate its power as an interactive technique. Related work includes [34] which used the NCut technique combined with anisotropic diffusion for unsupervised US segmentation.
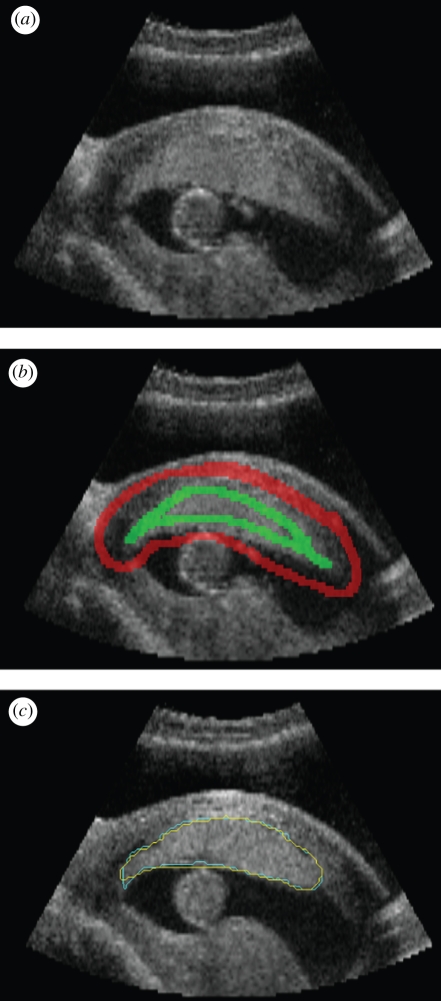
Figure 3.
(a) Image slice through a three-dimensional ultrasound scan of a placenta; (b) seeding for random walker algorithm. (c) Boundary found by automated algorithm (blue) and manual segmentation (yellow). (Result courtesy of Gordon Stevenson, University of Oxford, UK).
2.3. Ultrasound image registration
The field of image registration deals with the alignment of two or more images of the same imaging modality or different imaging modalities. The degree of difficulty in solving a given image registration problem is principally determined by the complexity of the geometric transformation between the two (or more) images (rigid, affine, non-rigid, etc.) and defining the matching criteria between the images, although other factors such as algorithm speed, capture range (sensitivity to initialization) and required accuracy of alignment are also key factors in selecting a method, particularly for real-time diagnosis and image-guided intervention applications. By far, the most popular method to US-based image registration uses the similarity-measure-based registration approach. For US–US registration, as used for instance in motion-tracking, free-hand US volume reconstruction and US mosaicing [35–39], classic similarity-measures such as normalized cross-correlation and normalized mutual information work well, although US-specific similarity measures based on a maximum likelihood framework have also been proposed [40,41].
US-X registration, where X is another imaging modality is more challenging. In this case, the similarity measure needs to capture some functional relationship between the intensities of the different imaging modalities. Unless pre-processing is done, this needs to be as insensitive as possible to US artefacts and insonification angle effects. Some groups have used mutual information [42], the generalized correlation-ratio [43], transformed an US image into a ‘vesselness’ image prior to registration [44] or proposed to define the similarity measure in terms of local-phase representation of each image rather than intensity [45,46]. Figure 4, for instance, shows an example of the method of Zhang et al. [46] applied to echocardiography-MR image registration. Here, the echocardiography image is shown as a green overlay on the corresponding MR image slice. Applications of US-X registration will be considered further in §3.
Figure 4.
Two-dimensional non-rigid alignment of echocardiography and cardiac MRI. (a) Cardiac MRI image, (b) ultrasound image with red, blue and yellow markers indicating points about which local alignment corrections are made at successive iterations of the registration algorithm. (c) The green overlay is the echocardiography image superimposed on the cardiac MR image slice. (Result courtesy of Weiwei Zhang, U. Oxford, UK).
Finally, US–US registration is the first step of fusion three-dimensional echocardiography a technique initially proposed in [47] for improving three-dimensional echocardiographic image segmentation by using multiple three-dimensional US scans, but which has been further improved (e.g. [48–51]), and demonstrated in clinical pilot studies to improve the field of view, anatomical definition and contrast-to-noise ratio relative to single scans for diagnostic purposes [52]. Fusion echocardiography (FE) is a software post-processing solution related to US spatial compounding [53] but it does not involve electronic beam steering of a transducer array (which has a restricted field of view). Rather than compounding (averaging) the intensity values the ‘best estimate of information content’ is taken by considering intensities from all scans. For instance, in figure 5 the wavelet-based fusion method described in [52] has been used.
Figure 5.
(a) Two-dimensional cut planes from a single three-dimensional echocardiography dataset: top left 4-chamber view, top right 2-chamber view, bottom left short axis view, bottom right 4-chamber view. Note the noisy recording with insufficient delineation of the endocardial borders. (b) Two-dimensional cut planes from a three-dimensional dataset where the dataset in figure 5(a) and three others of similar quality were combined together using the image fusion algorithm: top left 4-chamber view, top right 2-chamber view, bottom left short axis view, bottom right 4-chamber view. All cut planes show a clear delineation of the myocardium from the cavity in all segments.
3. Image-guided interventions
Image-guided interventions were traditionally mostly performed under X-ray fluoroscopic imaging. The first such procedures were in fact performed shortly after the invention of X-ray at the end of nineteenth century. With the introduction of US imaging into medicine in the second half of twentieth century, gradually it was also considered for its use in interventional procedures. Because of its non-ionizing characteristic and its cost effectiveness, its popularity has increased considerably in the last decades. US is the modality of choice for many clinical applications. US has also become a common imaging tool to support pain management during various interventional procedures. The large number of applications in which US is regularly used as the main modality for diagnosis and treatment is too large to be reviewed within this article. Here, we rather focus more on the recent medical imaging and computer-assisted intervention techniques, which enable multimodal imaging and propose novel techniques for integrating US into further clinical applications. In spite of its many advantages over other imaging modalities such as X-ray, CT and MRI in term of its real-time nature, its small size and possibility of further miniaturization, its suitability for integration into interventional procedures and its price, rapid increase of its interventional use has been hampered by several characteristics of this imaging modality. One of these is its high user dependency. This property could be considered by experts as an advantage and by trainees as a disadvantage. This results in two issues. First, the training becomes an important subject for interventional use of US imaging. Second, the success of interventional US imaging procedures is highly dependent on the experience level of its user and therefore difficult to assess. In this regard standard US procedures sometimes have difficulties in establishing themselves as an alternative or a replacement for traditional X-ray-, CT- or MRI-based image-guided procedures. This has pushed the scientific community towards many related research directions. On one side, US simulation has become an active subject of research with the clear objective of improving and accelerating the training of practitioners. On the other side, the registration of interventional US and other standard imaging modalities such as CT and MRI has been another focus of research, in this case with the objective of allowing the physician to transfer their diagnostic information and intervention plans into an US-based procedure, which in turn reduces the user dependency on interpretation of interventional US images and helps improve and standardize the procedures. In addition to these points which are the focus of our discussion, there are two other directions of active research in interventional US imaging. The first one is US mosaicing as well as compounding into three-dimensional and four-dimensional volumes. This allows for imaging and visualization of larger areas and volume, once again reducing the need for the physician to compound such dynamic information mentally [54–58]. In the following, we also emphasize the suitability of US as an enabling imaging technology in medical robotics.
