Abstract
Computational systems biology is concerned with the development of detailed mechanistic models of biological processes. Such models are often stochastic and analytically intractable, containing uncertain parameters that must be estimated from time course data. In this article, we consider the task of inferring the parameters of a stochastic kinetic model defined as a Markov (jump) process. Inference for the parameters of complex nonlinear multivariate stochastic process models is a challenging problem, but we find here that algorithms based on particle Markov chain Monte Carlo turn out to be a very effective computationally intensive approach to the problem. Approximations to the inferential model based on stochastic differential equations (SDEs) are considered, as well as improvements to the inference scheme that exploit the SDE structure. We apply the methodology to a Lotka–Volterra system and a prokaryotic auto-regulatory network.
Keywords: chemical Langevin equation, Markov jump process, pseudo-marginal approach, sequential Monte Carlo, stochastic differential equation, stochastic kinetic model
1. Introduction
Computational systems biology [1] is concerned with developing dynamic simulation models of complex biological processes. Such models are useful for developing a quantitative understanding of the process, for testing current understanding of the mechanisms, and to allow in silico experimentation that would be difficult or time-consuming to carry out on the real system in the laboratory. The dynamics of biochemical networks at the level of the single cell are well known to exhibit stochastic behaviour [2]. A major component of the stochasticity is intrinsic to the system, arising from the discreteness of molecular processes [3]. The theory of stochastic chemical kinetics allows the development of Markov process models for network dynamics [4], but such models typically contain rate parameters that must be estimated from imperfect time course data [5]. Inference for such partially observed nonlinear multivariate Markov process models is an extremely challenging problem [6]. However, several strategies for rendering the inference problems more tractable have been employed in recent years, and new methodological approaches have recently been developed that offer additional possibilities. This paper will review these methods, and show how they may be applied in practice to some low-dimensional but nevertheless challenging problems.
In §2, a review of the basic structure of the problem is presented, showing how the Markov process representation of a biochemical network is constructed, and introducing a diffusion approximation that greatly improves computational tractability. In §3, a Bayesian approach to the inferential problem is given, together with an introduction to methods of solution that are ‘likelihood-free’ in the sense that they do not require evaluation of the discrete time transition kernel of the Markov process. Unfortunately, most obvious inferential algorithms suffer from scalability problems as either the number of parameters or time points increase. In §4, it is shown how the recently proposed particle Markov chain Monte Carlo (pMCMC) algorithms [7] may be applied to this class of models, as these do not suffer from scalability problems in the same way as more naive approaches. It is also shown how the structure of stochastic differential equation (SDE) models may be exploited in order to adapt the basic pMCMC approach, departing from the likelihood-free paradigm, but leading to algorithms that are (relatively) computationally efficient, even in low/no measurement error scenarios where likelihood-free approaches tend to break down.
2. Stochastic chemical kinetics
For mass-action stochastic kinetic models (SKMs) [3], it is assumed that the state of the system at a given time is represented by the number of molecules of each reacting chemical ‘species’ present in the system at that time, and that the state of the system is changed at discrete times according to one or more reaction ‘channels’. So consider a biochemical reaction network involving u species 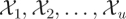 and v reactions ℛ1, ℛ2, … , ℛv, written using standard chemical reaction notation as
and v reactions ℛ1, ℛ2, … , ℛv, written using standard chemical reaction notation as
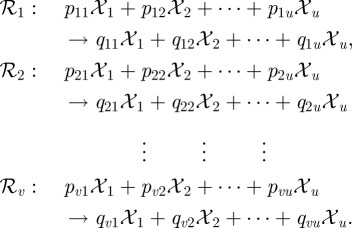 |
Let Xj,t denote the number of molecules of species 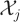 at time t, and let Xt be the u-vector Xt = (X1,t, X2,t, … , Xu,t)⊤. The v × u matrix P consists of the coefficients pij, and Q is defined similarly. The u × v stoichiometry matrix S is defined by
at time t, and let Xt be the u-vector Xt = (X1,t, X2,t, … , Xu,t)⊤. The v × u matrix P consists of the coefficients pij, and Q is defined similarly. The u × v stoichiometry matrix S is defined by
The matrices P, Q and S will typically be sparse. On the occurrence of a reaction of type i, the system state, Xt, is updated by adding the ith column of S. Consequently, if ΔR is a v-vector containing the number of reaction events of each type in a given time interval, then the system state should be updated by ΔX, where
The stoichiometry matrix therefore encodes important structural information about the reaction network. In particular, vectors in the left null-space of S correspond to conservation laws in the network. That is, any u-vector a that satisfies a⊤S = 0 has the property (clear from the above equation) that a⊤Xt remains constant for all t.
Under the standard assumption of mass-action stochastic kinetics, each reaction ℛi is assumed to have an associated rate constant, ci, and a propensity function, hi(Xt, ci), giving the overall hazard of a type i reaction occurring. That is, the system is a Markov jump process (MJP), and for an infinitesimal time increment dt, the probability of a type i reaction occurring in the time interval (t,t + dt] is hi(Xt, ci)dt. The hazard function for a particular reaction of type i takes the form of the rate constant multiplied by a product of binomial coefficients expressing the number of ways in which the reaction can occur, that is,
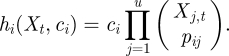 |
It should be noted that this hazard function differs slightly from the standard mass action rate laws used in continuous deterministic modelling, but is consistent (up to a constant of proportionality in the rate constant) asymptotically in the high concentration limit. Let c = (c1, c2, … , cv)⊤ and h(Xt,c) = (h1(Xt, c1),h2(Xt, c2), … , hv(Xt, cv))⊤. Values for c and the initial system state X0 = x0 complete specification of the Markov process. Although this process is rarely analytically tractable for interesting models, it is straightforward to forward-simulate exact realizations of this Markov process using a discrete event simulation method. This is due to the fact that if the current time and state of the system are t and Xt, respectively, then the time to the next event will be exponential with rate parameter
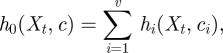 |
and the event will be a reaction of type ℛi with probability hi(Xt, ci)/h0(Xt, c) independently of the waiting time. Forwards simulation of process realizations in this way is typically referred to as Gillespie's direct method in the stochastic kinetics literature, after Gillespie [4]. See Wilkinson [3] for further background on stochastic kinetic modelling.
In fact, the assumptions of mass-action kinetics, as well as the one-to-one correspondence between reactions and rate constants may both be relaxed. All of what follows is applicable to essentially arbitrary v-dimensional hazard functions h(Xt, c).
The central problem considered in this paper is that of inference for the stochastic rate constants, c, given some time course data on the system state, Xt. It is therefore most natural to first consider inference for the earlier-mentioned MJP SKM. As demonstrated by Boys et al. [6], exact Bayesian inference in this setting is theoretically possible. However, the problem appears to be computationally intractable for models of realistic size and complexity, due primarily to the difficulty of efficiently exploring large integer lattice state space trajectories. It turns out to be more tractable (though by no means straightforward) to conduct inference for a continuous state Markov process approximation to the MJP model. Construction of this diffusion approximation, known as the chemical Langevin equation (CLE), is the subject of the next section.
2.1. The diffusion approximation
The diffusion approximation to the MJP can be constructed in a number of more or less formal ways. We will present here an informal intuitive construction, and then provide brief references to more rigorous approaches.
Consider an infinitesimal time interval, (t,t + dt]. Over this time, the reaction hazards will remain constant almost surely. The occurrence of reaction events can therefore be regarded as the occurrence of events of a Poisson process with independent realizations for each reaction type. Therefore, if we write dRt for the v-vector of the number of reaction events of each type in the time increment, it is clear that the elements are independent of one another and that the ith element is a Po(hi(Xt, ci)dt) random quantity. From this, we have that E(dRt) = h(Xt, c)dt and Var(dRt) = diag{h(Xt, c)} dt. It is therefore clear that
is the Itô SDE that has the same infinitesimal mean and variance as the true MJP (where dWt is the increment of a v-dimensional Brownian motion). Now since dXt = SdRt, we obtain
| 2.1 |
where now Xt and Wt are both u-vectors. Equation (2.1) is the SDE most commonly referred to as the CLE, and represents the diffusion process that most closely matches the dynamics of the associated MJP, and can be shown to approximate the SKM increasingly well in high concentration scenarios [8]. In particular, while it relaxes the assumption of discrete states, it keeps all of the stochasticity associated with the discreteness of state in its noise term. It also preserves many of the important structural properties of the MJP. For example, equation (2.1) has the same conservation laws as the original SKM. However, it should be noted that the approximation breaks down in low-concentration scenarios, and therefore should not be expected to work well for models involving species with very low copy-number. This is quite typical for many SKMs; yet the approximation often turns out to be adequate for inferential purposes in practice.
