Abstract
The concepts of functional equivalence classes and information control in living systems are useful to characterize downward (or top-down) causation by feedback information control in synthetic biology. Herein, we re-analyse published experiments of microbiology and synthetic biology that demonstrate the existence of several classes of functional equivalence in microbial organisms. Classes of functional equivalence from the bacterial operating system, which processes and controls the information encoded in the genome, can readily be interpreted as strong evidence, if not demonstration, of top-down causation (TDC) by information control. The proposed biological framework reveals how this type of causality is put in action in the cellular operating system. Considerations on TDC by information control and adaptive selection can be useful for synthetic biology by delineating the irreducible set of properties that characterizes living systems. Through a ‘retro-synthetic’ biology approach, these considerations could contribute to identifying the constraints behind the emergence of molecular complexity during the evolution of an ancient RNA/peptide world into a modern DNA/RNA/protein world. In conclusion, we propose TDCs by information control and adaptive selection as the two types of downward causality absolutely necessary for life.
Keywords: RNA world, RNA evolution, information selection, molecular complexity, convergence, convergent evolution
1. Introduction
Despite being at the core of natural sciences, the understanding of the nature of causation has attracted the attention of a rather small number of scientists. However, with the rise of systems biology and synthetic biology that aim at understanding biological systems in a more global, holistic way, causal concepts could be helpful to those fascinated by problems related to the emergence of complexity in life. Understanding the nature of causation in living systems can provide the necessary framework to characterize the relationships existing between different levels of biological complexity. As the perceptible world around us is based on matter, most scientists usually assume that causal effects stem from the bottom up and that the complexity of living systems could be explained in terms of simple ‘bottom-up’ and ‘same-level’ causations. However, this present view likely reflects only parts of the truth and can be put to the test by the fields of systems biology [1,2] and synthetic biology [3–6]. For instance, the recent area of synthetic biology aims at designing, synthesizing and engineering new biological functions and systems not found in nature. Therefore, one can get a better understanding of how new functions and systems should be integrated to fulfil the properties of life.
For most scientists interested by questions related to the problem of emergence of complexity, it is generally accepted that the development of emergent properties, which is clearly a bottom-up (or upward) causality, is influenced by top-down (or downward) causality [7–10]. While bottom-up causation is the ability of lower levels of reality to have causal power over higher levels, top-down causation (TDC) is typically defined as the ability of a higher hierarchic level to have causal power over lower levels. Therefore, while biological components (for example, proteins and RNA molecules) have causal effects on the functioning of a whole biological system, it is also apparent that the whole, as a context, can have an effect on the lower components through boundaries and constraints that determine the outcomes of lower level causations ([9,11] and references therein). Simple and concrete examples from biochemistry and molecular biology can demonstrate this phenomenon. For instance, a particular RNA sequence can fold into a functional three-dimensional conformation via an intricate network of hydrogen bonds between its constituent residues. In the context of the whole RNA sequence, the conformation of a set of residues is subject to the residues that surround it and can be energetically less favourable than the one that it might eventually adopt in the context of a smaller portion of the RNA sequence. Consequently, a complex RNA or protein illustrates how a network of structural interactions as a whole can affect the properties of its constitutive parts [12].
It was recently suggested by Ellis [11,13] that there are at least five different types of TDC taking place in natural sciences depending on the context: algorithmic TDC; TDC via non-adaptive information control; TDC via adaptive selection; TDC via adaptive information control; intelligent TDC. Recognizing these different forms of causation implies that causes other than those involving physical and chemical interactions exist in the real world [11,13]. Because of its key implication in living systems, TDC by information control was investigated more extensively by Auletta et al. [12] in order to propose an experimental and biological framework aimed at testing TDCs at the bacterial level. TDC by information control occurs when a higher level entity influences lower level entities so as to attain a specific functional outcome (goal) through feedback information control loops [12]. The feedback control system depends essentially on the flow of information, coupled to an evaluation of that information relative to the particular functional outcome.
In Auletta et al. [12], we proposed that the establishment of classes of equivalence in living organisms could potentially be used as objective criteria for demonstrating the existence of TDC by information control. A class of functional equivalence is defined by a functional outcome (goal) that is operated by lower level components that can be different as long as they produce the same outcome. By exemplifying the conservation of functions rather than the conservation of modes of operations, the existence of classes of equivalence strongly suggests that it is the biological system as a whole that defines the boundaries and constraints within which a particular class of functional equivalence is established by natural selection. In other words, it is a functional need developed by the whole biological system that defines the constraints within which a particular class of equivalence is established. However, without understanding their respective roles within the context of a whole cell, functional equivalence classes are more symptoms of TDC rather than being direct proofs of TDC. It is therefore of prime importance to clearly establish the controlling instances behind the existence of an equivalence class and to understand how this class is established by natural selection. To address this question, it is beneficial to look at bacterial cells because of their simpler organization. Presently, tool kits from synthetic biology allow the demonstration of the existence of functional equivalence classes in bacteria by experiments of complementation, which typically consist of the replacement of one functional gene by another one in a single organism [12] (see box 1 in electronic supplementary material, appendix). This can subsequently demonstrate TDC by information control if a good understanding of the controlling instances behind the existence of the class of equivalence studied can be provided.
In the present paper, we first define a bacterial cell based on the concepts of master functions, networks and flow of information. We then outline the concept of classes of functional equivalence and briefly describe the way they can be experimentally established in microbiology and synthetic biology. We then review several experiments published in the literature that present compelling experimental evidence for the existence of various functional equivalence classes in bacteria. By focusing on functional equivalence classes belonging to the cellular operating system (COS) of bacteria, we establish the information control instances behind their existence and propose that TDC by information control, in conjunction with TDC by adaptive selection, can—to some extent—explain how the functions from the COS came to be through evolution. As such, TDC by information control and adaptive selection are at the root of what characterizes living systems on the Earth. These considerations provide a helpful framework for future experiments in synthetic biology, which will facilitate the build-up of a minimal living system and will assist in the establishment of the higher level functions that characterize living systems of greater complexity than bacteria.
2. Defining bacterial micro-organisms
To better understand how TDC operates in bacteria, a basic knowledge of how a cell functions is essential. By taking the example of bacteria, one can view this biological system as a single cell, a homogeneous cellular colony or a more elaborate ecosystem involving a genetically heterogeneous cellular population. Controlling instances might be ascribed differently depending upon the context used as a reference. Herein, we consider the cell as a unit that is a complex intracellular network defined by chemical reactions, structural and functional parts, and the ensemble of their functional relationships with respect to one another.
2.1. Hierarchical functional networks and master functions
To fully comprehend the functional organization of a cell is possible only through the understanding of it as a cellular network [14–17]. While metabolic, genetic and regulatory sub-networks can be distinguished, it is the ensemble of these networks that constitute the fundamental system characterizing life. In a holistic way, it is the cell—as an irreducible ‘controlling’ unit—that determines the set of informational feedback control loops necessary for its survival. In the literature, the metabolic regulatory network is often distinguished from the genetic regulatory network, but it is clearly the interplay between the two that forms the controlling informational instances of the cell. The metabolite pools act as intracellular molecular signals that link the metabolic network to the genetic network through genetic regulations [18]. Despite their apparent daunting complexity, these biological networks are themselves very modular in their overall organization, with sub-domains and network motifs being easily distinguishable [15,19]. In fact, mathematical models suggest that biological networks are inherently simple with modular units that could virtually perform independently [16,19,20]. Nevertheless, much remains to be performed at the experimental level for understanding this modularity in terms of functions. This necessitates comprehending what is the essential functional network that defines the invariance for life and how these functional modules are wired to create the fundamental living network.