Many technical development ideas in image-guided interventions have first been developed and tested for brain surgery applications. It is no surprise that researchers are also looking at using US imaging to improve the performance of image-guided intervention techniques for brain surgical applications. A neuro-navigation system based on a custom-built three-dimensional US was proposed in [59,60]. They claimed that the use of three-dimensional US improves the results by providing real-time imaging feedback. Rasmussen et al. [61] used a commercial three-dimensional US system provided by the company SonoWand, for intra-operative recording of multiple three-dimensional US volumes and navigation through a co-registered pre-operative surgical plan. The registration and data visualization was based on external optical tracking and therefore did not include automatic estimation of brain shift. They, however, presented a post-operative study of a vessel-based co-registration of three-dimensional Power Doppler and segmented vessel trees from magnetic resonance angiography to evaluate its potential for brain shift estimation. Recently, Rasmussen et al. [62] presented an image-based method for registration of MRI to three-dimensional US acquired before the opening of the dura and provided promising results for four cases of brain tumour surgery. The real-time estimation of brain shift using intra-operative two-dimensional and three-dimensional US and its fusion with pre-operative MRI and electrophysiological data for surgical navigation is still an ongoing subject of research and development. We expect to see many exciting development in this domain in the following years.
US is probably one of the most suitable imaging modalities for supporting medical robotics applications. A visual servoing scheme, based on real-time two-dimensional US imaging, was proposed in [63] to control the insertion of a surgical forceps with four degrees of freedom into a beating heart within a minimally invasive procedure. Real-time three-dimensional US was employed in [64] to detect the instrument and track the tissue in order to synchronize the motion of instrument with the heart. Such work could enable advance robotics solution to be deployed in minimally invasive cardiac surgery simplifying the human control of the robot through US-based compensation of fast cardiac motion.
US-assisted cardiac interventional guidance is considered in [65]. The proposed system co-registered patient-specific models obtained from pre-operative data with intra-operative real-time US images within an advanced mixed reality visualization environment also integrating the three-dimensional virtual models of the tracked surgical instruments. The authors demonstrate the high accuracy of the system and its appropriateness for cardiac interventions.
Miniaturization of US sensors [66] has allowed their integration into catheters, endoscopes and laparoscopes. For example, US is systematically used for needle guidance in many procedures. Many systems also offer the visualization of a needle within the US B-mode plane by mechanical construction and pre-calibration of the US detector array and the needle insertion support. The use of such systems in transrectal US (TRUS) as well as endoscopic US has entered into routine practice in many clinical applications.
Since its introduction, intravascular imaging (IVUS) has been considered and used by many interventional cardiologists not only for analysis and characterization of plaques composition [67] but also for navigation during stent placement [68]. This has incited many academic and industrial researchers to focus on many different challenges required for the integration and use of this technology within vascular interventions. In general, IVUS catheters are navigated under X-ray fluoroscopy guidance and the existence of pre-operative CT angiography data in many cases encourages the registration of these modalities. In addition, the integration of IVUS with other intra-vascular modalities such as intravascular optical coherence tomography or the recently introduced intra-vascular near-infrared fluorescence imaging [69] could complement this imaging and increase its successful use for image-guided interventions.
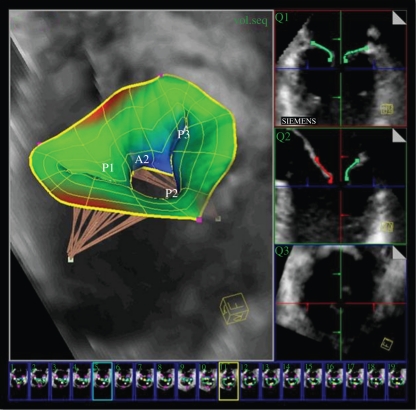
Transesophageal echocardiography (TEE) is routinely used in operating rooms for pre-operative analysis, intra-operative guidance and post-operative assessment of various minimal invasive valve repair and replacement procedures. A robust learning-based framework was introduced in [70] to estimate patient-specific models of the aortic and mitral valves morphology and function from four-dimensional TEE data. See figure 6. This work is extended in [71] by incorporating models of the left ventricle and atrium, to enable comprehensive hemodynamic analysis within the left side of the heart.
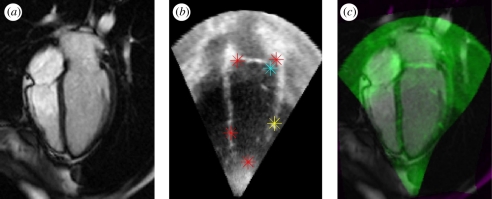
Figure 6.
(Top) Patient-specific model of the mitral valve with subvalvular apparatus out of four-dimensional transesophageal echocardiography images. (Bottom) The morphological and functional quantification of the mitral valve, based on the model. Courtesy of Razvan Ionasec, Siemens Corporate Research, Princeton, USA.
The fusion of intra-cardiac three-dimensional echocardiography with C-arm CT for treating atrial fibrillation using catheter ablation is investigated in [72]. In this work, an intracardiac real-time three-dimensional US catheter is presented and used. The authors develop methods for registration and visualization of CT reconstruction obtained by an angiographic C-arm and real-time three-dimensional US data acquired with the proposed US catheter.
Tracked B-mode US has also been frequently used for treatment of different cancers. For instance, different properties of US imaging were used in [73] to define a specific algorithm to co-register CT and US for its use in radiation therapy of head and neck cancer. A novel CT-US registration method was proposed in [74] satisfying both requirements of full automation and real-time computation. The method simultaneously optimized the linear combination of different ultrasonic effects simulated from CT, and the CT-US affine registration parameters. The method is aimed at assisting treatment of cancer in both the liver and kidney. Instead of a global rigid or affine registration of US with CT, Wein et al. [75] presented first results for a full deformable registration using the simulation-based approach on the graphics processing unit to better represent the real situation. However, this still does not achieve the real-time performance required for image-guided applications and could be considered as work in progress.
A recent approach is based on creation of a statistical model from CT or MR and its registration to two-dimensional or three-dimensional US. A statistical deformation model of the femur and pelvis was built in [76] and registered to a set of surface points obtained from intra-operative three-dimensional US data. A statistical shape model of a lumbar spine and interventional three-dimensional US were registered in [77] for image guidance in spinal needle injection. In [78], a patient-specific statistical motion model of the prostate was built from biomechanical simulations of a finite element model of the prostate obtained from pre-operative MR data. This was then registered to intra-operative three-dimensional TRUS images for its use in guiding prostate biopsy and interventions.
