Abstract
The Notch signaling pathway is highly conserved from Drosophila to humans and plays an important role in the regulation of cellular proliferation, differentiation and apoptosis.
Constitutive activation of Notch signaling has been shown to result in excessive cellular proliferation and a wide range of malignancies, including leukemia, glioblastoma and lung and breast cancers. Notch can also act as a tumor suppressor, and its inactivation has been associated with an increased risk of spontaneous squamous cell carcinoma. This minireview focuses on recent advances related to the mechanisms and roles of activated Notch1, Notch2, Notch3 and Notch4 signaling in human lymphocytic leukemia, myeloid leukemia and B cell lymphoma, as well as their significance, and recent advances in Notch-targeted therapies.
Review
Canonical and noncanonical activation of the Notch signaling pathway
The Notch gene was first described following the observation of Notches on the wings of fruit flies (Drosophila melanogaster) caused by partial loss of function of the Notch gene. Notch signaling is involved in many biological processes, ranging from embryonic development to cell proliferation and survival. It has been demonstrated that the Notch signaling pathway is involved in vascular formation and morphogenesis during vascular development. Notch1, Notch2 and Notch4 and its ligands (Jagged1, Jagged2, Dll1 and Dll4) are expressed in vascular endothelium, whereas Notch3 is expressed in vascular smooth muscle cells. Mutations in Notch3 are associated with CADASIL syndrome (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), the human degenerative vascular disease.
The human Notch family includes four receptors and five ligands [1,2]. All four Notch receptors are synthesized as a single transmembrane polypeptide in the endoplasmic reticulum and transported to the cell surface trough the trans-Golgi network. Notch receptors are expressed as heterodimeric proteins with extracellular, transmembrane and intracellular domains (Figure 1). When a ligand of the Delta/Serrate/LAG-2 family (located on the surface of neighboring cells) binds to the extracellular domain of the Notch receptor, it triggers proteolytic cleavage by a metalloprotease (a disintegrin and metalloprotease (ADAM)). ADAM cleavage produces a substrate for a second cleavage by the presenilin-containing γ-secretase complex, releasing the Notch intracellular domain (NICD) [2,3] (Figure 2). NCID corresponds to the activated form of Notch, which translocates to the nucleus and forms complexes with specific DNA-binding proteins (CBF1/Suppressor of Hairless/LAG-1 and Mastermind/SEL-8) and transcriptionally activates target genes [4] (Figure 2). In the absence of receptor activation and NICD, CBF1 acts as a transcriptional repressor through interactions with the corepressors SMRT (silencing mediator of retinoid and thyroid receptors), KyoT2, CIR (CBF1-interacting corepressor) and SHARP (SMRT/HDAC1 (histone deacetylase 1)-associated repressor protein) [5]. In addition to canonical intracellular signaling pathways, there are other types of noncanonical Notch signaling (Figure 3). The first one involves Notch ligation and translocation of activation signals independent of CBF1 (NICD-dependent), the second involves activation of Notch target genes that are independent of γ-secretase cleavage (NICD- and CBF1-independent) and the third involves CBF1-dependent gene activation without receptor cleavage and NICD release. Termination of Notch signaling can occur at or downstream of the Notch receptor. The Notch receptor can be degraded through the lysosomes by the ubiquitin ligase Itch/AIP4 [6] or another ubiquitin ligase, Nedd4 [7], which act together with Numb [8] and Itch/AIP4 to stimulate endocytosis and lysosomal degradation of the Notch receptor [9]. Finally, NICD1 phosphorylation by GSK3 regulates its interaction with the E3 ubiquitin ligase CDC4/FBW7, thereby controlling NICD1 ubiquitination and proteasome-mediated degradation [10]. This multifaceted control of Notch expression underscores its critical functions in cellular homeostasis.
