Abstract
Cerebral aneurysms are a multi-factorial disease with severe consequences. A core part of the European project @neurIST was the physical characterization of aneurysms to find candidate risk factors associated with aneurysm rupture. The project investigated measures based on morphological, haemodynamic and aneurysm wall structure analyses for more than 300 cases of ruptured and unruptured aneurysms, extracting descriptors suitable for statistical studies. This paper deals with the unique challenges associated with this task, and the implemented solutions. The consistency of results required by the subsequent statistical analyses, given the heterogeneous image data sources and multiple human operators, was met by a highly automated toolchain combined with training. A testimonial of the successful automation is the positive evaluation of the toolchain by over 260 clinicians during various hands-on workshops. The specification of the analyses required thorough investigations of modelling and processing choices, discussed in a detailed analysis protocol. Finally, an abstract data model governing the management of the simulation-related data provides a framework for data provenance and supports future use of data and toolchain. This is achieved by enabling the easy modification of the modelling approaches and solution details through abstract problem descriptions, removing the need of repetition of manual processing work.
Keywords: cerebral aneurysms, computational imaging and modelling, computational physiology, virtual physiological human
1. Introduction
Cerebral aneurysms are local pathological dilatations of arteries in the brain causing significant morbidity and mortality rates [1]. A large proportion (1–5%) of the population has unruptured aneurysms. Once an aneurysm ruptures, subarachnoid haemorrhage (SAH) usually occurs, and the impact is devastating. After SAH, patients have a 45 per cent 30 day mortality rate, and an estimated 30 per cent of survivors present moderate-to-severe disability. As a consequence, SAH induces a serious burden for patients, their relatives and society.
A considerable debate regarding the optimal treatment of patients harbouring unruptured intracranial aneurysms has developed in recent years, stemming largely from the influence of several factors. Among them stand out the impact of recent technological improvements on diagnostic and interventional imaging, the appearance of a new generation of therapeutic devices, and the increase in the number of large-scale, randomized, multi-institutional studies centred in the outcome evaluation of existing treatments [2] as well as the evolution of unruptured aneurysms [3]. The combination of these factors and the sometimes contradictory conclusions introduced by these studies have led to considerable uncertainty about the optimal treatment for patients harbouring unruptured intracranial aneurysms.
These efforts from the clinical community have caused an increasing interest in modelling the complex multi-factorial mechanisms thought to be involved in aneurysm genesis, growth and rupture, which include genetic, physical and environmental factors. This global approach to the disease links with the wide international interest in the construction of an integrated in silico human. The ‘IUPS Physiome’ [4,5] provides the framework for a hierarchy of models spanning the length scales from the molecule to the individual. Other activities exploring the development of multi-scale and multi-disciplinary physiological models up to specific organ level have resulted in the development of the virtual physiological human (VPH) [6–8], in the context of which we could frame this work.
1.1. The @neurIST project
This work was developed as part of the European project ‘Integrated Biomedical Informatics for the Management of Cerebral Aneurysms’ (@neurIST; http://www.aneurist.org) [9,10]. @neurIST was a 4 year project started in 2006 with a 17 million euro budget, gathering 28 public and private institutions of 12 European countries, including industrial, medical and academic institutions. Several organizations from the USA, New Zealand and Japan participated as external collaborators. The mission statement of the project was the following:
@neurIST will transform the management of cerebral aneurysms by providing new insights, personalized risk assessment, and methods for the design of improved medical devices and treatment protocols.
To fulfil this mission, four software suites (@neuRisk, @neuLink, @neuFuse and @neuEndo) and two infrastructure components (@neuCompute and @neuInfo) were developed to collaborate with each other [11–13]. @neuRisk provides personalized risk assessment and treatment guidelines to practitioners based on the analyses performed by the other three suites. @neuLink enables not only the study of the existing links between genetics and disease, but also provides links to the biomechanical indices extracted from @neuFuse. @neuFuse addresses the patient data fusion from different modalities and the extraction of such biomechanical indices. @neuEndo works closely to the other three suites facilitating the optimization and customization of medical devices used for patient treatment. The two infrastructure components allowing the interoperability and collaboration of the suites are @neuCompute, by providing high-performance-distributed computing capabilities, and @neuInfo, by enabling a homogeneous data access and storage to the heterogeneous data types handled in the suites. A more detailed description of the @neurIST system architecture is provided in Benkner et al. [14], where the authors introduce the developed service-oriented IT infrastructure supporting the seamless access to distributed medical data and computational resources in an easy and secure manner.
From the previous, one can understand the role that @neurIST has in tackling a disease in a global manner, a concept which directly links to the VPH. The developed multi-scale paradigm could be extended to other diseases.
1.2. Finding biomechanical risk factors: approach and challenges
This paper describes the @neurIST complex information processing toolchain approach to provide physical models of cerebral aneurysms and derive representative descriptors that could be associated with their rupture.
To create a comprehensive basis for investigating potential physical risk factors, the project aimed to compute for a large number of cases many different characterizations based on the analysis of shape, blood flow (haemodynamics) and wall mechanics. To enable statistical analyses, the typically continuous output of these analyses had to be reduced to a finite number of descriptors suitable for finding statistical associations that would help understanding the natural history and the risks of available treatments.
To this end, @neurIST collected extremely heterogeneous data that included patient's clinical history, images and blood samples, for more than a total of 1400 patients with ruptured and unruptured aneurysms. Our goal was then to compute the whole set of physical characterizations mentioned before for the nearly 500 patients having associated medical images.
To reach these goals, we had to face three major challenges. First, to determine the details of the most representative biomechanical analyses to be performed and select the most meaningful physical descriptors that could be derived (§2). Second, to enable performing multi-case, multi-operator and multi-site analyses guaranteeing consistency and repeatability among users and processing sites, and to reduce the manual work needed (§3). And third, to preserve the provenance of the derived descriptors, to link back to the clinical data, and to permit revisions of the modelling decisions, allowing for future variations and extensions of the analysis (§4).
Section 5 gives an account on our practical experience with using the toolchain. Among others, the work needed to synchronize between different processing teams and feedback collected from exposure to clinicians, i.e. non-experts with respect to the tools. Finally, we discuss how relevant these results are for clinical practice and VPH research in general.
2. Challenge 1: biomechanical modelling and descriptors
The first challenge faced while designing the @neurIST toolchain was to identify the set of most representative biomechanical descriptors suitable for performing risk assessment and association studies, and to identify the most appropriate computational model of the underlying physical problem. Along the lines set by Ma et al. [15] and Ma [16], three general analyses were chosen to be performed: morphological, haemodynamic and structural. These analyses have been linked in the literature to aneurysm genesis, growth and rupture [17–19]. Evidence indicating differences in morphology [20–23] and flow [18,24] between ruptured and unruptured aneurysms have been shown for reduced patient cohorts. Structural wall mechanics [25–28] has been used to justify the growth and remodelling happening at the aneurysm level.