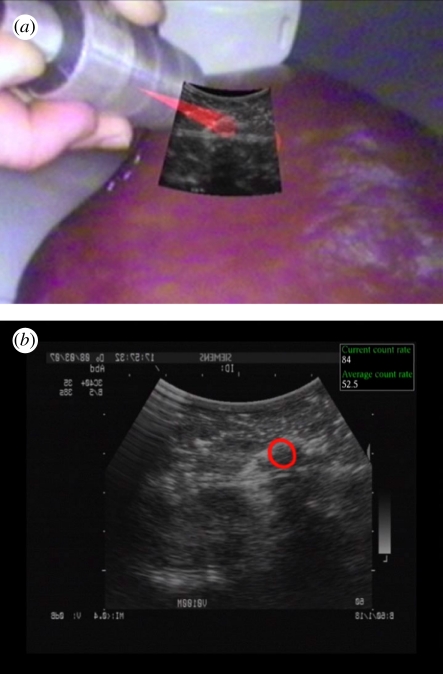
Another new trend is the use of tracked nuclear probes to get functional intra-operative information. Surgeons, however, wish to position such functional information within the context of anatomical imaging. The registration of intra-operative and dynamic functional imaging to pre-operative CT or MRI data requires the algorithms to compute complex deformation in real-time. Another alternative offered by two-dimensional and three-dimensional US imaging is the fusion and visualization of co-registered US and functional imaging data (see figure 7). A system for systematic co-registration of tracked nuclear gamma and B-mode US probes was proposed in [79]. The ease of integration of real-time US imaging and different functional imaging systems allows the development of novel techniques [80] and even hardware [81] and their integration into procedures in the near future. The real-time co-registered acquisition of US and functional data will also open the path for deformable registration of interventional functional data to pre-operative CT or MRI data, by taking advantages of US-CT and US-MR registration methods discussed earlier in this paper.
Figure 7.
(a) Augmented reality visualization of tracked US and gamma probe acquisition on a liver phantom with implanted malignant and benign tumours. (b) US imaging display augmented by the gamma information presented as a red circle where the probe is sensing high activity. The count rate of detected activity is provided by the gamma probe and is displayed within the upper right part of the display.
4. Clinical perspective
The driver for image analysis in clinical echocardiography for many years has been the desire to have automated quantification tools to compute measures such as the left ventricular (LV) ejection fraction, and local wall motion and myocardial thickening as indicators of LV dysfunction and ischaemia (e.g. [82]). Many of the advances described in §1 are motivated by this. Although three-dimensional echocardiography-based LV volumes are underestimated with respect to cardiac MR [83] three-dimensional echocardiography still offers a very convenient and cheaper way to assess LV function. Sophisticated commercial software packages for visualizing and manipulating three-dimensional echocardiography now exist, but few semi or fully automated quantification methods have been well-validated or commercialized [84,85] and none are used in routine clinical practice in hospitals. The limiting factor remains the wide variability of quality of patient data with the literature citing between 40 and 60 per cent of all routine clinical data having a ‘good or intermediate acoustic window’ meaning having a good enough quality for an image analysis method to work reliably on it. From a clinical perspective this means that it is not cost-effective to introduce automated image analysis into clinical workflow at the current time.
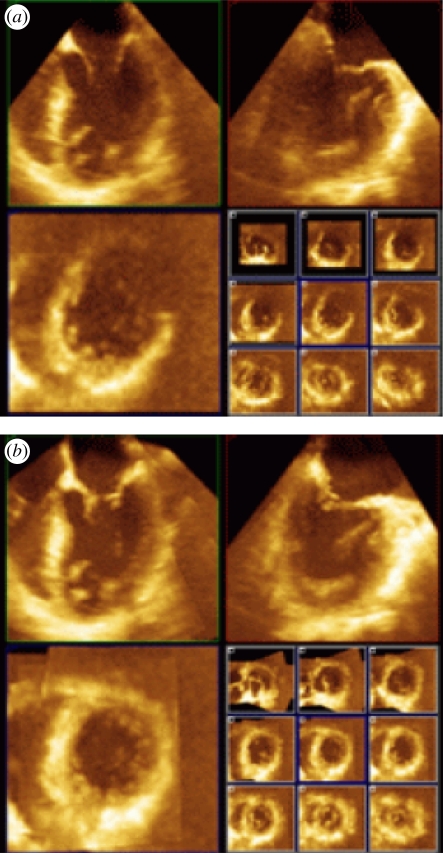
The technique of fusion three-dimensional echocardiography mentioned in §1 (see also figures 5 and 8) provides one way to improve image quality and the field of view, both limiting factors of current US technology. Here we briefly discuss some of the challenges in clinical three-dimensional echocardiography today from the clinical perspective and suggest how FE may help to overcome some of them. In particular, several aspects of fusion three-dimensional echocardiography make it particularly attractive for clinical echocardiography: besides better image quality and comprehensive datasets, three-dimensional FE has the potential for a major improvement in workflow in the clinical echo laboratory.
Figure 8.
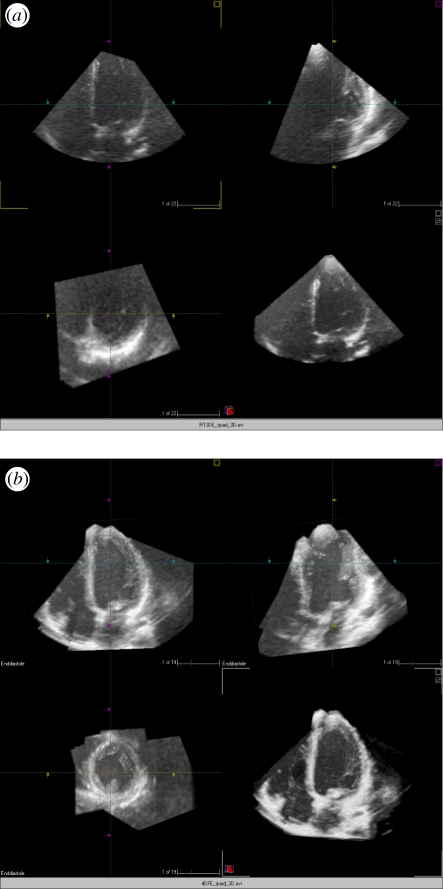
(a) Two-dimensional orthogonal cut planes from a single three-dimensional transoesophageal echocardiography dataset with 9 short-axis cross-sections shown in the lower right quadrant. (b) Equivalent for a fused three-dimensional echocardiography reconstruction clearly indicating superior image quality. (Result courtesy of Daniel Augustine, University of Oxford, UK).