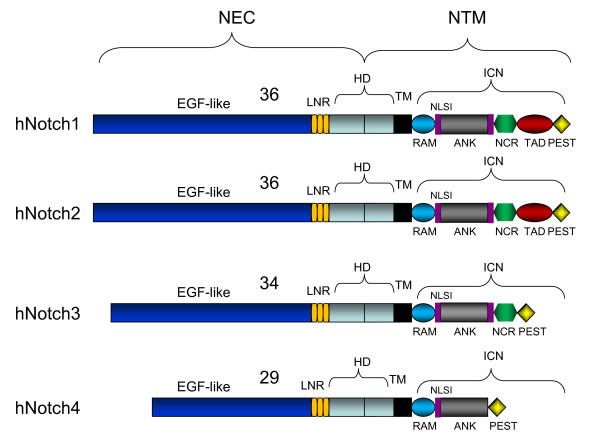
Figure 1.
Structure of the four human Notch receptors. NEC: extracellular subunit; NTM: transmembrane subunit; EGF: epidermal growth factor; HD: heterodimerization domain; ICN: intracellular domain; LNR: cysteine-rich LNR repeats; TM: transmembrane domain; RAM: RAM domain; NLS: nuclear localizing signals; ANK: ankyrin repeat domain; NCR: cysteine response region; TAD: transactivation domain; PEST: region rich in proline (P), glutamine (E), serine (S) and threonine (T) residues.
Figure 2.
The Notch signaling pathway. The initiation of the Notch signaling pathway begins when the Notch ligand binds to the Notch receptor. This action triggers two proteolytic cleavages by ADAM-type protease (S2) and γ-secretase (S3), respectively. Following cleavages, the activated form of Notch is released (NICD) and is translocated to the nucleus, where NICD forms complexes with specific DNA-binding proteins (CBF1/Suppressor of Hairless/LAG-1 and Mastermind/SEL-8). Afterward the transcriptional process of target genes is initiated. MAML1: Mastermind-like 1 protein; CBF1: DNA-binding transcription factor.
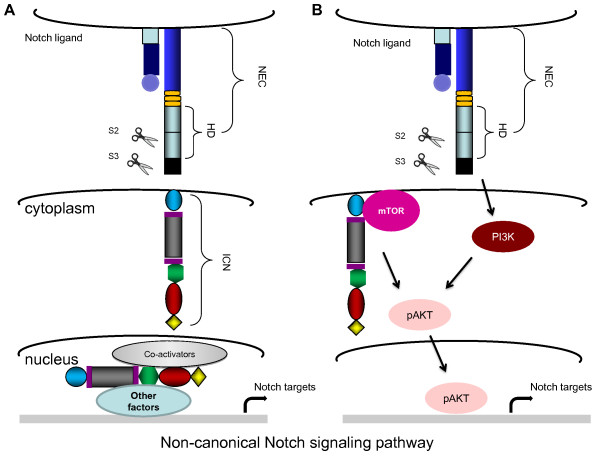
Figure 3.
Noncanonical Notch signaling pathway. (A) NICD-dependent, CBF1-independent transcriptional activation by NICD, coactivators and other undefined factors (B) Interaction of NICD with components of other signaling pathways to activate Notch targets or tissue-specific factors.
The role of the microenvironment in the activation of Notch in leukemia is increasingly recognized. Recently, cis-inhibition of Notch signaling by the DLL1 ligand has been described in Drosophila and mice [11]. These investigations have suggested that while expression of ligands on neighboring cells stimulates Notch activation, expression on the same cell as the Notch receptor may have an inhibitory effect [11]. Along these lines, activation of Notch signaling in B-cell malignancies might result from interactions between tumor cells as well as between the tumor cell and the microenvironment. There is evidence suggesting the importance of Notch signaling in the cross-talk between multiple myeloma (MM) cells and their environment. Bone marrow stromal cells express both Notch ligands, Jagged and δ, and are able to activate Notch signaling in MM cells [12,13].