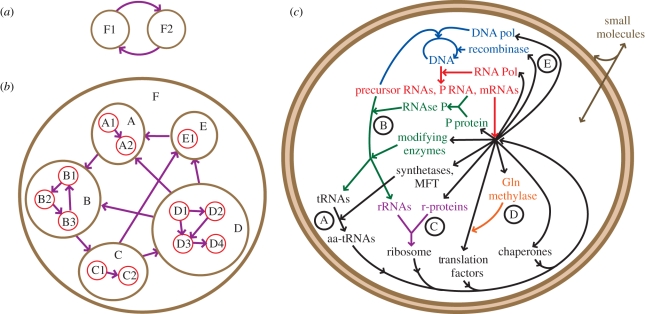
Regardless of the earlier-mentioned considerations, a living cell can be described at an organic level as an autonomous functional entity, while its constitutive parts cannot. The way we approach the cell as a network is to look at it as a hierarchical system of functions in informational relationships with one another where master functions are the functions essential for life. These master functions can be defined as higher order functional cyclic networks composed of multiple sub-functions (figure 1). They unify and integrate the system into a single autonomously behaving and responding, temporarily persistent, identifiably acting entity. A master function can be reduced to a minimal set of sub-functions, which itself is typically irreducible. Building up from von Neumann's [31] idea of self-reproducing automata, Danchin [32] proposed that a bacterial cell could be described as a biological computer that is split into a machine (cellular machinery) and a program (genome). The COS is the link between the program and the cell machinery (figure 1c). By involving multiple operations such as replication, transcription, translation and regulation, the COS is at the root of the two main master functions of bacterial life: the replication of the genome and reproduction of the cell machinery. The intimate informational network that results from the symbiotic association of these two master functions is what characterizes cellular life. As such, ‘cellular life’ could be seen as the higher level master function. For a cell, its outcome is the formation of two daughter cells from one. Albeit reductive, this view is useful for describing the phenomenon of TDC by information control.
Figure 1.
The concept of master functions and networks in bacterial cells. (a) Two functions can be linked to one another by informational relationships (arrows) resulting from sharing, exchanging information or action on one another to form a functional cyclic network. (b) A master function F can be defined as a higher order functional network of lower functional entities (A–E) that can themselves involve multiple functional operations (e.g. A1, C2, D4, etc.) in informational relationship with one another. (c) Example of a functional cyclic network: the minimal cellular operating system (COS) network comprises biological macromolecules and pathways supposed necessary and sufficient for replication (and reproduction) from small molecule nutrients diffusing through the bilayer lipid vesicle (figure adapted from Forster & Church [21]). Biomolecules and chemical reactions are coloured according to the biochemical pathway which they belong to: DNA replication (blue), RNA transcription (red), RNA processing (green), ribosome assembly (violet), protein translation (black) and post-translational processing (orange). MFT: methionyl-tRNAfMet formyltransferase. Circled letters correspond to steps subjected to complementation experiments described in table 1 (A [22,23]; B [24,25]; C [26,27]; D [28,29]; E [30]).
2.2. The cell as a flow of information
While there are different kinds of information, it can generally be defined as that which brings about a reduction of uncertainty or indeterminacy [33,34]. An anticipated characteristic of all master functions is that they are functional cyclic networks. Note that the ‘chemoton’ proto-cell described by Szathmary & Smith [35] fits the present description of a cell as a functional cyclic network [36,37]. Because of their cyclic organization, these networks can also be seen as informational networks with inbuilt informational links acting as feedback control loops. As such, an informational link can be broadly defined as any type of physical and/or chemical interaction existing between two functions. This link has both a functional and informational meaning in contrast to random molecular interactions that are part of the noise. Within the context of a cyclic network, an informational link can include a chemical reaction resulting in the transformation of one molecule into another, the operation of an enzyme on a substrate or the operation of a molecular effector acting as a signal on another molecule.
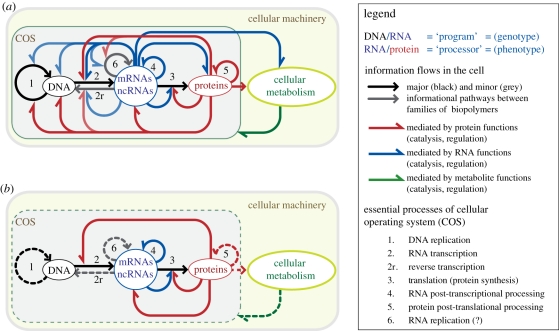
The flow of information pertaining to the various biomolecular processes of the COS is shown schematically in figure 2 within the context of the present ‘dogma’ of molecular biology of the cell. It exemplifies the relations between the three major categories of informational polymers: DNA, RNA and proteins. The control of informational flows contributes to cellular homeostasis. Within the COS, informational flows mediated by protein, RNA and metabolite functions locally form numerous cycles. The COS is therefore characterized by multiple feedback loops of information. As such, the COS has an inbuilt control of the information it processes. It contributes to the selection of the ‘quality’ of the information carried by the informational molecules and regulates the level of expression of the information.
Figure 2.
Flow of information within the central dogma of a cell. The diagram in (a) conceptualizes and summarizes the different pathways and feedback loops that relate to the three major categories of informational polymers involved in the COS. ncRNAs, non-coding RNAs; mRNAs, messenger RNAs (coding for proteins). RNA replication (step 6) is a process that occurs in eukaryotic cells [38,39]. It might also occur in bacteria. The cellular metabolism can theoretically have an informational effect on any of the COS (this is indicated by a green arrow directed from the cellular metabolism to the COS). (b) Simplified diagram with information feedback loops involved in the processing of ncRNAs that directly participate in translation (e.g. rRNA, tRNA). Dashed arrows indicate steps that are not essential to the process. The gene corresponding to the ncRNA is transcribed (step 2) into a ncRNA precursor that might require post-transcriptional processing (step 4), so it can be used by the translational apparatus for synthesizing proteins (step 3). Translation (step 3) is possible only when the information encoded at the level of the ncRNA gene is valid. The quality of the information carried by the ncRNA is necessary for the production of the enzymes responsible for the production of the mature ncRNA, like the RNA polymerase (step 2), the proteins of the translational apparatus (step 3) and the proteins involved in the maturation (step 4). This feedback loop controls the quality of the informational properties of the ncRNA.
In a functional network, it is possible that the flow of information forms a self-replicative cyclical operational network, also called a hypercycle [40]. However, functional cycles are not all self-replicative and could also take place at the level of lower constituent functional entities (figure 1b).
3. The cell and top-down causation by information control and adaptive selection
The sub-functions that constitute a master function emerge from the bottom up. A master function as complex as the ‘modern day’ replication of the cellular programme is however extremely improbable to have emerged by mere ‘chance’ (figure 1c). Even in the simplest modern cell, a master function ‘ought to come’ from simpler molecular systems that nevertheless keep the intrinsic properties and characteristics of the master function conserved through the process of evolution. Therefore, a master function creates the functional constraints and boundaries within which the lower functions can evolve, diversify and eventually become more complex. It is important to realize here that it is the COS that operates the reproduction and replication of the cell. The various molecular components of the COS can interact, recombine and diversify to lead to further complexification and differentiation, offering new capabilities and potentialities. Thus, the COS does constitute, in the philosophical sense, the system formal cause [12]. This characterizes a top-down causal effect by the whole system (defined by master functions) on the molecular parts (defined by the functions of lower levels that operate the master functions). Considering the master functions of a cell, the functional constraints are informational constraints. Each sub-function that has evolved within the master function is constantly selected for the quality of its information. Therefore, it is the master function that ultimately ‘decides’ whether the information will be kept or not. This is characterizing TDC by information control [12]. As the sub-functions are intimate parts of the master function, the acceptance or rejection of the new information carried by the new sub-functionalities leads either to the survival or to the death of the cellular system. However, within the context of evolution, death as an outcome is not an issue. There are typically millions of cells that can proofread the quality of the information and function in parallel (like parallel processors of information). To be fully operational, the cell therefore needs to perform within a space of possibilities. The process of selection for valid information is operated ‘in a blind way’ by multiple copies of identical (or quasi-identical) cells. This is characteristic of TDC by adaptive selection. Bacterial cells can therefore be characterized by TDC by information control in conjunction with TDC by adaptive selection.
In summary, this TDC framework implies that the cell can be operated by different modes of operations or functions of lower levels as long as the properties defined by the master functions are conserved [12] (figure 1). The direct philosophical implication of this framework is as follows: through TDC by information control, the master functions affect the outcome of their functional parts that contribute to the conservation of the properties of the master functions that define the whole system. Additionally, the master functions define the boundaries within which the lower level functions evolve by natural selection. Aspects relative to the emergence of the functional properties for cellular life are discussed at a latter stage in this paper. Within an evolutionary process (that takes advantage of adaptive selection by definition), it is because of information control from the top down that the phenomenon of convergence of functions can possibly take place.
The concept of classes of functional equivalence can be extremely useful for providing experimental biological evidences in favour of this framework. Indeed, the demonstration that different modalities can operate similar or identical functions within the cell and that these functions are under information feedback loop control would constitute a rather eloquent proof of TDC by information control.