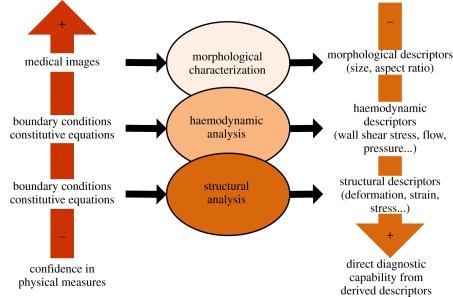
The balance between merits and demerits of each analysis is sketched in figure 1. The theoretical level of confidence in the physical measures on which each of the analyses are based and the direct diagnostic capability derived from them varies inversely depending on the analysis. As an example, morphological characterizations have the least requirements in terms of the physical measures, only depending on the anatomical information extracted from the medical image. This is opposed to the difficulties in obtaining the physical measures needed to perform haemodynamic and structural analyses in this order of complexity. In the case of haemodynamics, the exact boundary conditions (inlet, outlet and wall movement) are usually unknown. For structural analysis, the situation is worse, as lack of information on the aneurysm wall (thickness, individual material properties) limits the analysis to provide qualitative results only. In contrast, the theoretical diagnostic capability would certainly be highest for a quantitatively correct wall mechanics analysis, as stresses exceeding the strength of the wall could be more directly associated with the mechanical processes inducing growth and remodelling phenomena and, therefore, linked to the event of rupture. Nevertheless, practically, morphological characterizations might currently have the highest predictive capabilities with respect to the other analyses.
Figure 1.
The theoretical diagnostic power (right) versus the practical reliability of physical measures (left) of the different analysis modes are inversely related [29].
Once these analyses were chosen, based on prior evidences found in the literature, and especially in the case of the haemodynamic and structural analyses, the immediate need was to decide what to simulate and how to perform such simulations. Unfortunately, there is not a clear answer to this problem. Each of the modelling choices presented trade-offs [30] and needed to be judged in the light of the available input data and intended purpose of the output.
To address these issues, an analysis protocol was developed in two stages. During the first stage, an initial version of the protocol was created and where the fundamental alternatives were identified, the core processing steps and data entities were specified and the topics that needed further exploration were highlighted. As an example of the last, the need for performing sensitivity studies was discussed. An external scientific advisory board of experts reviewed the protocol and raised several questions, including the need of performing linear versus nonlinear wall mechanics studies, using moving versus fixed wall boundary conditions for haemodynamics, employing coupled or isolated models (fluid structure interaction being an option) and using a personalized versus generic systemic model.
As a result, a second version of the protocol was written [29] providing a high-level tool-independent description of the processing chain, clearly describing each major operation with the specification of inputs and outputs. In addition, it included references to several of the studies carried out by the project to evaluate aspects such as the sensitivity of flow simulations to the imaging modality [31] and inlet boundary conditions setting [32], and the relationship between patient-specific and generic boundary conditions [33].
3. Challenge 2: implementation of the toolchain
The second challenge was to enable multi-case, multi-user and multi-site analyses, which guaranteed consistency and repeatability among operators and geographically disperse processing sites, while minimizing the amount of required user manual intervention. However, some steps had to remain under user control and considerable efforts had to be devoted to synchronize the different operators in the various processing teams. In the current section, the actual implementation of the toolchain is discussed, highlighting the efforts made to automate or semi-automate it. The toolchain encompassed the whole process from reading a medical image to the extraction of the morphological, structural and haemodynamic descriptors selected in the analysis protocol. These were combined with information available from the patient record and stored within a structured data repository, as described in §4.
3.1. Vascular geometry creation
All three analyses specified in the analysis protocol get as an input a surface mesh. Therefore, the first stage in the toolchain proceeds to create a vascular geometry from a three-dimensional rotational angiographic image (3DRA). This imaging modality is currently considered the gold standard [34,35] for aneurysm detection owing to its superior spatial and contrast resolution. A three-dimensional triangulated surface mesh is obtained using an automatic segmentation method based on geodesic active regions (GAR) in combination with an image standardization technique [36,37], recently presented and validated by Bogunovic et al. [38]. The method eliminates most of the dependency on the operator, and on the specific imaging protocols and equipment used. The only possible choice for the operator is the selection of the region of interest to be segmented as opposed to other methods such as the manual iso-intensity surface extraction (ISE), where in addition the selection of a threshold is left to the operator. While the impact of the selection of the region of interest for both methods has shown to be negligible, GAR outperformed ISE qualitatively and quantitatively for 3DRA and time of flight magnetic resonance angiographic images obtained from clinical routine when compared with manual measurements performed by clinicians. The dependency on the equipment used to image the patient is removed, thanks to the use of a training set and standardized images. The impact of the selection of images to build a training set has also shown to be minimal when compared with the inter-observer variability among clinicians when performing manual measurements. In terms of execution time, it was able to segment a region of interest with a size of 2563 voxels in 17 ± 4 min (average ± standard deviation) on a standard personal computer with an Intel quad-core 2.4 GHz processor and 4GB of memory.
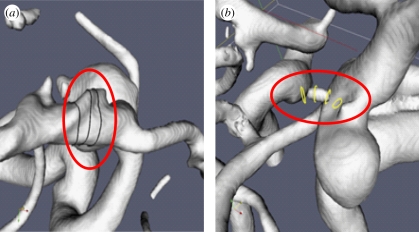
Once the segmented surface mesh is available, it is manually manipulated to remove some of the artefacts not belonging to the cerebral vasculature or not relevant for the subsequent analyses (figure 2). A skeletonization algorithm [39] is used to extract the skeleton (medial axis) of this geometry. The skeleton acts as a reference to perform the perpendicular cuts where the flow boundary conditions need to be set, and as the linking point to the three-dimensional model of the human systemic circulation [40]. This first step in the toolchain is critical because the resulting vascular geometry is used as the basis for all further analyses. The impact of the chosen imaging modality and the quality of the image might have a strong influence in quantitative parameters extracted from flow simulations [31]. Therefore, the role of the operator in accepting or rejecting the images based on their quality before they are processed is fundamental for the accuracy of subsequent analyses. In our experience in @neurIST, the approximate discard rate of 3DRA images for all clinical centres involved in the data collection was between 30 and 40 per cent.
Figure 2.
Examples of typical topological problems introduced during the segmentation because of low image quality: (a) touching vessels and (b) missing vessels.