Mechanisms leading to constitutive activation of Notch signaling
Notch1 was discovered in humans through a t(7;9)(q34;q34.3) chromosomal translocation observed in some patients with T-cell acute lymphoblastic leukemia (T-ALL) [14,15]. However, a direct role of Notch activation in T-ALL remained obscure, since only 1% to 3% of patients with T-ALL were found to carry this translocation. It was only after the discovery of a high rate of activating mutations that it became clear that Notch1 expression is linked to the development of T-ALL [16]. Activating mutations identified in the T-ALL cluster at the heterodimerization domain (HD) and the proline, glutamine, serine and threonine (PEST) domain led to ligand-independent cleavage of the Notch receptor and a reduced degradation of NICD1, respectively. Recently, activating mutations in Notch were identified in more than 30% of human T-lymphotropic virus type I (HTLV-I)-associated adult T-cell leukemia (ATL) patients, suggesting an important role for Notch signaling in HTLV-I-associated ATL [17]. Activating mutations found in ATL patients are different from those previously reported in patients with T-ALL and mostly involve single-substitution mutations in the PEST domain that do not create an early stop codon, but rather lead to reduced CDC4/Fbw7-mediated degradation and stabilization of NICD1 [17].
Additional mechanisms have also been reported to lead to increased Notch expression in cancer cells. Mutations and internal duplication insertions in exon 28 of NICD [18], as well as mutations in CDC4/Fbw7 [10,19], have been reported, but they seem to occur at a very low frequency. The Wnt/β-catenin and Notch1 signaling pathways play an important role in a variety of biological processes, including cell proliferation and survival. Studies have shown that β-catenin can regulate the level and transcriptional activity of Notch1 [20]. β-catenin can prevent NICD degradation, possibly by competing with CDC4/Fbw7-dependent degradation [20]. Moreover, β-catenin increases the transcriptional activity of NICD, and the effects of β-catenin on Notch1 are noticeably reduced by overexpression of the lymphocyte enhancer-binding factor 1, LEF1.
Implication of Notch signaling in various hematological disorders
Notch1
Notch1 has been reported to play a role in T-ALL and ATL, with approximate mutation rates of 50% and 30%, respectively [16,17,21,22]. Notch1 is required for the proliferation and survival of leukemia cells, and its role has been described in recent reviews [23-26]. The high prevalence of activating mutations found in Notch1 in T-ALL and ATL patients (Table 1) might suggest that this event plays a role in promoting the emergence of a particular subclone. Whether a mutation in Notch is a primary or secondary event in tumor cells is unclear. Mansour et al. [27] reported low-level Notch1 despite high blast counts in T-ALL patients, suggesting that these mutations were acquired as a secondary event in a preselected subclone. On the other hand, it has also been reported that mutation of Notch1 can be an early or initiating event in T-ALL arising prenatally, to be complemented by a postnatal SIL-TAL1 fusion gene and emergence of tumor clones [28].
Table 1.
Mutation of Notch1 and Notch2 in human leukemia and/or lymphomaa
| Leukemia and/or lymphoma type | Notch mutations in PEST/HD domain, % | Studies |
|---|---|---|
| Notch1 | ||
| T-ALL | 50% PEST/HD | [16,49] |
| B-CLL | 4.6% PEST | [50,51] |
| CLL | 12.2% PEST | [52] |
| AML | 8.3% PEST | [53] |
| T-NHL | 42.9% PEST/HD | [49] |
| ATL | 30% PEST | [17] |
| Notch2 | ||
| DLBCL | 8% PEST | [31] |
| MZL | 5% PEST | [32] |
aPEST: proline, glutamine, serine, and threonine; HD: heterodimerization domain; T-ALL: T-cell acute lymphoblastic leukemia; B-CLL: B-cell chronic lymphocytic leukemia; CLL: chronic lymphocytic leukemia; AML: acute myeloid leukemia; T-NHL: T-cell non-Hodgkin lymphoma; ATL: adult T-cell leukemia; DLBCL: diffuse large B-cell lymphoma; MZL: marginal zone lymphoma.
