Abstract
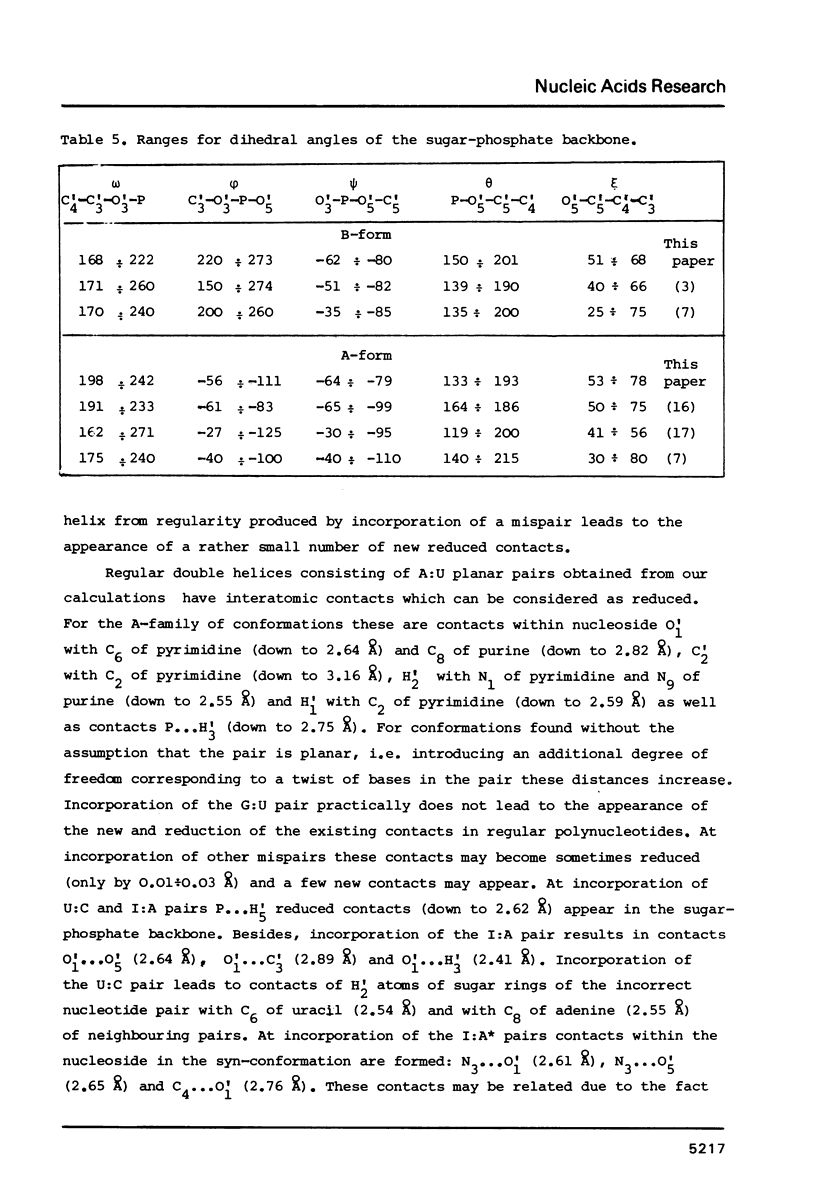
Theoretical conformational analysis using classical potential functions has shown the possibility of incorporation of nucleotide mispairs with the bases in normal tautomeric forms into the DNA double helix. Incorrect purine-pyrimidine, purine-purine and pyrimidine-pyrimidine pairs can be incorporated into the double helix existing both in A- and B-conformations. The most energy favourable conformations of fragments containing a mispair have all the dihedral angles of the sugar-phosphate backbone within the limits characteristic of double helices consisting of Watson-Crick nucleotide pairs. Incorporation of mispairs is possible practically without the appearance of reduced interatomic contacts. Mutual position of bases in the incorporated mispair does not differ much from their position at the energy minimum of the corresponding isolated base pairs. Conformational parameters of irregular regions of double-stranded polynucleotides containing G:U, I:A, I:A* (syn) and U:C pairs are presented. Distortion of the sugar-phosphate backbone is the least upon incorporation of the G:U pair. Formation of mispairs in the processes of nucleic acid biosynthesis and spontaneous mutagenesis is discussed.