4.1. Improving three-dimensional echocardiography image quality
Over the last 10 years there has been some improvement of three-dimensional echocardiography image quality but still a large proportion of three-dimensional studies are not good enough to be used instead of standard two-dimensional echocardiography. This means that there are parts of the left ventricle and other heart chambers in which the border between the heart muscle and the blood pool cannot be tracked visually or by an automated contour finding system. In the first clinical trial using three-dimensional FE only approximately 70 per cent of three-dimensional non-fused (i.e. native) datasets were graded as having intermediate or good image quality [51]. Note that this study included a high number of healthy younger adult individuals—in a typical clinical echo laboratory one would expect a higher number of patients with insufficient image quality for quantitative analysis. The study went on to show that three-dimensional FE increases the proportion of participants with general image quality graded as good or intermediate quality from 70 per cent with standard three-dimensional echocardiography to 97 per cent with three-dimensional FE. This difference should translate into improved diagnostic potential for even small areas of wall motion abnormality. The improvements in image quality seen with three-dimensional FE are much more impressive than the progress obtained from newer transducers.
Even for the fetal heart getting consistent, shadow free, data remain problematic due to the uncontrollable nature of foetal orientation. The utility of image fusion has been demonstrated in a recent clinical study [86] where it was shown that it reduced the variability of volume estimates by about 50 per cent relative to measurement on a single scan.
The few clinical studies validating fusion three-dimensional echocardiography to-date have focused on the left ventricle. However, there is also a need to improve the border definition of the other heart chambers. In particular, there is a need to improve the display of the right ventricle. Because of the geometry of the right ventricle two-dimensional echocardiography for assessment of volumes and function is challenging and three-dimensional echocardiography has been shown to provide good agreement with cardiac MRI. We hypothesize that three-dimensional FE of the right ventricle will result in a similar improvement as observed in three-dimensional fusion of the left ventricle and are currently performing a study with MRI as the gold standard.
4.2. Three-dimensional datasets including the entire organ
At present abnormally large hearts cannot be displayed in a single three-dimensional echocardiography dataset. Three-dimensional FE provides a comprehensive dataset, in which the sonographer can navigate and follow structures without reviewing different loops which are not aligned. With three-dimensional FE it is possible to maintain the highest volume rate and line density. In particular, three-dimensional TEE suffers from limited field of view due to the limited size of the transducer. The image quality is less an issue because of the proximity of the transducer to the heart. Although three-dimensional TEE has become a valuable tool for guiding cardiac surgery and interventions such as transcutaneous closure of shunts and paravalvular leaks, it is often difficult to assess the exact topography of pathology in separate three-dimensional datasets. The availability of a comprehensive dataset displaying the entire heart would facilitate planning and guiding interventional procedures. Early results on three-dimensional FE applied to TEE are reported in [87], see figure 8.
4.3. Simplified image acquisition: a vision
Echocardiographic imaging requires the skills of an experienced sonographer to find the optimal acoustic window and adjustment of the probe. Unfortunately, often the optimal acoustic window is missed. With three-dimensional FE there is the potential to become less dependent on the sonographer's experience: a first step is using a simple protocol of consecutive acquisitions in the vicinity of where a single dataset is normally acquired and then the fusion process is started. The optimal cut-planes can then be found in the high quality three-dimensional FE reconstruction and analysed by using already available programs such as Qlab (Philips) or four-dimensional LV analysis (TomTec) [88]. With faster implementations of three-dimensional FE, it might be possible to scan and fuse consecutive heart beats in near real-time. Recently, transducers have become available that can acquire datasets within a single heart beat and do not require breath holding. Eventually the image acquisition may only need the simple placement of a new type of probe comparable to putting electrocardiogram patches and then further acquisition is automatic. That would have major financial implications as the cost of a sonographer and the scan time per patient would be greatly diminished.
5. Conclusion
In this article, we have outlined how US has evolved in the fields of medical image analysis and image-guided interventions over the past 15–20 years. In spite of strong competition from MRI and CT in particular, there is no question that US will continue to play a key role in multi-modality clinical decision making in the future and if anything its role will increase as three-dimensional US imaging becomes more widely available as a cost-effective three-dimensional imaging modality. A key barrier to more widespread use of automated analysis methods remains the high level of expertise required to acquire good quality data but as we have seen, providing solutions to this is an active area of research at the current time. Today, US acquisition and image analysis are generally seen as two separate tasks but in the future we can expect to see the boundaries between the two blurred with the ‘intelligence’ of image analysis software driving the advanced capabilities of US machines.
Acknowledgements
J.A.N. acknowledges the financial support of the UK Engineering and Physical Sciences Research Council (EPSRC) (and particularly EP/G030 693/1 shared jointly with H.B.), the NIHR funded Oxford Biomedical Research Centre Imaging Theme, and the Oxford Centre of Excellence in Personalized Healthcare funded by the Wellcome Trust and EPSRC under grant number WT 088 877/Z/09.
