Abstract
We propose a Bayesian multivariate model in which a single linear combination of the covariates predict multiple outcomes simultaneously. The single linear combination is a data-derived score along the lines of the Apache or Charlson index scores for critically ill patients, the Karnofsky or Eastern Cooperative Oncology Group score for cancer patients or Euro-score for cardiac patients that may be used to predict multiple outcomes. Outcomes may be discrete or continuous and we use a composition of generalized linear models for the marginal distribution for each outcome. We explain how to set the prior distribution and we use Markov chain Monte Carlo methods to calculate the posterior distribution. We propose two types of expanded models to diagnose whether each outcome indeed has predictor effects common with the other outcomes, and whether a particular predictor is commonly predictive for all outcomes. We determine a final model based on the diagnostic models. The method is applied to a study yielding multiple psychometric outcomes of mixed type measured in young people living with human immunodeficiency virus.
Keywords: Bayesian Wald test, human immunodeficiency virus, index construction, multivariate regression, single index model
1. Introduction
Multiple outcomes are common in medical, psychological and sociological studies. Joint analysis of continuous multiple outcomes is common in recent decades [1–18]. Difficulty arises when the outcomes comprise a mix of continuous, binary and non-negative integer variables. In recent years, a number of models have been developed to handle mixed outcomes, e.g. [19–24].
Little & Schluchter [19] and Gueorguieva & Agresti [24] have proposed maximum-likelihood procedures for analysing mixed continuous and binary data with missing values. Little & Schluchter model q categorical variables defining a q-way contingency table with C = ∏Ii cells where the ith categorical variable has Ii levels. They assumed a multinomial distribution over the C cells of the contingency table. Given the contingency table cells, the continuous outcomes follow a multivariate normal distribution. Sparse data can make inference problematic. Gueorguieva & Agresti proposed an underlying multivariate probit model for the binary variables that correlate directly with the continuous normal variables.
Sammel et al. [20] proposed a latent variable model for mixed discrete and continuous outcomes with the multiple outcomes correlated through subject-specific latent variables. Conditional on the latent variables, outcomes were assumed independent with each outcome arising from a one parameter exponential family, with its mean a function of the latent variable and other covariates. An expectation-maximization (EM) algorithm was used to fit the model.
Arminger & Kusters [21] assumed each outcome had a latent variable and jointly, the latent variables follow a multivariate normal distribution with expectation potentially dependent on covariates. Dunson [23] proposed a Bayesian latent variable model for clustered mixed outcomes. It generalized Arminger & Kusters' work to accommodate non-normal latent variables for multi-level data. It allowed nonlinear relationships between the covariates and latent variables and used multiple latent variables for each type of outcome and covariate-dependent modification of the relationship between the latent and covariates.
In this paper, we propose a double link common predictor effect (COPE) model in which a single linear combination of the covariates is used to predict all outcomes simultaneously. Generalized linear models are used to model the marginal distribution for each outcome. Outcome-specific double link functions connect each outcome's mean to the single linear predictor, where a double link refers to the composition of one link function in a second link function. Correlations among outcomes are modelled by latent variables. Markov chain Monte Carlo (MCMC) algorithms are used to estimate the posterior distribution of the parameters and the latent variables.
Compared with the model proposed by Sammel et al. [20] in which each outcome has an outcome-specific set of fixed effect coefficients, our model furnishes a parsimonious common set of fixed effects for all outcomes, thus we have greater efficiency in our parameter estimates. Compared with Dunson's model, in which all outcomes have a common set of fixed effects, our model returns greater flexibility for modelling COPE through the double link function between the linear predictor and outcomes. In general, the usual multiple outcome model is the saturated model, which has a separate set of coefficients for each outcome. Our model, when appropriate, is much more efficient as there are many fewer coefficients.
Frequentist methods are based on asymptotic maximum-likelihood theory and the standard errors and confidence interval are based on asymptotic normality assumptions. In Bayesian inference, posterior distribution of parameters and latent variables can be estimated by MCMC methods, and means and variances for parameters can be appropriately estimated without requiring a large sample size. For our proposed model, a key inference involves the product of parameters; using a maximum-likelihood approach requires the delta method to estimate point estimates and variances of the product of parameters. With the Bayesian approach, inferences can be drawn directly from posterior samples of the product of the parameters.
Index scores for evaluating disease severity are common in medicine. Examples include the Apache [25,26] and Charlson index scores [27,28] for critically ill patients, the Karnofsky and Eastern Cooperative Oncology Group scores for cancer patients [29,30] and EuroScore [31,32] for cardiac patients. These scores are often used for predicting multiple outcomes beyond the outcome used for the initial derivation. The single linear combination estimated by our model is a principled data-derived score along the lines of these scores that may be used to predict multiple outcomes. Our Bayesian methodology using MCMC methodology makes it easy to produce both (i) index calculations for an individual complete with uncertainty intervals and (ii) predictions for single or multiple outcomes. Construction of online calculators is straightforward using MCMC methodology.
This paper is organized as following: §2 proposes the double link COPE model for mixed outcomes. Section 3 explains the prior specification. In §4, we relax our insistence on only one linear combination of the covariates for predicting all outcomes, and we describe two types of extended models both as model diagnostics and as potentially valuable models in their own right. Section 5 applies the method to multiple, mixed-type psychometric outcomes measured on young people living with human immunodeficiency virus (YPLH) and the paper concludes with a discussion in §6.
2. Common predictor effect model specification
We observe k1 continuous outcomes, k2 binary outcomes, k3 count outcomes and L covariates on subject i, i = 1, …, n. Let yik denote the kth outcome for subject i, k = 1, …, K, K = k1 + k2 + k3, and define the ith observation yi = (yi1, …, yiK)′; let xil denote the lth predictor, l = 1, …, L, for subject i with xi = (xi1, …, xiL)′, where xi does not include an intercept. Covariates are the same for all outcomes and a single linear predictor, meaning a single linear combination of covariates, xi′α, will be used to predict all outcomes.
