Abstract
Enzymatic catalysis and protein signaling are dynamic processes that involve local and/or global conformational changes that occur across a broad range of time scales. 1H-15N relaxation NMR provides a comprehensive understanding of protein backbone dynamics both in the apo (unliganded) and ligand-bound conformations enabling both fast and slow internal motions of individual amino acid residues to be observed. We recently reported the structure and nucleotide binding properties of the STAS domain of Rv1739c, a SulP anion transporter protein of M. tuberculosis. In this report we present 1H-15N NMR backbone dynamics measurements (T1, T2 and steady-state ({1H}-15N) heteronuclear NOE) of the Rv1739c STAS domain, in the absence and presence of saturating concentrations of GTP and GDP. Analysis of measured relaxation data and estimated dynamic parameters indicated distinct features differentiating binding of GTP and GDP to Rv1739c STAS. The 9.55 nsec overall rotational correlation time (τm) of Rv1739c STAS increased to 10.48 nsec in the presence of GTP, and to 13.25 nsec in the presence of GDP, indicating significant nucleotide-induced conformational changes. These conformational changes were accompanied by slow time scale (μsec-msec) motions in discrete regions of the protein, reflected in guanine nucleotide-induced changes in relaxation parameters. The observed nucleotide-specific alterations in relaxation properties of individual STAS residues reflect increased molecular anisotropy and/or the emergence of conformational equilibria governing functional properties of the STAS domain.
Keywords: SLC26, SulP, GTP, GDP, relaxation rates, order parameter, reduced spectral density function
Introduction
SulP/SLC26 proteins constitute a phylogenetically ancient superfamily of anion transporters with an N-terminal polytopic transmembrane domain and a C-terminal cytoplasmic STAS (sulfate transporter and anti-sigma factor antagonist) domain [1–3]. Among the 10 SLC26 genes of the human genome, loss-of-function mutations identified to date underlie familial recessive syndromes of deafness (SLC26A4, SLC26A5), goiter (SLC26A4), chondrodysplasia (SLC26A2), and chloride-diarrhea (SLC26A3). Disruption of additional Slc26 genes in mice leads to nephrolithiasis (Slc26a1, Slc26a6), distal renal tubular acidosis (Slc26a7), gastric hypochlorhydria and dysplasia (Slc26a7, Slc26a9), and male infertility (Slc26a8). Slc26 genes or closely adjacent loci have also been genetically or epigenetically linked to increased risk inflammatory bowel disease [4] and colon cancer [5], among other disorders. These phenotypes likely result from tissue-specific loss of transport of chloride, bicarbonate, iodide, sulfate, or oxalate by a mechanism of anion exchange and/or (for Slc26a7 and Slc26a9) chloride conductance [6]. Among the many SLC26 gene disease mutations characterized to date are numerous missense mutations of their STAS domains.
Slc26-like sulP genes are expressed in all metazoan phyla examined to date, as well as in unicellular eukaryotes and bacteria [7], and most contain STAS domains. The STAS domains of yeast sulfate transporters Sult1 and Sult2 proteins are essential for plasmalemmal targeting [8]. The ychM protein of E. coli is associated with bicarbonate uptake, and binding of acyl carrier protein by ychM STAS domain appears to contribute to fatty acid elongation reactions [9]. The bicA protein of Synechococcus likely mediates Na+-dependent bicarbonate uptake as part of the organism’s carbon fixation process [10, 11], but the function of its STAS domain has not been determined.
The structure of SulP/SLC26 transmembrane domains is unknown, but C-terminal alkaline phosphatase and β-lactamase fusion studies of bicA predict 12 transmembrane spans [1], consistent with cytoplasmic localization of both N- and C-termini of mammalian SLC26 polypeptides [12, 13]. The X-ray crystal structures of STAS domains have been solved for a centrally deleted form of rat prestin [14] and for E. coli ychM in spontaneous complex with acyl carrier protein liganded with malonyl-coA [9]. The NMR structural analyses of the engineered central deletion variant of rat prestin corroborated the prestin crytstal structure.