References
- 1.Tao Z., Tagare H. D., Beaty J. D. 2006. Evaluation of four probability distribution models for speckle in clinical cardiac ultrasound. IEEE Trans. Med. Imag. 25, 1483–1491. 10.1109/TMI.2006.881376 ( 10.1109/TMI.2006.881376) [DOI] [PubMed] [Google Scholar]
- 2.Xiao G., Brady M., Noble J. A., Zhang Y. 2002. Segmentation of ultrasound B-mode images with intensity inhomogeneity correction. IEEE Trans. Med. Imag. 21, 48–57. 10.1109/42.981233 ( 10.1109/42.981233) [DOI] [PubMed] [Google Scholar]
- 3.Boukerroui D., Noble J. A., Robini M. C., Brady M. 2001. Enhancement of contrast regions in suboptimal ultrasound images with application to echocardiography. Ultrasound Med. Biol. 27, 1583–1594. 10.1016/S0301-5629(01)00478-1 ( 10.1016/S0301-5629(01)00478-1) [DOI] [PubMed] [Google Scholar]
- 4.Slabaugh G., Unal G., Fang T., Wels M. 2006. Ultrasound-specific segmentation via decorrelation and statistical region-based active contours. IEEE Computer Society Conf. on Computer Vision and Pattern Recognition (CVPR'06), New York City, NY, vol. 1, pp. 45–53. IEEE Computer Society. [Google Scholar]
- 5.Loupas T., McDicken W., Allan P. 1989. An adaptive weighted median filter for speckle suppression in medical ultrasound image. IEEE Trans. Circuits Syst. 36, 129–135. 10.1109/31.16577 ( 10.1109/31.16577) [DOI] [Google Scholar]
- 6.Lee J. S. 1980. Digital image enhancement and noise filtering by use of local statistics. IEEE Trans. Pattern Anal. Mach. Intell. 2, 165–168. 10.1109/TPAMI.1980.4766994 ( 10.1109/TPAMI.1980.4766994) [DOI] [PubMed] [Google Scholar]
- 7.Frost V., Stiles J. A., Shanmugan K. S., Holtzman J. C. 1982. A model for radar images and its application to adaptive digital filtering of multiplicative noise. IEEE Trans. Pattern Anal. Mach. Intell. 4, 157–166. 10.1109/TPAMI.1982.4767223 ( 10.1109/TPAMI.1982.4767223) [DOI] [PubMed] [Google Scholar]
- 8.Karaman M., Kutay M. A., Bozdagi G. 1995. An adaptive speckle suppression filter for medical ultrasonic imaging. IEEE Trans. Med. Imag. 14, 283–292. 10.1109/42.387710 ( 10.1109/42.387710) [DOI] [PubMed] [Google Scholar]
- 9.Yu Y., Acton S. T. 2002. Speckle reducing anisotropic diffusion. IEEE Trans. Image Process. 11, 1260–1270. 10.1109/TIP.2002.804276 ( 10.1109/TIP.2002.804276) [DOI] [PubMed] [Google Scholar]
- 10.Krissian K., Westin C. F., Kikinis R., Vosburgh K. G. 2007. Oriented speckle reducing anisotropic diffusion. IEEE Trans. Image Process. 16, 1412–1424. 10.1109/TIP.2007.891803 ( 10.1109/TIP.2007.891803) [DOI] [PubMed] [Google Scholar]
- 11.Zong X., Laine A. F., Geiser E. A. 1998. Speckle reduction and contrast enhancement of echocardiograms via multiscale nonlinear processing. IEEE Trans. Med. Imag. 17, 532–540. 10.1109/42.730398 ( 10.1109/42.730398) [DOI] [PubMed] [Google Scholar]
- 12.Achim A., Berzerianos A., Tsakalides P. 2001. Novel Bayesian multi-scale method for speckle removal in medical ultrasound images. IEEE Trans. Med. Imag. 20, 772–783. 10.1109/42.938245 ( 10.1109/42.938245) [DOI] [PubMed] [Google Scholar]
- 13.Coupe P., Hellier P., Kervrann C., Barillot C. 2009. Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans. Image Process. 18, 2221–2229. 10.1109/TIP.2009.2024064 ( 10.1109/TIP.2009.2024064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axel L., Montillo A., Kim D. 2005. Tagged magnetic resonance imaging of the heart: a survey. Med. Image Anal. 9, 376–393. 10.1016/j.media.2005.01.003 ( 10.1016/j.media.2005.01.003) [DOI] [PubMed] [Google Scholar]
- 15.Crosby J., Amundsen B. H., Hergum T., Remme E. W., Langland S., Torp H. 2009. 3-D speckle tracking for assessment of regional left ventricular function. Ultrasound Med. Biol. 35, 458–471. 10.1016/j.ultrasmedbio.2008.09.011 ( 10.1016/j.ultrasmedbio.2008.09.011) [DOI] [PubMed] [Google Scholar]
- 16.Noble J. A. 2010. Ultrasound image segmentation and tissue characterization. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 224, 307–316. 10.1243/09544119JEIM604 ( 10.1243/09544119JEIM604) [DOI] [PubMed] [Google Scholar]
- 17.Noble J. A., Boukerroui D. 2006. Ultrasound image segmentation: a survey. IEEE Trans. Med. Imag. 25, 987–1010. 10.1109/TMI.2006.877092 ( 10.1109/TMI.2006.877092) [DOI] [PubMed] [Google Scholar]
- 18.Kyriacou E. C., Pattichis C., Pattichis M., Loizou C., Christodoulou C., Kakkos S. K., Nicolaides A. 2010. A review of noninvasive ultrasound image processing methods in the analysis of carotid plaque morphology for the assessment of stroke risk. IEEE Trans. Inform. Technol. Biomed. 14, 1027–1038. 10.1109/TITB.2010.2047649 ( 10.1109/TITB.2010.2047649) [DOI] [PubMed] [Google Scholar]
- 19.Gong L., Pathak S. D., Haynor D. R., Cho P. S., Kim Y. 2004. Parametric shape modeling using deformable superellipses for prostate segmentation. IEEE Trans. Med. Imag. 23, 340–349. 10.1109/TMI.2004.824237 ( 10.1109/TMI.2004.824237) [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y., Papademetris X., Sinusas A. J., Duncan J. S. 2009. A dynamical shape prior for LV segmentation from RT3D echocardiography. Medical Image Computing and Computer-assisted Intervention – MICCAI 2009, Lecture Notes in Computer Science, vol. 5761, pp. 206–213. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikic I., Krucinski S., Thomas J. D. 1998. Segmentation and tracking in echocardiographic sequences: active contours guided by optimal flow estimates. IEEE Trans. Med. Imag. 17, 274–284. 10.1109/42.700739 ( 10.1109/42.700739) [DOI] [PubMed] [Google Scholar]
- 22.Jacob G., Noble J. A., Behrenburch C., Kelion A. D., Banning A. P. 2008. A shape-space based approach to tracking myocardial thickening and quantifying regional left ventricular function. IEEE Trans. Med. Imag. 21, 226–238. [DOI] [PubMed] [Google Scholar]
- 23.Bosch J. G., Mitchell S. C., Lelieveldt B. P. F., Nijland F., Kamp O., Sonka M., Reiber J. H. C. 2002. Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE Trans. Med. Imag. 21, 1374–1383. 10.1109/TMI.2002.806427 ( 10.1109/TMI.2002.806427) [DOI] [PubMed] [Google Scholar]