An outcome-specific double link function is needed for each outcome in our proposed model: the usual link function gk(·) for a generalized linear model that connects the expected value of subject i outcome k, E(Yik) = θik, to the natural parameter ηik; and a location-scale link function ψk(·) that connects its natural parameter with the single linear predictor. In our model, we take ψk(·) to be linear, 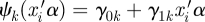 , which allows the COPE for each outcome to be modified by a scale parameter. The usual link functions for generalized linear models are the identity link for continuous outcomes ηik = gk(θik) = θik; the logit link for binary outcomes ηik = gk(θik) = logit(θik) = log(θik(1−θik)−1), where logit(x) = log(x(1 − x)−1) and its inverse, called expit, expit(x) = exp(x)(1 + exp(x))−1; and the log link for count outcomes ηik = gk(θik) = log(θik). We model the correlations across the multivariate outcomes on the linear predictor scale through a random effect variable βik,
, which allows the COPE for each outcome to be modified by a scale parameter. The usual link functions for generalized linear models are the identity link for continuous outcomes ηik = gk(θik) = θik; the logit link for binary outcomes ηik = gk(θik) = logit(θik) = log(θik(1−θik)−1), where logit(x) = log(x(1 − x)−1) and its inverse, called expit, expit(x) = exp(x)(1 + exp(x))−1; and the log link for count outcomes ηik = gk(θik) = log(θik). We model the correlations across the multivariate outcomes on the linear predictor scale through a random effect variable βik, 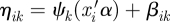 .
.
The double link COPE model for mixed outcomes is
| 2.1 |
| 2.2a |
| 2.2b |
and
| 2.2c |
where α is an L × 1 vector of fixed-effect regression coefficients for all outcomes, γ0k and γ1k are the intercept and scale parameter in the link for outcome k, respectively, γ0 = (γ01, … , γ0K)′, γ1 = (γ11, … , γ1K)′, βi = (βi1, …, βiK)′ follows a multivariate normal distribution with mean 0 and covariance Σ(ξ), βi ∼ NK(0,Σ(ξ)), where Σ(ξ) models the correlations among the multiple outcomes on the linear predictor scale, and ξ is a vector of unknown parameters for the components of the correlation matrix.
As a whole,  can be estimated in model (2.1), however there is an identifiability problem if we want to estimate α and γ1 individually. For example, α and γ1 can be replaced by
can be estimated in model (2.1), however there is an identifiability problem if we want to estimate α and γ1 individually. For example, α and γ1 can be replaced by  and
and 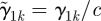 for an arbitrary constant c ≠ 0 and we still have the same prediction. We need to constrain α and/or γ1 to solve the problem. The solution we propose is to divide the regression coefficients by their Euclidean norm ‖α‖ = (α′α)1/2 and fix the sign of the regression coefficient for one pre-specified predictor whose regression coefficient is significantly different from 0, without lack of generality, say αL. We modify ηik in model (2.1) as
for an arbitrary constant c ≠ 0 and we still have the same prediction. We need to constrain α and/or γ1 to solve the problem. The solution we propose is to divide the regression coefficients by their Euclidean norm ‖α‖ = (α′α)1/2 and fix the sign of the regression coefficient for one pre-specified predictor whose regression coefficient is significantly different from 0, without lack of generality, say αL. We modify ηik in model (2.1) as
| 2.3 |
3. Prior specification
We specify prior distributions for the parameters in our proposed model to complete the Bayesian modelling. Many researchers have discussed different ways to specify priors [33–36]. Prior information may come from previous studies, published results from similar studies, scientific reasoning about the measured variables, other related data or expert opinions. For any model, it is helpful to have an automatic prior that requires relatively little in the way of substantive input to allow data and model exploration to begin. Our own preference is to add scientific/statistical reasoning on top of that to allow for sensible informative proper priors particularly for parameters not of primary scientific interest. In contrast, specifying a fully subjective prior requires a large investment in time and training of subject matter specialists and is not an activity that can be lightly undertaken.
For models (2.2a)–(2.3), we specify the prior distributions for common regression coefficients α, intercepts γ0, scale parameters γ1 and the covariance Σ of the latent variables βi. We first standardize each continuous predictor by subtracting its mean and dividing by its standard deviation, the resulting continuous predictor has mean 0 and standard deviation 1. We assign a normal prior to α, α ∼ N(μα, Σα), where μα is a vector of location parameters and Σα is a covariance matrix. We set μα = 0L, an L-vector of 0s, and α's to be independent, 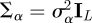 ,
, 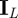 is the L × L identity matrix, and we give
is the L × L identity matrix, and we give  an arbitrary value c. This is a flat prior for α /‖α‖ on the unit ball in ℜL. The sampling distribution of Y is not affected by the length ‖α‖ of α and the posterior of ‖α‖ is equal to its prior.
an arbitrary value c. This is a flat prior for α /‖α‖ on the unit ball in ℜL. The sampling distribution of Y is not affected by the length ‖α‖ of α and the posterior of ‖α‖ is equal to its prior.
We discuss priors for the intercept γ0k and scale parameters γ1k separately for each type of outcome: Bernoulli, count and continuous. For Bernoulli outcomes, a wide range of probabilities of a response equal to 1 is from 0.001 to 0.999 (0.001 < P(yik = 1) < 0.999), which gives a range after logit transformation of −6.9 to 6.9. We assign a normal prior to γ0k with mean 0 and variance ((6.9 − (−6.9))/4)2 =11.9 for binary outcomes, which is the square of the range divided by 4, inspired by the idea that in the prior, the mean ± 2 s.d. should cover 95 per cent of the prior range. For count outcomes, we take the mean of the response to be somewhere between 0.01 and 100. After a log transformation, we have a range of the linear predictor of −4.6 to 4.6. We assign a normal prior to γ0k with mean 0 and variance ((4.6 − (−4.6))/4)2 = 5.3 for count outcomes. For continuous outcomes, we assign a normal prior to γ0k with mean 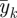 and variance
and variance 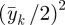 . We consider these priors to be vague and not particularly informative although proper. The prior for continuous outcome uses the data, but the information in the prior is limited. Another approach for continuous outcomes is to guess at the range = (max−min) of each outcome and use the mid-range, (max + min)/2, as prior mean and ((max−min)/4)2 as prior variance.
. We consider these priors to be vague and not particularly informative although proper. The prior for continuous outcome uses the data, but the information in the prior is limited. Another approach for continuous outcomes is to guess at the range = (max−min) of each outcome and use the mid-range, (max + min)/2, as prior mean and ((max−min)/4)2 as prior variance.