We recently reported the NMR solution structure of the cytoplasmic C-terminal STAS domain of M. tuberculosis SulP protein Rv1739c [15, 16], a probable sulfate transporter or sulfate transport regulator [17]. These data also demonstrate binding of Rv1739c to the guanine nucleotides GTP and GDP [15]. The anti-sigma factor antagonist SpoIIAA of B. subtilis is a GTPase [18], and the copurification of GTPase activity with Rv1739c STAS domain [15] suggests that Rv1739c STAS may also possess intrinsic GTPase activity. The interaction of specific guanine nucleotides with Rv1739c STAS was characterized by photo-affinity labeling, steady state intrinsic fluorescence quench, and residue-specific 1HN, 15N chemical shift perturbation [15].
Protein-ligand interactions are governed by dynamic processes and interaction mechanisms that vary greatly with respect to rate constants across a wide range time scales. NMR relaxation parameters reveal intrinsic atomic level dynamic properties of proteins alone as well as in complex. These dynamic properties of proteins and protein-ligand interactions determine protein tumbling behavior in solution, identify local or global protein regions affected by ligand binding, and may correlate to protein function. We therefore measured 15N- backbone relaxation parameters, T1, T2 and steady-state ({1H}-15N) heteronuclear NOE of Rv1739c STAS in the absence and presence of GTP and GDP. The relaxation data were analyzed by model-free formalism [19–21] and reduced spectral density mapping [22, 23] to define dynamic properties of the Rv1739c STAS domain in unliganded and liganded forms. The results demonstrate that nucleotide binding significantly altered motional properties of defined residues of the STAS domain. Nucleotide-specific relaxation properties of select residues or segments exhibiting internal motions of varied time scales suggest molecular regions important to this nucleotide-STAS complex.
Results
15N backbone dynamics of Rv1739c STAS domain
(a) relaxation NMR data
To study dynamics of the Rv1739c STAS domain, the backbone 15N-1HN resonance-based 15N longitudinal (T1) and transverse (T2) relaxation times as well as {1H}-15N heteronuclear NOEs were collected and measured. Well-resolved 1H-15N crosspeaks, as seen in 2D 1H-15N HSQC [16], allowed us to record T1 and T2 data for each of 112 unambiguously assigned resonances from a total of 119 non-proline residues. {1H}-15N NOEs were obtained for 108 unambiguously assigned resonances (Supplementary Table S1). {1H}-15N NOEs could not be obtained for loop residues D3 (L1), I4 (L1), Q47 (L3), and G106 (L8) (the bracketed L represents the loop structure as numbered from the N-terminus; this convention, used throughout the present study, also extends to the symbols α for helix and β for strand) as a result of resonance degeneracy in either NOE or NONOE experiment. The 7 unidentified residues in T1, T2, and {1H}-15N heteronuclear NOE experiments are those previously unassigned [16]. Longitudinal relaxation rates (R1), transverse relaxation rates (R2), {1H}-15N heteronuclear NOE, and the ratio R2/R1 are plotted with their estimated error values for protein backbone 15N-1HN atoms as a function of Rv1739c STAS amino acid sequence (Figure 1). Average values of these 15N-relaxation parameters for Rv1739c STAS were: < R1 > = 1.36 ± 0.05 s−1; < R2 > = 14.62 ± 0.70 s−1; and < NOE > = 0.79 ± 0.04.