4. Classes of functional equivalence in biology
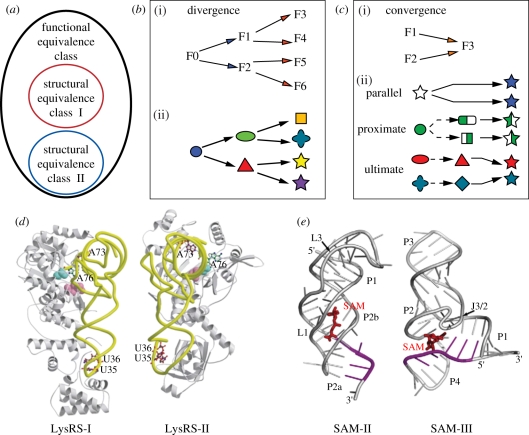
In biology, a class of functional equivalence is defined by different modes of operations (typically defined by a set of molecules) that have the same function or lead to the same functional outcome [12]. The underlying concept of functions is therefore crucial for asserting without ambiguity whether two distinct modes of operations are equivalent or not in living organisms. Potential molecular candidates to a particular class of functional equivalence are often identified by comparative genomic sequence analysis of different species. This approach mainly allows identification of homologous functional molecules that result from evolutionary divergence (figure 3b) and that operate by similar conserved mechanisms (e.g. RNA polymerases, ribosomes or RNase P RNAs from different organisms). However, equivalent modes of operations are not necessarily related by homology as they can result from ultimate evolutionary convergence (figure 3c). Two functionally analogous sets of biomolecular operations may have completely different unrelated structural features, different mechanisms of catalysis, different modalities of recognizing substrates and may even involve a different number of molecular components. Still, they can have similar, if not identical, functional outcomes (figure 3d,e). Consequently, molecular candidates analogous to a particular functional equivalence class require extensive experimental characterization of their function. This can partially explain why the number of examples of molecular functional convergence reported in the literature is still scarce.
Figure 3.
Evolutionary divergence, convergence and classes of functional equivalence. (a) A functional equivalence class can be formed of different structural equivalence classes. (b) By evolutionary divergence, a functional molecular system can lead to new molecules with different functionalities (i) and significant sequence and structural variations (ii). Some of these molecules can still retain the same function and belong to the same functional class. Molecular divergence implies that the divergent molecules are evolutionarily related as they have a common ancestor. (c) Two different unrelated molecules can evolve towards the same function through evolutionary convergence (i) [41]. At the molecular level (ii), one can distinguish parallel convergence (where the same molecular system in two different organisms evolves independently the same way), proximate convergence (where a molecular system that has diverged significantly in two lineages converges towards the same features) and ultimate convergence (where two different evolutionarily unrelated molecular systems converge towards the same functional features) [42]. In contrast to ultimate convergence, parallel and proximate convergences are phenomena that pertain to evolutionarily related molecules. (d–e) Classes of functional equivalence with molecules resulting from ultimate convergence. (d) Functional convergence of class I and class II Lysyl-tRNA synthetases (LysRS). These two enzyme classes have different structural topologies with different modalities of recognition of Lysyl-tRNAs (adapted from Terada et al. [43]). They are distinct structural equivalence classes that belong to the same class of functional equivalence defined by Lysyl-tRNA aminoacylation functionalities. (e) Functional convergence of class II and class III of S-adenosyl-methionine (SAM) riboswitches [44]. These two distinct structural classes have different modalities of recognizing SAM [45,46]. Figure adapted from Serganov [47].
The concept of functional equivalence classes allows the dismissal of evolutionary concerns such as homology or analogy because what really matters when comparing two modes of operation is their degree of structural and functional similarities, not their evolutionary relationship per se. Nevertheless, within a class of functional equivalence, evolutionary considerations are useful to identify its most compelling members, which should be structurally very different (different shapes and structure topologies) and most likely result from evolutionary convergence [41,42].
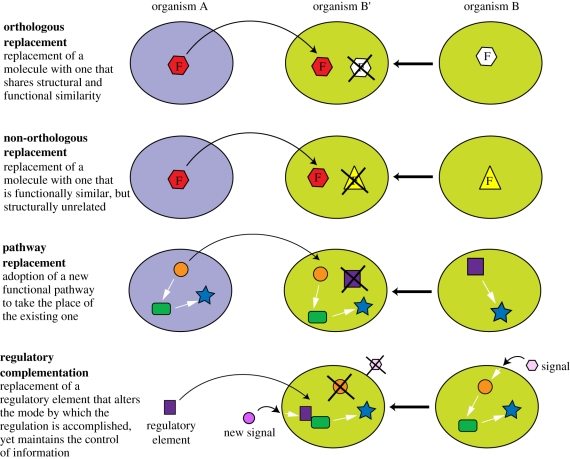
Complementation experiments are the most straightforward methods for establishing the functional equivalence of different biomolecules or pathways in organisms (figure 4 and box 1 in electronic supplementary material, appendix). They are part of the toolkits of synthetic biology. By replacing a set of operations, defined by one or more molecules with a particular functional outcome, for another one, it is possible to verify whether the new set of operations is able to recover the initial function within an organism. Because of the remarkable functional modularity of living organisms, numerous classes of functional equivalence could potentially be defined in this way. It is, however, important to keep in mind that the demonstration of functional equivalence by complementation is dependent upon the initial experimental conditions. In some optimal conditions, two sets of operations might look functionally alike (equivalent) while in more discriminative conditions, they might show distinctive functional behaviour. Grouping two different sets of operations into the same class of equivalence can therefore require a certain degree of coarse graining. Moreover, if the complex interconnectivity between components of a sub-function is not fully understood (i.e. not all of its operations have been characterized), then replacing only part of its operations might appear to have no functional equivalence.
Figure 4.
Examples of four different categories of complementation experiments. Organism A is the donor of the operation of function F, and organism B′, resulting from organism B after knock out of the operation of function F in organism B, is the recipient. Molecular entities are represented by small, coloured geometric symbols. Selected references corresponding to each category of complementation: orthologous replacement [25], non-orthologous replacement [22,24], pathway replacement [23,48] and regulatory complementation [49,51]. Additional examples are provided in table 1.
From an evolutionary point of view, complementation experiments are good models for horizontal gene transfer (HGT), also called lateral gene transfer. In bacteria, it is not uncommon for genes to be transferred horizontally from one organism to another. HGT allows bacteria to acquire new functional modules and can therefore be seen as contributing to the innovation and potential emergence of new functions. It is also through this modality of genetic exchange that functional convergence of different modes of operations can potentially be established [50]. For instance, the quasi-universality of the genetic code shared by all living organisms might be resulting from such unifying processes [50].
5. Experimental evidence for classes of functional equivalence
Several examples of complementation experiments published in the literature demonstrate the functional interchangeability of different molecules or even different molecular pathways that are involved in similar cellular processes (table 1). They establish in vivo the existence of various classes of functional equivalence within the OS genetic network [22–24] and the metabolic networks [52,53] in bacteria, archaea and lower eukaryotes (e.g. yeast [53]). Many of these complementation experiments demonstrate that functions from eukaryotes could be similar or at least partly similar to those found in bacteria, indicating strong conservation of functionalities throughout the three different branches of life [53]. With the exception of the work by Wegscheid et al. [25], none of the reported complementation experiments has ever been interpreted in the literature within a conceptual TDC framework.
Table 1.
Examples of complementation experiments demonstrating the existence of functional equivalence classes in the genetic, metabolic and regulatory networks. The listed examples only represent a small subset of experiments of complementation or gene replacements. They were identified in PubMed with the key words: complementation, gene, function replacement (or displacement), orthologous, parallel, non-orthologous (or non-orthologous) and heterologous. The experiment by Wegscheid et al. [25] has been described in Auletta et al. [12]. Experiments referenced [22–25,51] are described in the text.