More formal approaches to the construction of the CLE usually revolve around the Kolmogorov forward equations for the Markov processes. The Kolmogorov forward equation for the MJP is usually referred to in this context as the chemical master equation. A second-order Taylor approximation to this system of differential equations can be constructed, and compared with the corresponding forward equation for an SDE model (known in this context as the Fokker–Planck equation). Matching the second-order approximation to the Fokker–Planck equation leads to the same CLE (2.1), as presented earlier. See Gillespie [8,9] for further details.
3. Bayesian inference
Suppose that the MJP X = {Xt|1 ≤ t ≤ T} is not observed directly, but observations (on a regular grid) y = {yt|t = 1, 2, … ,T} are available and assumed conditionally independent (given X), with conditional probability distribution obtained via the observation equation
| 3.1 |
Here, we take Yt to be a length-p vector, F is a constant matrix of dimension u × p and ɛt is a length-p Gaussian random vector. This flexible setup allows for observing only a subset of components of Xt, and taking F to be the u × u identity matrix corresponds to the case of observing all components of Xt (subject to error). Note that in the case of unknown measurement error variance, the parameter vector c can be augmented to include the elements of Σ. Bayesian inference may then proceed through the posterior density
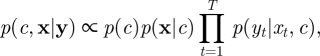 |
3.2 |
where p(c) is the prior density ascribed to c, p(x|c) is the probability of the MJP and p(yt|xt, c) is the observation density constructed from equation (3.1), which we let depend explicitly on c for the purposes of generality. Since the posterior in equation (3.2) will typically be unavailable in closed form, samples are usually generated from p(c,x|y) through a suitable MCMC scheme.
3.1. Likelihood-free/plug-and-play methods
One of the problems with standard approaches to using MCMC for inference in realistic data-poor scenarios is the difficulty of developing algorithms to explore a huge (often discrete) state space with a complex likelihood structure that makes conditional simulation difficult. Such problems arise frequently, and in recent years, interest has increasingly turned to methods that avoid some of the complexity of the problem by exploiting the fact that we are easily able to forward-simulate realizations of the process of interest. Methods such as likelihood-free MCMC (LF-MCMC) [10] and approximate Bayesian computation [11] are now commonly used to tackle Bayesian inference problems, which would be extremely difficult to solve otherwise. Although not the main focus of this paper, it is of course possible to develop similar computationally intensive algorithms from a non-Bayesian standpoint, where such likelihood-free approaches are sometimes termed ‘plug-and-play’; see Ionides et al. [12], He et al. [13] and Wood [14] for further details.
A likelihood-free approach to this problem can be constructed as follows. Suppose that interest lies in the posterior distribution p(c,x|y). A Metropolis–Hastings (MH) scheme can be constructed by proposing a joint update for c and x as follows. Supposing that the current state of the Markov chain is (c,x), first sample a proposed new value for c, c★, by sampling from some (essentially) arbitrary proposal distribution q(c★|c). Then, conditional on this newly proposed value, sample a proposed new sample path, x★, by forwards simulation from the model p(x★|c★). Together, the newly proposed pair (c★,x★) is accepted with probability min{1,A}, where
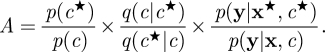 |
Crucially, the potentially problematic likelihood term, p(x|c), does not occur in the acceptance probability, owing to the fact that a sample from it was used in the construction of the proposal. Note that choosing an independence proposal of the form q(c★|c) = p(c★) leads to the simpler acceptance ratio
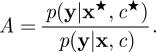 |
This ‘canonical’ choice of proposal will not be ‘optimal’, but lends itself to more elaborate schemes, as we will consider shortly.
3.2. Sequential likelihood-free Markov chain Monte Carlo
The basic LF-MCMC scheme discussed earlier might perform reasonably well, provided that y is not high-dimensional, and there is sufficient ‘noise’ in the measurement process to make the probability of acceptance non-negligible. However, in practice y is often of sufficiently large dimension that the overall acceptance rate of the scheme is intolerably low. In this case, it is natural to try and ‘bridge’ between the prior and the posterior with a sequence of intermediate distributions. There are several ways to do this, but here it is most natural to exploit the Markovian nature of the process and consider the sequence of posterior distributions obtained as each additional time point is observed. Define the data up to time t as yt = {y1, … , yt}. Also, define sample paths xt ≡{xs|t − 1< s ≤ t}, t = 1, 2, … , so that x = {x1, x2, … }. The posterior at time t can then be computed inductively as follows.
Assume at time t that we have a (large) sample from p(c,xt|yt) (for t = 0, initialize with sample from the prior, p(c)p(x0|c)).
- Run an MCMC algorithm that constructs a proposal in two stages:
- First sample (c★, xt★) ∼ p(c,xt|yt) by picking at random and perturbing c★ slightly (sampling from a kernel density estimate of the distribution).
- Next sample xt+1★ by forward simulation from p(xt+1★|c★, xt★).
- Accept/reject (c★, xt+1★) with probability min {1,A}, where
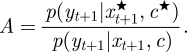
Output the sample from p(c,xt+1|yt+1), put t := t + 1, return to step 2.
Consequently, for each observation yt, an MCMC algorithm is run that takes as input the current posterior distribution prior to observation of yt and outputs the posterior distribution given all observations up to yt. As yt is typically low-dimensional, this strategy usually leads to good acceptance rates.
This algorithm has been applied successfully to biochemical network inference [15], but suffers from two different problems as the problem size increases. First, as the number of parameters (the dimension of c) increases, the algorithm suffers from the usual ‘curse of dimensionality’ as it becomes increasingly difficult to effectively cover the parameter space with a Monte Carlo sample. Second, as the number of time points increases, the method suffers from the ‘particle degeneracy’ problem that is well known to affect sequential Monte Carlo (SMC) algorithms targeting fixed model parameters [16]. The jittering approach used in step 2(a) can alleviate the problem somewhat [17] but does not completely overcome it. Both of these problems can be addressed by using particle MCMC methods [18], and by the particle marginal Metropolis–Hastings (PMMH) algorithm, in particular.
4. Particle marginal Metropolis–Hastings
Suppose that interest is in the marginal parameter posterior
where p(c,x|y) is given in equation (3.2). If p(c|y) can be evaluated (up to proportionality), then an MH scheme with arbitrary proposal density q(c★|c) has an acceptance ratio of the form
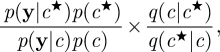 |
where we have used the standard decomposition p(c|y) ∝ p(y|c)p(c). Of course, in practice the marginal-likelihood term p(y|c) is difficult to compute exactly. Some progress can be made by considering the pseudo-marginal MH method described in Beaumont [19] and Andrieu & Roberts [20]. Here, the intractable term p(y|c) in the acceptance probability is replaced with a Monte Carlo estimate 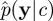 to become
to become
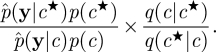 |
4.1 |
Provided that the corresponding estimator is unbiased (or has a constant bias that does not depend on c), it is possible to verify that the method targets the marginal p(c|y). By considering 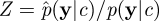 and augmenting the state space of the chain to include Z, it is straightforward to rewrite the acceptance ratio in equation (4.1) to find that the chain targets the joint density
and augmenting the state space of the chain to include Z, it is straightforward to rewrite the acceptance ratio in equation (4.1) to find that the chain targets the joint density
Marginalizing over Z then gives
Clearly, if E(Z|c) is a constant independent of c, then the chain targets the required marginal.
A closely related approach is the PMMH scheme of Andrieu et al. [7,18]. This method uses an SMC estimate of marginal likelihood 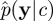 in the acceptance ratio (4.1). The PMMH scheme targets the correct marginal p(c|y) since the SMC scheme can be constructed to give an unbiased estimate of the marginal likelihood p(y|c) under some fairly mild conditions involving the resampling scheme [21]. It should also be noted that the PMMH scheme can be used to sample the joint posterior p(c,x|y). Essentially, a proposal mechanism of the form
in the acceptance ratio (4.1). The PMMH scheme targets the correct marginal p(c|y) since the SMC scheme can be constructed to give an unbiased estimate of the marginal likelihood p(y|c) under some fairly mild conditions involving the resampling scheme [21]. It should also be noted that the PMMH scheme can be used to sample the joint posterior p(c,x|y). Essentially, a proposal mechanism of the form 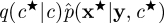 , where
, where 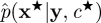 is an SMC approximation of p(x★|y,c★), is used. The resulting MH acceptance ratio is given by equation (4.1). Full details of the PMMH scheme including a proof establishing that the method leaves the target p(c,x|y) invariant can be found in Andrieu et al. [18].
is an SMC approximation of p(x★|y,c★), is used. The resulting MH acceptance ratio is given by equation (4.1). Full details of the PMMH scheme including a proof establishing that the method leaves the target p(c,x|y) invariant can be found in Andrieu et al. [18].