3.2. Morphological characterization chain
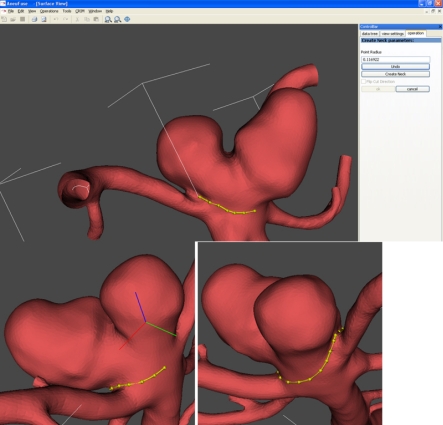
A collection of morphological descriptors of various complexity was selected among the wide variety available in the literature [21,22]. Basic size indices such as aspect ratio [20], non-sphericity index [22], aneurysm volume and surface area are calculated for the aneurysm sac. Aspect ratio relates the aneurysm depth and neck width, while non-sphericity index relates the aneurysm volume and surface area. To isolate the aneurysm sac, the aneurysm neck is manually delineated as shown in figure 3. A set of more sophisticated descriptors called three-dimensional Zernike moment invariants [23,41] are also used to characterize the aneurysm shape and a portion of the surrounding feeding vessels using a fixed cutting protocol at every inlet and outlet. Three-dimensional Zernike moments provide a complete three-dimensional morphological characterization of objects, and their invariants remove scale and orientation dependency. These have shown for a small heterogeneous population promising rupture prediction rates [42]. Finally, inlet and outlet vessel diameters are also calculated at the cut planes used to isolate the geometry for which the Zernike moment invariants are computed. All of these descriptors are automatically computed in real time from the isolated geometries of the sac and aneurysm plus a portion of the surrounding feeding vessels.
Figure 3.
Example of the manual delineation of the aneurysm neck implemented in the @neurIST toolchain.
3.3. Haemodynamics analysis chain
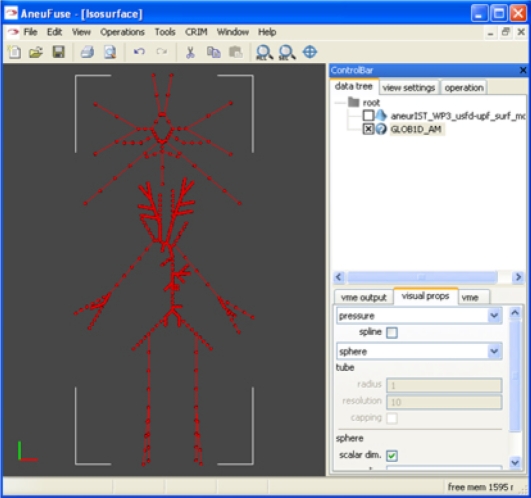
Two complementary haemodynamic analyses are carried out as part of this chain: a global one and a local one, with an optional uni-directional coupling between them. The global one is based on a one-dimensional model of the human systemic circulation [40], including the main arteries of a complete circle of Willis, and predicts flow rate and pressure waveforms at predefined points (figure 4). The local analysis uses the full three-dimensional geometry near the region of interest, obtained by clipping the three-dimensional geometry at appropriate places [29,32].
Figure 4.
A view of the @neurIST one-dimensional model.
As no patient-specific measurements of flow properties at the openings defined by the clipping are generally available, the results of the one-dimensional global analysis can be used to obtain boundary conditions for the local three-dimensional analysis. Using the skeletonized representation of the three-dimensional vasculature in the region of interest calculated during the vascular geometry extraction stage, the operator manually defines perpendicular clipping planes and links the appropriate nodes of the one-dimensional circulation model to the corresponding points of the three-dimensional inlet and outlet boundaries, thus defining the boundary conditions for the three-dimensional model (figure 4).
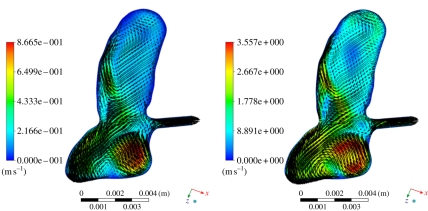
The set of qualitative and quantitative descriptors (candidate risk factors) defined by the analysis protocol were extracted from the flow solution automatically, except a few qualitative ones such as flow stability and intra-aneurysmal flow type (figure 5) requiring expert judgement. Quantitative volumetric measures were performed within the aneurysm and at the neck. These included the maximum and mean velocities defined at peak systole and averaged over the cardiac cycle. Similarly, other measures were computed on the model surface, including areas of elevated pressure, impingement jet location, maximum static pressure on the wall at peak systole and the wall shear stress and derivates such as the oscillatory shear index. Some of these measures are highly dependent on the location of the aneurysm neck, which unfortunately had to be manually defined as already mentioned in the morphological analysis. There have been several efforts in the literature aimed to solve this problem in an automated way, but to date none has been able to extract the aneurysm neck reliably in the general case [43–46].
Figure 5.
Example of unstable flow from (a) end diastole and (b) peak systole.
The analysis protocol chose to model the transient blood flow by the Navier–Stokes equations under the assumption of rigid walls and a simple Newtonian rheology model with blood density of 1060 kg m−3 and viscosity of 0.0035 Pa s.
The production runs were performed using a commercial three-dimensional flow solver ANSYS CFX (Ansys Inc., Canonsburg, PA, USA), which is a finite-volume computational fluid dyamics (CFD) code solving for the Navier–Stokes flow equations across an unstructured three-dimensional discretization (known as mesh) of the vasculature volume. Such mesh was generated by the ANSYS ICEM 3D (Ansys Inc.) mesh generator that labelled the aneurysmal interior in a completely automatic manner.
3.4. Structural analysis chain
Of all types of analyses in figure 1, an ‘accurate’ structural analysis would have the largest predictive capabilities. Nevertheless, in practice it also is the analysis requiring the most assumptions and simplifications. Still it can be hoped to gain information about risk factors [28]. In @neurIST, and owing to the lack of patient-specific data, the assumptions being made included averaged material properties and aneurysmal wall thickness and spatially and temporally constant pressure, for simplicity assumed at diastole. However, the membrane code FEANOR [47], developed specifically for @neurIST, computes the zero-load state of the aneurysm under the above assumptions, which is a first step towards more realistic wall mechanics simulation.
The thickness of the aneurysm wall was set to a constant value of 86 µm, the thickness of the vessel wall to 500 µm, and was linearly interpolated in a transition region (figure 6a). Owing to this thin wall, computing the zero-load state is a numerically delicate process (figure 6) and great effort was required to make this numerically stable and hence automatic.
Figure 6.
Computing the zero-load state leads to considerable wrinkling and is numerically delicate. (a) Computed zero-load configuration (with assumed thickness) and (b) stresses at 120 mmHg computed by using the nonlinear material model.