| function | molecular systems | type of complementation (evolutionary significance) |
reference | |
|---|---|---|---|---|
| recipient(s) | donor(s) | |||
| genetic operating system network | ||||
| tRNAlys aminoacylation | class I (Bacillus subtilis) and class II (Borrelia burgdorferi) Lysyl-tRNA synthetases | non-orthologous replacement (convergence) | [22] | |
| B. subtilis (Bact.) | B. burgdorferi (Bact.) | |||
| tRNAgln aminoacylation | Gln-tRNAgln direct (Escherichia coli) and indirect (B. subtilis) aminoacylation pathways | pathway replacement (convergence) | [23] | |
| E. coli (Bact.) | B. subtilis (Bact.) | |||
| tRNA processing | type A RNase P RNA (E. coli) and MRORP1 protein (Arabidopsis) | non-orthologous replacement (convergence) | [24] | |
| E. coli (Bact.) | Arabidopsis (Euk.) | |||
| TMP synthesis | ThyA (E. coli) and ThyX (Bo. burgdorferi) Thymidylate synthases | non-orthologous replacement (convergence) | [28] | |
| E. coli (Bact.) | Bo. burgdorferi (Bact.) | |||
| protein folding | Rpl25 (Saccharomyces cerevisiae) and TF (E. coli) ribosomal protein chaperones | non-orthologous replacement (convergence) | [26] | |
| E. coli (Bact.) | S. cerevisiae (Euk.) | |||
| tRNA processing | type A (E. coli) and type B (B. subtilis) RNase P RNAs | orthologous replacement (divergence) | [25] | |
| E. coli (Bact.) | B. subtilis (Bact.) | |||
| B. subtilis (Bact.) | E. coli (Bact.) | |||
| ribosome assembly | rRNA/r-protein operons | orthologous replacement (divergence) | [27] | |
| E. coli (Bact.) | Salmonella typhimurium (Bact.) | |||
| Proteus vulgaris (Bact.) | ||||
| DNA recombination repair | RAD54 (S. cerevisiae) and AtRAD54 (Arabipdopsis) repair proteins | orthologous replacement (divergence) | [29] | |
| S. cerevisiae (Euk.) | Arabidopsis (Euk.) | |||
| post-translational processing | alg7 (S. cerevisiae) and mv1751 (Methanococcus voltae) N-glycosylation proteins | orthologous replacement (divergence) | [30] | |
| S. cerevisiae (Euk.) | M. voltae (Arch.) | |||
| metabolic network | ||||
| lipid-linked oligosaccharides translocation | ABC type Wzx (E. coli) and non-ABC-type WlaB (Campylobacter jejuni) flippases | non-orthologous replacement (convergence) | [52] | |
| E. coli (Bact.) | C. jejuni (Bact.) | |||
| inorganic pyrophosphate hydrolysis | soluble (S. cerevisiae) and membrane-bound H+-translocating (Arabidopsis) inorganic pyrophosphatases | non-orthologous replacement (convergence) | [53] | |
| S. cerevisiae (Euk.) | Arabidopsis (Euk.) | |||
| Chloroflexus aurantiacus (Bact.) | ||||
| antibiotic resistance | low Mg2+ (Salmonella enterica) and high Mg2+ (E. coli) Polymyxin B resistance pathways | pathway replacement (divergence) | [48] | |
| E. coli (Bact.) | S. enterica (Bact.) | |||
| molecular transport | YopB/YopD (Pseudomonas aeruginosai) and PopB/PopD (Yersinia pestis) proteins | orthologous replacement (divergence) | [54] | |
| P. aeruginosai (Bact.) | Y. pestis (Bact.) | |||
| regulatory network | ||||
| bacteria motility | pseudotaxis pathway (dependent on a theophylline-riboswitch) and natural chemotaxis pathway (E. coli) | regulatory complementation (convergence) | [51] | |
| E. coli (Bact.) | synthetic parts | |||
| cellular and hormonal regulation | lower and higher eukaryote calmodulins | orthologous replacement (divergence) | [55] | |
| S. cerevisiae (Euk.) | Xenopus laevis (Euk.) | |||
| transcriptional regulation | piD261/Bud32 (S. cerevisiae) and PRPK (human) kinase proteins | partial orthologous replacement (divergence) | [56] | |
| S. cerevisiae (Euk.) | Homo sapiens (Euk.) | |||
The complementation experiments involving functions carried out by amino-acyl tRNA synthetases (aaRS) [22,23] (figure 3d), RNase P [24], flippases [52] and pyrophosphatases [53] demonstrate without ambiguity that very different modes of molecular operations could be substituted by one another as long as their function remains the same (table 1 and figure 5). In all cases, the molecular instructions that are exchanged have likely originated by ultimate convergent evolution [41,42] because they do not share any apparent structural similarities. The best example of all is the complementation experiment in Escherichia coli where the RNase P function, which is essentially based on an RNA in bacteria, is replaced by a purely proteinaceous RNase P from plant organelles [24]. By substituting a function carried by an RNA with one carried by a protein, one cannot argue that the two molecular systems are swappable simply because they share conserved structural features. In this experiment and others [22–24,52,53], the only feature that the exchanged molecular systems share is their functional outcome. These experiments are therefore in much stronger support of TDC than that [25] originally mentioned by Auletta et al. [12]. Indeed, in Wegscheid et al. [25], the RNase P RNAs that were exchanged are still phylogenetically related despite their significant biophysical differences.
Figure 5.
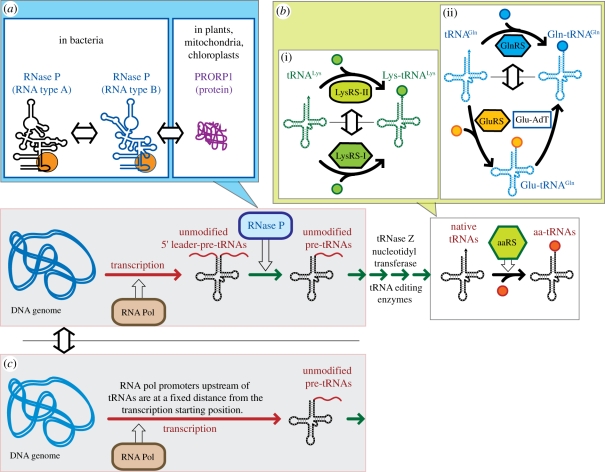
Examples of functional equivalence classes within the process of tRNA synthesis and activation from the bacterial operating system. (a) In Escherichia coli, the endogenous type A RNase P RNA can be substituted by a type B RNase P RNA [25]. It can also be substituted by PROP1, a proteinaceous RNase P from plant mitochondria (Arabidopsis thaliana) [24]. (b) (i) Lysyl-tRNA synthetases (LysRS) have been found in different organisms and are categorized as either class I or class II. Substituting a class II bacterial LysRS by a class I archaeal LysRS still allows the bacteria to operate the COS [22]. (ii) The direct pathway of amino acylation of glutaminyl tRNA found in E. coli can potentially be substituted by an indirect pathway involving first mischarging of tRNAgln with Glu by glutamyl-tRNA synthetase (GluRS) to form glutamyl tRNAGln. Then, glutamyl tRNAGln is converted into glutaminyl tRNAgln by Glu-tRNAGln amidotransferase (Glu-AdT) [23]. (c) The process of maturation of 5′-leader pre-tRNA by RNase P does not exist anymore in Nanoarchaeum equitans. Instead, the tRNA genes code for tRNA precursors without 5′ leader because transcription starts now at the level of the mature 5′ end [57]. With respect to mature tRNAs production for translation, the Nanoarchaeum process is expected to be equivalent to the RNase P-dependent one. However, this remains to be tested in vivo. Colour code for reaction processes is the same as in figure 1c.
On the basis of the cellular framework presented in figure 2, the classes of functional equivalence identified in the COS readily provide strong clues, if not proof, of TDC by information control. As shown in figure 5, both tRNA aminoacylation (involving aaRS) and tRNA maturation (involving RNase P) are essential processes for the proper implementation of the basic protocol of the genetic code. If tRNAs are not able to be properly amino-acylated or matured, the enzymes responsible for tRNA production (RNA polymerases, aaRS and RNase P protein) cannot be accurately synthesized by the translational apparatus (see also electronic supplementary material, figure S1). Therefore, the very existence of the multiple feedback control loops at the level of the COS offers several possibilities to verify the quality of the operations necessary for replication and cellular reproduction (figure 2b).
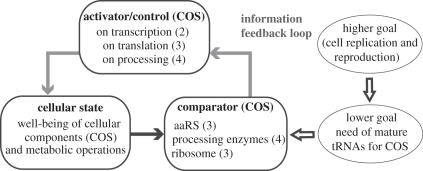
In these examples, we can clearly establish that the information selection defining the operational elements of the class is conserved despite the differences in lower level variables. This is demonstrating TDC by information control. A more philosophical framework is also shown in figure 6b.
Figure 6.
Philosophical interpretation of top-down causation by information control in the cellular operating system. Numbers correspond to the steps indicated in figure 2.
Presently, it is apparent that regulatory elements, which control the expression of gene operons in bacteria, can have very different modes of operations for regulating similar gene operons in different bacteria [58,59]. At least three different categories of attenuation mechanisms (ribosome-mediated, protein-mediated and uncharged-tRNA-mediated) are involved in the regulation of the tryptophan operon in bacteria [58,60]. Another remarkable example is given by the very distinct structural classes of S-adenosyl-methionine riboswitches that seem to be interchangeable for controlling—via an attenuation mechanism—the same type of genes in different bacteria species [44,61] (figure 3e). These examples are far from being isolated cases and would require a thorough investigation. At the molecular level, functional convergence is likely more common than it was initially thought to be. By looking at cellular functions that are linked to the intrinsic regulatory mechanisms of the bacterial cell as well as the extrinsic physical and chemical environment in which the bacteria live [6], other interesting conserved functional features under TDC by information control are likely in existence, especially at the level of regulatory controlling elements. In the future, it will also be of prime importance to look at functional networks in an evolutionary context to ultimately understand their modularity and possibly how their parts came to be [19].