The general PMMH algorithm is described in appendix A.1 and we use the special case in which only samples from the marginal parameter posterior are required, as the basis for our inference scheme. A key ingredient of the PMMH algorithm is the construction of an SMC estimate of p(y|c). We therefore outline this approach in detail for some specific models.
4.1. Particle marginal Metropolis–Hastings for discrete stochastic kinetic models
Implementation of the PMMH scheme requires an SMC approximation of p(x|y,c) and the filter's estimate of the marginal likelihood p(y|c). Note that if interest is only in the marginal posterior p(c|y), then it suffices that we can calculate the SMC estimate of p(y|c). We now give a brief account of the SMC scheme and refer the reader to Doucet et al. [16] for further details.
An SMC scheme targeting p(x|y,c) can be constructed in the following way. At time t + 1, we observe yt+1 and our goal is to generate a sample from p(xt+1|yt+1, c) where, as before, we define xt+1 = {xs|t < s ≤ t + 1} and yt+1 = {ys|s = 1, 2, … , t + 1}. We have (up to proportionality),
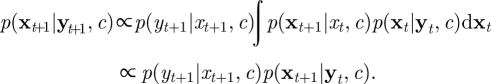 |
4.2 |
However, for general problems of interest, p(xt|yt, c) does not have analytic form. The SMC scheme therefore approximates p(xt|yt, c) by the cloud of points or ‘particles’ {xt1, … , xtN} with each particle xti having probability mass wti = 1/N. Hence the predictive density p(xt+1|yt, c) is approximated by
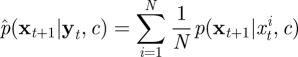 |
4.3 |
and equation (4.2) is replaced with
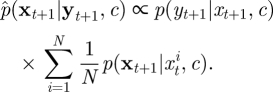 |
4.4 |
This approximation can be sampled for example via MCMC, using an algorithm similar to the one described in §3.2. We note, however, that the importance resampling strategy described by Gordon et al. [22] is simple to implement and permits straightforward calculation of an unbiased estimate of marginal likelihood, p(y| c). The basic idea is to use the (approximate) predictive 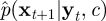 as an importance density, and this task is made easy by the ability to simulate from p(xt+1|xti, c) using the Gillespie algorithm with initial condition xti. The weights required for the resampling step at time t + 1, wt+1i, are proportional to p(yt+1|xt+1i, c), where each xt+1i is the final component of xt+1i generated from equation (4.3). We resample with replacement among the new particle set {xt+11, … , xt+1N} using the corresponding (normalized) weights as probabilities. Hence, an approximate sample of size N can be generated from (4.2) for each t = 1, 2, … ,T, after initializing the scheme with a sample from the initial density p(x1). Once all T observations have been assimilated, the filter's estimate of p(x|y,c) can be sampled (if required) by drawing uniformly from the set {x1, … , xN}. Note that to this end, it is necessary to keep track of the genealogy of particles over time. The algorithmic form of this simple SMC algorithm is given in appendix A.2.
as an importance density, and this task is made easy by the ability to simulate from p(xt+1|xti, c) using the Gillespie algorithm with initial condition xti. The weights required for the resampling step at time t + 1, wt+1i, are proportional to p(yt+1|xt+1i, c), where each xt+1i is the final component of xt+1i generated from equation (4.3). We resample with replacement among the new particle set {xt+11, … , xt+1N} using the corresponding (normalized) weights as probabilities. Hence, an approximate sample of size N can be generated from (4.2) for each t = 1, 2, … ,T, after initializing the scheme with a sample from the initial density p(x1). Once all T observations have been assimilated, the filter's estimate of p(x|y,c) can be sampled (if required) by drawing uniformly from the set {x1, … , xN}. Note that to this end, it is necessary to keep track of the genealogy of particles over time. The algorithmic form of this simple SMC algorithm is given in appendix A.2.
After assimilating the information in all T observations, the filter's estimate of marginal likelihood p(y|c) is obtained as
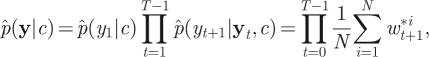 |
4.5 |
where w*it+1=p(yt+1|xit+1,c). Perhaps surprisingly, it turns out that this estimator of marginal likelihood is exactly unbiased [23]. It is then straightforward to implement the PMMH scheme, using the estimate of marginal likelihood given by equation (4.5) in the acceptance probability in equation (4.1). It should be noted that unlike the sequential LF-MCMC scheme described in §3.2, PMMH does not suffer from the particle degeneracy problem. As discussed by Andrieu et al. [18], performance of the PMMH scheme should be roughly constant even as T increases, provided that N increases linearly with T. The mixing of the PMMH scheme, however, is likely to depend on the number of particles used in the SMC scheme. While the method can be implemented using just N = 1 particle (reducing exactly to the simple LF-MCMC scheme given in §3.1), the corresponding estimator of marginal likelihood will be highly variable, and the impact of this on the PMMH algorithm will be a poorly mixing chain. Using a large value of N is therefore desirable but will come at a greater computational cost, since every iteration of the PMMH scheme requires a run of the SMC scheme. A discussion of the relationship between the variability of the estimator of marginal likelihood and the efficiency of the pseudo-marginal approach can be found in Andrieu & Roberts [20].
We therefore consider using the PMMH algorithm in conjunction with an approximation to the SKM, namely, the CLE. This is the subject of the next section. Improvements to the efficiency of the SMC scheme (and therefore the PMMH scheme in turn) are considered in the context of the CLE in §4.3.
4.2. Particle marginal Metropolis–Hastings for the chemical Langevin equation
Consider the CLE (2.1) and write it as
where
We refer to α(Xt, c) as the drift and β(Xt, c) as the diffusion coefficient. Since the transition density associated with the process will typically be analytically intractable, performing parameter inference in this setting is non-trivial. Attempts to overcome this problem have included the use of estimating functions [24], simulated maximum-likelihood estimation [25,26] and Bayesian imputation approaches [27–29]. These methods are summarized by Sørensen [30]. A related approach to the work considered here can be found in Stramer & Bognar [31]. The latter use a pseudo-marginal approach to infer parameters in stochastic volatility type models. Monte Carlo methods that are both exact (and avoid the error associated with imputation approaches) and computationally efficient have been proposed by Beskos et al. [32]. A summary of this work and some extensions can be found in Sermaidis et al. [33]. Such methods are attractive but at present cannot be readily applied to the general CLE described here.
We therefore follow a Bayesian imputation approach by working with the Euler–Maruyama approximation with density p(· |x,c) such that
As before, suppose that we have observations on a regular grid, y = {yt|t = 1, 2, … , T}. To allow sufficient accuracy of the Euler–Maruyama approximation, we augment observed data by introducing m − 1 latent values between every pair of observations. At this point, it is helpful to redefine X = {Xt|t = 1, 1 + Δt, 1 + 2Δt, …, T} where Δt = 1/m. The joint posterior density for parameters and latent states is then given (up to proportionality) as
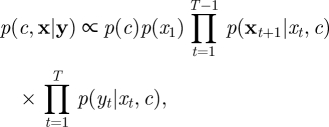 |
4.6 |
where
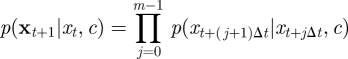 |
4.7 |
and we redefine xt+1 = {xs|s = t + Δt,t + 2Δt, … , t + 1} to be the skeleton path on (t,t + 1]. The PMMH scheme can be used to sample p(c, x|y) and requires an SMC scheme targeting p(x|y, c). A simple SMC strategy is to follow the approach described in §4.1—that is, we recursively sample 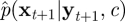 whose form is given by equation (4.4), with p(xt+1|xti, c) replaced by equation (4.7). Essentially, the Euler approximation is used to generate new values of the latent path, X, inside an SMC scheme. An estimate of marginal likelihood
whose form is given by equation (4.4), with p(xt+1|xti, c) replaced by equation (4.7). Essentially, the Euler approximation is used to generate new values of the latent path, X, inside an SMC scheme. An estimate of marginal likelihood 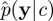 can be calculated via equation (4.5).
can be calculated via equation (4.5).