Two different protocols are supported. The first one uses a nonlinear material model and computation of a zero-load state, and the second uses a linear material model assuming zero stresses at diastole. Given the essential uncertainties in quantitatively estimating the model parameters, the characteristic measures include only relative measures like ratio of areas exceeding certain thresholds of maximum stress and strain, as these are expected to be less sensitive to variations of the unknown patient-specific values of aneurysm wall thickness and material properties.
4. Challenge 3: A model for @neurIST simulation data
4.1. Motivation and overview
The third challenge was to preserve the provenance of the derived descriptors, to be able to link back to the clinical data and to permit future revisions of the modelling decisions, allowing for future variations and extensions of the analysis. This concern was partially originated by the huge amount of manual work going into the toolchain and into the data processing, despite the level of automation and semi-automation achieved during the implementation of the toolchain. Moreover, feeding the results into the sophisticated data mining machinery of @neuLink [11] meant that various subsets and combinations of the result data were possibly needed, in order to select statistically meaningful groups of cases and to provide the greatest flexibility in the search for patterns.
The answer was to design a systematic model of the different data entities involved to facilitate meeting these requirements. Relationships, mutual dependencies and history (provenance) needed to be clearly captured. In contrast to the general approaches for provenance recording described in Simmhan et al. [48], our approach is geared towards the specific needs of the @neurIST project. According to the taxonomy of Simmhan et al. [48], we use a coarse-grained and somewhat abstracted replication recipe (see also below §4c on abstract problem descriptions (APDs)). At the same time, to maximize opportunities for future use, the entities in this model had to be represented in a sufficiently general way in order to distinguish from details specific to concrete tools and applications. That is, to abstract from information which will become obsolete sooner or later. This application-neutrality was already required within the @neurIST complex information toolchain, as it was designed to support different solvers for the same computational problem (e.g. ANSYS CFX and a Lattice–Boltzmann code for haemodynamics).
Our solution thus consisted of the following ingredients:
— A relational data model (using structured query language (SQL) as data definition language) linking the important classes of simulation (or ‘derived’) data to the clinical data, hence ‘derived data model’ (DDM).
— A set of tools automatically translating the abstract, application-neutral description of the DDM into complete input specifications for concrete simulation applications.
— A strategy for storing and retrieving bulk data, like raw simulation results, images, etc. While most of the bulk data is application-specific and can be reproduced using the high-level data and the two tools just mentioned, it may be worthwhile to keep it for a certain period of time, e.g. for visualization and review purposes.
4.2. A relational data model for derived data
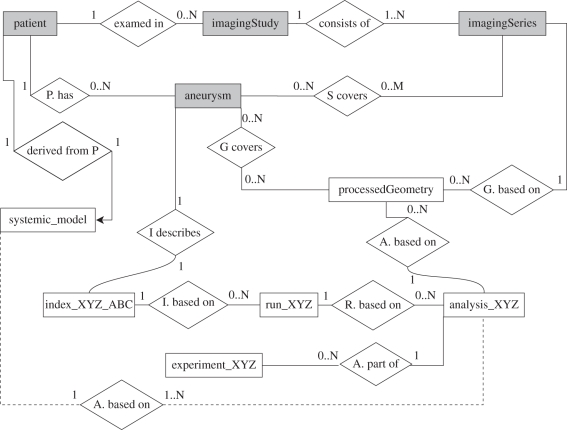
The DDM forms the core of these developments. A high-level overview in the form of an entity-relationship (ER) diagram is shown in figure 7. The DDM covered the following major entities.
— Aneurysm. The anatomical structure in the focus of interest.
— Image (split in imagingStudy and imagingSeries). A medical image of the region of interest.
— Processed geometry. A valid geometric representation of one or more aneurysms with anatomically correct topology.
— Analysis. A high-level description of a computational model, e.g. blood flow through the vasculature surrounding an aneurysm.
— Run. An actual computation of an analysis with a simulation programme.
— Index. A discrete1 characterization of an aneurysm, e.g. aspect ratio, collection of three-dimensional Zernike moment invariants or maximal wall shear stress inside the aneurysm.
— Experiment. A group of similar analyses suitable for finding statistical correlations.
Figure 7.
An entity-relationship diagram of the @neurIST derived data model [29]. Grey entities link to their counterparts in the clinical reference information model (CRIM) developed in the project, white ones to derived (i.e. computed) data. The diamond-shaped boxes describe the relationship of two entities, and the numbers give their multiplicities. For example, a processed geometry is based on exactly one imaging series, and each imaging series is the basis for 0 up to N processed geometries. The suffix XYZ is a placeholder for the different types of analysis.
Some of these entities (namely aneurysm and image) contained references to their counterparts in clinical databases that may be enhanced with additional information, such as image quality. The clinical databases containing the anonymized patient's clinical record use their own data model developed in the project, the @neurIST clinical reference information model (CRIM). The DDM was kept deliberately separate from the CRIM to minimize dependencies.
The central use case of the DDM in our context of finding risk indicators is the ability to retrieve answers to questions (known as queries) like ‘for a set of aneurysms, retrieve rupture status and values of biomechanical characterizations A, B, and C’. This query could easily be refined by restriction to aneurysms in certain locations, or by excluding cases with certain irregularities (like bad image quality), and so on.
An important means to form meaningful groups of comparable analyses is the experiment entity. For instance, we used two different experiments for the wall mechanics analyses, one using a linear material model, and one using a non-linear model with computation of a zero-load state. Another example of use of experiments is for different conventions for geometry clipping in shape analysis. Clearly, it does not make sense to mix analyses pertaining to different experiments in a statistical evaluation.
4.3. Handling bulk data
The storage of the actual bulk data (meshes, pictures and full simulation results) can be handled in a quite flexible way, as the data model provides a placeholder for each such data entities, with the necessary meta-data as to where and how it is stored. Smaller data like surface meshes can even be stored directly in a data base management system (DBMS) as binary large object (BLOB). Data stored outside the DBMS can be referenced by URLs, or most generally, by endpoint references (EPR), containing the details for addressing a service delivering the data.
An important criterion to support decisions about whether to store a particular piece of data, such as raw simulation results or processed geometries, is the cost associated to (re-)generate them. This can be computational costly as well as, and more importantly, the amount of manual work needed to create them. For the case of the @neurIST toolchain, this differentiates processed geometries, possibly requiring a lot of manual work, see §3a, from simulation results, which can be generated automatically from their APDs, a concept we will discuss in more detail next.
4.4. An important concept: abstract problem descriptions
An APD is a high-level definition of a computational biomechanical problem, like a blood flow analysis through an aneurysm and surrounding vasculature. Despite their high-level nature, they contain or reference all information needed to generate input for any concrete simulation application run computing the analysis. There could be different runs using different applications for the same analysis, which should yield the ‘same’ results (up to some limit of accuracy). Sets of different runs could be used for verification of the solver-independence of results.