- 24.Leung K. Y. E., Danilouchkine M. G., van Stralen M., de Jong N., van der Steen A. F. W., Bosch J. G. 2011. Left ventricular border tracking using cardiac motion models and optical flow. Ultrasound Med. Biol. 37, 605–616. [DOI] [PubMed] [Google Scholar]
- 25.Lin N., Yu W., Duncan J. S. 2002. Combinative multi-scale level set framework for echocardiographic image segmentation. In Medical Image Computing and Computer-assisted Interventions – MICCAI 2002, Lecture Notes in Computer Science, vol. 2488, pp. 682–689. Springer. [DOI] [PubMed] [Google Scholar]
- 26.Paragios N. 2003. A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans. Med. Imag. 22, 773–776. 10.1109/TMI.2003.814785 ( 10.1109/TMI.2003.814785) [DOI] [PubMed] [Google Scholar]
- 27.Leung K. Y. E., Stralen M., van Burken G., van Jong N., de Bosch J. G. 2010. Automatic active appearance model segmentation of 3D echocardiograms. Proc. 2010 IEEE Int. Symp. Biomedical Imaging: from Nano to Macro, Rotterdam, The Netherlands, April 14–17, 2010, pp. 320–323. IEEE. [Google Scholar]
- 28.Carneiro G., Georgescu B., Good S., Comanicui D. 2008. Detection of fetal abnormalities from ultrasound images using a constrained probabilistic boosting tree. IEEE Trans. Med. Imag. 27, 1342–1355. 10.1109/TMI.2008.928917 ( 10.1109/TMI.2008.928917) [DOI] [PubMed] [Google Scholar]
- 29.Pujol O., Rosales M., Radeva P., Nofrerias-Fernandez E. 2003. Intravascular ultrasound images vessel characterization using Adaboost. In Proc. Functional imaging and modelling of the heart, Lyon, France, pp. 242–251. Springer. [Google Scholar]
- 30.Yaqub M., Mahon P., Javaid M. K., Cooper C., Noble J. A. 2010. Weighted voting in 3D random forest segmentation. In Proc. Medical Image Understanding and Analysis, Warwick, UK, July 2010 (eds Bhalerao A. H., Rajpoot N. M.), pp. 261–266. [Google Scholar]
- 31.Yang L., Georgescu B., Zheng Y., Meer P., Comaniciu D. 2008. 3D ultrasound tracking of the left ventricle using one-step forward prediction and data fusion of collaborative trackers. IEEE Conf. on Computer Vision and Pattern Recognition, pp. 1–8, 10.1109/CVPR.2008.4587518. [DOI] [Google Scholar]
- 32.Wang Y., Georgescu B., Comaniciu D., Houle H. 2010. Learning-based 3D myocardial motion flow estimation using high frame rate volumetric ultrasound data, Int. Symp. on Biomedical Imaging: from Nano to Macro, Rotterdam, The Netherlands, April 14–17, 2010, pp. 1097–1100. IEEE. [Google Scholar]
- 33.Grady L. 2006. Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1768–1783. 10.1109/TPAMI.2006.233 ( 10.1109/TPAMI.2006.233) [DOI] [PubMed] [Google Scholar]
- 34.Archip N., Rohling R., Cooperberg P., Tahmasebpour H. 2005. Ultrasound image segmentation using spectral clustering. Ultrasound Med. Biol. 31, 1485–1497. 10.1016/j.ultrasmedbio.2005.07.005 ( 10.1016/j.ultrasmedbio.2005.07.005) [DOI] [PubMed] [Google Scholar]
- 35.Ledesma-Carbayo M. J., Kybic J., Desco M., Santos A., Suhling M., Hunziker P. R., Unser M. 2005. Spatio-temporal nonrigid registration for ultrasound cardiac motion estimation. IEEE Trans. Med. Imag. 24, 1113–1126. 10.1109/TMI.2005.852050 ( 10.1109/TMI.2005.852050) [DOI] [PubMed] [Google Scholar]
- 36.Shekar R., Zagrodsky V. 2002. Mutual information-based rigid and nonrigid registration of ultrasound volumes. IEEE Trans. Med. Imag. 21, 9–22. 10.1109/42.981230 ( 10.1109/42.981230) [DOI] [PubMed] [Google Scholar]
- 37.Xiao G., Brady J. M., Noble J. A., Burcher M., English R. 2002. Nonrigid registration of 3-D free-hand ultrasound images of the breast. IEEE Trans. Med. Imag. 21, 405–412. 10.1109/TMI.2002.1000264 ( 10.1109/TMI.2002.1000264) [DOI] [PubMed] [Google Scholar]
- 38.Gee A. H., Treece G. M., Prager R. W., Cash C. J. C., Berman L. 2003. Rapid registration for wide field-of-view freehand three-dimensional ultrasound. IEEE Trans. Med. Imag. 22, 1344–1357. 10.1109/TMI.2003.819279 ( 10.1109/TMI.2003.819279) [DOI] [PubMed] [Google Scholar]
- 39.Wachinger C., Wein W., Navab N. 2008. Registration strategies and similarity measures for three-dimensional ultrasound mosaicing. Acad. Radiol. 15, 1404–1415. 10.1016/j.acra.2008.07.004 ( 10.1016/j.acra.2008.07.004) [DOI] [PubMed] [Google Scholar]
- 40.Cohen B., Dinstein I. 2002. New maximum likelihood motion estimation schemes for noisy ultrasound images. Pattern Recogn. 35, 455–463. 10.1016/S0031-3203(01)00053-X ( 10.1016/S0031-3203(01)00053-X) [DOI] [Google Scholar]
- 41.Myronenko A., Song X., Sahn D. J. 2009. Maximum likelihood motion estimation in 3D echocardiography through non-rigid registration in spherical coordinates. In Functional Imaging and Modeling of the Heart 2009, Lecture Notes in Computer Science, vol. 5528, pp. 427–436. [Google Scholar]
- 42.Huang X., Hill N. A., Ren J., Guiraudon G., Boughner D. R., Peters T. M. 2005. Dynamic 3D ultrasound and MR image registration of the beating heart. In Medical Image Computing and Computer-assisted Interventions – MICCAI 2005, Lecture Notes in Computer Science, vol. 3750, pp. 171–178. Springer. [DOI] [PubMed] [Google Scholar]
- 43.Roche A., Pennec X., Malandain G., Ayache N. 2001. Rigid registration of 3D ultrasound with US images: a new approach combining intensity and gradient information. IEEE Trans. Med. Imag. 20, 1038–1049. 10.1109/42.959301 ( 10.1109/42.959301) [DOI] [PubMed] [Google Scholar]
- 44.Penney G., Blackall J., Hamady M., Sabharwal T., Adam A., Hawkes D. 2004. Registration of freehand 3D ultrasound and magnetic resonance liver images. Med. Image Anal. 8, 81–91. 10.1016/j.media.2003.07.003 ( 10.1016/j.media.2003.07.003) [DOI] [PubMed] [Google Scholar]
- 45.Mellor M., Brady M. 2005. Phase mutual information as a similarity measure for registration. Med. Image Anal. 9, 330–343. 10.1016/j.media.2005.01.002 ( 10.1016/j.media.2005.01.002) [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., Noble J. A., Brady J. M. 2007. Adaptive non-rigid registration of real time 3D ultrasound to cardiovascular MR images. Proc. 20th Int. Conf. Information Processing in Medical Imaging, Kerkade, The Netherlands, Lecture Notes in Computer Science, vol. 4585, pp. 50–61. Springer. [DOI] [PubMed] [Google Scholar]
- 47.Ye X., Atkinson D., Noble J. A. 2002. 3D freehand echocardiography for automatic LV reconstruction and analysis based on multiple acoustic windows. IEEE Trans. Med. Imag. 21, 1051–1058. [DOI] [PubMed] [Google Scholar]