Note that the PMMH scheme permits a joint update of the parameters c and latent path X. This is particularly useful when working with the CLE owing to dependence between X and parameters in the diffusion coefficient β(Xt, c). This dependence is highlighted as a problem in Roberts & Stramer [28] and can result in poor mixing of Gibbs sampling strategies (such as those considered in Eraker [29] and Golightly & Wilkinson [34]) for large values of m corresponding to fine discretizations. Updating both c and X in a single block effectively side-steps this issue [31,35].
Choosing the value of m to balance accuracy and computational cost remains an open research problem. The error due to discretization is of order 1/m and it is therefore desirable to choose a sufficiently large value of m to allow the Euler approximation to be accurate. As discussed in Stramer & Bognar [31], one method is to look at the marginal parameter posteriors for increasing m. The authors suggest choosing a value m* such that (estimated) marginal posteriors are approximately the same for m ≥ m*.
4.3. Particle marginal Metropolis–Hastings using diffusion bridges
The mixing of the PMMH scheme will depend on the variability of the estimator of marginal likelihood, which in turn depends on the efficiency of the SMC scheme, which will in turn depend on the degree of noise in the measurement process. Methods based on blind forward simulation from the model will break down as measurement error decreases to zero. Although this is not a major problem in many practical applications, it is a matter of general concern. We therefore consider an improved SMC strategy where we use a discretization of a conditioned diffusion in the weighted resampling procedure. It should be noted that other improvements to the SMC scheme that do not rely on exploiting the structure of the CLE are possible. For example, residual and stratified resampling methods can reduce variance introduced by the resampling step [36,37]. A review of these methods can be found in the study of Doucet & Johansen [38].
Recall that the SMC approximation of p(xt+1|yt, c) is given (up to proportionality) as
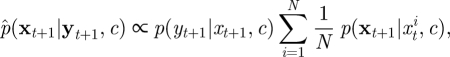 |
where p(xt+1|xt, c) is given in equation (4.7). Now note that
Although p(yt+1|xt, c) and p(xt+1|xt, yt+1, c) are typically intractable under the nonlinear structure of the CLE, the above equation can be exploited to reduce the variability of the unnormalized weights and in turn improve the estimator of marginal likelihood required in the PMMH scheme. A fully adapted auxiliary particle filter aims to sample xti with probability proportional to p(yt+1|xti, c) and simulate the latent path via p(xt+1|xti, yt+1, c), giving a corresponding normalized weight of wt+1i = 1. This approach was introduced by Pitt & Shephard [39] in the context of (nonlinear) state space models. See Pitt et al. [23] for a discussion of the use of an auxiliary particle filter inside an MH scheme. Naturally, the aim is to get as close to full adaption as possible. Here, we focus on the task of approximating p(xt+1|xt, yt+1, c) by a tractable Gaussian density. Denote the approximation by 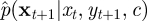 , which can be factorized as
, which can be factorized as
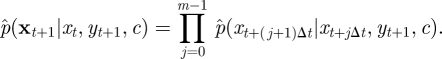 |
The approximation we require is derived in appendix A.3 and takes the form
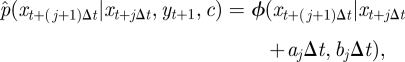 |
4.8 |
where ϕ(·|μ, Σ) denotes the Gaussian density with mean μ, variance Σ and aj, bj are given in appendix A.3 by equations (A 1) and (A 2). We use the density in equation (4.8) recursively to sample the latent skeleton path xt+1 conditional on xt and xt+1. We provide details of an SMC scheme that uses this construct in appendix A.3.
The PMMH algorithm can then be implemented by using the SMC scheme of appendix A.3 targeting p(x|y,c). If 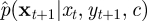 is close to p(xt+1|xt,yt+1, c), then the variance of the weights (relative to the variance of the weights obtained under the forward simulation approach described in §4.2) should be reduced. In turn, we would expect a reduction in the variance of the estimator of the marginal likelihood p(y|c).
is close to p(xt+1|xt,yt+1, c), then the variance of the weights (relative to the variance of the weights obtained under the forward simulation approach described in §4.2) should be reduced. In turn, we would expect a reduction in the variance of the estimator of the marginal likelihood p(y|c).
In the case of full observation and no error, aj and bj reduce to
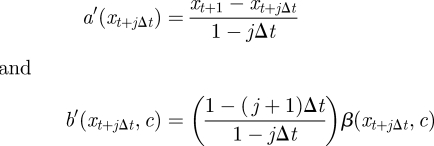 |
giving the form of the modified diffusion bridge (MDB) construct, which was first derived explicitly by Durham & Gallant [26]. Now consider the process {Xt′} satisfying the MDB discretization
This can be regarded as a discrete-time approximation of an SDE with limiting form [40]
which has the same diffusion coefficient as the conditioned SDE satisfied by {Xt}. Consequently, the law of one process is absolutely continuous with respect to the other [41]. The MDB construct therefore provides an efficient and appealing way of sampling the latent values within the SMC scheme. In particular, in situations involving low or no measurement error, use of the MDB as a proposal mechanism inside the SMC scheme ensures that as Δt → 0, the (unnormalized) weights approach a finite, non-zero limit. Sampling the latent values via the Euler approximation cannot be recommended in low noise situations. In this case, using a proposal mechanism which fails to condition on the observations results in an extremely inefficient sampling scheme. Examples in which the measurement error is varied are considered in §5. Finally, it should be noted that we preclude an observation regime where the dynamics of {X′t} between two consecutive measurements are dominated by the drift term α(Xt, c). It is likely that the MDB construct will perform poorly in such scenarios, as a′(Xt′) is independent of the drift term α(Xt, c). In this case, a better approach might be to use a mixture of the MDB construct and Euler forward simulator. See, for example, Fearnhead [42] for further details and a suggested approach.
5. Applications
In order to illustrate the inferential methods considered in §4, we consider the stochastic Lotka–Volterra model examined by Boys et al. [6] and a simple prokaryotic auto-regulatory model introduced in Golightly & Wilkinson [34]. We examine parameter inference in data-poor scenarios. Note that we eschew the sequential MCMC scheme described in §3.2 in favour of the numerically stable PMMH scheme.
5.1. Lotka–Volterra
We consider a simple model of predator and prey interaction comprising three reactions:
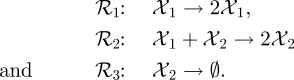 |
Denote the current state of the system by X = (X1, X2)⊤ where we have dropped dependence of the state on t for notational simplicity. The stoichiometry matrix is given by
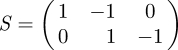 |
and the associated hazard function is
The diffusion approximation can be calculated by substituting equations (5.1) and (5.2) into the CLE (2.1) to give respective drift and diffusion coefficients of
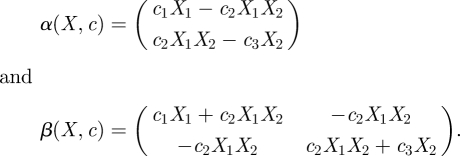 |
5.1.1. Results
We consider two synthetic datasets  and
and  , consisting of 50 observations at integer times on prey and predator levels simulated from the SKM using the Gillespie algorithm and corrupted with zero mean Gaussian noise. The observation equation (3.1) is therefore
, consisting of 50 observations at integer times on prey and predator levels simulated from the SKM using the Gillespie algorithm and corrupted with zero mean Gaussian noise. The observation equation (3.1) is therefore
where Xt = (X1,t, X2,t)⊤, ɛt ∼ N(0,σ2). We took σ2 = 10 to construct the first dataset 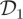 and σ2 = 200 to construct
and σ2 = 200 to construct 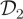 . In both cases, we assume σ2 to be known. True values of the rate constants (c1, c2, c3)⊤ were taken to be 0.5, 0.0025 and 0.3 following Boys et al. [6]. Initial latent states x1,0 and x2,0 were taken to be 100.
. In both cases, we assume σ2 to be known. True values of the rate constants (c1, c2, c3)⊤ were taken to be 0.5, 0.0025 and 0.3 following Boys et al. [6]. Initial latent states x1,0 and x2,0 were taken to be 100.
We ran the PMMH scheme for the SKM, CLE and CLE using the bridging strategy for 5 × 105 iterations. A Metropolis random walk update with variance tuned from a short pilot run was used to propose log(c). Independent proper Uniform U(−7,2) priors were taken for each log(ci). The SMC scheme employed at each iteration of PMMH used N = 100 particles. After discarding a number of iterations as burn-in and thinning the output, a sample of 10 000 draws with low auto-correlations was obtained for each scheme. When working with the CLE, we must specify a value of m to determine the size of the Euler time step Δt. We report results for m = 5 only (and therefore Δt = 0.2) but note that these are not particularly sensitive to m > 5.