In equation (2.3), scale parameter γ1k is the slope of the regression of ηik on 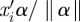 . When the linear predictor is perfectly predictive, the absolute value of the slope can be estimated as the range (max − min) of ηik divided by the range (max − min) of
. When the linear predictor is perfectly predictive, the absolute value of the slope can be estimated as the range (max − min) of ηik divided by the range (max − min) of 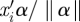 , with an appropriate sign either positive or negative. When the linear predictor is not predictive at all, γ1k = 0. We assign a normal prior with mean 0 and variance the square of the range of the natural parameter divided by 2 times the range of single linear predictor
, with an appropriate sign either positive or negative. When the linear predictor is not predictive at all, γ1k = 0. We assign a normal prior with mean 0 and variance the square of the range of the natural parameter divided by 2 times the range of single linear predictor
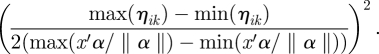 |
3.1 |
We previously discussed the range of the natural parameters for binary, Poisson and continuous outcomes when specifying priors for the intercept. We now discuss the range of 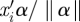 . We deal with continuous predictors and binary predictors separately. The standardized continuous predictors, the norm of the fixed effect regression coefficients equal to 1 implies that the absolute value of each regression coefficient αl /‖α‖ is less than 1; an approximate range of xil αl /‖α‖ is then at most roughly −2 to 2. For binary predictors, we have
. We deal with continuous predictors and binary predictors separately. The standardized continuous predictors, the norm of the fixed effect regression coefficients equal to 1 implies that the absolute value of each regression coefficient αl /‖α‖ is less than 1; an approximate range of xil αl /‖α‖ is then at most roughly −2 to 2. For binary predictors, we have
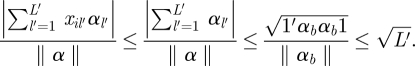 |
3.2 |
Here l′ indexes the binary predictors, L′ is the total number of binary predictors, 1 is a vector of 1 with length L′ and αb /‖ α ‖ is the vector of regression coefficients for the binary predictors. The range of x′α /‖ α ‖ is therefore roughly 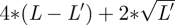 , where L − L′ is the number of continuous predictors. We substitute this range in place of the denominator of equation (3.1) to calculate the prior variances for γ1k.
, where L − L′ is the number of continuous predictors. We substitute this range in place of the denominator of equation (3.1) to calculate the prior variances for γ1k.
We use a Wishart prior Σ−1 ∼ Wishart(ν, Λ) for precision matrix Σ−1, with degrees of freedom ν and precision Λ, where Σ−1 is the inverse of covariance matrix Σ in model (2.3). We set ν to be a number somewhat bigger than K, the dimension of Λ, and we set Λ to be a diagonal matrix. We set diagonal element Λkk to be ν − K − 1 divided by a prior estimate of the variance of ηik. For continuous outcomes, we use the prior-estimated range divided by 4 as our prior estimate of the variance of ηik; for binary outcomes, we use ((6.9–(−6.9))/4)2 = 11.9 and for count outcomes, we use ((4.6–(−4.6))/4)2 = 5.3.
4. Diagnostic models
A priori, outcomes are chosen to share a single linear predictor. In practice, we may choose outcomes which we hope are appropriate. Similarly, we may have chosen predictors that are generally predictive of all outcomes, however some of them may not belong to a single-shared linear predictor. The COPE model assumes that all outcomes are predicted by the single linear predictor and all predictors are commonly predictive of all outcomes. There are models whose number of fixed effects parameters is in between the COPE model and the saturated model, and we propose two extended models to check the COPE model assumptions. Model A checks whether one pre-specified outcome belongs with other outcomes, and model B checks whether one pre-specified predictor is commonly predictive. We apply model A to all outcomes in turn. Any outcomes that do not belong to the COPE model are removed and are fit separately barring some value to continuing to fit a multivariate model. We then apply model B to all predictors in turn. In contrast to outcomes, predictors often can not be arbitrarily dropped for statistical reasons and must be kept in the model for scientific reasons. Thus, we may actually prefer the model B version of the COPE model—a kind of relaxed COPE model where most covariates belong to a single linear predictor, but a few covariates are not required to be part of the COPE linear predictor.
4.1. Extended model A: one outcome has a separate set of predictor effects
For a pre-specified outcome s, we modify the linear predictor in model (2.3) by adding an extra set of regression coefficients for outcome s on top of the common regression coefficients, to allow the fixed effects for outcome s to differ from the common effects while keeping the linear predictor for other outcomes the same as in model (2.3)
| 4.1 |
where αs = (αs1, …, αsL) is the extra set of regression coefficients, αsl is the departure from the common effect for all outcomes of the effect of predictor l on outcome s, l = 1,…, L. We assign αs a normal prior with mean 0L and variance IL.
Models (2.2a)–(4.1) have an identifiability problem, γ1s and αs can be replaced by 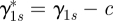 and
and 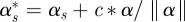 for an arbitrary constant c. To solve the problem, we use the Gram–Schmidt process to split αs into two components, one component
for an arbitrary constant c. To solve the problem, we use the Gram–Schmidt process to split αs into two components, one component 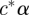 is parallel to α, where c* = αs′α/α′α and the other component, denoted α*s, α*s = αs − c*α, is perpendicular to α, α*′s α = 0. We use MCMC sampling for computations and we calculate α*s for each posterior sample of αs.
is parallel to α, where c* = αs′α/α′α and the other component, denoted α*s, α*s = αs − c*α, is perpendicular to α, α*′s α = 0. We use MCMC sampling for computations and we calculate α*s for each posterior sample of αs.
Having fit the model, we test the hypothesis H0: α*s = 0L against the alternative hypothesis HA:α*s ≠ 0L using a Bayesian Wald test. The test statistic is calculated as
| 4.2 |
where 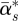 is the posterior mean of α*s, the posterior variance Var(α*s) is estimated by
is the posterior mean of α*s, the posterior variance Var(α*s) is estimated by 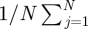
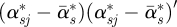 , N is the total number of posterior samples, j is the index of jth posterior sample, and Wα is compared with a χ2-distribution with degree of freedom L. If Wα > χL2(0.95), we conclude that the sth outcome does not have regression parameters in common with other outcomes.
, N is the total number of posterior samples, j is the index of jth posterior sample, and Wα is compared with a χ2-distribution with degree of freedom L. If Wα > χL2(0.95), we conclude that the sth outcome does not have regression parameters in common with other outcomes.
4.2. Extended model B: one predictor has a distinct effect for each outcome
Suppose we have K* outcomes remaining in our model. For a pre-specified predictor l, we modify model (2.3) to allow the lth predictor xl to have a separate coefficient for each outcome k. We modify ηik for outcome k in model (2.3)
| 4.3 |
where ϕk, k = 1, … , K*, is the departure from the common effect of the fixed effect of predictor l on outcome k and xil is the lth component of xi for the ith subject; also recall that αl is the lth component of α. Define ϕ = (ϕ1, … , ϕK*)′ and γ1 = (γ11, … , γ1K*)′. We assign ϕ a normal prior with mean 0K* and variance IK*.
As with model B, we have an identifiability problem because αl and ϕ can be replaced by αl* = αl * c and ϕ* = ϕ+ (γ1αl (1 − c))/‖ α‖ for an arbitrary constant c, thus γ1l and ϕl are confounded. We again use the Gram–Schmidt process to split ϕ into two components, one component c*γ1 is parallel to γ1, where c* = ϕ′γ1/γ1′γ1 and the other component ϕ* = ϕ− c*γ1 are perpendicular to γ1.