- 48.Grau V., Noble J. A. 2005. Adaptive multiscale ultrasound compounding using phase information. In Medical Image Computing and Computer-assisted Interventions – MICCAI 2005, Palm Springs, CA, Lecture Notes in Computer Science, vol. 3749, pp. 589–596. Springer. [DOI] [PubMed] [Google Scholar]
- 49.Grau V., Becher H., Noble J. A. 2007. Registration of multi-view real time three dimensional echocardiographic sequences. IEEE Trans. Med. Imag. 26, 1154–1165. 10.1109/TMI.2007.903568 ( 10.1109/TMI.2007.903568) [DOI] [PubMed] [Google Scholar]
- 50.Rajpoot K., Grau V., Becher H., Noble J. A. 2009. Multiview RT3D echocardiography image fusion. Proc. 5th Int. Conf. Modelling and Imaging of the Heart, Nice, France, Lecture Notes in Computer Science, vol. 5528, pp. 134–143. Springer. [Google Scholar]
- 51.Rajpoot K., Szmigielski C., Grau V., Becher H., Noble J. A. 2011. The evaluation of single-view and multi-view fusion 3D echocardiography using image-driven segmentation and tracking. Med. Image Anal. (in press) ( 10.1016/j.media.2011.02.007) [DOI] [PubMed] [Google Scholar]
- 52.Szmigielski C., Rajpoot K., Grau V., Myerson S. G., Holloway C., Noble J. A., Kerber R., Becher H. 2010. Real-time 3D fusion echocardiography. JACC Cardiovasc Imaging. 3, 682–690. 10.1016/j.jcmg.2010.03.010 ( 10.1016/j.jcmg.2010.03.010) [DOI] [PubMed] [Google Scholar]
- 53.Entrekin R. R., Porter B. A., Sillesen H. H., Wong A. D., Cooperberg P. L., Fix C. H. 2001. Real-time spatial compound imaging: application to breast, vascular and musculoskeletal ultrasound. Seminars in ultrasound, CT and MRI 22, 50–64. 10.1016/S0887-2171(01)90018-6 ( 10.1016/S0887-2171(01)90018-6) [DOI] [PubMed] [Google Scholar]
- 54.Poon T., Rohling R. 2005. Three-dimensional extended field-of-view ultrasound. Ultrasound Med. Biol. 32, 356–369. [DOI] [PubMed] [Google Scholar]
- 55.Wachinger C., Wein W., Navab N. 2007. Three-dimensional ultrasound mosaicing. In Medical Image Computing and Computer-assisted Interventions – MICCAI 2007, Brisbane, Australia, Lecture Notes in Computer Science, vol. 4792, pp. 327–335. Springer. [DOI] [PubMed] [Google Scholar]
- 56.Kutter O., Wein W., Navab N. 2009. Multi-modal registration based ultrasound mosaicing. In Medical Image Computing and Computer-assisted Intervention – MICCAI 2009, London, UK, Lecture Notes in Computer Science, vol. 5761, pp. 763–770. Springer. [DOI] [PubMed] [Google Scholar]
- 57.Yao C., Simpson J. M., Schaeffter T., Penney G. P. 2010. Spatial compounding of large numbers of multi-view 3D echocardiography images using feature consistency. IEEE Int. Symp. on Biomedical Imaging: from Nano to Macro, Rotterdam, The Netherlands, April 14–17, 2010, pp. 968–971. [Google Scholar]
- 58.Wachinger C., Yigitsoy N M., Navab N. 2010. Manifold learning for image-based breathing gating with application to 4D ultrasound. In Medical Image Computing and Computer-assisted Intervention – MICCAI 2010, Beijing, China, Lecture Notes in Computer Science, vol. 6362, pp. 26–33. Springer. [DOI] [PubMed] [Google Scholar]
- 59.De Nigris D., Mercier L., Del Maestro R., Collins D. L., Arbel T. 2010. Hierarchical multimodal image registration based on adaptive local mutual information. In Medical Image Computing and Computer-assisted Intervention – MICCAI 2010, Beijing, China, Lecture Notes in Computer Science, vol. 6362, pp. 643–651. Springer. [DOI] [PubMed] [Google Scholar]
- 60.Lindner D., Trantakis C., Renner C., Arnold S., Schmitgen A., Schneider J., Meixensberger J. 2006. Application of intraoperative 3D ultrasound during navigated tumor resection. Invasive Neurosurg. 49, 197–202. 10.1055/s-2006-947997 ( 10.1055/s-2006-947997) [DOI] [PubMed] [Google Scholar]
- 61.Letteboer M. M., Willems P. W., Viergever M. A., Niessen W. J. 2005. Brain shift estimation in image-guided neurosurgery using 3-D ultrasound. IEEE Trans. Biomed. Eng. 52, 268–276. 10.1109/TBME.2004.840186 ( 10.1109/TBME.2004.840186) [DOI] [PubMed] [Google Scholar]
- 62.Rasmussen I. A., Lindseth F., Rygh O. M., Berntsen E. M., Selbekk Xu T. J., Hernes T. A. N., Harg E., Håberg A., Unsgaard G. 2007. Functional neuronavigation combined with intra-operative 3D ultrasound: initial experiences during surgical resections close to eloquent brain areas and future directions in automatic brain shift compensation of preoperative data. Acta Neurochirurgia (Wien) 149, 365–378. 10.1007/s00701-006-1110-0 ( 10.1007/s00701-006-1110-0) [DOI] [PubMed] [Google Scholar]
- 63.Vitrani M., Mitterhofer H., Morel G., Bonnet N. 2007. Robust ultrasound-based visual servoing for beating heart intracardiac surgery. In IEEE Int. Conf. on Robotics and Automation—ICRA, Rome, Italy, pp. 3021–3027. IEEE. [Google Scholar]
- 64.Yuen S. G., Vasilyev N. V., del Nido P. J., Howe R. D. 2011. Robotic Tissue Tracking for Beating Heart Mitral Valve Surgery. Med. Image Anal. (in press) ( 10.1016/j.media.2010.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linte C. A., Moore J., Wedlake C., Peters T. M. 2010. Evaluation of model-enhanced ultrasound-assisted interventional guidance in a cardiac phantom. IEEE Trans. Biomed. Eng. 57, 2209–2218. 10.1109/TBME.2010.2050886 ( 10.1109/TBME.2010.2050886) [DOI] [PubMed] [Google Scholar]
- 66.Khuri-Yakub B. T., et al. 2010. Miniaturized ultrasound imaging probes enabled by CMUT arrays with integrated frontend electronic circuits. Proc. Conf. IEEE Eng. Med. Biol. Soc. pp. 5987–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colombo A., Hall P., Nakamura S., Almagor Y., Maiello L., Martini G., Gaglione A., Goldberg S. L., Tobis J. M. 1995. Intracoronary stenting without anticoagulation accomplished with intravascular ultrasound guidance. Circulation 91, 1676–1688. [DOI] [PubMed] [Google Scholar]
- 68.Nair A., Kuban B. D., Tuzcu E. M., Schoenhagen P., Nissen S. E., Vince D. G. 2002. Coronary plaque classification with intravascular ultrasound radiofrequency data analysis. Circulation 106, 2200–2206. 10.1161/01.CIR.0000035654.18341.5E ( 10.1161/01.CIR.0000035654.18341.5E) [DOI] [PubMed] [Google Scholar]