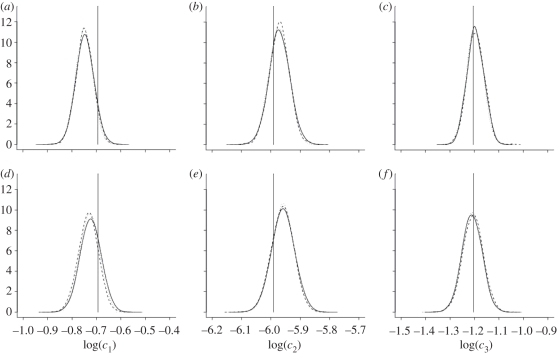
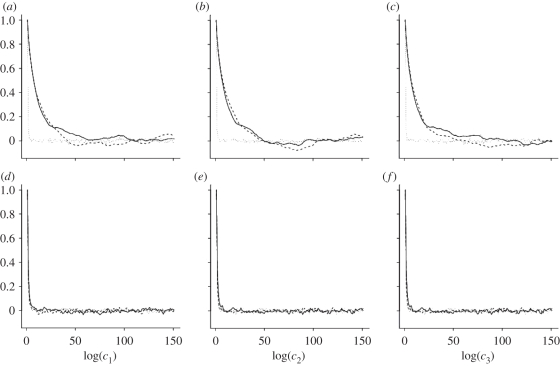
Figure 1 and table 1 summarize the output of the PMMH scheme when applied to the SKM, CLE and CLE with the bridge proposal mechanism. Inspection of the kernel density estimates of the marginal densities of each log(ci) given in figure 1 shows little difference in posteriors obtained under the two inferential models (SKM and CLE) for both synthetic datasets. Auto-correlations are reported in figure 2. Not surprisingly, for dataset 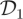 , which represents high signal to noise σ2 = 10, failure to condition on the observations in the proposal mechanism when applying the PMMH strategy to the SKM and CLE results in comparatively poorly mixing chains. Mixing is much improved when using the bridging strategy for the CLE. Of course, as the signal to noise is weakened, as in dataset
, which represents high signal to noise σ2 = 10, failure to condition on the observations in the proposal mechanism when applying the PMMH strategy to the SKM and CLE results in comparatively poorly mixing chains. Mixing is much improved when using the bridging strategy for the CLE. Of course, as the signal to noise is weakened, as in dataset 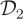 with σ2 = 200, there is fairly little to be gained in terms of mixing, by conditioning on the observations in the proposal mechanism.
with σ2 = 200, there is fairly little to be gained in terms of mixing, by conditioning on the observations in the proposal mechanism.
Figure 1.
Marginal posterior distributions based on the output of the PMMH scheme for the SKM (solid), CLE (dashed) and CLE with bridging strategy (dotted) using synthetic data generated from the Lotka–Volterra model with (a–c) σ2 = 10 and (d–f) σ2 = 200. Values of each log(ci) that produced the data are indicated.
Table 1.
Marginal posterior means and standard deviations for log(ci) from the output of the PMMH scheme under three different models using synthetic data generated from the Lotka–Volterra SKM. The ESS rows show effective sample size of each chain per 1000 iterations. The adjusted effective sample size is shown in the ESSadj rows.
| log(c1) | log(c2) | log(c3) | log(c1) | log(c2) | log(c3) | |
|---|---|---|---|---|---|---|
| SKM (σ2 = 10) | SKM (σ2 = 200) | |||||
| mean | −0.746 | −5.970 | −1.195 | −0.723 | −5.958 | −1.210 |
| s.d. | 0.035 | 0.034 | 0.034 | 0.040 | 0.037 | 0.040 |
| ESS | 51.377 | 52.191 | 54.924 | 459.193 | 432.287 | 417.818 |
| ESSadj | 0.856 | 0.867 | 0.915 | 7.653 | 7.205 | 6.964 |
| CLE (σ2 = 10) | CLE (σ2 = 200) | |||||
| mean | −0.747 | −5.971 | −1.194 | −0.733 | −5.961 | −1.206 |
| s.d. | 0.035 | 0.034 | 0.035 | 0.040 | 0.037 | 0.041 |
| ESS | 50.453 | 47.200 | 47.917 | 611.758 | 585.064 | 604.966 |
| ESSadj | 3.604 | 3.371 | 3.423 | 43.687 | 41.790 | 43.212 |
| CLE (bridge, σ2 = 10) | CLE (bridge, σ2 = 200) | |||||
| mean | −0.750 | −5.973 | −1.198 | −0.728 | −5.960 | −1.210 |
| s.d. | 0.034 | 0.033 | 0.034 | 0.040 | 0.037 | 0.040 |
| ESS | 823.902 | 814.871 | 852.608 | 606.988 | 666.097 | 676.050 |
| ESSadj | 24.967 | 24.693 | 25.837 | 18.394 | 20.185 | 20.304 |
Figure 2.
Auto-correlations based on the output of the PMMH scheme for the SKM (solid), CLE (dashed) and CLE with bridging strategy (dotted) using synthetic data generated from the Lotka–Volterra model with (a–c) σ2 = 10 and (d–f) σ2 = 200.
All algorithms are coded in C and implemented on a desktop personal computer with a 2.83 GHz clock speed. Computational cost of implementing the PMMH scheme for the SKM, CLE and CLE (bridge) scales roughly as 4.29 : 1 : 2.36 and it takes approximately 14 s to perform 1000 iterations of the CLE scheme. Naturally, computational cost of the PMMH scheme applied to the SKM depends on the values of the rate constants and latent states that are consistent with the data, as these determine the number of reaction events per simulation. To compare each scheme, we report the effective sample size (ESS) [43] in table 2. We see that when using 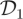 with σ2 = 10, the CLE (bridge) algorithm clearly outperforms SKM and CLE in terms of ESS. When using
with σ2 = 10, the CLE (bridge) algorithm clearly outperforms SKM and CLE in terms of ESS. When using 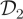 with σ2 = 200, the performance of the CLE approach is comparable to that of the CLE (bridge) in terms of ESS. We also provide an adjusted effective sample size ESSadj by taking the ratio of ESS to CPU time. In terms of ESSadj, the CLE (bridge) approach outperforms both the CLE and SKM methods when using dataset
with σ2 = 200, the performance of the CLE approach is comparable to that of the CLE (bridge) in terms of ESS. We also provide an adjusted effective sample size ESSadj by taking the ratio of ESS to CPU time. In terms of ESSadj, the CLE (bridge) approach outperforms both the CLE and SKM methods when using dataset 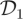 , with typical ESSadj values 27 times larger than those obtained under the SKM and around seven times larger than those under the CLE approach. For the low signal-to-noise case (
, with typical ESSadj values 27 times larger than those obtained under the SKM and around seven times larger than those under the CLE approach. For the low signal-to-noise case (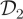 ), there is little to be gained by conditioning on the observations in the proposal mechanism, and the CLE approach is therefore to be preferred.
), there is little to be gained by conditioning on the observations in the proposal mechanism, and the CLE approach is therefore to be preferred.
Table 2.
Marginal posterior means and standard deviations for log(ci) from the output of the PMMH scheme under three different models using synthetic data generated from the prokaryotic auto-regulatory network. The ESS rows show effective sample size of each chain per 1000 iterations. The adjusted effective sample size is shown in the ESSadj rows.