6. Discussion
One of the important questions relative to living systems is to identify the core functional properties that characterize life. TDC by information control and adaptive selection could be particularly insightful for explaining the emergence of novel functions in living systems. These considerations could also be useful for proposing new experiments of synthetic biology.
6.1. The emergence of the core functional properties of life
Among the definitions of life, many share the notion that cellular life is associated to the emerging properties of a replicating informational molecular system able to mutate [32,62,63]. While the informational template should be allowed to change for exploring a space of possibilities for new functions, it still needs to retain the ability to replicate effectively. Exploration of novel functions is possible because of imperfections in the replication process, which creates the constraints for this exploration. Like the pre-existing information, the new resulting molecular information requires selection for its viability within the system. In other words, the new information is expressed and controlled for its ability to operate within the system. The control of the ‘quality’ of the information (through feedback control) is embedded within the replicating system and can be seen as a necessary underlying property associated to the function of replication. However, the issue here is not merely to replicate but to also properly segregate the new information resulting from replication so that what is essential to the system is kept while what is deleterious (lethal) to the system is disregarded. Therefore, the replicating system requires compartmentalization with selective reproduction of the molecular sub-functions responsible for DNA replication and cell reproduction, as well as the novel functions that are not detrimental to the cellular master functions. While there is a drive here for perpetuating the informational master properties within the system through time (a drive for life that allows exploration of novelty), there is also a drive for eliminating parts of the molecular information that do not fulfil the master cellular functions (therefore a drive towards decay and death). The main selection drive behind perpetuation of the informational properties is to retain among others, the novel functions that allow intake of the sources of chemical energy, production of the molecular building blocks necessary for the synthesis of the biomolecules that support the master functions of the cell as well as their repair, etc. The main selection drive behind the elimination of undesirable information is to retain, for example, the functions that favour degradation processes and the segregation of undesirable molecules from the correct ones as well as elimination of the cellular systems that have aged, etc. [64]. Because of TDC by information control and adaptive selection, one can perceive how unrelated molecular systems could converge towards similar or identical functions during evolution.
The master functions of the cell (replication and reproduction) are the properties that characterize the cellular unit as a whole. They are defined at the molecular level by a set of relationships that abstractly define the COS network and absolutely need to be conserved (figure 2). Through the COS, the cell selects the information that it can process, which is also the information that constitutes parts of the COS itself. The COS is, by definition, built up for working via feedback through information control. Any information that it processes, if disruptive, is ultimately eliminated because it induces the destruction of the whole cell. By contrast, if selected, the COS can operate with it and the replication of the information and the reproduction of the cell can occur. The cell therefore operates through TDC by information control; the higher level master functions have causal power on the set of lower level functions or operations that are causally effective for controlling all the information processed by the cell in order to assure conservation of the master functions.
Interestingly, the constitutive lower level functions of replication and cellular reproduction can be carried out by different modalities of operations. This is particularly well exemplified by the COS of bacteria, archaea and eukaryotes. Despite sharing similar overall mechanisms and numerous similar macromolecular machineries for replication, transcription and translation (e.g. RNA polymerases, ribosomes, DNA replisomes), they can nevertheless present significant differences at the level of the molecular operations leading to these functional outcomes. As long as the master functions are conserved, the intracellular network of molecules and their associated interactions can significantly vary as long as the overall process is maintained. For instance, different classes of enzymes are structurally highly divergent or even unrelated at the level of their structures (e.g. class I and class II LysRS, figure 3d) but essentially carry the same function at the level of the COS (see earlier text).
The phenomenon of TDC by information control through adaptive selection can explain how living systems could possibly have some of their parts evolving through time from an RNA/peptide world to a DNA/RNA/protein world [65] (figure 7). As long as the basic master functions of the cell are conserved, the cellular system can increase in complexity by creating new functions that fit the goal of the master function. The optimization of the COS is essentially driven by the need to efficiently process a greater amount of information. The fitness of the cell is defined here in terms of being able to efficiently operate the master functions.
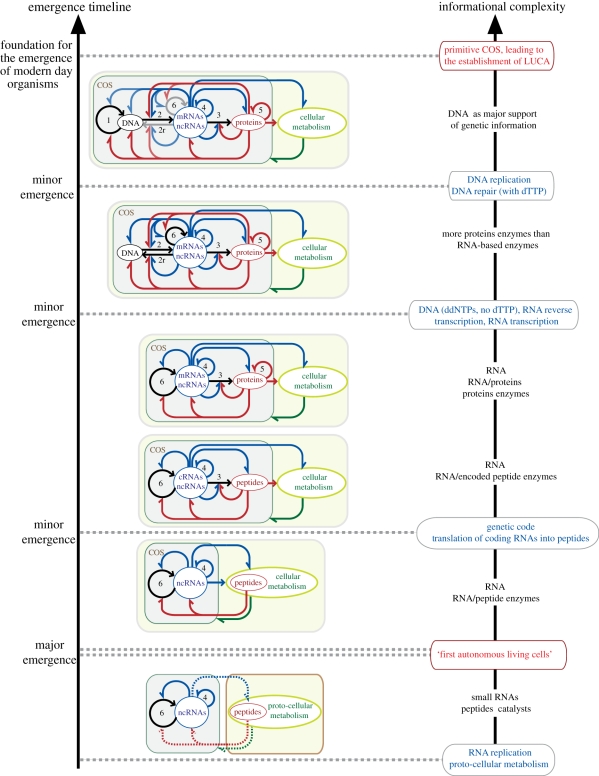
Figure 7.
Possible evolutionary scenario for the optimization and increase in complexity of the cellular operating system (COS) through top-down causation by information control and adaptive selection. This leads to the foundational ‘modern’ core of information pathways belonging to the last universal common ancestor (LUCA) of all living organisms on the Earth. While this scenario delineates clear transition steps, some of the optimization steps could overlap in time. The major emergence of the first autonomous living cells corresponds to the emergence of the regulatory network linking RNA replication with cellular reproduction. See also legend of figure 2.
Within our TDC framework, we propose a scenario explaining how simple replicating and reproducing cellular systems, based on COSs involving a limited set of functionalities, could have developed into modern COSs (figure 7). What is making this scenario possible is the existence of classes of functional equivalence with modes of operations able to be interchanged between different cellular entities through HGT [66,67]. It is therefore with TDC by information control and adaptive selection that the phenomenon of convergence towards a unique COS can take place. Several important emergence events might have occurred. First, coupling between RNA replication and cellular metabolic reproduction led to the first autonomous living cells. This coupling corresponded to the emergence of the first regulatory network associated to the COS. Then, the natural drive of informational molecules to increase in size led to an increase of complexity that required improvement in the processing of this information. This could have taken place in multiple steps, which likely overlapped in time, each of these steps improving the accuracy and speed of the COS. Key emergence events were (i) the invention of the universal genetic code with the translation of coding RNA into encoded peptides allowing the COS to rely more and more on proteins to carry important catalytic functions and followed by (ii) the invention of DNA as a superior support of the genetic information. As such, DNA genomes offered the advantage to be more easily repaired than RNA genomes. Additionally, other essential functions, which paved the way towards efficient production of chemical energy, communication between cells and their environments as well as between different cells, emerged very early in conjunction with the emergence of the common ‘modern’ COS of all living organisms [66]. This point in the history of cellular evolution, which can be traced back from modern day organisms, gave rise to LUCA, the last universal common ancestor (figure 7). It is only from that point that cellular life was ready for the next evolutionary step leading to speciation through division of labour and multi-cellular organization [66]. This point, called the Darwinian threshold, is therefore at the origin of the major division between eukaryotic, bacterial and archaeal cells. However, note that the process of optimization of the COS has continued to take place to a minor extent after that point of history.
Our discussion here pertains only to the core master functions of cellular life. During evolution, new master functions can emerge with a selection drive towards higher order functions (such as those pertaining to differentiated multicellular living systems). However, any new set of emerging master functions cannot work against the master properties of lower levels but have to be built upon them. Good examples are parasitic- or symbiotic-living cellular systems. The genomes of these obligate systems might have lost some of the lower level operations necessary for their autonomy outside their host. However, they rely on the fundamental lower level properties provided by their host to operate their master functions. In all cases, any new master functions will create new functional boundaries for TDC by information control and adaptive selection. Therefore, TDC can enable new classes of equivalence to appear through an explosion of diverse life forms and species [68] that are themselves subjected to new phenomena of convergence [69].