The conversion of an APD into a concrete simulation input and the subsequent run is, in principle, a completely automatic task, as the APD contained all relevant information; all necessary pieces of data needing human intervention are generated and stored before creating the APD. In practice, the feasibility of automation is certainly dependent on the application; in the case of @neurIST, we fully automated this step for the supported simulation packages. An exception to this rule are some qualitative characterizations that inevitably required human intervention.
The precise but application-neutral representation of biomechanical problems in the APDs enables their long-term storage, making them insensitive to the rise and fall of simulation applications, or just incompatibilities between versions. For instance, we integrated two fundamentally different types of CFD solvers. First, ANSYS CFX based on a finite volume scheme on unstructured meshes, and, second, a Lattice–Boltzmann solver, based on regular structured grids (the latter was not used for production runs). Basically, this meant implementing specific preprocessors translating the APD into either ANSYS CFX or Lattice–Boltzmann input. The abstractness of the representation also facilitates automatic transformation of analyses to yield variants implementing different experiments. For instance, it would be easy to implement features like changing the generation of boundary conditions or using a different material model. Such automatic transformations pave the way to practical future uses of the results, like doing parametric studies or setting up completely new modelling approaches.
5. Practical experience
Even if the degree of automation accomplished in the implementation of the complex information toolchain is beyond the state of the art, it was not sufficient to achieve consistent results across the different processing teams. To achieve this, training was fundamental, and verification of the level of understanding of the case processing rules was mandatory. To evaluate the overall impact of the operator decisions on the final-derived descriptors, six patient images were chosen as demonstrators and were fully processed by eight representatives of the processing teams. The objective was to synchronize the results of the operators for the shape, structural and haemodynamic analyses.
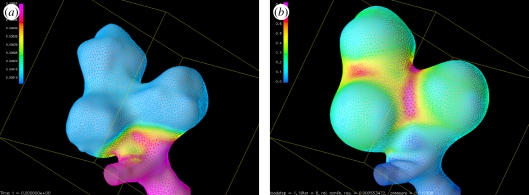
Several rounds of this synchronization were needed before a common agreement was reached and a set of accurate guidelines could be distilled. This process was simpler in the case of the morphological analysis and only two rounds were needed. As an example of the importance of this process, see in figure 8 the surprising variability in the results for some operators after the first synchronization iteration. In the case of the haemodynamic analysis, the synchronization took longer because of the elevated complexity of this chain as well as the impact that operator decisions could have on the final simulation results. Still, agreement was reached in the end and the @neurIST case processing was started.
Figure 8.
Results for the first synchronization round showing the differences among six of the eight operators during the manual isolation of the region of interest. Operators 1, 2 and 4 showed significant discrepancies with the correct geometries isolated by operators 3, 5 and 6.
In total, 15 operators from four case processing centres were involved and fully processed images from five hospitals producing descriptors for more than 300 aneurysms. The essential data items created during this process were stored using the mechanism described in §4 and are thus available for future work. Statistical associations are currently being analysed between these descriptors and the aneurysm rupture status information, among others.
In addition, we also wanted to evaluate the suitability of our toolchain for clinical practice. In this spirit, and to gain useful feedback, the toolchain has been officially exposed to more than 260 neurointerventionists, neurosurgeons and neurologists for their evaluation through hands-on workshops at different public events, where the participants worked under guidance through a simple case, from medical image to a three-dimensional blood flow simulation. They achieved results comparable to those obtained by experts users, with similar performance.
The main venues have been the 1st and 2nd European Society for Minimally Invasive Neurovascular Treatment (ESMINT) Teaching Courses, respectively, held in Lisbon, Portugal (7–12 September 2008) and Barcelona, Spain (13–18 December 2009), the 4th International Conference on Computational Bioengineering, Bertinoro, Italy (16–18 September 2009), the XIV World Congress of Neurological Surgery, Boston, USA (30 August–4 September 2009) and the Simposio Internacional de Neuroradiología Intervencional, Santiago de Chile, Chile (26–27 November 2009).
The collected feedback was used to further improve the subsequent releases of the toolchain. Some of the conclusions reached during the 1st ESMINT Teaching Course were reported and described in Singh et al. [49]. In general, clinicians recognized shortcomings in current management of cerebral aneurysms and the need for better understanding of the disease and the identification of novel and more efficient risk descriptors. Although most clinicians believe that physical characterization (mainly haemodynamics at that workshop) may offer better diagnostic value, there is a clear lack of awareness concerning the role of haemodynamics in the aetiopathogenesis of cerebral aneurysms and the use of CFD in this context.
6. Discussion
A number of unique challenges were associated with the large-scale multi-case, multi-operator and multi-site biomechanical processing undertaken by @neurIST. The solutions we have developed to meet these challenges are, we believe, of interest not only for the field of aneurysm research, but also beyond that for the larger VPH community studying other diseases.2
The toolchain itself was developed and automated to a degree which proved sufficient to process more than 300 aneurysms minimizing and quantifying the impact of human operators, either by ensuring synchronization among operators through training or by performing several sensitivity studies. Also, more than 260 clinicians ran full blood flow simulations starting from medical images, using our toolchain during internationally renowned hands-on events. Their feedback convinced us that these kinds of tools will find their way into clinical practice. All together, the amount of practical usage, testing and continual improvement based on user feedback of this toolchain certainly surpasses that of most similar research-oriented efforts. There is, however, still room for improvements.
Our analysis protocol [29], with its detailed specifications and ample discussion of limitations versus modelling alternatives, is an ideal starting point for continuing and extending our efforts. It can also serve as a blueprint for transferring the approach to other diseases, which should be straightforward for vascular diseases.
The analysis of the biomechanical characterizations with respect to their diagnostic value is still ongoing. Some preliminary works [42,50–53] have already used the toolchain on a reduced number of cases. However, it can be expected that the larger the number of cases, the statistically more reliable conclusions will be achieved compared with similar prior works [53], but always under the strong and arguable assumption that aneurysms do not change over time.
Using the toolchain to simulate flow in the presence of virtual endovascular devices such as stents [54] currently allows evaluating the performance of endovascular devices from a haemodynamic point of view [55,56], benefitting from the ability of setting boundary conditions based on a one-dimensional model of the systemic circulation [32,33,40]. The inclusion in the project of genetics data has rendered already relevant results [57] to the scientific community in identifying three new risk loci, which still need to be linked to biomechanical factors through @neuLink [12].
The results of our processing are organized using the DDM and are available for future extensions of the our work, both by adding new cases or by varying the modelling approach. In this case, it might be interesting to enable the toolchain to investigate effects of certain drugs on blood viscosity and hence on the computed flow characterizations. The prospect of being able to run large-scale computational experiments with these many cases without having to invest a lot of human effort into manual case-by-case model adaptation is exciting.