| log(c1) | log(c2) | log(c3) | log(c4) | log(c7) | log(c8) | |
|---|---|---|---|---|---|---|
| SKM | ||||||
| mean | −1.888 | −0.437 | −4.789 | −0.867 | −1.742 | −3.630 |
| s.d. | 1.000 | 0.767 | 0.933 | 0.630 | 0.725 | 0.368 |
| ESS | 16.501 | 27.31 | 25.599 | 29.321 | 28.862 | 55.148 |
| ESSadj | 0.220 | 0.364 | 0.341 | 0.390 | 0.384 | 0.734 |
| CLE | ||||||
| mean | −2.624 | −1.176 | −4.644 | −1.215 | −1.995 | −3.775 |
| s.d. | 0.695 | 0.610 | 1.158 | 0.962 | 0.780 | 0.755 |
| ESS | 17.842 | 37.450 | 20.531 | 16.941 | 25.547 | 24.209 |
| ESSadj | 0.317 | 0.666 | 0.513 | 0.301 | 0.454 | 0.430 |
| CLE (bridge) | ||||||
| mean | −2.574 | −1.214 | −4.718 | −1.102 | −1.982 | −3.812 |
| s.d. | 0.682 | 0.554 | 1.014 | 0.777 | 0.729 | 0.736 |
| ESS | 61.091 | 115.412 | 57.74 | 40.791 | 98.741 | 61.790 |
| ESSadj | 0.469 | 0.886 | 0.443 | 0.313 | 0.758 | 0.474 |
5.2. Prokaryotic auto-regulation
Genetic regulation is a notoriously complex biochemical process, especially in eukaryotic organisms [44]. Even in prokaryotes, there are many mechanisms used, not all of which are fully understood. However, one commonly used mechanism for auto-regulation in prokaryotes that has been well studied and modelled is a negative feedback mechanism whereby dimers of a protein repress its own transcription. The classic example of this is the λ repressor protein cI of phage λ in Escherichia coli, originally modelled stochastically by Arkin et al. [45]. Here we consider a simplified model for such a prokaryotic auto-regulation, based on this mechanism of dimers of a protein coded for by a gene repressing its own transcription. The full set of reactions in this simplified model are as follows:
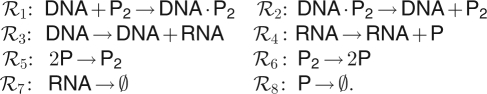 |
See Golightly & Wilkinson [34] for further explanation. We order the variables as X = (RNA, P, P2, DNA · P2, DNA)⊤, giving the stoichiometry matrix for this system:
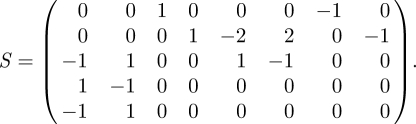 |
The associated hazard function is given by
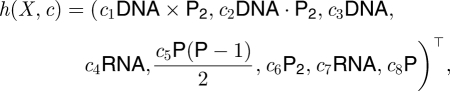 |
using an obvious notation.
Like many biochemical network models, this model contains conservation laws leading to rank degeneracy of the stoichiometry matrix, S. The diffusion bridge method considered in §4.3 is the simplest to implement in the case of models of full rank. This is without loss of generality, as we can simply strip out redundant species from the rank-deficient model. Here, there is just one conservation law,
where k is the number of copies of this gene in the genome. We can use this relation to remove DNA · P2 from the model, replacing any occurrences of DNA · P2 in rate laws with k − DNA. This leads to a reduced full-rank model with species X = (RNA, P, P2, DNA)⊤, stoichiometry matrix
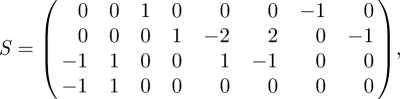 |
5.1 |
and associated hazard function
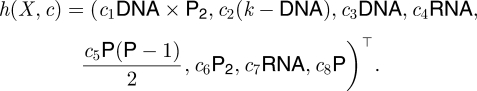 |
5.2 |
We can then obtain the diffusion approximation by substituting equations (5.1) and (5.2) into the CLE (2.1). Although greatly simplified relative to our current understanding of the full biological process, this model captures the key interactions and feedbacks characteristic of the real system being modelled.
5.2.1. Results
We consider a challenging data-poor scenario by analysing a synthetic dataset consisting of 100 observations at integer times of total protein numbers simulated from the SKM using the Gillespie algorithm. The observation equation (3.1) becomes
where Xt = (RNAt, Pt, (P2)t, DNAt)⊤, ɛt ∼ N(0,σ2) and we take σ2 = 4 to be known. Hence, we have observations on Pt + 2(P2)t subject to error. Again, this is a simplification of a real single-cell measurement scenario, but captures many important features. First, it captures the partial nature of the observation process; this is a very data-poor scenario, typical of real measurement systems. Second, it attempts to model a reporter mechanism where the protein of interest is tagged to form a fusion with a fluorescent reporter, such as GFP, where it is not possible to distinguish monomers from dimers in the measured signal. True values of the rate constants (c1, … , c8)⊤ were taken to be 0.1, 0.7, 0.35, 0.2, 0.1, 0.9, 0.3 and 0.1 with the reversible dimerization rates c5 and c6 assumed known. Note that we take the conservation constant (that is, the number of copies of the gene on the genome) and the initial latent state x1 to be known with respective values of 10 and (8,8,8,5)⊤. Simulations from the SKM with these settings give inherently discrete time series of each species. This scenario should therefore be challenging for the CLE when used as an inferential model.
We ran the PMMH scheme for the SKM, CLE and CLE using the bridging strategy for 3 × 106 iterations. A Metropolis random walk update with variance tuned from a short pilot run was used to propose log(c) and independent proper Uniform U(−7,2) priors were taken for each log(ci). The SMC scheme employed at each iteration of PMMH used N = 100 particles. After discarding a number of iterations as burn-in and thinning the output, a sample of 10 000 draws with low auto-correlations was obtained for each scheme. As before, we must specify a value of m to determine the size of the Euler time step Δt to use in the numerical solution of the CLE. We took m = 5 (and therefore Δt = 0.2) to limit computational cost.
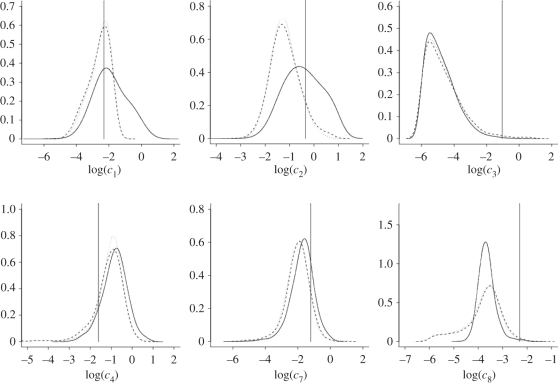
Figure 3 and table 2 summarize the output of the PMMH scheme for each inferential model. Kernel density estimates of the marginal densities of each log(ci) are given in figure 3. We see that sampled values of each parameter are consistent with the true values used to produce the synthetic data. In addition, it appears that little is lost by ignoring the inherent discreteness in the data. The output of the PMMH scheme when using the CLE is largely consistent with that obtained under the SKM (see also marginal posterior means and standard deviations in table 2). Not surprisingly, kernel densities obtained under the CLE and CLE (bridge) methods match up fairly well as both schemes are designed to sample the same invariant distribution.
Figure 3.
Marginal posterior distributions based on the output of the PMMH scheme for the SKM (solid), CLE (dashed) and CLE with bridging strategy (dotted) using synthetic data generated from the prokaryotic auto-regulatory network. Values of each log(ci) that produced the data are indicated.
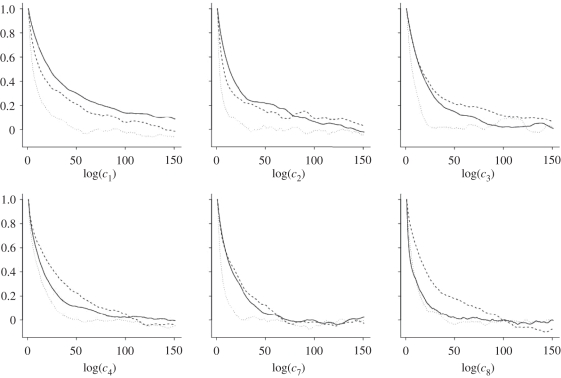
Computational cost of implementing the PMMH scheme for the SKM, CLE and CLE (bridge) scales as 1 : 0.75 : 1.73 with 1000 iterations of the CLE approach requiring approximately 56 s of CPU time. However, it is clear from the auto-correlation plots in figure 4 that the mixing of the chains under the CLE (bridge) method is far better than that obtained under the SKM and CLE. Table 2 gives the ESS for which the CLE (bridge) algorithm clearly outperforms SKM and CLE. Again, adjusted ESSs are provided. In terms of computational performance, the CLE approach outperforms SKM for four out of six parameters and the CLE (bridge) approach outperforms its vanilla counterpart for five out of six.
Figure 4.
Auto-correlations based on the output of the PMMH scheme for the SKM (solid), CLE (dashed) and CLE with bridging strategy (dotted) using synthetic data generated from the prokaryotic auto-regulatory network.