Notch2
Increasing evidence suggests that Notch2 may play a role in leukemia and lymphoma. Early studies showed that feline leukemia virus recombinant genomes isolated from lymphomas captured Notch2, which included the intracellular ankyrin repeat functional domain in the envelope gene [29]. Later it was found that Notch2 plays a role in CD8 thymocyte maturation and that enforced expression of activated Notch2 invariably resulted in T-cell leukemia in mice [30]. Table 1 shows that approximately 8% of diffuse large B-cell lymphomas (DLBCLs) have Notch2 mutations [31]. Similar to observations in Notch1, mutations in DLCBL affected the PEST domain or a single-amino acid substitution at the C terminus and resulted in Notch2-reduced turnover [31]. These observations suggest Notch2 gain-of-function mutations in a subset of B-cell lymphomas. In fact, Notch2 is involved in the development of B1 and marginal zone B cells, and Notch2 is overexpressed in some marginal zone lymphomas (MZLs) [32]. Potential activating mutations of human Notch2 presented in Table 1 were also detected in 5% of MZL patients [32]. Notch2 may also play an indirect role in chronic B-cell lymphocytic leukemia (B-CLL) through upregulated expression of CD23 [33,34].
Notch3
A possible role of Notch3 in leukemia was postulated in studies in which transgenic mice expressing the constitutively active intracellular domain of Notch3 in thymocytes and T cells developed early and aggressive T-cell neoplasia [35]. Importantly, these results were validated in humans, and examination of T-ALL patients demonstrated high expression of Notch3 and pTα transcripts, whereas the expression of these genes was considerably reduced in or absent from patients in remission [36]. pTα and Notch3 interactions are essential for distribution of the E3 ligase protein, c-Cbl, to the lipid rafts. This is important in the development of leukemogenesis, since in the absence of pTα, c-Cbl targets Notch3 for proteasome degradation [37]. Moreover, the NF-κΒ pathway may be involved in the development of Notch3-dependent T-cell lymphoma in humans, and there is genetic and biochemical evidence that Notch3 triggers multiple NF-κΒ activation pathways [38]. Recently, Notch3 was found to control expression of mitogen-activated protein kinase phosphatase 1 and plays a role in the survival of T-ALL cells [39].
Notch4
Notch4 is expressed in human bone marrow cells and in CD34+ and CD34- populations [40]. Notch4 intracellular domain-transduced cord cells transplanted into mice showed remarkably elevated levels of engraftment of an immature T-cell population, while B-cell development was inhibited. Taken together, these results suggest that activation of Notch4 leads to enhanced stem cell activity, reduced differentiation and altered lymphoid development [41].
Clinical relevance and therapeutic approaches aimed at targeting Notch signaling
The molecular pathogenesis of Notch has recently been reviewed [24]. Targeting Notch receptor cleavage through γ-secretase inhibitors (GSIs) is an attractive approach, as GSI treatment inhibits proliferation of T-ALL and ATL tumor cells in vitro and in vivo [42,43]. The use of GSIs, however, poses several challenges. Current GSIs have been shown to have significant intestinal toxicity in patients because of the dual inhibition of Notch1 and Notch2. In addition, high levels of IL-6 and IL-8 have been reported to abrogate or significantly reduce the efficacy of the GSI (RO4929097) [44]. GSIs may not be useful in patients with a Notch1 mutation in the HD, as this results in weakened association or complete dissociation of the receptor subunits and thus leads to ligand-independent activation. Finally, recent studies have also shown that T-ALL patients with loss of PTEN are resistant to GSI effects [45]. Together these results highlight the need for new targeted therapies.