We test the hypothesis H0 : ϕ* = 0k* against the alternative hypothesis HA : ϕ* ≠ 0K*. The Wald test statistic is
| 4.4 |
here 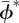 is the posterior mean of ϕ*, posterior variance Var(ϕ*) is estimated by
is the posterior mean of ϕ*, posterior variance Var(ϕ*) is estimated by 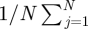
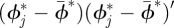 and Wϕ is compared with a χ2-distribution with degree of freedom K*. If
and Wϕ is compared with a χ2-distribution with degree of freedom K*. If 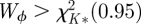 , we conclude that the lth predictor is not commonly predictive for all outcomes.
, we conclude that the lth predictor is not commonly predictive for all outcomes.
4.3. Final model
We fit extended model A for each outcome in turn and calculate each Wald test. We remove the outcomes with significant Wald test results from our COPE model. We then fit extended model B with the remaining outcomes for each predictor in turn and calculate each Wald test statistic as well. We remove predictors with significant Wald tests from the common set of predictors and these predictors are allowed to have separate coefficients for each outcome. Suppose, we have K* outcomes left for the COPE model, L* predictors are common and L − L* predictors have outcome-specific regression coefficients. Without loss of generality, suppose the first L* predictors are common for all outcomes, denoted as x′i− = (xi1, … , xiL*). Then our final model is
 |
4.5 |
where α− = (α1, … , αL*) is the common regression coefficient vector, αmk is the regression coefficient of predictor m, m = L* + 1, … , L, for outcome k, k = 1, … , K*, γ0k, γ1k and βik are the same as in model (2.3). If the Wald test for every outcome is significant, the simple solution is to not use the COPE model, though some B-type diagnostic model might still be appropriate. If all predictors are significant, we get the usual saturated model.
5. Data analysis: teens linked to care
We illustrate the methodology by examining baseline observations from the teens linked to care (TLC) study on YPLH, which was conducted in eight adolescent clinical care sites in Los Angeles, New York, and San Francisco from 1995 to 1996 [37,38]. In this paper, we study outcomes that are subscales from the psychometric measure brief symptom inventory (BSI). The BSI consists of 53 items covering nine symptom dimensions: somatization, obsessive-compulsive disorder (OCD), interpersonal sensitivity, depression, anxiety, hostility, phobic anxiety, paranoid ideation and psychoticism. Each item is given an integer-valued score ranging from 0 to 4.
We generate continuous, binary and Poisson variables from the 53 items by the following rules: continuous subscale scores are calculated by summing the values for the items included in that dimension and dividing by the number of items; binary variables are calculated from a single item score by setting the binary variable to 0 if the original item score is 0 else setting the binary variable to 1 for an item score of 1–4; and Poisson variables are calculated by summing the score of the items included in that dimension.
These are all commonly used distributions for items and subscales except for the Poisson distribution, which we think merits greater consideration than it has received in the past. For example, many items on the BSI are skewed with respondents choosing mostly 0, occasionally 1 and rarely 2 to 4. For an item scored on a 0 to 4 scale, a Poisson random variable with mean less than 1.27 has less than a 1 per cent chance of being 5 or larger, and thus the Poisson approximation to the sampling distribution has the potential to be surprisingly accurate; this must necessarily be confirmed for each item in the dataset. Summing several approximately Poisson-distributed random variables then produces a random variable that, in practice, is indistinguishable from a Poisson, even in the presence of modest correlation. Poisson regression is a commonly accepted model for non-negative integer-valued data; our model introduces the random effect to model both the correlation and the overdispersion, for when a non-negative integer outcome being modelled as Poisson has variance bigger than its mean.
We then apply the proposed COPE model on a set of generated outcomes that include continuous, binary and Poisson variables. Higher scores on every scale indicate a worse psychological state.
We use data collected at baseline interview in our analysis and start with a candidate model with eight outcomes, two treated as continuous subscales: depression and OCD; two treated as Poisson subscales: anxiety and somatization; and four dichotomized as binary items: BS11 (poor appetite), BS13 (temper uncontrollable), BS21 (people are unfriendly) and BS31 (avoid things that frighten you). No items are common between BSI subscales and the four binary items belong to subscales not otherwise used in this analysis. The two continuous outcomes have long right tails, we transform each outcome as log2 (x + c), where c is the smallest non-zero value for that outcome, in our example, c is 1/6 for both outcomes. For response variables analysed on the log scale, the exponentiated regression coefficients can be interpreted as the multiplicative increase in the unlogged response per unit increase in the predictor.
The predictors are gender (73% male); age standardized to have mean 0 and standard deviation 1 by subtracting the mean 20.8 and then dividing by the standard deviation 2.1; food (87% yes), yes means that a subject can get enough food easily daily; finance (66% yes), yes means that a subject has the necessities to live comfortably and no means that the subject is struggling to survive and has difficulty paying bills; marijuana use in the past three months (79% yes); acquired immune deficiency syndrome (AIDS) symptoms (37% yes), yes means that the subject has physical health symptoms resulting from HIV infection; hard drug use (62% yes), yes means that the subject used at least one type of hard drug from stimulants, LSD, inhalants, coke, crack and heroin in the past three months; and attempted suicide (41% yes), yes means that the subject attempted suicide at least once prior to being enrolled in this study.
We complete a Bayesian prior specification of the model by following the recommendations from §3. We assign a normal prior with mean 08 and variance 9I8 to α, α ∼ N(0,9I8), here 9 can be changed to any arbitrary value. For the intercepts γ0k, for continuous outcomes, we use the centre of the range (max + min)/2 as the prior mean and 1/4 of the range (max−min)/4 as the prior standard deviation; for binary outcomes, we use 0 as the prior mean and 3.4 as the prior standard deviation; for Poisson data, we use 0 as the prior mean and 2.3 as the prior standard deviation. A guess of the ranges of log depression and OCD are both from −2.58 to 2.06. The centre of the range is −0.26 and the range divided by 4 is 1.16. Therefore, we set normal independent priors for γ0 with means (−0.26,−0.26,0,0,0,0,0,0) and standard deviations (1.16,1.16,3.4,3.4,3.4,3.4,2.3,2.3).