Full text
PDF






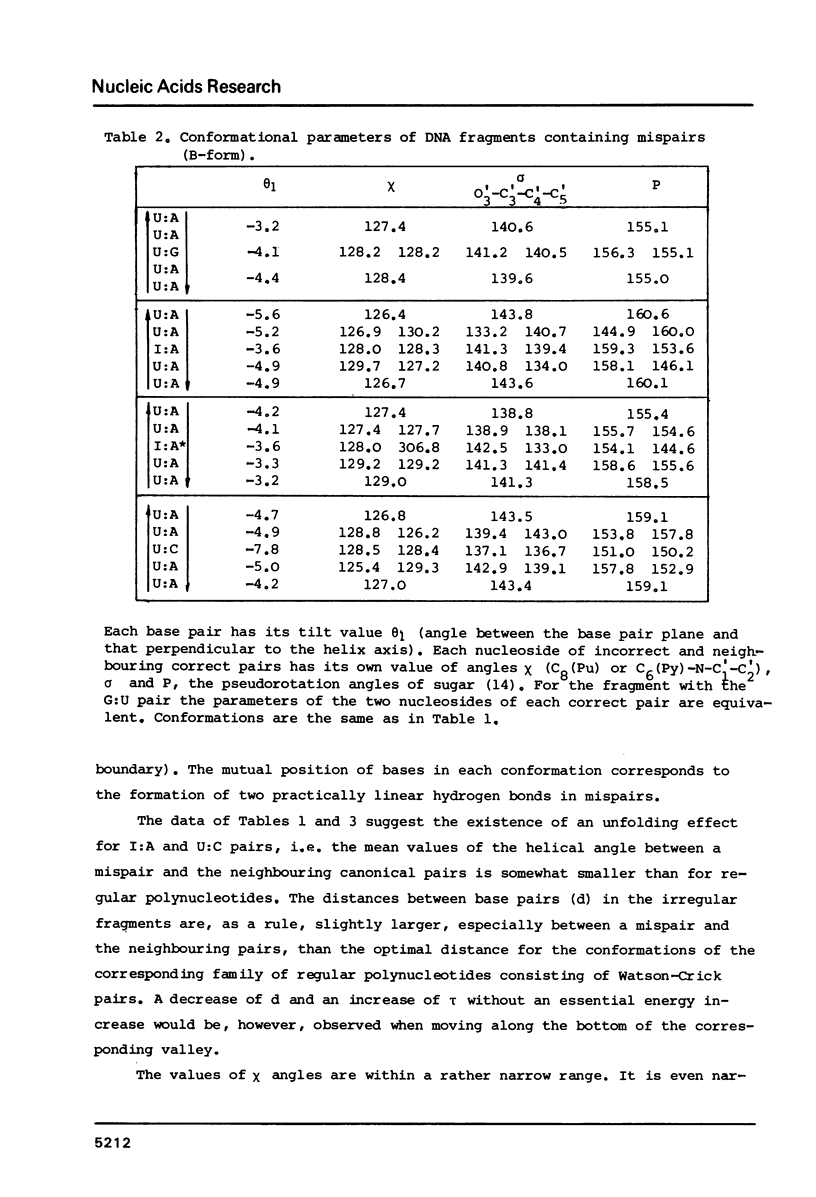
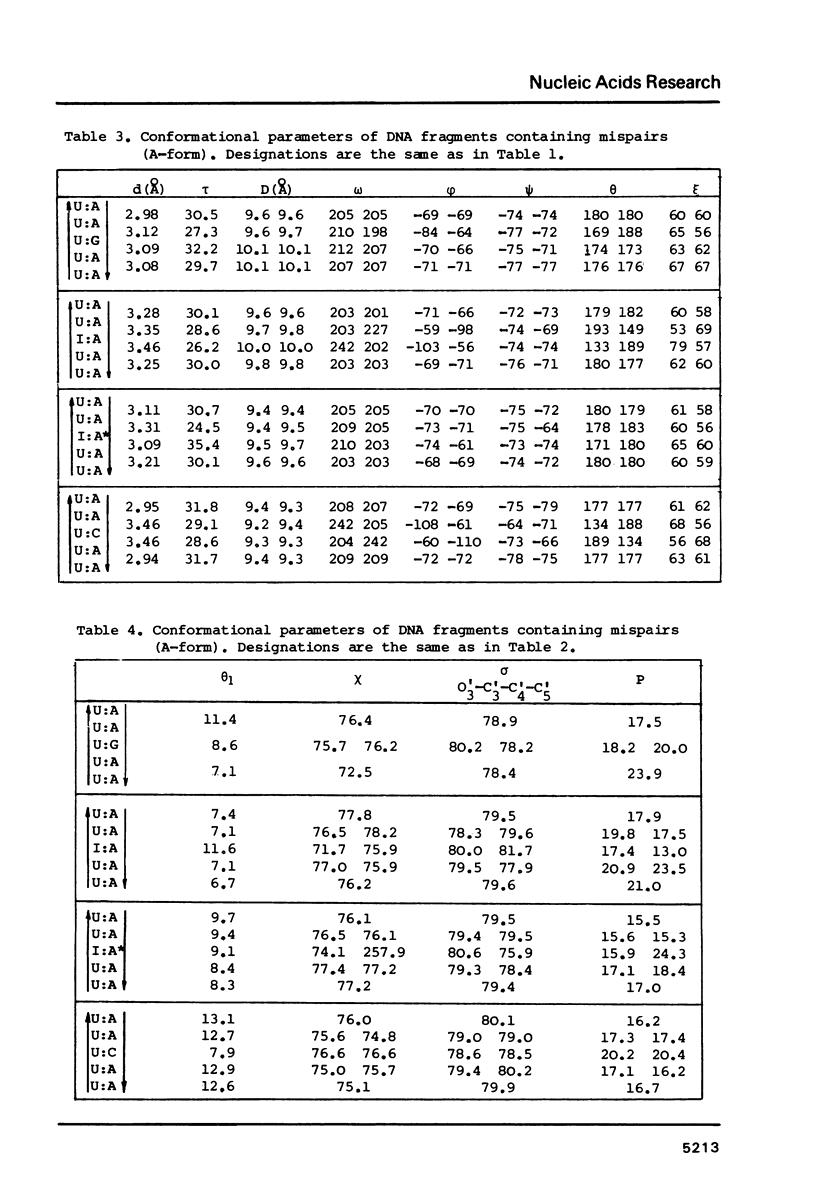

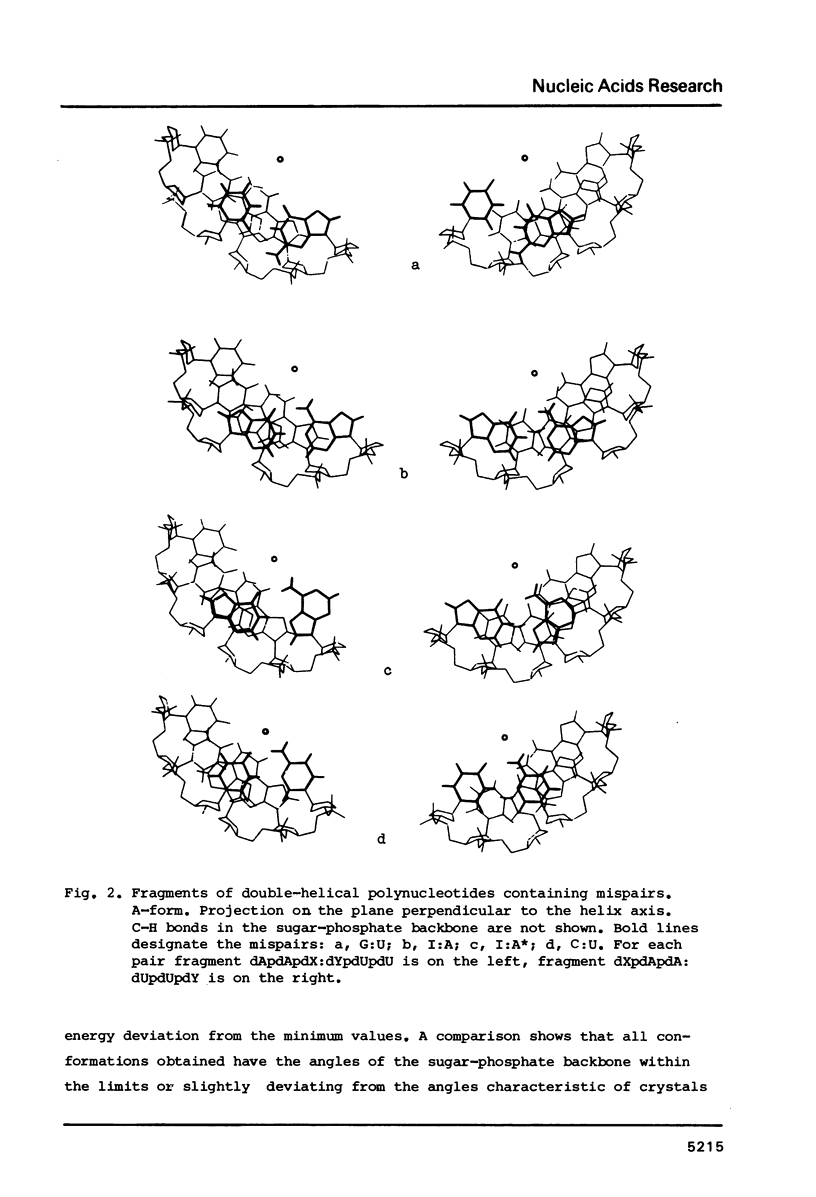
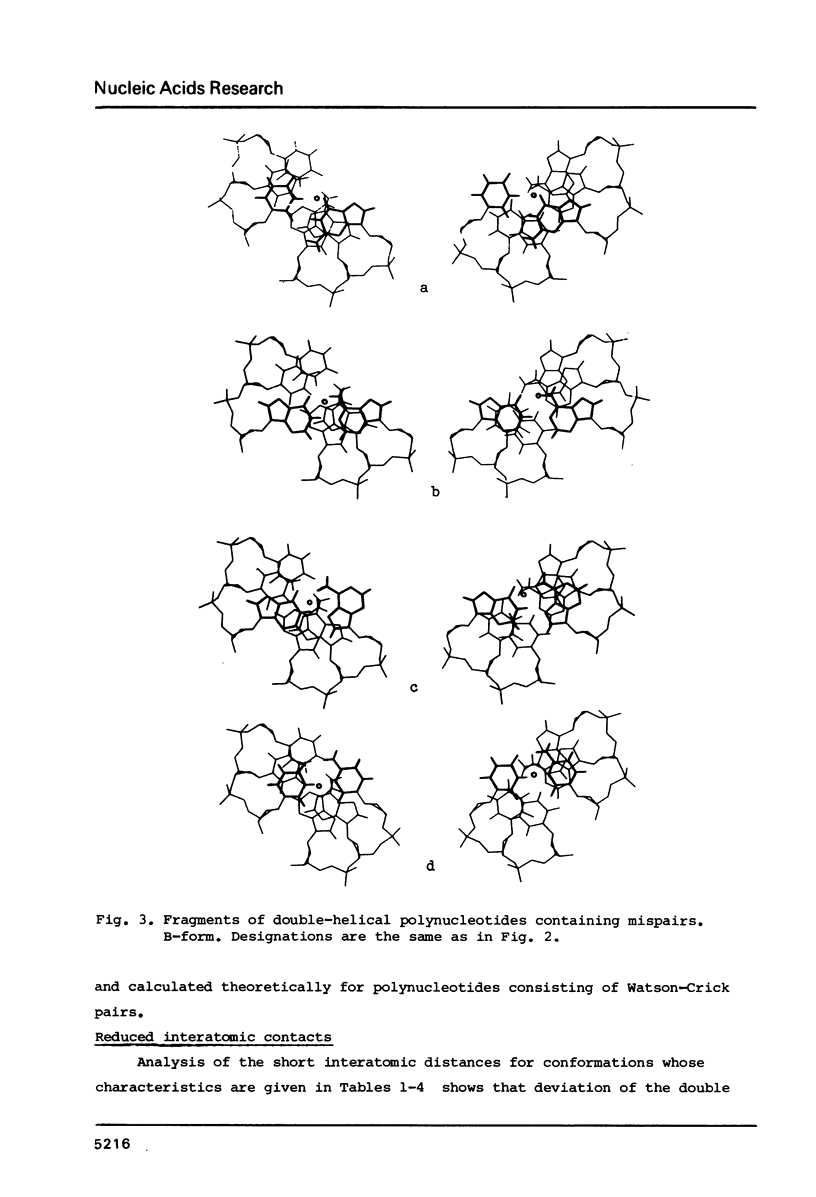





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann T., Gramlich V., Klump H., Knäble T., Schmid E. D., Seliger H., Stulz J. Demonstration of G . U wobble base pairs by Raman and IR spectroscopy. Biophys Chem. 1979 Nov;10(3-4):231–238. doi: 10.1016/0301-4622(79)85011-5. [DOI] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Bruskov V. I., Poltev V. I. Uznavanie fermentami komplementarnykh par azotistykh osnovanii i usilenie spetsifichnosti vzaimodeistvii v protsessakh matrichnogo sinteza. Dokl