6. Discussion and conclusions
This paper has considered the problem of inference for the parameters of a very general class of Markov process models using time course data, and the application of such techniques to challenging parameter estimation problems in systems biology. We have seen how it is possible to develop extremely general ‘likelihood-free’ pMCMC algorithms based only on the ability to forward simulate from the Markov process model. Although these methods are extremely computationally intensive, they work very reliably, provided that the process is observed with a reasonable amount of measurement error. Exact likelihood-free approaches break down in low/no measurement error scenarios, but in this case it is possible to use established methods for sampling diffusion bridges in order to carry out inference for the parameters of a diffusion approximation to the true MJP model. Diffusion approximations to SKMs are useful for at least two reasons. The first reason is computational speed. As the complexity of the true model increases, exact simulation becomes intolerably slow, whereas (approximate, but accurate) simulation from the diffusion approximation remains computationally efficient. The second reason is tractability. There are well-established, computationally intensive methods for simulating from diffusion bridges, and these techniques allow the development of much more efficient pMCMC algorithms, especially in low/no measurement error scenarios. These more efficient schemes work well even in very challenging multivariate settings, and we expect that they will scale up to problems of relatively high dimension (i.e. 20 parameters and 50 species) without encountering serious problems.
Despite the relative efficiency of the pMCMC algorithms discussed here, the computational expense of pMCMC algorithms for complex models remains a limiting factor, and the development of software toolkits that make it easy to fully exploit the speed of modern processors and the parallelism of modern computing architectures is a high priority [46].
Acknowledgements
The second author (D.J.W.) was supported by a fellowship from the Biotechnology and Biological Sciences Research Council (BBSRC), grant no. BBF0235451, and the Statistical and Applied Mathematical Sciences Institute (SAMSI) programme on the Analysis of Object Data. The authors would also like to thank three anonymous referees, whose comments helped to greatly improve the clarity of this paper.
Appendix A
A.1. Particle marginal Metropolis–Hastings scheme
The PMMH scheme has the following algorithmic form.
1. Initialization, i = 0,
set c(0) arbitrarily and
run an SMC scheme targeting p(x|y,c(0)), sample
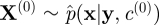 from the SMC approximation and let
from the SMC approximation and let 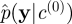 denote the marginal likelihood estimate.
denote the marginal likelihood estimate.
2. For iteration i ≥ 1,
sample c★ ∼ q(· |c(i−1)),
run an SMC scheme targeting p(x|y,c★), sample
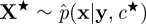 , let
, let 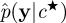 denote the marginal-likelihood estimate, and
denote the marginal-likelihood estimate, and- with probability min{1,A} where
accept a move to c★ and X★ otherwise store the current values.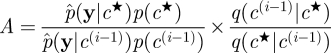
If interest lies purely in the marginal posterior p(c|y) then the scheme can be simplified by noting that in this case there is no requirement to sample or store draws of the latent states X.
A.2. Sequential Monte Carlo for the stochastic kinetic model
An SMC scheme based on the bootstrap filter of Gordon et al. [22] can be stated as follows.
1. Initialization.
Generate a sample of size N, {x11, … , x1N} from the initial density p(x1).
- Assign each x1i a (normalized) weight given by
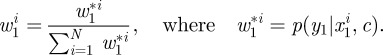
- Construct and store the currently available estimate of marginal likelihood,
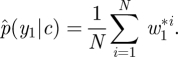
Resample N times with replacement from {x11, … , x1N} with probabilities given by {w11, … , w1N}.
2. For times t = 1, 2, … , T − 1.
For i = 1, … , N: draw Xt+1i ∼ p(xt+1|xti,c) using the Gillespie algorithm.
- Assign each xt+1i a (normalized) weight given by
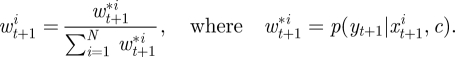
- Construct and store the currently available estimate of marginal likelihood,
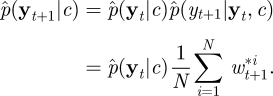
Resample N times with replacement from {xt+11, … , xt+1N} with probabilities given by {wt+11, … , wt+1N}.
A.3. Sequential Monte Carlo using diffusion bridges
We consider first the task of deriving the form of the density 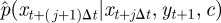 , which, when sampled recursively, gives the skeleton of a diffusion bridge. Following Wilkinson & Golightly [47], we derive the required density by constructing a Gaussian approximation to the joint density of Xt+(j+1)Δt and Yt+1 (conditional on Xt+jΔt and c). We have that
, which, when sampled recursively, gives the skeleton of a diffusion bridge. Following Wilkinson & Golightly [47], we derive the required density by constructing a Gaussian approximation to the joint density of Xt+(j+1)Δt and Yt+1 (conditional on Xt+jΔt and c). We have that
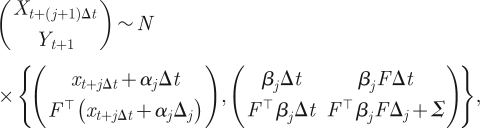 |
where Δj = 1 − jΔt and we use the short-hand notation αj = α (xt+jΔt, c) and βj = β (xt+jΔt, c). Conditioning on Yt+1 = yt+1 yields
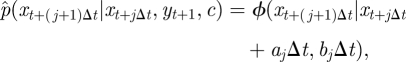 |
where
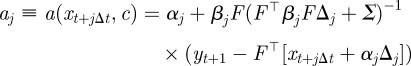 |
A 1 |
and
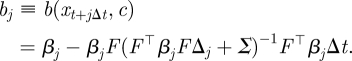 |
A 2 |
This density can be sampled recursively to produce a latent skeleton path xt+1 conditional on xt and xt+1. An SMC strategy that uses this construct can be obtained by replacing step 2 of the SMC algorithm described in appendix A.2 with the following.
2. For times t = 1, 2, … , T − 1.
For i = 1, … , N: draw
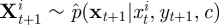 using equation (4.8) recursively.
using equation (4.8) recursively.- Assign each xt+1i a (normalized) weight given by
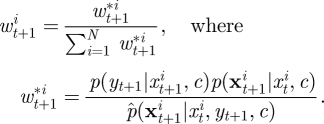
- Construct and store the currently available estimate of marginal likelihood,
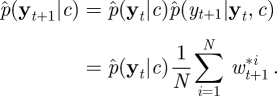
Resample N times with replacement from {xt+11, … , xt+1N} with probabilities given by {wt+11, … , wt+1N}.
Note that theorem 1 of Pitt et al. [23] establishes that the auxiliary particle filter (of which the algorithm presented above is a special case) gives an unbiased estimator of marginal likelihood.