We have one standardized continuous predictor and seven binary predictors, the range of xi′ α /‖ α ‖ is then from  to
to  which is −4.6 to 4.6 and the length of the range is 9.2. A guess of the range of ηik is 5.2 for continuous outcomes after logarithm transformation, 13.8 for binary outcomes and 9.2 for Poisson outcomes. We calculate the prior standard deviation for scale parameters by dividing the range of ηik by 4 times the range of x′α /‖α‖ and we get 0.14 for continuous outcomes, 0.38 for binary outcomes and 0.25 for count outcomes. We set normal independent priors for scale parameters γ1 with mean (0,0,0,0,0,0,0,0,0,0) and standard deviations (0.28,0.28,0.76,0.76,0.76,0.76,0.50,0.50).
which is −4.6 to 4.6 and the length of the range is 9.2. A guess of the range of ηik is 5.2 for continuous outcomes after logarithm transformation, 13.8 for binary outcomes and 9.2 for Poisson outcomes. We calculate the prior standard deviation for scale parameters by dividing the range of ηik by 4 times the range of x′α /‖α‖ and we get 0.14 for continuous outcomes, 0.38 for binary outcomes and 0.25 for count outcomes. We set normal independent priors for scale parameters γ1 with mean (0,0,0,0,0,0,0,0,0,0) and standard deviations (0.28,0.28,0.76,0.76,0.76,0.76,0.50,0.50).
We set a Wishart prior for the inverse of the covariance matrix for the random effects βi with degrees of freedom 20 and we calculate the kth diagonal element by dividing ν − K − 1 = 20 − 8 − 1 = 11 by a guess of the variance of ηik, which are (5.2/4)2 = 1.7 for continuous outcomes, (13.8/4)2 = 11.9 for binary outcomes and (9.2/4)2 = 5.3 for count outcomes. We get the prior precision matrix Λ = diag(6.5,6.5,0.9,0.9,0.9,0.9,2.1,2.1).
We run the proposed candidate model in WinBUGS [39] and generate 500 000 posterior samples, discarding the first 4000 samples as a burn-in. We examine autocorrelation plots and find that the autocorrelations for most parameters are close to 0 after 300 lags. The posterior density plots show that the posterior distributions for α, γ0 and γ1 are bell-shaped and unimodal.
We diagnose the candidate model by running extended model A, models (2.2a)–(4.1), for each outcome in turn to test whether the outcome has a separate set of regression coefficients. This results in eight extended models corresponding to the eight outcomes. We calculate the Bayesian Wald test statistic for each model and compare it with χ82(0.95) = 15.51, where the degree of freedom is equal to the number of predictors in the model. Results show that the Wald test statistic is significant for anxiety (p-value = 0.048), but not for any of the other outcomes. We exclude the outcome anxiety then run extended model B, models (2.2a)–(2.2c) and (4.3) for each predictor in turn to test whether the predictor is not commonly predictive for all outcomes, but has a different regression coefficient for each outcome, resulting in another eight extended models corresponding to the eight predictors. We calculate the Bayesian Wald test statistic for each model and compare it with χ72(0.95) = 14.07, where the degree of freedom is the number of outcomes in the model, which is 7 now because we excluded anxiety. Results show that the Wald test statistic is significant for suicide attempt (p-value = 0.040) but not for the other predictors. The Bayesian Wald test statistics and p-values for these two sets of extended models are shown in table 1.
Table 1.
Model diagnostics for checking whether an outcome or a predictor belongs in the COPE model. Left three columns: the Bayesian Wald statistic Ws in extended model A is used to test whether an outcome has a separate set of regression coefficients different from other outcomes. The p-value next to Ws comes from comparing Ws with χ82, a χ2-distribution with 8 d.f. Anxiety is the only outcome whose Wald statistic is significant. We exclude anxiety from the COPE model. Right three columns: Wald statistics Wϕ are from extended model B and are used to test whether a predictor has a different regression coefficient for each outcome. The p-value next to Wϕ comes from comparing Wϕ with a χ72 distribution. Suicide attempt is the only predictor whose Wald statistic is significant.
| outcome | Ws | p-value | predictor | Wϕ | p-value |
|---|---|---|---|---|---|
| depression | 10.2 | 0.25 | male | 8.5 | 0.29 |
| obsessive compulsive | 9.8 | 0.28 | age | 4.2 | 0.76 |
| BS11 (poor appetite) | 5.5 | 0.70 | food | 8.1 | 0.33 |
| BS13 (temper uncontrol) | 10.3 | 0.24 | finance | 4.0 | 0.78 |
| BS21 (people not friendly) | 5.4 | 0.72 | marijuana | 6.2 | 0.52 |
| BS31 (avoid frighten) | 9.0 | 0.34 | hard drugs | 9.6 | 0.21 |
| anxiety | 15.6 | 0.05 | AIDS symptoms | 6.8 | 0.45 |
| somatization | 13.7 | 0.09 | suicide attempt | 14.7 | 0.04 |
Our final model has anxiety by itself in a separate model and a relaxed COPE model for the other seven outcomes, and eight predictors among which suicide attempt has an outcome-specific effect and the other seven predictors constitute a single linear predictor commonly predictive for the seven remaining outcomes. Posterior summaries for the seven common regression coefficients α and eight scale parameters γ1 are shown in table 2. Having enough food and a good financial situation imparts negative effects on all seven outcomes. Using marijuana and having AIDS symptoms have positive effects on all seven outcomes. The magnitude of the effects is modified by the scale parameter for each outcome. The scale parameter for the depression and OCD subscales is 0.58 and 0.71, respectively, which means that 1 unit change in the single linear predictor would associate with a change of 20.58 = 1.49 units in depression and a change of 20.71 = 1.64 for the OCD subscale because we used a base 2 logarithm transformation for these two continuous variables prior to analysis. The scale parameters for binary outcomes B11, B13, B21 and B31 are 1.42, 0.47, 0.87 and 0.71, respectively, which means that 1 unit change in the single linear predictor would associated with e1.42 =4.13, e0.47 =1.59, e0.87 =2.39 and e0.71 = 2.04 odds ratio change for B11, B13, B21 and B31. The scale parameter for Poisson outcome somatization is 0.71, indicting a one unit change in linear predictor is associated with e0.71 = 2.04 unit change in somatization.
Table 2.