Akad Nauk SSSR. 1974 Nov 1;219(1):231–234. [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. DNA polymerase accuracy and spontaneous mutation rates: frequencies of purine.purine, purine.pyrimidine, and pyrimidine.pyrimidine mismatches during DNA replication. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4251–4255. doi: 10.1073/pnas.78.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Poltev V. I., Bruskov V. I. O molekuliarnykh mekhanizmakh spontannykh transversiii i tranzitsii. Mol Biol (Mosk) 1977 May-Jun;11(3):661–670. [PubMed] [Google Scholar]
- Poltev V. I., Bruskov V. I. On molecular mechanisms of nucleic acid synthesis fidelity aspects. 1. Contribution of base interactions. J Theor Biol. 1978 Jan 7;70(1):69–83. doi: 10.1016/0022-5193(78)90303-x. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Hughes D. W., Grégoire R. J., Bell R. A., Neilson T. Effects of internal nonbonded bases and a G.U base pair on the stability of a short ribonucleic acid helix. Biochemistry. 1979 Nov 13;18(23):5109–5116. doi: 10.1021/bi00590a013. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953 May 30;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B., Poltev V. I., Florent'ev V. L. Atom-atomnye potentsial'nye funktsii dlia konformatsionnykh raschetov nukleinovykh kislot. Mol Biol (Mosk) 1980 Sep-Oct;14(5):1116–1130. [PubMed] [Google Scholar]