Recently, the use of therapeutic antibodies that selectively block Notch1 receptor signaling has been reported to inhibit tumor growth in mouse models [46]. Rather than targeting receptor processing, alternative strategies may also focus on blocking NICD functions and transcriptional activities. Along these lines, the use of a stapled peptide to inhibit the Notch transcription factor complex has been reported to result in Notch-specific antiproliferative effects in cultured cells and in a mouse model of Notch1-driven T-ALL [47].
Conclusions
Increased Notch signaling is linked to hematological malignancies. Overexpression of activated the Notch1, Notch2 or Notch3 genes in bone marrow progenitor cells reliably induces T-ALL at high frequency in murine models. Activating mutations in Notch1 is frequent in both T-ALL and ATL. It has been proposed that a fraction of T-ALLs that present as leukemia without evidence of thymic involvement may originate from bone marrow progenitors that have acquired a Notch1 mutation, suggesting that constitutive active Notch1 could, in some cases, be an initial event driving tumor development. On the other hand, animal model studies have indicated that Notch can also play a secondary role during T-ALL development. The role played by Notch1 in other human leukemias is less clear, as is the potential role of other Notch genes. Although inhibition of Notch signaling in xenograft tumors in animal models effectively prevents tumor cell growth, clinical outcomes in humans do not seem to be significantly associated with Notch status, which has an impact on early response to treatment but not on general outcomes in pediatric patients with T-ALL [48]. This may be related to acquisition of additional mutations in patients who have received several therapies and may relieve tumor cells from Notch1 addiction. As is the case with many targeted therapies, resistance to single drugs emerges rapidly, suggesting that a multidrug chemotherapy targeting Notch and connected pathways is needed.
Abbreviations
IL: interleukin; NF-κΒ: nuclear factor κΒ.
Competing interests
CN is a section editor for BMC Cancer.
Authors' contributions
JP created the figures and wrote the manuscript. CN wrote the manuscript. Both authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Joanna Pancewicz, Email: jpancewicz@kumc.edu.
Christophe Nicot, Email: cnicot@kumc.edu.
Acknowledgements
This work was supported by National Cancer Institute (NCI) grant CA106258 (to CN). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI. The authors thank Brandi Miller for editorial assistance.
References
- D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Bernard F. Notch targets and their regulation. Curr Top Dev Biol. 2010;92:253–275. doi: 10.1016/S0070-2153(10)92008-5. [DOI] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israël A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DAP, Cornell M, Baron M. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- del Álamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21:R40–R47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19:781–791. doi: 10.1016/S1074-7613(03)00325-X. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- Reynolds TC, Smith SD, Sklar J. Analysis of DNA surrounding the breakpoints of chromosomal translocations involving the β T cell receptor gene in human lymphoblastic neoplasms. Cell. 1987;50:107–117. doi: 10.1016/0092-8674(87)90667-2. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Pancewicz J, Taylor JM, Datta A, Baydoun HH, Waldmann TA, Hermine O, Nicot C. Notch signaling contributes to proliferation and tumor formation of human T-cell leukemia virus type 1-associated adult T-cell leukemia. Proc Natl Acad Sci USA. 2010;107:16619–16624. doi: 10.1073/pnas.1010722107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, Barnes K, Zuurbier L, Meijerink JP, Ferrando AA. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Kim H, Ki H, Yang I, Yang N, Lee KY, Kim N, Park HS, Kim K. β-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim Biophys Acta. 2009;1793:290–299. doi: 10.1016/j.bbamcr.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Erbilgin Y, Sayitoglu M, Hatirnaz O, Dogru O, Akcay A, Tuysuz G, Celkan T, Aydogan G, Salcioglu Z, Timur C, Yuksel-Soycan L, Ure U, Anak S, Agaoglu L, Devecioglu O, Yildiz I, Ozbek U. Prognostic significance of NOTCH1 and FBXW7 mutations in pediatric T-ALL. Dis Markers. 2010;28:353–360. doi: 10.3233/DMA-2010-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Sulis ML, Duke V, Foroni L, Jenkinson S, Koo K, Allen CG, Gale RE, Buck G, Richards S, Paietta E, Rowe JM, Tallman MS, Goldstone AH, Ferrando AA, Linch DC. Prognostic implications of NOTCH1 and FBXW7 mutations in adults with T-cell acute lymphoblastic leukemia treated on the MRC UKALLXII/ECOG E2993 protocol. J Clin Oncol. 2009;27:4352–4356. doi: 10.1200/JCO.2009.22.0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002;1:466–476. doi: 10.4161/cbt.1.5.162. [DOI] [PubMed] [Google Scholar]
- Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2011;25:83–90. doi: 10.1016/j.blre.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay PA. Notch signaling in pediatric malignancies. Curr Oncol Rep. 2008;10:459–468. doi: 10.1007/s11912-008-0071-2. [DOI] [PubMed] [Google Scholar]
- Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, Gale RE, Linch DC. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- Eguchi-Ishimae M, Eguchi M, Kempski H, Greaves M. NOTCH1 mutation can be an early, prenatal genetic event in T-ALL. Blood. 2008;111:376–378. doi: 10.1182/blood-2007-02-074690. [DOI] [PubMed] [Google Scholar]
- Rohn JL, Lauring AS, Linenberger ML, Overbaugh J. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CM, Hurez V, Swindle CS, Hamada Y, Klug CA. Activated Notch2 potentiates CD8 lineage maturation and promotes the selective development of B1 B cells. Mol Cell Biol. 2003;23:8637–8650. doi: 10.1128/MCB.23.23.8637-8650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G, Nannya Y, Suzuki R, Ota S, Ota Y, Izutsu K, Sakata-Yanagimoto M, Hangaishi A, Yagita H, Fukayama M, Seto M, Kurokawa M, Ogawa S, Chiba S. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100:920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trøen G, Wlodarska I, Warsame A, Hernández Llodrà S, De Wolf-Peeters C, Delabie J. NOTCH2 mutations in marginal zone lymphoma. Haematologica. 2008;93:1107–1109. doi: 10.3324/haematol.11635. [DOI] [PubMed] [Google Scholar]
- Hubmann R, Düchler M, Schnabl S, Hilgarth M, Demirtas D, Mitteregger D, Hölbl A, Vanura K, Le T, Look T, Schwarzmeier JD, Valent P, Jäger U, Shehata M. NOTCH2 links protein kinase C δ to the expression of CD23 in chronic lymphocytic leukaemia (CLL) cells. Br J Haematol. 2010;148:868–878. doi: 10.1111/j.1365-2141.2009.08024.x. [DOI] [PubMed] [Google Scholar]
- Hubmann R, Schwarzmeier JD, Shehata M, Hilgarth M, Duechler M, Dettke M, Berger R. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.V99.10.3742. [DOI] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Checquolo S, Balestri A, Biondi A, Cazzaniga G, Lendahl U, Fehling HJ, Hayday AC, Frati L, von Boehmer H, Gulino A, Screpanti I. Combined expression of pTα and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci USA. 2002;99:3788–3793. doi: 10.1073/pnas.062050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutour P, Estienne MH, Davi F, Landman-Parker J, Quartier P, Buzyn A, Delabesse E, Valensi F, Macintyre E. Analysis of TCR, pTα, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood. 2003;101:2693–2703. doi: 10.1182/blood-2002-08-2438. [DOI] [PubMed] [Google Scholar]