References
- 1.Kitano H. 2002. Computational systems biology. Nature 420, 206–210 10.1038/nature01254 (doi:10.1038/nature01254) [DOI] [PubMed] [Google Scholar]
- 2.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. 2002. Stochastic gene expression in a single cell. Science 297, 1183–1186 10.1126/science.1070919 (doi:10.1126/science.1070919) [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson D. J. 2006. Stochastic modelling for systems biology. Boca Raton, FL: Chapman & Hall/CRC Press [Google Scholar]
- 4.Gillespie D. T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 10.1021/j100540a008 (doi:10.1021/j100540a008) [DOI] [Google Scholar]
- 5.Wilkinson D. J. 2009. Stochastic modelling for quantitative description of heterogeneous biological systems. Nat. Rev. Genet. 10, 122–133 10.1038/nrg2509 (doi:10.1038/nrg2509) [DOI] [PubMed] [Google Scholar]
- 6.Boys R. J., Wilkinson D. J., Kirkwood T. B. L. 2008. Bayesian inference for a discretely observed stochastic kinetic model. Stat. Comput. 18, 125–135 10.1007/s11222-007-9043-x (doi:10.1007/s11222-007-9043-x) [DOI] [Google Scholar]
- 7.Andrieu C., Doucet A., Holenstein R. 2009. Particle Markov chain Monte Carlo for efficient numerical simulation. In Monte Carlo and quasi-Monte Carlo methods 2008 (eds L'Ecuyer P., Owen A. B.), pp. 45–60 Berlin/Heidelberg, Germany: Springer [Google Scholar]
- 8.Gillespie D. T. 2000. The chemical Langevin equation. J. Chem. Phys. 113, 297–306 10.1063/1.481811 (doi:10.1063/1.481811) [DOI] [Google Scholar]
- 9.Gillespie D. T. 1992. A rigorous derivation of the chemical master equation. Phys. A 188, 404–425 10.1016/0378-4371(92)90283-V (doi:10.1016/0378-4371(92)90283-V) [DOI] [Google Scholar]
- 10.Marjoram P., Molitor J., Plagnol V., Tavare S. 2003. Markov chain Monte Carlo without likelihoods. Proc. Natl Acad. Sci. USA 100, 15 324–15 328 10.1073/pnas.0306899100 (doi:10.1073/pnas.0306899100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaumont M. A., Zhang W., Balding D. J. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ionides E. L., Breto C., King A. A. 2006. Inference for nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 103, 18 438–18 443 10.1073/pnas.0603181103 (doi:10.1073/pnas.0603181103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He D., Ionides E. L., King A. A. 2010. Plug-and-play inference for disease dynamics: measles in large and small populations as a case study. J. R. Soc. Interface 7, 271–283 10.1098/rsif.2009.0151 (doi:10.1098/rsif.2009.0151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood S. N. 2010. Statistical inference for noisy nonlinear ecological dynamic systems. Nature 466, 1102–1104 10.1038/nature09319 (doi:10.1038/nature09319) [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson D. J. 2011. Parameter inference for stochastic kinetic models of bacterial gene regulation: a Bayesian approach to systems biology (with discussion). In Bayesian statistics, vol. 9 (ed. Bernardo J. M.). Oxford, UK: Oxford Science Publications [Google Scholar]
- 16.Doucet A., de Freitas N., Gordon N. 2001. Sequential Monte Carlo methods in practice. Statistics for Engineering and Information Science New York, NY: Springer [Google Scholar]
- 17.Liu J., West M. 2001. Combined parameter and state estimation in simulation-based filtering. In Sequential Monte Carlo methods in practice (eds Doucet A., de Freitas N., Gordon N.), pp. 197–224 New York, NY: Springer [Google Scholar]
- 18.Andrieu C., Doucet A., Holenstein R. 2010. Particle Markov chain Monte Carlo methods (with discussion). J. R. Stat. Soc. B 72, 269–342 10.1111/j.1467-9868.2009.00736.x (doi:10.1111/j.1467-9868.2009.00736.x) [DOI] [Google Scholar]
- 19.Beaumont M. A. 2003. Estimation of population growth or decline in genetically monitored populations. Genetics 164, 1139–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrieu C., Roberts G. O. 2009. The pseudo-marginal approach for efficient computation. Ann. Stat. 37, 697–725 10.1214/07-AOS574 (doi:10.1214/07-AOS574) [DOI] [Google Scholar]
- 21.Del Moral P. 2004. Feynman-Kac formulae: genealogical and interacting particle systems with applications. New York, NY: Springer [Google Scholar]
- 22.Gordon N. J., Salmond D. J., Smith A. F. M. 1993. Novel approach to nonlinear/non-Gaussian Bayesian state estimation. IEE Proc. F 140, 107–113 [Google Scholar]
- 23.Pitt M. K., Silva R. S., Giordani P., Kohn R. Auxiliary particle filtering within adaptive Metropolis–Hastings sampling. 2011 See http://arxiv.org/abs/1006.1914. [Google Scholar]
- 24.Bibby B. M., Sørensen M. 1995. Martingale estimating functions for discretely observed diffusion processes. Bernouilli 1, 17–39 10.2307/3318679 (doi:10.2307/3318679) [DOI] [Google Scholar]
- 25.Pedersen A. 1995. A new approach to maximum likelihood estimation for stochastic differential equations based on discrete observations. Scand. J. Stat. 22, 55–71 [Google Scholar]
- 26.Durham G. B., Gallant R. A. 2002. Numerical techniques for maximum likelihood estimation of continuous time diffusion processes. J. Bus. Econ. Stat. 20, 279–316 10.1198/073500102288618397 (doi:10.1198/073500102288618397) [DOI] [Google Scholar]
- 27.Elerian O., Chib S., Shephard N. 2001. Likelihood inference for discretely observed non-linear diffusions. Econometrika 69, 959–993 10.1111/1468-0262.00226 (doi:10.1111/1468-0262.00226) [DOI] [Google Scholar]
- 28.Roberts G. O., Stramer O. 2001. On inference for non-linear diffusion models using Metropolis–Hastings algorithms. Biometrika 88, 603–621 10.1093/biomet/88.3.603 (doi:10.1093/biomet/88.3.603) [DOI] [Google Scholar]
- 29.Eraker B. 2001. MCMC analysis of diffusion models with application to finance. J. Bus. Econ. Stat. 19, 177–191 10.1198/073500101316970403 (doi:10.1198/073500101316970403) [DOI] [Google Scholar]
- 30.Sørensen H. 2004. Parametric inference for diffusion processes observed at discrete points in time. Int. Stat. Rev. 72, 337–354 10.1111/j.1751-5823.2004.tb00241.x (doi:10.1111/j.1751-5823.2004.tb00241.x) [DOI] [Google Scholar]
- 31.Stramer O., Bognar M. 2011. Bayesian inference for irreducible diffusion processes using the pseudo-marginal approach. Bayesian Anal. 6, 231–258 10.1214/11-BA608 (doi:10.1214/11-BA608) [DOI] [Google Scholar]
- 32.Beskos A., Papaspiliopoulos O., Roberts G. O., Fearnhead P. 2006. Exact and computationally efficient likelihood-based estimation for discretely observed diffusion processes. J. R. Stat. Soc. B 68, 333–382 10.1111/j.1467-9868.2006.00552.x (doi:10.1111/j.1467-9868.2006.00552.x) [DOI] [Google Scholar]
- 33.Sermaidis S., Papaspiliopoulos O., Roberts G. O., Beskos A., Fearnhead P. 2011. Markov chain Monte Carlo for exact inference for diffusions. See http://arxiv.org/abs/1102.5541. [Google Scholar]
- 34.Golightly A., Wilkinson D. J. 2005. Bayesian inference for stochastic kinetic models using a diffusion approximation. Biometrics 61, 781–788 10.1111/j.1541-0420.2005.00345.x (doi:10.1111/j.1541-0420.2005.00345.x) [DOI] [PubMed] [Google Scholar]
- 35.Golightly A., Wilkinson D. J. 2006. Bayesian sequential inference for nonlinear multivariate diffusions. Stat. Comput. 16, 323–338 10.1007/s11222-006-9392-x (doi:10.1007/s11222-006-9392-x) [DOI] [Google Scholar]
- 36.Kitagawa G. 1996. Monte Carlo filter and smoother for non-Gaussian nonlinear state space models. J. Comput. Graphical Stat. 5, 1–25 10.2307/1390750 (doi:10.2307/1390750) [DOI] [Google Scholar]
- 37.Liu J. S. 2001. Monte Carlo strategies in scientific computing. New York, NY: Springer [Google Scholar]
- 38.Doucet A., Johansen A. M. 2011. A tutorial on particle filtering and smoothing: fifteen years later. In The Oxford handbook of nonlinear filtering (eds Crisan D., Rozovsky B.), pp. 656–704 Oxford, UK: Oxford University Press [Google Scholar]
- 39.Pitt M. K., Shephard N. 1999. Filtering via simulation: auxiliary particle filters. J. Am. Stat. Assoc. 446, 590–599 10.2307/2670179 (doi:10.2307/2670179) [DOI] [Google Scholar]
- 40.Stramer O., Yan J. 2007. Asymptotics of an efficient Monte Carlo estimation for the transition density of diffusion processes. Methodol. Comput. Appl. Probab. 9, 483–496 10.1007/s11009-006-9006-2 (doi:10.1007/s11009-006-9006-2) [DOI] [Google Scholar]
- 41.Delyon B., Hu Y. 2006. Simulation of conditioned diffusion and application to parameter estimation. Stoch. Process. Appl. 116, 1660–1675 [Google Scholar]
- 42.Fearnhead P. 2008. Computational methods for complex stochastic systems: a review of some alternatives to MCMC. Stat. Comput. 18, 151–171 10.1007/s11222-007-9045-8 (doi:10.1007/s11222-007-9045-8) [DOI] [Google Scholar]
- 43.Plummer M., Best N., Cowles K., Vines K. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News 6, 7–11 [Google Scholar]
- 44.Latchman D. 2002. Gene regulation: a eukaryotic perspective, 4th edn. New York, NY: Garland Science [Google Scholar]
- 45.Arkin A., Ross J., McAdams H. H. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149, 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson D. J. 2005. Parallel Bayesian computation. In Handbook of parallel computing and statistics (ed. Kontoghiorghes E. J.), pp. 481–512 New York, NY: Marcel Dekker/CRC Press; 10.1201/9781420028683.ch16 (doi:10.1201/9781420028683.ch16) [DOI] [Google Scholar]
- 47.Wilkinson D. J., Golightly A. 2010. Markov chain Monte Carlo algorithms for SDE parameter estimation. In Learning and inference in computational systems biology (eds Lawrence N., Girolami M., Rattray M., Sanguinetti G.), pp. 253–275 Cambridge, MA: MIT Press [Google Scholar]