We believe that our approach has the potential to serve as a model for organizing large-scale multi-case simulation in the VPH context in general, as it supports looking both into the past, by providing provenance information, and into the future, by providing a means to modify toolchain details (like replacing specific solvers) and to transform problem descriptions to support alternative modelling choices. Currently, the model only supports the coupling between the systemic and the local three-dimensional haemodynamics analyses; a particularly promising path for future evolution is to add explicit support for coupling between different analyses on different scales.
Acknowledgements
This work was supported by the @neurIST Integrated Project (co-financed by the European Commission through the contract no. IST-027703). The authors would also like to thank for their contribution to the processing of cases as well as to the implementation of the toolchain the following researchers: C. Valencia, H. Tahir, H. Morales, A. J. Geers, M. Aguilar, M. Kim, C. Bludszuweit, D. Capdeferro, P. Watton, M. Giacomoni, H. Bogunovic, A. Radaelli and K. McCormack.
Footnotes
I.e. a small, finite set of values, as opposed to a continuum like a pressure field, which is not usable in a statistical clustering analysis.
Parts of the toolchain are available through the @neurIST web site http://www.aneurist.org/, as well as the data model SQL definition and the final analysis protocol document. Currently, there are efforts in the direction of making derived data available through the European Society of Minimally Invasive Neurological Therapy (ESMINT) society http://www.esmint.eu/.
One contribution of 17 to a Theme Issue ‘The virtual physiological human’.
References
- 1.Brisman J., Song J., Newell D. 2006. Medical progress: cerebral aneurysms. New Engl. J. Med. 355, 928–939. 10.1056/NEJMra052760 ( 10.1056/NEJMra052760) [DOI] [PubMed] [Google Scholar]
- 2.Molyneux A. J.& the International Subarachnoid Aneurysm Trial (ISAT) Collaborative Group 2002. International subarachnoid aneurysm trial (ISAT) of neurological clipping versus endovascular coiling in 2143 patients with ruptured intracranial aneurysms: a randomised trial. Lancet 360, 1267–1274. 10.1016/S0140-6736(02)11314-6 ( 10.1016/S0140-6736(02)11314-6) [DOI] [PubMed] [Google Scholar]
- 3.Wiebers D.& the International Study of the Unruptured Intracranial Aneurysms Investigators 2003. Unruptured intracranial aneurysms: natural history, clinical outcome, and risks of surgical and endovascular treatment. Lancet 362, 103–110. 10.1016/S0140-6736(03)13860-3 ( 10.1016/S0140-6736(03)13860-3) [DOI] [PubMed] [Google Scholar]
- 4.Hunter P., Robbins P., Noble D. 2002. The IUPS human physiome project. Eur. J. Physiol. 445, 1–9. 10.1007/s00424-002-0890-1 ( 10.1007/s00424-002-0890-1) [DOI] [PubMed] [Google Scholar]
- 5.Hunter P., Borg T. 2003. Integration from proteins to organs: the physiome project. Nature 4, 237–243. 10.1038/nrm1017 ( 10.1038/nrm1017) [DOI] [PubMed] [Google Scholar]
- 6.STEP Consortium. 2007. Seeding the EuroPhysiome: a roadmap to the virtual physiological human. See http://www.europhysiome.org/roadmap. [Google Scholar]
- 7.Fenner J., et al. 2008. The EuroPhysiome, STEP and a roadmap for the virtual physiological human. Proc. R. Soc. A 366, 2979–2999. 10.1098/rsta.2008.0089 ( 10.1098/rsta.2008.0089) [DOI] [PubMed] [Google Scholar]
- 8.Viceconti M., Clapworthy G., Van Sint Jan S. 2008. The virtual physiological human—a European initiative for in silico human modelling. J. Physiol. Sci. 58, 441–447. 10.2170/physiolsci.RP009908 ( 10.2170/physiolsci.RP009908) [DOI] [PubMed] [Google Scholar]
- 9.Iavindrasana J., et al. 2008. The @neurIST project. Stud. Health Technol. Inform. 138, 161–164. [PubMed] [Google Scholar]
- 10.Dunlop R., et al. 2008. @neurIST—chronic disease management through integration of heterogeneous data and computer-interpretable guideline services. Stud. Health Technol. Inform. 138, 173–177. [PubMed] [Google Scholar]
- 11.Arbona A., Benkner S., Engelbrecht G., Fingberg J., Hofmann M., Kumpf K., Lonsdale G., Woehrer A. 2007. A service-oriented grid infrastructure for biomedical data and compute services. IEEE Trans. Nanobiosci. 6, 136–141. 10.1109/TNB.2007.897438 ( 10.1109/TNB.2007.897438) [DOI] [PubMed] [Google Scholar]
- 12.Friedrich C. M., Dach H., Gattermayer T., Engelbrecht G., Benkner S., Hofmann-Apitius M. 2008. @neuLink: a service-oriented application for biomedical knowledge discovery. Stud. Health Technol. Inform. 138, 165–172. [PubMed] [Google Scholar]
- 13.Hofmann-Apitius M., et al. 2008. Knowledge environments representing molecular entities for the virtual physiological human. Phil. Trans. R. Soc. A 366, 3091–3110. 10.1098/rsta.2008.0099 ( 10.1098/rsta.2008.0099) [DOI] [PubMed] [Google Scholar]
- 14.Benkner S., et al. 2010. @neurist—infrastructure for advanced disease management through integration of heterogeneous data, computing, and complex processing services. IEEE Trans. Info. Technol. BioMed. 14, 1365–1377. 10.1109/TITB.2010.2049268 ( 10.1109/TITB.2010.2049268) [DOI] [PubMed] [Google Scholar]
- 15.Ma B., Harbaugh R., Lu J., Raghavan M. 2004. Modeling the geometry, hemodynamics and tissue mechanics of cerebral aneurysms. In Proc. of IMECE: ASME Int. Mechanical Engineering Congress and Exposition, Advances in Bioengineering, pp. 397–398. Washington, DC: ASME 10.1115/IMECE2004-60024 ( 10.1115/IMECE2004-60024) [DOI] [Google Scholar]
- 16.Ma B. 2004. Modeling the geometry, hemodynamics, and tissue mechanics of cerebral aneurysms. Ph.D. thesis, University of Iowa, USA. [Google Scholar]