- Checquolo S, Palermo R, Cialfi S, Ferrara G, Oliviero C, Talora C, Bellavia D, Giovenco A, Grazioli P, Frati L, Gulino A, Screpanti I. Differential subcellular localization regulates c-Cbl E3 ligase activity upon Notch3 protein in T-cell leukemia. Oncogene. 2010;29:1463–1474. doi: 10.1038/onc.2009.446. [DOI] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, Gaetano C, Ruco L, Hoffman ES, Hayday AC, Lendahl U, Frati L, Gulino A, Screpanti I. Constitutive activation of NF-κΒ and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero M, Minuzzo S, Pusceddu I, Moserle L, Persano L, Agnusdei V, Tosello V, Basso G, Amadori A, Indraccolo S. Notch3-mediated regulation of MKP-1 levels promotes survival of T acute lymphoblastic leukemia cells. Leukemia. 2011;25:588–598. doi: 10.1038/leu.2010.323. [DOI] [PubMed] [Google Scholar]
- Karanu FN, Yuefei L, Gallacher L, Sakano S, Bhatia M. Differential response of primitive human. Leukemia. 2003;17:1366–1374. doi: 10.1038/sj.leu.2402973. [DOI] [PubMed] [Google Scholar]
- Vercauteren SM, Sutherland HJ. Constitutively active Notch4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood. 2004;104:2315–2322. doi: 10.1182/blood-2004-01-0204. [DOI] [PubMed] [Google Scholar]
- Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, Tsaparikos K, Jani JP, Hosea N, Sands M, Randolph S, Smeal T. Evaluation of selective γ-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- Luistro L, He W, Smith M, Packman K, Vilenchik M, Carvajal D, Roberts J, Cai J, Berkofsky-Fessler W, Hilton H, Linn M, Flohr A, Jakob-Røtne R, Jacobsen H, Glenn K, Heimbrook D, Boylan JF. Preclinical profile of a potent γ-secretase inhibitor targeting Notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009;69:7672–7680. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Luistro L, Carvajal D, Smith M, Nevins T, Yin X, Cai J, Higgins B, Kolinsky K, Rizzo C, Packman K, Heimbrook D, Boylan JF. High tumor levels of IL6 and IL8 abrogate preclinical efficacy of the γ-secretase inhibitor, RO4929097. Mol Oncol. 2011;5:292–301. doi: 10.1016/j.molonc.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zúñiga-Pflücker JC, Dominguez M, Ferrando AA. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G, Kaltenbach S, Yakouben K, Mazingue F, Robert A, Boutard P, Plantaz D, Rohrlich P, van Vlierberghe P, Preudhomme C, Otten J, Speleman F, Dastugue N, Suciu S, Benoit Y, Bertrand Y, Cavé H. EORTC-CLG. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24:2023–2031. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- Park MJ, Taki T, Oda M, Watanabe T, Yumura-Yagi K, Kobayashi R, Suzuki N, Hara J, Horibe K, Hayashi Y. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. 2009;145:198–206. doi: 10.1111/j.1365-2141.2009.07607.x. [DOI] [PubMed] [Google Scholar]
- Sportoletti P, Baldoni S, Cavalli L, Del Papa B, Bonifacio E, Ciurnelli R, Bell AS, Di Tommaso A, Rosati E, Crescenzi B, Mecucci C, Screpanti I, Marconi P, Martelli MF, Di Iannini M, Falzetti F. NOTCH1 PEST domain mutation is an adverse prognostic factor in B-CLL. Br J Haematol. 2010;151:404–406. doi: 10.1111/j.1365-2141.2010.08368.x. [DOI] [PubMed] [Google Scholar]
- Di Ianni M, Baldoni S, Rosati E, Ciurnelli R, Cavalli L, Martelli MF, Marconi P, Screpanti I, Falzetti F. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br J Haematol. 2009;146:689–691. doi: 10.1111/j.1365-2141.2009.07816.x. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, Bassaganyas L, Baumann T, Juan M, López-Guerra M, Colomer D, Tubío JM, López C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernández JM, Puente DA, Freije JM, Velasco G, Gutiérrez-Fernández A, Costa D, Carrió A, Guijarro S, Enjuanes A. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Kogoshi H, Nara N, Tohda S. NOTCH1 mutations are rare in acute myeloid leukemia. Leuk Lymphoma. 2006;47:2400–2403. doi: 10.1080/10428190600773339. [DOI] [PubMed] [Google Scholar]