6.2. Future perspectives for synthetic biology
With synthetic biology, new biological functions and systems not found in nature can be created through a combination of in vivo, in vitro and in silico techniques. One can envision the engineering of new living organisms by modification of their genomes from a top-down approach [70,71] or by integration of artificial molecular parts from the bottom up [70,72,73]. For the past few years, bootstrapping experiments with synthetic bacterial genomes derived from natural ones have been underway for recreating minimal living bacterial organisms from Mycoplasma [74–76] and, very recently, some of the technical challenges have been overcome [77]. For instance, the genesis of new Mycoplasma mycoides cells has been demonstrated by transplantation of a synthetic genome into a M. capricolum recipient cell [77].
We believe that our present TDC framework could be useful for planning future experiments of synthetic biology in order to unravel the minimal set of functions that characterize living systems [21,78] and providing insight about the way some of the essential cellular functions came to be. For instance, using the concept of classes of equivalence, it might be possible to substitute some of the existing cellular processes by simpler ones, once a class of functional equivalence is established. As it has been shown in the archaeon Nanoarchaeum equitan [57], the production of mature tRNA does not require RNase P as long as the tRNA genes are organized at the level of the genome such that all pre-tRNA transcripts are produced without 5′-leader sequences (figure 5c). A deeper understanding of the different classes of functional equivalence and their modularity could be essential for the integration of new functionalities that would be more deeply rooted in the cellular regulatory network of the cell. Key factors will be the proper identification of the modality of regulations and flow of information at the level of these minimal living systems that are likely to present some degree of hierarchical organization. The minimal set of cellular functions identified in Mycoplasma organisms is likely to be an interesting starting point for investigating the process of emergence of new functions and properties at the level of these organisms.
While it is unlikely that the new M. mycoides strain will be able to be (de)-evolved into a form of life that does not require the translational apparatus, it is however anticipated that the determination of the important characteristics at the root of the master functions of life through the top-down approach will be of some help to those interested by the bottom-up approach. It might be possible to substitute a certain number of operations of lower functions naturally carried out by proteins into some carried by artificial RNA molecules, creating a new cellular ‘RNA world’. These experiments could be seen as ‘retro-synthetic’ biology. In this exercise, several hypothetical evolutionary pathways could be investigated for their potential to have led to modern day cellular systems. A better understanding of the necessary emerging requirements for cellular life might come from giant viruses such as mimiviruses [79–81]. These obligate parasitic systems offer the intriguing possibility to be engineered into biological systems that could express a COS (with a fully operational translational apparatus) that might allow them to function independently from their cellular host. However, these experiments come with possible ethical issues that should not be neglected.
Synthetic biology has presently demonstrated that it is possible to design novel metabolic pathways with new specific synthetic goals (systemically ‘needed’ outcome or products) in bacteria [71,82,83] or to express orthologous pathways that can work in parallel with existing pathways in order to express new genes to create artificial metabolic systems [84,85]. For instance, it has been demonstrated that a synthetically evolved orthogonal ribosome system can function within E. coli to generate protein with unnatural amino acids in parallel to the ‘normal’ ribosomal system [85]. It is possible to create new regulatory pathways by triggering specific ‘behaviour’ (e.g. bacterial motility) in response to specific molecular signals (e.g. theophylline or the herbicide atrazine) [49,86]. In other words, at the microbial level, an outcome can be reached by ‘re-programming’ the cell with artificial genes that could lead to this outcome with different pathways and different molecular triggers. Most of these approaches are based on the present understanding of how natural regulatory pathways work in bacteria [18,19,71]. Because of the high modularity and rather simple regulatory pathways in bacteria (especially when compared with those from eukaryotes), artificial regulatory pathways can be built, and existing functions and molecular parts can be easily exchanged with the new ones (http://partsregistry.org) [87].
Within the context of synthetic biology, complementation experiments are extremely useful tools for experimentally unravelling the real nature of the modularity behind the various functions characterizing living organisms. Determination of this functional modularity allows identification of classes of functional equivalence that could potentially be hierarchically organized based on their degree of importance for the bacteria in well-defined experimental conditions.
7. Conclusion
In conclusion, TDC by information control and adaptive selection are at the root of converging forces that shape the evolution of living biosystems from the simplest to the most complex levels. Living systems could therefore be defined as self-reproducing systems that function via TDC by information control and adaptive selection. The functions of the COS that control cellular reproduction and DNA replication are maintained through TDC by information control leading to a converging driving force. This is apparent within the COS as it can have different modes of operations with the same functions. It is anticipated that functional convergence at the molecular level might not be as rare as initially thought. With the development of synthetic biology, it is expected that functional testing by complementation of newly identified molecules could lead to the discovery of a greater number of examples of ultimate functional convergence that are indicative of TDC. As long as the fundamental functions of reproduction and replication are kept, emergence of novel functions from the bottom-up is possible. This is, however, under the dependency of TDC by information control and adaptive selection. Indeed, whatever emerges from the bottom up still must work within the context of the living system. Darwinian evolutionary processes in living systems are therefore not only ruled from the bottom up but also by fundamental emerging organizational principles that are hierarchically built up and impose necessary constraints from the top down. These principles are the key for defining organic life.
Acknowledgements
We warmly thank George Ellis and Bill Stoeger for their input on a preliminary version of this paper. This work was funded through a STARS grant from the Centre for Theology and the Natural Sciences, Berkeley, CA (www.ctns.org). L.J. wishes to dedicate this paper to St Thomas Aquinas and Roger Bacon.
References
- 1.Bard J. 2010. A systems biology view of evolutionary genetics: network-driven processes incorporate much more variation than evolutionary genetics can handle. This variation is hard to formalise but allows fast change. Bioessays 32, 559–563 10.1002/bies.200900166 (doi:10.1002/bies.200900166) [DOI] [PubMed] [Google Scholar]
- 2.Martins dos Santos V. A., Damborsky J. 2010. Systems biology at work. Curr. Opin. Biotechnol. 21, 498–501 10.1016/j.copbio.2010.08.001 (doi:10.1016/j.copbio.2010.08.001) [DOI] [PubMed] [Google Scholar]
- 3.Khalil A. S., Collins J. J. 2010. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379 10.1038/nrg2775 (doi:10.1038/nrg2775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lorenzo V. 2010. Synthetic biology: something old, something new. Bioessays 32, 267–270 10.1002/bies.200900194 (doi:10.1002/bies.200900194) [DOI] [PubMed] [Google Scholar]
- 5.Danchin A. 2009. Information of the chassis and information of the program in synthetic cells. Syst. Synth. Biol. 3, 125–134 10.1007/s11693-009-9036-5 (doi:10.1007/s11693-009-9036-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young E., Alper H. 2010. Synthetic biology: tools to design, build, and optimize cellular processes. J. Biomed. Biotechnol. 2010, 130781. 10.1155/2010/130781 (doi:10.1155/2010/130781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luisi P. L. 2002. Toward the engineering of minimal living cells. Anat. Rec. 268, 208–214 10.1002/ar.10155 (doi:10.1002/ar.10155) [DOI] [PubMed] [Google Scholar]