- 17.Humphrey J., Taylor C. 2008. Intracranial and abdominal aortic aneurysms: similarities, differences, and need for a new class of computational models. Annu. Rev. Biomed. Eng. 10, 221–246. 10.1146/annurev.bioeng.10.061807.160439 ( 10.1146/annurev.bioeng.10.061807.160439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cebral J. R., Castro M. A., Appanaboyina S., Putman C. M., Millan D., Frangi A. F. 2005. Efficient pipeline for image-based patient-specific analysis of cerebral aneurysm hemodynamics: technique and sensitivity. IEEE Trans. Med. Imag. 24, 457–467. 10.1109/TMI.2005.844159 ( 10.1109/TMI.2005.844159) [DOI] [PubMed] [Google Scholar]
- 19.Villa-Uriol M. C., et al. 2010. Toward integrated management of cerebral aneurysms. Phil. Trans. R. Soc. A 368, 2961–2982. 10.1098/rsta.2010.0095 ( 10.1098/rsta.2010.0095) [DOI] [PubMed] [Google Scholar]
- 20.Ujiie H., et al. 1999. Effects of size and shape (aspect ratio) on the hemodynamics of saccular aneurysms: a possible index for surgical treatment of intracranial aneurysms. Neurosurgery 45, 119–130. 10.1097/00006123-199907000-00028 ( 10.1097/00006123-199907000-00028) [DOI] [PubMed] [Google Scholar]
- 21.Ma B., Harbaugh R. E., Raghavan M. L. 2004. Three-dimensional geometrical characterization of cerebral aneurysms. Ann. Biomed. Eng. 32, 264–273. 10.1023/B:ABME.0000012746.31343.92 ( 10.1023/B:ABME.0000012746.31343.92) [DOI] [PubMed] [Google Scholar]
- 22.Raghavan M. L., Ma B., Harbaugh R. E. 2005. Quantified aneurysm shape and rupture risk. J. Neurosurg. 102, 355–362. 10.3171/jns.2005.102.2.0355 ( 10.3171/jns.2005.102.2.0355) [DOI] [PubMed] [Google Scholar]
- 23.Millan R., Dempere-Marco L., Pozo J. M., Cebral J. R., Frangi A. F. 2007. Morphological characterization of intracranial aneurysms using 3-D moment invariants. IEEE Trans. Med. Imag. 26, 1270–1282. 10.1109/TMI.2007.901008 ( 10.1109/TMI.2007.901008) [DOI] [PubMed] [Google Scholar]
- 24.Steinman D., Milner J., Norley C., Lownie S., Holdsworth D. 2003. Image-based computational simulation of flow dynamics in a giant intracranial aneurysm. Am. J. Neuroradiol. 24, 559–566. [PMC free article] [PubMed] [Google Scholar]
- 25.Stehbens W. 1963. Histopathology of cerebral aneurysms. Arch. Neurol. 8, 272–285. [DOI] [PubMed] [Google Scholar]
- 26.Stehbens W. 1981. Arterial structure at branches and bifurcations with reference to physiological and pathological processes, including aneurysm formation. In Structure and function of the circulation, vol. 2 (eds Schwarz C. J., Werthessen N. T., Wolf S. G.), pp. 667–693. New York, NY: Plenum Press. [Google Scholar]
- 27.Seshaiyer P., Hsu F., Shah A., Kyriacou S., Humphrey J. 2001. Multiaxial mechanical behavior of human saccular aneurysms. Comput. Methods Biomech. Biomed. Eng. 4, 281–289. 10.1080/10255840108908009 ( 10.1080/10255840108908009) [DOI] [Google Scholar]
- 28.Ma B., Lu J., Harbaugh R., Raghavan M. 2007. Nonlinear anisotropic stress analysis of anatomically realistic cerebral Aneurysms. Trans. ASME J. Biomech. Eng. 129, 88–96. 10.1115/1.2401187 ( 10.1115/1.2401187) [DOI] [PubMed] [Google Scholar]
- 29.Berti G., Hose R., Marzo A., Villa-Uriol M. C., Singh P., Lawford P. 2010 Analysis protocols version 2. See http://www.aneurist.org/UserFiles/File/PUBLIC_DELIVERABLES/D23v2_v1.2_final.pdf . [Google Scholar]
- 30.Taylor C., Humphrey J. 2009. Open problems in computational vascular biomechanics: hemodynamics and arterial wall mechanics. Comput. Methods Appl. Mech. Eng. 198, 3514–3523. 10.1016/j.cma.2009.02.004 ( 10.1016/j.cma.2009.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geers A. J., Larrabide I., Radaelli A. G., Bogunovic H., Kim M., van Andel H. A. F. G., Majoie C. B., VanBavel E., Frangi A. F. 2011. Patient-specific computational hemodynamics of intracranial aneurysms from 3DRA and CTA: an in vivo reproducibility study. Am. J. Neuroradiol. ( 10.3174/ajnr.A2306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzo A., Singh P., Reymond P., Stergiopulos N., Patel U., Hose D. R. 2009. Influence of inlet boundary conditions on the local haemodynamics of intracranial aneurysms. Comput. Methods Biomech. Biomed. Eng. 12, 431–444. 10.1080/10255840802654335 ( 10.1080/10255840802654335) [DOI] [PubMed] [Google Scholar]
- 33.Marzo A., et al. 2010. Computational haemodynamics in cerebral aneurysms: the effects of modelled versus measured boundary conditions. Ann. Biomed. Eng. 2, 884–896 ( 10.1007/s10439-010-0187-z) [DOI] [PubMed] [Google Scholar]
- 34.McKinney A., Palmer C., Truwit C., Karagulle A., Teksam M. 2008. Detection of aneurysms by 64-section multidetector CT angiography in patients acutely suspected of having an intracranial aneurysm and comparison with digital subtraction and 3D rotational angiography. Am. J. Neuroradiol. 29, 594–602. 10.3174/ajnr.A0848 ( 10.3174/ajnr.A0848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rooij W., Sprengers M., de Gast A., Peluso J., Sluzewski M. 2008. 3D rotational angiography: the new gold standard in the detection of additional intracranial aneurysms. Am. J. Neuroradiol. 29, 976–979. 10.3174/ajnr.A0964 ( 10.3174/ajnr.A0964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez M., Frangi A. F. 2007. Non-parametric geodesic active regions: method and evaluation for cerebral aneurysms segmentation in 3DRA and CTA. Med. Image Anal. 11, 224–241. 10.1016/j.media.2007.01.002 ( 10.1016/j.media.2007.01.002) [DOI] [PubMed] [Google Scholar]
- 37.Bogunović H., Radaelli A., de Craene M., Delgado D., Frangi A. F. 2008. Image intensity standardization in 3D rotational angiography and its application to vascular segmentation. In SPIE Medical Imaging 2008: Image Processing, San Diego, CA, USA, vol. 6914 (eds Reinhardt J., Pluim J.), p. 691419 Bellingham, WA: Society of Photo-Optical Instrumentation Engineers; 10.1117/12.770564 ( 10.1117/12.770564) [DOI] [Google Scholar]