Posterior summaries for common predictors α, scale parameters γ1 and intercept γ0 in final model. Age is standardized with mean 0 and standard deviation 1, other covariates are 0–1 variables; mean and standard deviation are the posterior sample mean and standard deviation for each parameter, respectively. Column headings 2.50% and 97.50% are the 2.5 percentile and 97.5 percentile, respectively, and % ≥ 0 = P(parameter ≥ 0| Y) is an estimate of the posterior probability that the parameter is positive.
| name | parm | mean | s.d. | 2.50% | 97.50% | % ≥ 0 |
|---|---|---|---|---|---|---|
| predictor | ||||||
| male | α1 | 0.05 | 0.20 | −0.34 | 0.44 | 0.59 |
| age | α2 | 0.01 | 0.07 | −0.14 | 0.15 | 0.54 |
| food | α3 | −0.58 | 0.16 | −0.84 | −0.22 | 0 |
| finance | α4 | −0.34 | 0.16 | −0.64 | −0.03 | 0.02 |
| marijuana | α5 | 0.17 | 0.17 | −0.19 | 0.49 | 0.83 |
| hard drugs | α6 | 0.05 | 0.21 | −0.33 | 0.45 | 0.58 |
| AIDS symptoms | α7 | 0.58 | 0.13 | 0.31 | 0.81 | 1.00 |
| scale parameter | ||||||
| depression | γ11 | 0.58 | 0.16 | 0.27 | 0.89 | 1.00 |
| obsessive compulsive | γ12 | 0.71 | 0.16 | 0.39 | 1.03 | 1.00 |
| BS11 (poor appetite) | γ13 | 1.42 | 0.37 | 0.71 | 2.18 | 1.00 |
| BS13 (temper uncontrol) | γ14 | 0.47 | 0.36 | −0.24 | 1.17 | 0.91 |
| BS21 (people not friendly) | γ15 | 0.87 | 0.34 | 0.22 | 1.56 | 1.00 |
| BS31 (avoid frighten) | γ16 | 0.71 | 0.38 | −0.05 | 1.45 | 0.97 |
| somatization | γ17 | 0.71 | 0.15 | 0.42 | 1.02 | 1.00 |
| intercept | ||||||
| depression | γ01 | −0.35 | 0.17 | −0.67 | 0 | 0.03 |
| obsessive compulsive | γ02 | −0.07 | 0.20 | −0.44 | 0.34 | 0.37 |
| BS11 (poor appetite) | γ03 | −0.10 | 0.42 | −0.88 | 0.76 | 0.39 |
| BS13 (temper uncontrol) | γ04 | −0.22 | 0.27 | −0.69 | 0.37 | 0.19 |
| BS21 (people not friendly) | γ05 | 0.13 | 0.31 | −0.43 | 0.79 | 0.64 |
| BS31 (avoid frighten) | γ06 | −0.80 | 0.31 | −1.38 | −0.17 | 0.01 |
| somatization | γ07 | 1.11 | 0.20 | 0.73 | 1.51 | 1.00 |
Table 3 shows posterior summaries for the regression coefficients of each predictor for each outcome. We fit a separate model for anxiety and a relaxed COPE model for the other seven outcomes. For the first seven common predictors, we calculate αlγ1k from the posterior samples of αl and γ1k, then draw inference from the calculated posterior samples of αlγ1k. We have the outcome-specific regression coefficient for suicide attempt, the one predictor that has a separate regression coefficient for each outcome. Suicide attempt has significant positive effects on depression, OCD, B31 and somatization. Food and finance have significant negative effects on anxiety while hard drugs, AIDS symptoms and suicide attempt have significant positive effects. We compare the standard deviation of the regression coefficients in our final relaxed COPE model with that in the saturated model and find that on average, the standard deviation in our final model is 29 per cent less than that in the saturated model. This is evidence that compared with the saturated model, the COPE model has greater efficiency.
Table 3.
Regression coefficients. There are three columns for each outcome. Columns give the posterior mean, posterior standard deviation and percentage of positive posterior samples. We fit anxiety separately and use a relaxed COPE model for the other 7 outcomes. In the COPE model, For the first 7 common predictors, we calculate αlγ1k from the posterior samples of αls and γ1k then draw inference from the calculated posterior samples of αlγ1k and we have the outcome-specific regression coefficient for suicide attempt, the one predictor that has a separate regression coefficient for each outcome.
| depression |
OCD |
BS11 |
BS13 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pred | mean | s.d. | % ≥ 0 | mean | s.d. | % ≥ 0 | mean | s.d. | % ≥ 0 | mean | s.d. | % ≥ 0 |
| male | 0.03 | 0.12 | 0.59 | 0.03 | 0.14 | 0.59 | 0.08 | 0.29 | 0.59 | −0.01 | 0.12 | 0.52 |
| age | 0 | 0.04 | 0.54 | 0.01 | 0.05 | 0.54 | 0.01 | 0.10 | 0.54 | 0.01 | 0.04 | 0.54 |
| food | −0.34 | 0.14 | 0 | −0.42 | 0.16 | 0 | −0.84 | 0.36 | 0 | −0.28 | 0.24 | 0.10 |
| financial | −0.19 | 0.10 | 0.02 | −0.24 | 0.12 | 0.02 | −0.47 | 0.23 | 0.02 | −0.15 | 0.14 | 0.11 |
| marijuana | 0.10 | 0.11 | 0.83 | 0.12 | 0.13 | 0.83 | 0.23 | 0.26 | 0.83 | 0.06 | 0.11 | 0.74 |
| hard drugs | 0.03 | 0.12 | 0.58 | 0.04 | 0.15 | 0.58 | 0.06 | 0.29 | 0.58 | 0.05 | 0.13 | 0.62 |
| AIDS symptoms | 0.33 | 0.11 | 1.00 | 0.41 | 0.12 | 1.00 | 0.81 | 0.26 | 1.00 | 0.28 | 0.22 | 0.91 |
| suicide attempt | 0.61 | 0.13 | 1.00 | 0.26 | 0.14 | 0.97 | 0.24 | 0.30 | 0.80 | 0.22 | 0.28 | 0.78 |
| B21 | B31 | som | anx | |||||||||
| pred | mean | s.d. | % ≥ 0 | mean | s.d. | % ≥0 | mean | s.d. | % ≥0 | mean | s.d. | % ≥ 0 |
| male | 0.04 | 0.19 | 0.59 | 0.05 | 0.17 | 0.60 | 0.04 | 0.14 | 0.59 | 0.12 | 0.06 | 0.98 |
| age | 0 | 0.07 | 0.54 | 0.01 | 0.06 | 0.54 | 0 | 0.05 | 0.54 | −0.01 | 0.03 | 0.29 |
| food | −0.51 | 0.25 | 0.01 | −0.40 | 0.25 | 0.03 | −0.42 | 0.15 | 0 | −0.30 | 0.06 | 0 |
| financial | −0.30 | 0.19 | 0.02 | −0.25 | 0.19 | 0.04 | −0.24 | 0.11 | 0.02 | −0.16 | 0.05 | 0 |
| marijuana | 0.15 | 0.18 | 0.82 | 0.11 | 0.15 | 0.80 | 0.12 | 0.13 | 0.83 | 0.01 | 0.07 | 0.58 |
| hard drugs | 0.03 | 0.19 | 0.58 | 0.02 | 0.17 | 0.57 | 0.03 | 0.15 | 0.58 | 0.26 | 0.06 | 1.00 |
| AIDS symptoms | 0.50 | 0.22 | 1.00 | 0.42 | 0.24 | 0.97 | 0.41 | 0.13 | 1.00 | 0.34 | 0.05 | 1.00 |
| suicide attempt | 0.20 | 0.28 | 0.77 | 0.91 | 0.31 | 1.00 | 0.35 | 0.13 | 1.00 | 0.32 | 0.05 | 1.00 |
6. Discussion
We began with a candidate COPE model that assumed all outcomes were similar and shared one common linear predictor. We then fit flexible diagnostic models to identify outcomes that did not belong to the COPE model, and which were then excluded from the final model. The model with one linear predictor is a one cluster model. When we have a large number of outcomes, more than one linear predictor may be needed to predict all outcomes. We may expand our one cluster model to a multi-cluster model by clustering outcomes and introducing cluster-specific regression coefficients for each cluster [40].