- 8.Luisi P. L. 2006. The emergence of life: from chemical origins to synthetic biology. Cambridge, UK: Cambridge University Press [Google Scholar]
- 9.Clayton P., Davies P. C. W. 2006. The re-emergence of emergence. Oxford, UK: Oxford University Press [Google Scholar]
- 10.Murphy N., Stoeger W. R. 2007. Evolution and emergence: systems, organisms, persons. Oxford, UK: Oxford University Press [Google Scholar]
- 11.Ellis G. F. 2008. On the nature of causation in complex systems. Trans. R. Soc. S. Afr. 63, 1–16 10.1080/00359190809519211 (doi:10.1080/00359190809519211) [DOI] [Google Scholar]
- 12.Auletta G., Ellis G. F., Jaeger L. 2008. Top-down causation by information control: from a philosophical problem to a scientific research programme. J. R. Soc. Interface 5, 1159–1172 10.1098/rsif.2008.0018 (doi:10.1098/rsif.2008.0018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis G. F. 2009. Top-down causation and the human brain. In Downward causation and the neurobiology of free will (eds Murphy N., Ellis G. F., O'Connor T.), pp. 63–81 Berlin/Heidelberg, Germany: Springer [Google Scholar]
- 14.Barabasi A. L., Oltvai Z. N. 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 10.1038/nrg1272 (doi:10.1038/nrg1272) [DOI] [PubMed] [Google Scholar]
- 15.Kashtan N., Alon U. 2005. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA 102, 13 773–13 778 10.1073/pnas.0503610102 (doi:10.1073/pnas.0503610102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alon U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 10.1038/nrg2102 (doi:10.1038/nrg2102) [DOI] [PubMed] [Google Scholar]
- 17.Silva-Rocha R., de Lorenzo V. 2010. Noise and robustness in prokaryotic regulatory networks. Annu. Rev. Microbiol. 64, 257–275 10.1146/annurev.micro.091208.073229 (doi:10.1146/annurev.micro.091208.073229) [DOI] [PubMed] [Google Scholar]
- 18.Goelzer A., Bekkal Brikci F., Martin-Verstraete I., Noirot P., Bessieres P., Aymerich S., Fromion V. 2008. Reconstruction and analysis of the genetic and metabolic regulatory networks of the central metabolism of Bacillus subtilis. BMC Syst. Biol. 2, 20. 10.1186/1752-0509-2-20 (doi:10.1186/1752-0509-2-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spirin V., Gelfand M. S., Mironov A. A., Mirny L. A. 2006. A metabolic network in the evolutionary context: multiscale structure and modularity. Proc. Natl Acad. Sci. USA 103, 8774–8779 10.1073/pnas.0510258103 (doi:10.1073/pnas.0510258103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer R., Danos V., Feret J., Krivine J., Fontana W. 2010. Intrinsic information carriers in combinatorial dynamical systems. Chaos 20, 037108. 10.1063/1.3491100 (doi:10.1063/1.3491100) [DOI] [PubMed] [Google Scholar]
- 21.Forster A. C., Church G. M. 2006. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2, 45. 10.1038/msb4100090 (doi:10.1038/msb4100090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jester B. C., Levengood J. D., Roy H., Ibba M., Devine K. M. 2003. Nonorthologous replacement of lysyl-tRNA synthetase prevents addition of lysine analogues to the genetic code. Proc. Natl Acad. Sci. USA 100, 14 351–14 356 10.1073/pnas.2036253100 (doi:10.1073/pnas.2036253100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baick J. W., Yoon J. H., Namgoong S., Soll D., Kim S. I., Eom S. H., Hong K. W. 2004. Growth inhibition of Escherichia coli during heterologous expression of Bacillus subtilis glutamyl-tRNA synthetase that catalyzes the formation of mischarged glutamyl-tRNA1 Gln. J. Microbiol. 42, 111–116 [PubMed] [Google Scholar]
- 24.Gobert A., Gutmann B., Taschner A., Gossringer M., Holzmann J., Hartmann R. K., Rossmanith W., Giege P. 2010. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 17, 740–744 10.1038/nsmb.1812 (doi:10.1038/nsmb.1812) [DOI] [PubMed] [Google Scholar]
- 25.Wegscheid B., Condon C., Hartmann R. K. 2006. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 7, 411–417 10.1038/sj.embor.7400641 (doi:10.1038/sj.embor.7400641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauch T., Hundley H. A., Pfund C., Wegrzyn R. D., Walter W., Kramer G., Kim S. Y., Craig E. A., Deuerling E. 2005. Dissecting functional similarities of ribosome-associated chaperones from Saccharomyces cerevisiae and Escherichia coli. Mol. Microbiol. 57, 357–365 10.1111/j.1365-2958.2005.04690.x (doi:10.1111/j.1365-2958.2005.04690.x) [DOI] [PubMed] [Google Scholar]
- 27.Asai T., Zaporojets D., Squires C., Squires C. L. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA 96, 1971–1976 10.1073/pnas.96.5.1971 (doi:10.1073/pnas.96.5.1971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong J., Skouloubris S., Dai Q., Myllykallio H., Barbour A. G. 2006. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J. Bacteriol. 188, 909–918 10.1128/JB.188.3.909-918.2006 (doi:10.1128/JB.188.3.909-918.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klutstein M., Shaked H., Sherman A., Avivi-Ragolsky N., Shema E., Zenvirth D., Levy A. A., Simchen G. 2008. Functional conservation of the yeast and Arabidopsis RAD54-like genes. Genetics 178, 2389–2397 10.1534/genetics.108.086777 (doi:10.1534/genetics.108.086777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shams-Eldin H., Chaban B., Niehus S., Schwarz R. T., Jarrell K. F. 2008. Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementation in Saccharomyces cerevisiae. J. Bacteriol. 190, 2217–2220 10.1128/JB.01778-07 (doi:10.1128/JB.01778-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Neumann J. 1958. The computer and the brain. New Haven, CT: Yale University Press [Google Scholar]
- 32.Danchin A. 2009. Bacteria as computers making computers. FEMS Microbiol. Rev. 33, 3–26 10.1111/j.1574-6976.2008.00137.x (doi:10.1111/j.1574-6976.2008.00137.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coffman J. A. 2011. Information as a manifestation of development. Information 2, 102–116 10.3390/info2010102 (doi:10.3390/info2010102) [DOI] [Google Scholar]
- 34.Coffman J. A. 2011. On causality in nonlinear complex systems: the developmentalist perspective. In Handbook of the philosophy of science: philosophy of complex systems, vol. 10 (ed. Hooker C.), pp. 214–221 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 35.Szathmary E., Smith J. M. 1995. The major evolutionary transitions. Nature 374, 227–232 10.1038/374227a0 (doi:10.1038/374227a0) [DOI] [PubMed] [Google Scholar]
- 36.Fontana W., Buss L. W. 1994. What would be conserved if ‘the tape were played twice’? Proc. Natl Acad. Sci. USA 91, 757–761 10.1073/pnas.91.2.757 (doi:10.1073/pnas.91.2.757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvestre D. A., Fontanari J. F. 2008. The information capacity of hypercycles. J. Theor. Biol. 254, 804–806 10.1016/j.jtbi.2008.07.023 (doi:10.1016/j.jtbi.2008.07.023) [DOI] [PubMed] [Google Scholar]
- 38.Kapranov P., et al. 2010. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature 466, 642–646 10.1038/nature09190 (doi:10.1038/nature09190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maida Y., Masutomi K. 2011. RNA-dependent RNA polymerases in RNA silencing. Biol. Chem. 392, 299–304 10.1515/BC.2011.035 (doi:10.1515/BC.2011.035) [DOI] [PubMed] [Google Scholar]
- 40.Eigen M., Biebricher C. K., Gebinoga M., Gardiner W. C. 1991. The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry 30, 11 005–11 018 10.1021/bi00110a001 (doi:10.1021/bi00110a001) [DOI] [PubMed] [Google Scholar]
- 41.Arendt J., Reznick D. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 10.1016/j.tree.2007.09.011 (doi:10.1016/j.tree.2007.09.011) [DOI] [PubMed] [Google Scholar]
- 42.Leander B. S. 2008. A hierarchical view of convergent evolution in microbial eukaryotes. J. Eukaryot. Microbiol. 55, 59–68 10.1111/j.1550-7408.2008.00308.x (doi:10.1111/j.1550-7408.2008.00308.x) [DOI] [PubMed] [Google Scholar]
- 43.Terada T., Nureki O., Ishitani R., Ambrogelly A., Ibba M., Soll D., Yokoyama S. 2002. Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nat. Struct. Biol. 9, 257–262 10.1038/nsb777 (doi:10.1038/nsb777) [DOI] [PubMed] [Google Scholar]
- 44.Roth A., Breaker R. R. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 10.1146/annurev.biochem.78.070507.135656 (doi:10.1146/annurev.biochem.78.070507.135656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert S. D., Rambo R. P., Van Tyne D., Batey R. T. 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 15, 177–182 10.1038/nsmb.1371 (doi:10.1038/nsmb.1371) [DOI] [PubMed] [Google Scholar]