- 69.Calfon M. A., Vinegoni C., Ntziachristos V., Jaffer F. A. 2010. Intravascular near-infrared fluorescence molecular imaging of atherosclerosis: toward coronary arterial visualization of biologically high-risk plaques. J. Biomed. Opt. 15, 011107. 10.1117/1.3280282 ( 10.1117/1.3280282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ionasec R., Voigt I., Georgescu B., Wang Y., Houle H., Vega-Higuera F., Navab N., Comaniciu D. 2010. Patient-specific modeling and quantification of the aortic and mitral valves from 4-D cardiac CT and TEE. IEEE Trans. Med. Imag. 29, 1636–1651. [DOI] [PubMed] [Google Scholar]
- 71.Voigt I., Mansi T., Mihalef V., et al. Patient-specific model of left heart anatomy, dynamics and hemodynamics from 4D TEE: a first validation study. Proc. 6th Int. Conf. Functional Modelling and Imaging of the Heart, New York City, NY, Lecture Notes in Computer Science, vol. 6666, pp. 341–349. Springer. [Google Scholar]
- 72.Wein W., Camus E., John M., Diallo M., Duong C., Al-Ahmad A., Fahrig R., Khamene A., Xu C. Towards guidance of electrophysiological procedures with real-time 3D intracardiac echocardiography fusion to C-arm CT. In Medical Image Computing and Computer-assisted Intervention – MICCAI 2009, Lecture Notes in Computer Science, vol. 5761/2009, pp. 9–16. Springer. [DOI] [PubMed] [Google Scholar]
- 73.Wein W., Röper B., Navab N. 2007. Integrating diagnostic B-mode ultrasonography into CT-based radiation treatment planning. IEEE Trans. Med. Imag. 26, 866–879. 10.1109/TMI.2007.895483 ( 10.1109/TMI.2007.895483) [DOI] [PubMed] [Google Scholar]
- 74.Wein W., Brunke S., Khamene A., Callstrom M. R., Navab N. 2008. Automatic CT-ultrasound registration for diagnostic imaging and image-guided intervention. Med. Image Anal. 12, 577–585. 10.1016/j.media.2008.06.006 ( 10.1016/j.media.2008.06.006) [DOI] [PubMed] [Google Scholar]
- 75.Wein W., Kutter O., Aichert A., Zikic D., Kamen A., Navab N. Automatic non-linear mapping of pre-procedure CT volumes to 3D ultrasound. In IEEE Int. Symp. on Biomedical Imaging (ISBI), April 14–17, 2010, Rotterdam, The Netherlands. [Google Scholar]
- 76.Barrett D. C., Chan C. S. K., Edwards P. J., Penney G. P., Slomczykowski M., Carter T. J., Hawkes D. J. 2008. Instantiation and registration of statistical shape models of the femur and pelvis using 3D ultrasound imaging. Med. Image Anal. 12, 358–374. 10.1016/j.media.2007.12.006 ( 10.1016/j.media.2007.12.006) [DOI] [PubMed] [Google Scholar]
- 77.Abolmaesumi P., Khallaghi S., Borshneck D., Fichtinger G., Pichora D., Mousavi P. 2010. Registration of a statistical shape model of the lumbar spine to 3D ultrasound images. Medical Image Computing and Computer-assisted Intervention – MICCAI 2010, Beijing, China, Lecture Notes in Computer Science, vol. 6362, pp. 68–75. Springer. [DOI] [PubMed] [Google Scholar]
- 78.Hu Y., Ahmed H. U., Allen C., Pendsé D., Sahu M., Emberton M., Hawkes D. J., Barratt D. C. 2009. MR to Ultrasound Image Registration for Guiding Prostate Biopsy and Interventions. Medical Image Computing and Computer-assisted Intervention – MICCAI 2009, London, UK, Lecture Notes in Computer Science, vol. 5761, pp. 787–794. Springer. [DOI] [PubMed] [Google Scholar]
- 79.Wendler T., Feuerstein M., Traub J., Lasser T., Vogel J., Daghighian F., Ziegler S. I., Navab N. 2007. Real-time fusion of ultrasound and gamma probe for navigated localization of liver metastases. In Proc. of Medical Image Computing and Computer-assisted Intervention – MICCAI 2007, Brisbane, Australia, Lecture Notes in Computer Science, vol. 47922, pp. 909–917. Springer. [DOI] [PubMed] [Google Scholar]
- 80.Wendler T., Lasser T., Traub J., Ziegler S. I., Navab N. 2009. Freehand SPECT/ultrasound fusion for hybrid image-guided resection. Proc. of Annu. Congr. of the European Association of Nuclear Medicine (EANM 2009), Barcelona. [Google Scholar]
- 81.Pani L., et al. In press Dual modality ultrasound-SPECT detector for molecular imaging; 12th topical seminar on innovative particle and radiation detectors Proc. 12th Topical Seminar on Innovative Particle and Radiation Detectors (IPRD10), Siena, Italy, 7–10 June 2010. Abstract available at http://www.bo.infn.it/sminiato/sm10/abstract/r-pani-ecorad-abstract.pdf) [Google Scholar]
- 82.Bermejo J., et al. 2008. Objective quantification of global and regional left ventricular systolic function by endocardial tracking of contrast echocardiographic sequences. Int. J. Cardiol. 124, 47–56. 10.1016/j.ijcard.2006.12.091 ( 10.1016/j.ijcard.2006.12.091) [DOI] [PubMed] [Google Scholar]
- 83.Mor-Avi V., et al. 2008. Real-time 3-dimensional echocardiographic quantification of left ventricular volumes: multicenter study for validation with magnetic resonance imaging and investigation of sources of error. JACC Cardiovasc. Imag. 1, 413–423. 10.1016/j.jcmg.2008.02.009 ( 10.1016/j.jcmg.2008.02.009) [DOI] [PubMed] [Google Scholar]
- 84.Hansegard J., Urheim S., Lunde K., Malm S., Rabben S. I. 2009. Semi-automated quantification of left ventricular volumes and ejection fraction by real-time three-dimensional echocardiography. Cardiovasc. Ultrasound 7, 18. 10.1186/1476-7120-7-18 ( 10.1186/1476-7120-7-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leung K. Y. E., Bosch J. G. 2010. Automated border detection in three-dimensional echocardiography: principles and promises. Eur. J. Echocardiogr. 11, 97–108. 10.1093/ejechocard/jeq005 ( 10.1093/ejechocard/jeq005) [DOI] [PubMed] [Google Scholar]
- 86.Gooding M. J., Rajpoot K., Mitchell S., Chamberlain P., Kennedy S. H., Noble J. A. 2010. Investigation into the fusion of multiple 4D fetal echocardiography images to improve image quality. Ultrasound Med. Biol. 36, 957–966. 10.1016/j.ultrasmedbio.2010.03.017 ( 10.1016/j.ultrasmedbio.2010.03.017) [DOI] [PubMed] [Google Scholar]
- 87.Rajpoot K., Augustine D., Basagiannis C., Noble J. A., Becher H., Leeson P. 2011. 3D fusion transoeosphageal echocardiography improves LV assessment. Proc. 6th Int. Conf. Functional Modelling and Imaging of the Heart, New York City, NY, Lecture Notes in Computer Science, vol. 6666, pp. 61–62. Springer. [Google Scholar]
- 88.Hascoet S., Brierre G., Coudron G., Cardin C., Bongard V., Acar P. 2010. Assessment of left ventricular volumes and function by real time three dimensional echocardiography in a pediatric population: a TomTec versus QLAB comparison. Echocardiography 27, 1263–1273. 10.1111/j.1540-8175.2010.01235.x ( 10.1111/j.1540-8175.2010.01235.x) [DOI] [PubMed] [Google Scholar]