We can use a graphical method to identify which outcomes belong to the same cluster and how many clusters we shall have. We first run a univariate generalized linear model for each outcome yk and get estimates 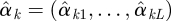 of regression coefficients omitting the intercept. We normalize
of regression coefficients omitting the intercept. We normalize 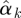 by dividing by its Euclidean norm,
by dividing by its Euclidean norm, 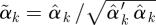 , so that
, so that 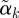 has length 1 and can be compared across outcomes. We plot a profile plot of the
has length 1 and can be compared across outcomes. We plot a profile plot of the 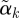 , plotting points (j,
, plotting points (j, 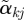 ) and drawing line segments connecting point
) and drawing line segments connecting point 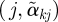 to
to 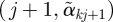 , and inspect the plot to identify potential outcome clusters.
, and inspect the plot to identify potential outcome clusters.
Finally, we can adapt the COPE model to longitudinal mixed outcome data by introducing two latent variables, one to model the correlation among outcomes at the same time and the other to model the correlation among outcomes over time. Consideration of the COPE model when it cannot be written as a linear model, as in model (2.3), could be an area of future work.
In this paper, we proposed a Bayesian Wald test to determine whether one outcome or one predictor belongs to the COPE model. There are alternative methods for model selection. The Bayes factor is a widely used statistic in which prior and posterior information are combined in a ratio that provides evidence in favour of one model versus another. We can calculate a Bayes factor as the marginal likelihood of the candidate model divided by the marginal likelihood of a diagnostic type A model or type B model, where the marginal likelihood of a model is the probability of the data with all the model parameters integrated out. We can also use Akaike information criterion (AIC), Bayesian information criterion (BIC) or deviance information criterion (DIC) to do model selection. In a Bayesian framework, DIC is easily calculated from the samples generated by a MCMC simulation while AIC and BIC require calculating the likelihood.
Acknowledgements
Weiss was supported in part by the Center for HIV Identification, Prevention and Treatment Services, NIH/NIMH P30MH58107. Suchard was supported in part by a John Simon Guggenheim Memorial Foundation Fellowship and a research gift from Google.
References
- 1.Travison T. G., Brookmeyer R. 2007. Global effects estimation for multidimensional outcomes. Stat. Med. 26, 4845–4859 10.1002/sim.2983 (doi:10.1002/sim.2983) [DOI] [PubMed] [Google Scholar]
- 2.Weiss R. E. 2005. Modeling longitudinal data. New York, NY: Springer [Google Scholar]
- 3.Molenberghs G., Verbeke G. 2005. Models for discrete longitudinal data. New York, NY: Springer [Google Scholar]
- 4.Beckett L. A., Tancredi D. J., Wilson R. S. 2004. Multivariate longitudinal models for complex change processes. Stat. Med. 23, 231–239 10.1002/sim.1712 (doi:10.1002/sim.1712) [DOI] [PubMed] [Google Scholar]
- 5.Dubin J. A., Müller H. G. 2005. Dynamical correlation for multivariate longitudinal data. J. Am. Stat. Assoc. 100, 872–881 10.1198/016214504000001989 (doi:10.1198/016214504000001989) [DOI] [Google Scholar]
- 6.Diggle P. J., Heagerty P. J., Liang K. Y., Zeger S. L. 2002. Analysis of longitudinal data, 2nd edn New York, NY: Oxford University Press [Google Scholar]
- 7.Davis C. S. 2002. Statistical methods for the analysis of repeated measurements. New York, NY: Springer [Google Scholar]
- 8.McCulloch C. E., Searle S. R. 2001. Generalized, linear, and mixed models. New York, NY: Wiley [Google Scholar]
- 9.Nummi T., Mottonen J. 2000. On the analysis of multivariate growth curve. Metrika, 52 77–89 10.1007/s001840000063 (doi:10.1007/s001840000063) [DOI] [Google Scholar]
- 10.Lin X., Ryan L., Sammel M., Zhang D., Padungtod C., Xu X. 2000. A scaled linear model for multiple outcomes. Biometrics, 56 593–601 10.1111/j.0006-341X.2000.00593.x (doi:10.1111/j.0006-341X.2000.00593.x) [DOI] [PubMed] [Google Scholar]
- 11.Verbeke G., Molenberghs G. 2000. Linear mixed models for longitudinal data. New York, NY: Springer [Google Scholar]
- 12.Fieuws S., Verbeke G. 2006. Pairwise fitting of mixed models for the joint modeling of multivariate longitudinal profiles. Biometrics, 62 424–431 10.1111/j.1541-0420.2006.00507.x (doi:10.1111/j.1541-0420.2006.00507.x) [DOI] [PubMed] [Google Scholar]
- 13.Gray S. M., Brookmeyer R. 2000. Multidimensional longitudinal data: estimating a treatment effect from continuous, discrete or time-to-event response variables. J. Am. Stat. Assoc. 95, 396–406 10.2307/2669376 (doi:10.2307/2669376) [DOI] [Google Scholar]
- 14.Brown H., Prescott R. 1999. Applied mixed models in medicine. New York, NY: Wiley [Google Scholar]
- 15.Gray S. M., Brookmeyer R. 1998. Estimating a treatment effect from multidimensional longitudinal data. Biometrics 54, 976–988 10.2307/2533850 (doi:10.2307/2533850) [DOI] [PubMed] [Google Scholar]
- 16.Littel R. C., Milliken G. A., Stroup W. A., Wolfinger R. D. 1996. The SAS system for mixed models. Cary, NC: SAS Institute Inc [Google Scholar]