- 46.Lu C., Smith A. M., Fuchs R. T., Ding F., Rajashankar K., Henkin T. M., Ke A. 2008. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat. Struct. Mol. Biol. 15, 1076–1083 10.1038/nsmb.1494 (doi:10.1038/nsmb.1494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serganov A. 2009. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 19, 251–259 10.1016/j.sbi.2009.02.002 (doi:10.1016/j.sbi.2009.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winfield M. D., Groisman E. A. 2004. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl Acad. Sci. USA 101, 17 162–17 167 10.1073/pnas.0406038101 (doi:10.1073/pnas.0406038101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha J., Reyes S. J., Gallivan J. P. 2010. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 6, 464–470 10.1038/nchembio.369 (doi:10.1038/nchembio.369) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Vetsigian K., Woese C., Goldenfeld N. 2006. Collective evolution and the genetic code. Proc. Natl Acad. Sci. USA 103, 10 696–10 701 10.1073/pnas.0603780103 (doi:10.1073/pnas.0603780103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Topp S., Gallivan J. P. 2007. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 129, 6807–6811 10.1021/ja0692480 (doi:10.1021/ja0692480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alaimo C., Catrein I., Morf L., Marolda C. L., Callewaert N., Valvano M. A., Feldman M. F., Aebi M. 2006. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25, 967–976 10.1038/sj.emboj.7601024 (doi:10.1038/sj.emboj.7601024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Castineira J. R., Lopez-Marques R. L., Villalba J. M., Losada M., Serrano A. 2002. Functional complementation of yeast cytosolic pyrophosphatase by bacterial and plant H + -translocating pyrophosphatases. Proc. Natl Acad. Sci. USA 99, 15 914–15 919 10.1073/pnas.242625399 (doi:10.1073/pnas.242625399) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Frithz-Lindsten E., Holmstrom A., Jacobsson L., Soltani M., Olsson J., Rosqvist R., Forsberg A. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol. Microbiol. 29, 1155–1165 10.1046/j.1365-2958.1998.00994.x (doi:10.1046/j.1365-2958.1998.00994.x) [DOI] [PubMed] [Google Scholar]
- 55.Davis T. N., Thorner J. 1989. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc. Natl Acad. Sci. USA 86, 7909–7913 10.1073/pnas.86.20.7909 (doi:10.1073/pnas.86.20.7909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Facchin S., Lopreiato R., Ruzzene M., Marin O., Sartori G., Gotz C., Montenarh M., Carignani G., Pinna L. A. 2003. Functional homology between yeast piD261/Bud32 and human PRPK: both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS Lett. 549, 63–66 10.1016/S0014-5793(03)00770-1 (doi:10.1016/S0014-5793(03)00770-1) [DOI] [PubMed] [Google Scholar]
- 57.Randau L., Schroder I., Soll D. 2008. Life without RNase P. Nature 453, 120–123 10.1038/nature06833 (doi:10.1038/nature06833) [DOI] [PubMed] [Google Scholar]
- 58.Merino E., Jensen R. A., Yanofsky C. 2008. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 11, 78–86 10.1016/j.mib.2008.02.005 (doi:10.1016/j.mib.2008.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vitreschak A. G., Mironov A. A., Lyubetsky V. A., Gelfand M. S. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14, 717–735 10.1261/rna.819308 (doi:10.1261/rna.819308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanofsky C. 2007. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 13, 1141–1154 10.1261/rna.620507 (doi:10.1261/rna.620507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poiata E., Meyer M. M., Ames T. D., Breaker R. R. 2009. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15, 2046–2056 10.1261/rna.1824209 (doi:10.1261/rna.1824209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luisi P. L. 1998. About various definitions of life. Orig. Life Evol. Biosph. 28, 613–622 10.1023/A:1006517315105 (doi:10.1023/A:1006517315105) [DOI] [PubMed] [Google Scholar]
- 63.Joyce G. F. 1989. RNA evolution and the origins of life. Nature 338, 217–224 10.1038/338217a0 (doi:10.1038/338217a0) [DOI] [PubMed] [Google Scholar]
- 64.Danchin A. 2009. Natural selection and immortality. Biogerontology 10, 503–516 10.1007/s10522-008-9171-5 (doi:10.1007/s10522-008-9171-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellington A. D., Chen X., Robertson M., Syrett A. 2009. Evolutionary origins and directed evolution of RNA. Int. J. Biochem. Cell Biol. 41, 254–265 10.1016/j.biocel.2008.08.015 (doi:10.1016/j.biocel.2008.08.015) [DOI] [PubMed] [Google Scholar]
- 66.Woese C. R. 2002. On the evolution of cells. Proc. Natl Acad. Sci. USA 99, 8742–8747 10.1073/pnas.132266999 (doi:10.1073/pnas.132266999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogan A. A., Higgs P. G. 2011. The advantages and disadvantages of horizontal gene transfer and the emergence of the first species. Biol. Direct 6, 1. 10.1186/1745-6150-6-1 (doi:10.1186/1745-6150-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gould S. J. 1989. Wonderful life: the burgess shale and the nature of history. New York, NY: W. W. Norton & Company [Google Scholar]
- 69.Conway Morris S. 1998. The crucible of creation: the burgess shale and the rise of animals. New York, NY: Oxford University Press [Google Scholar]
- 70.Pereto J., Catala J. 2007. The renaissance of synthetic biology. Biol. Theory 2, 128–130 10.1162/biot.2007.2.2.128 (doi:10.1162/biot.2007.2.2.128) [DOI] [Google Scholar]
- 71.Keasling J. D. 2008. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 10.1021/cb7002434 (doi:10.1021/cb7002434) [DOI] [PubMed] [Google Scholar]
- 72.Szostak J. W., Bartel D. P., Luisi P. L. 2001. Synthesizing life. Nature 409, 387–390 10.1038/35053176 (doi:10.1038/35053176) [DOI] [PubMed] [Google Scholar]
- 73.Schrum J. P., Zhu T. F., Szostak J. W. 2010. The origins of cellular life. Cold Spring Harb. Perspect. Biol. 2, a002212. 10.1101/cshperspect.a002212 (doi:10.1101/cshperspect.a002212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lartigue C., Glass J. I., Alperovich N., Pieper R., Parmar P. P., Hutchison C. A., III, Smith H. O., Venter J. C. 2007. Genome transplantation in bacteria: changing one species to another. Science 317, 632–638 10.1126/science.1144622 (doi:10.1126/science.1144622) [DOI] [PubMed] [Google Scholar]
- 75.Gibson D. G., et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 10.1126/science.1151721 (doi:10.1126/science.1151721) [DOI] [PubMed] [Google Scholar]
- 76.Kaiser J. 2007. Synthetic biology. Attempt to patent artificial organism draws a protest. Science 316, 1557. 10.1126/science.316.5831.1557 (doi:10.1126/science.316.5831.1557) [DOI] [PubMed] [Google Scholar]
- 77.Gibson D. G., et al. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 10.1126/science.1190719 (doi:10.1126/science.1190719) [DOI] [PubMed] [Google Scholar]
- 78.Jewett M. C., Forster A. C. 2010. Update on designing and building minimal cells. Curr. Opin. Biotechnol. 5, 697–703 10.1016/j.copbio.2010.06.008 (doi:10.1016/j.copbio.2010.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Claverie J. M., Abergel C. 2010. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 26, 431–437 10.1016/j.tig.2010.07.003 (doi:10.1016/j.tig.2010.07.003) [DOI] [PubMed] [Google Scholar]
- 80.Forterre P. 2010. Giant viruses: conflicts in revisiting the virus concept. Intervirology 53, 362–378 10.1159/000312921 (doi:10.1159/000312921) [DOI] [PubMed] [Google Scholar]
- 81.Boyer M., et al. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl Acad. Sci. USA 106, 21 848–21 853 10.1073/pnas.0911354106 (doi:10.1073/pnas.0911354106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 10.1038/nature08721 (doi:10.1038/nature08721) [DOI] [PubMed] [Google Scholar]
- 83.Peralta-Yahya P. P., Keasling J. D. 2010. Advanced biofuel production in microbes. Biotechnol. J. 5, 147–162 10.1002/biot.200900220 (doi:10.1002/biot.200900220) [DOI] [PubMed] [Google Scholar]
- 84.An W., Chin J. W. 2009. Synthesis of orthogonal transcription-translation networks. Proc. Natl Acad. Sci. USA 106, 8477–8482 10.1073/pnas.0900267106 (doi:10.1073/pnas.0900267106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J. W. 2010. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 10.1038/nature08817 (doi:10.1038/nature08817) [DOI] [PubMed] [Google Scholar]
- 86.Gallivan J. P. 2007. Toward reprogramming bacteria with small molecules and RNA. Curr. Opin. Chem. Biol. 11, 612–619 10.1016/j.cbpa.2007.10.004 (doi:10.1016/j.cbpa.2007.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson J. C., Dueber J. E., Leguia M., Wu G. C., Goler J. A., Arkin A. P., Keasling J. D. 2010. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4, 1. 10.1186/1754-1611-4-1 (doi:10.1186/1754-1611-4-1) [DOI] [PMC free article] [PubMed] [Google Scholar]