- 38.Bogunovic H., et al. 2011. Automated segmentation of cerebral vasculature with aneurysms in 3DRA and TOF-MRA using geodesic active regions: an evaluation study. Med. Phys. 38, 210–222. 10.1118/1.3515749 ( 10.1118/1.3515749) [DOI] [PubMed] [Google Scholar]
- 39.Mellado X., Larrabide I., Hernandez M., Frangi A. F. 2007. Flux driven medial curve extraction. Insight J. [Google Scholar]
- 40.Reymond P., Merenda F., Perren F., Rüfenacht D., Stergiopulos N. 2009. Validation of a one-dimensional model of the systemic arterial tree. Am. J. Physiol. Heart Circ. Physiol. 297, H208–H222. 10.1152/ajpheart.00037.2009 ( 10.1152/ajpheart.00037.2009) [DOI] [PubMed] [Google Scholar]
- 41.Pozo J. M., Villa-Uriol M., Frangi A. F. 2011. Efficient 3D Geometric and Zernike moments computation from unstructured surface meshes. IEEE Trans. Pattern Anal. Machine Intell. 33, 471–484. 10.1109/TPAMI.2010.139 ( 10.1109/TPAMI.2010.139) [DOI] [PubMed] [Google Scholar]
- 42.Valencia C., Villa-Uriol M. C., Pozo J. M., Frangi A. F. 2010. Morphological descriptors as rupture indicators in middle cerebral artery aneurysms. In Proc. IEEE EMBS. EMBC, pp. 6046–6049. Buenos Aires, Argentina. Piscataway, NJ: IEEE Press 10.1109/IEMBS.2010.5627610 ( 10.1109/IEMBS.2010.5627610) [DOI] [PubMed] [Google Scholar]
- 43.Lauric A., Miller E., Frisken S., Malek A. M. 2010. Automated detection of intracranial aneurysms based on parent vessel 3D analysis. Med. Image Anal. 14, 149–159. 10.1016/j.media.2009.10.005 ( 10.1016/j.media.2009.10.005) [DOI] [PubMed] [Google Scholar]
- 44.Ford M., Hoi Y., Piccinelli M., Antiga L., Steinman D. 2009. An objective approach to digital removal of saccular aneurysms: technique and applications. Bri. J. Radiol. 82, S55–S61. 10.1259/bjr/67593727 ( 10.1259/bjr/67593727) [DOI] [PubMed] [Google Scholar]
- 45.Sgouritsa E., Mohamed A., Morsi H., Shaltoni H., Mawad M., Kakadiaris I. 2010. Neck localization and geometry quantification of intracranial aneurysms. In IEEE Int. Symp. Biomed. Imag., pp. 1057–1060. Piscataway, NJ: IEEE Press. Rotterdam, The Netherlands 10.1109/ISBI.2010.5490172 ( 10.1109/ISBI.2010.5490172) [DOI] [Google Scholar]
- 46.Larrabide I., Villa-Uriol M. C., Cardenes R., Pozo J. M., Hose D., Frangi A. F. 2010. Automated intracranial aneurysm isolation and quantification. In Proc. IEEE EMBS. EMBC, pp. 2841–2844. Argentina: Buenos Aires; Piscataway, NJ: IEEE Press 10.1109/IEMBS.2010.5626075 ( 10.1109/IEMBS.2010.5626075) [DOI] [PubMed] [Google Scholar]
- 47.Schmidt J. G. 2010. The FEANOR source code. See http://www.rheinahrcampus.de/∼medsim. [Google Scholar]
- 48.Simmhan Y. L., Plale B., Gannon D. 2005. A survey of data provenance in e-science. SIGMOD Record 34, 31–36. 10.1145/1084805.1084812 ( 10.1145/1084805.1084812) [DOI] [Google Scholar]
- 49.Singh P., et al. 2009. The role of computational fluid dynamics in the management of unruptured intracranial aneurysms: a clinicians' view. Comput. Intell. Neurosci. 2009, 1–12. 10.1155/2009/760364 ( 10.1155/2009/760364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radaelli A. G., Sola Martínez T., Vivas Díaz E., Mellado X., Castro M. A., Putman C. M., Guimaraens L., Cebral J. R., Frangi A. F. 2007. Combined clinical and computational information in complex cerebral aneurysms: application to mirror cerebral aneurysms. In SPIE Medical Imaging: Physiology, Function, and Structure from Medical Images, San Diego, CA, USA (eds Manduca A., Hu X. P.). Lecture Notes on Computer Science 6511, p. 65111F Bellingham, WA: Society of Photo-Optical Instrumentation Engineers; 10.1117/12.708955 ( 10.1117/12.708955) [DOI] [Google Scholar]
- 51.Singh P., et al. 2010. Effects of smoking and hypertension on wall shear stress and oscillatory shear index at the site of intracranial aneurysm formation. Clin. Neurol. Neurosurg. 112, 306–313. 10.1016/j.clineuro.2009.12.018 ( 10.1016/j.clineuro.2009.12.018) [DOI] [PubMed] [Google Scholar]
- 52.Singh P., et al. 2010. The effects of aortic coarctation on cerebral hemodynamics and its importance in the etiopathogenesis of intracranial aneurysms. J. Vasc. Interv. Neurol. 3, 17–30. [PMC free article] [PubMed] [Google Scholar]
- 53.Cebral J., Mut F., Raschi M., Scrivano E., Ceratto R., Lylyk P., Putman C. 2010. Aneurysm rupture following treatment with flow-diverting stents: computational hemodynamics analysis of treatment. Am. J. Neuroradiol. 32, 27–33. 10.3174/ajnr.A2398 ( 10.3174/ajnr.A2398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larrabide I., Kim M., Augsburger L., Villa-Uriol M., Rüfenacht D., Frangi A. 2011. Fast virtual deployment of self-expandable stents: method and in vitro evaluation for intracranial aneurysmal stenting. Med. Image Anal. ( 10.1016/j.media.2010.04.009) [DOI] [PubMed] [Google Scholar]
- 55.Radaelli A., et al. 2008. Reproducibility of haemodynamical simulations in a subject-specific stented aneurysm model: A report on the Virtual Intracranial Stenting Challenge 2007. J. Biomech. 41, 2069–2081. 10.1016/j.jbiomech.2008.04.035 ( 10.1016/j.jbiomech.2008.04.035) [DOI] [PubMed] [Google Scholar]
- 56.Kim M., Larrabide I., Villa-Uriol M. C., Frangi A. F. 2009. Hemodynamic alterations of a patient-specific intracranial aneurysm induced by virtual deployment of stents in various axial orientation. In IEEE Int. Symp. Biomed. Imag., Boston, MA, USA, pp. 1215–1218. Piscataway, NJ: IEEE Press; 10.1109/ISBI.2009.5193280 ( 10.1109/ISBI.2009.5193280) [DOI] [Google Scholar]
- 57.Yasuno K., et al. 2010. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat. Genet. 42, 420–425. 10.1038/ng.563 ( 10.1038/ng.563) [DOI] [PMC free article] [PubMed] [Google Scholar]