- 17.Mickey R. M., Shema S. J., Vacek P. M., Bell D. Y. 1994. Analysis of multiple outcome variables measured longitudinally. Comput. Stat. Data Anal. 17, 17–33 10.1016/0167-9473(92)00059-Z (doi:10.1016/0167-9473(92)00059-Z) [DOI] [Google Scholar]
- 18.O’Brien P. C. 1984. Procedures for comparing samples with multiple endpoints. Biometrics 40, 1079–1087 10.2307/2531158 (doi:10.2307/2531158) [DOI] [PubMed] [Google Scholar]
- 19.Little R. J. A., Schluchter M. D. 1985. Maximum likelihood estimation for mixed continuous and categorical data with missing. Biometrika 72, 497–512 10.1093/biomet/72.3.497 (doi:10.1093/biomet/72.3.497) [DOI] [Google Scholar]
- 20.Sammel M., Ryan L. M., Legler J. M. 1997. Latent variable models for mixed discrete and continuous outcomes. J. R. Stat. Soc. Ser. B 59, 667–678 10.1111/1467-9868.00090 (doi:10.1111/1467-9868.00090) [DOI] [Google Scholar]
- 21.Arminger G., Kusters U. 1988. Latent trait models with indicators of mixed measurement level. In Latent trait and latent class models (eds Langeheine R., Rost J.). New York, NY: Plenum [Google Scholar]
- 22.Fitzmaurice G. M., Laird N. M. 1997. Regression models for mixed discrete and continuous responses with potentially missing values. Biometrics 53, 110–122 10.2307/2533101 (doi:10.2307/2533101) [DOI] [PubMed] [Google Scholar]
- 23.Dunson D. B. 2000. Bayesian latent variable models for clustered mixed outcomes. J. R. Stat. Soc. Ser. B 62, 355–366 10.1111/1467-9868.00236 (doi:10.1111/1467-9868.00236) [DOI] [Google Scholar]
- 24.Gueorguieva R. V., Agresti A. 2001. A correlated probit model for joint modeling of clustered binary and continuous responses. J. Am. Stat. Assoc. 96, 1102–1112 10.1198/016214501753208762 (doi:10.1198/016214501753208762) [DOI] [Google Scholar]
- 25.Knaus W., Draper E., Wagner D., Zimmerman J. 1985. APACHE II: a severity of disease classification system. Crit. Care Med. 13, 818–829 10.1097/00003246-198510000-00009 (doi:10.1097/00003246-198510000-00009) [DOI] [PubMed] [Google Scholar]
- 26.Knaus W. A., et al. 1991. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest 100, 1619–1636 10.1378/chest.100.6.1619 (doi:10.1378/chest.100.6.1619) [DOI] [PubMed] [Google Scholar]
- 27.Charlson M. E., Pompei P., Ales K. L., MacKenzie C. R. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J. Chronic Dis. 40, 373–383 10.1016/0021-9681(87)90171-8 (doi:10.1016/0021-9681(87)90171-8) [DOI] [PubMed] [Google Scholar]
- 28.Hall W. H., Ramachandran R., Narayan S., Jani A. B., Vijayakumar S. 2004. An electronic application for rapidly calculating Charlson comorbidity score. BMC Cancer 4, 94. 10.1186/1471-2407-4-94 (doi:10.1186/1471-2407-4-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnofsky D., Burchenal J. 1949. The clinical evaluation of chemotherapeutic agents in cancer. In Evaluation of chemotherapeutic agents (ed. Macleod C.), pp. 191–205, 2nd edn New York, NY: Columbia University Press. [Google Scholar]
- 30.Oken M. M., Creech R. H., Tormey D. C., Horton J., Davis T. E., McFadden E. T., Carbone P. P. 1982. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am. J. Clin. Oncol. 5, 649–655 10.1097/00000421-198212000-00014 (doi:10.1097/00000421-198212000-00014) [DOI] [PubMed] [Google Scholar]
- 31.Nashef S. A. M, Roques F., Michel P., Gauducheau E., Lemeshow S., Salamon R. EuroSCORE study group 1999. European system for cardiac operative risk evaluation (EuroSCORE). Eur. J. Cardio-thorac. Surg. 16, 9–13 10.1016/S1010-7940(99)00134-7 (doi:10.1016/S1010-7940(99)00134-7) [DOI] [PubMed] [Google Scholar]
- 32.Roques F., Michel P., Goldstone A. R., Nashef S. A. M. 2003. Letter to the editor: the logistic EuroSCORE. Eur. Heart J. 24, 1–2 10.1016/S0195-668X(02)00799-6 (doi:10.1016/S0195-668X(02)00799-6) [DOI] [PubMed] [Google Scholar]
- 33.Dongen S. V. 2006. Prior specification in Bayesian statistics: three cautionary tales. J. Theor. Biol. 242, 90–100 10.1016/j.jtbi.2006.02.002 (doi:10.1016/j.jtbi.2006.02.002) [DOI] [PubMed] [Google Scholar]
- 34.Kass R. E., Wasserman L. 1996. The selection of prior distributions by formal rules. J. Am. Stat. Assoc. 91, 1343–1370 10.2307/2291752 (doi:10.2307/2291752) [DOI] [Google Scholar]
- 35.Clarke B., Wasserman L. 1993. Noninformative priors and nuisance parameters. J. Am. Stat. Assoc. 88, 1427–1432 10.2307/2291287 (doi:10.2307/2291287) [DOI] [Google Scholar]
- 36.Akaike H. 1980. The interpretation of improper prior distributions as limits of data dependent proper prior distributions. J. R. Stat. Soc. Ser. B 42, 46–52 See http://www.jstor.org/stable/2984737 [Google Scholar]
- 37.Rotheram-Borus M. J., Murphy D. A., Swendeman D., Chao B., Chabon B., Zhou S., Birnbaum J., O'Hara P. 1999. Substance use and its relationship to depression, anxiety, and isolation among youth living with HIV. Int. J. Behav. Med. 6, 293–311 10.1207/s15327558ijbm0604_1 (doi:10.1207/s15327558ijbm0604_1) [DOI] [PubMed] [Google Scholar]
- 38.Rotheram-Borus M. J., Murphy D. A., Wight R. G., Lee M. B., Lightfoot M., Swendeman D., Birnbaum J. M., Wright W. 2001. Improving the quality of life among young people living with HIV. Eval. Program Plann. 24, 227–237 See http://ideas.repec.org/a/eee/epplan/v24y2001i2p227-237.html [Google Scholar]
- 39.Lunn D. J., Thomas A., Best N., Spiegelhalter D. 2000. WinBUGS—a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 10, 325–337 10.1023/A:1008929526011 (doi:10.1023/A:1008929526011) [DOI] [Google Scholar]
- 40.Jia J., Weiss R. E. 2009. Common predictor effects for multivariate longitudinal data. Stat. Med. 28, 1793–1804 10.1002/sim.3589 (doi:10.1002/sim.3589) [DOI] [PMC free article] [PubMed] [Google Scholar]