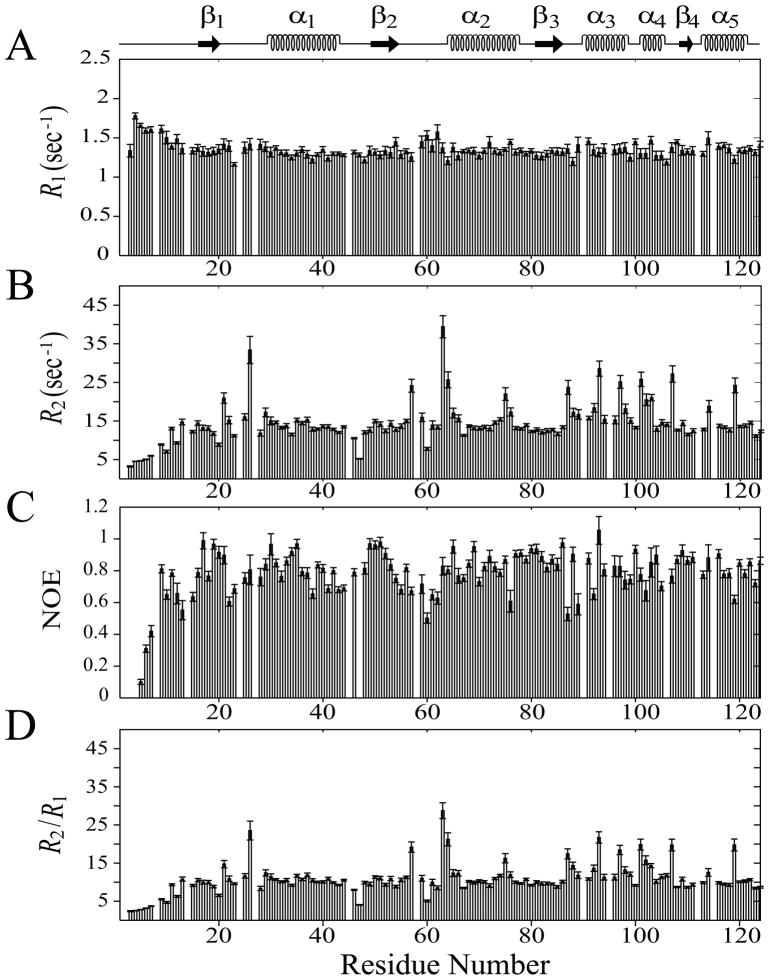
Fig. 1.
15N-relaxation data of Rv1739c STAS domain (~0.5 mM in 50 mM Na phosphate, pH 7.2, 275 mM NaCl) recorded at 600.13 MHz, 300 K. (A) longitudinal relaxation rates (R1), (B) transverse relaxation rates (R2), (C) steady-state heteronuclear NOEs, and (D) R2/R1 ratios are plotted as a function of Rv1739c STAS domain amino acid sequence. STAS secondary structure deduced from the NMR solution tertiary structure is shown at top.
R1 values are homogeneously distributed along the sequence (panel A, Figure 1). Residues localized within secondary structures demonstrated lower R1 (1.33 s−1), in contrast to the residues in the loop regions (Table 1 and Supplementary Table S1). These data show that residues in structured regions exhibit relatively restricted motions compared to those of loop regions. The high flexibility of the Rv1739c STAS N-terminus [15], (L1, first 15 aa) is reflected by their average R1 value of 1.52 s−1, which is higher than the overall R1 of the STAS domain (Supplementary Table S1). The structured region α1-β3 exhibits a tighter distribution, with the exception of the few residues within and near loop region L4.
Table 1.
15N relaxation rates R1 (sec−1), R2 (sec−1), τm (nsec),{1H}-15N heteronuclear NOEs recorded at 600.13 MHz, 300 K, and reduced spectral density function parameters {J(0) (ns/rad), J(ωN) (ns/rad), and J(0.87ωH) (ps/rad)} of secondary structural elements of Rv1739c STAS in the absence or presence of 20 mM GTP or GDP.
| parameter | structural segment | STAS | STAS+GTP | STAS+GDP |
|---|---|---|---|---|
| R1 | α-helices | 1.34 ± 0.05 | 1.27 ± 0.06 | 1.06 ± 0.04 |
| β-strands | 1.33 ± 0.05 | 1.29 ± 0.06 | 1.03 ± 0.04 | |
| loops | 1.36 ± 0.05 | 1.30 ± 0.05 | 1.06 ± 0.04 | |
|
| ||||
| R2 | α-helices | 16.13 ± 0.80 | 16.52 ± 0.69 | 18.63 ± 1.00 |
| β-strands | 12.65 ± 0.49 | 14.46 ± 0.45 | 17.23 ± 0.63 | |
| loops | 15.02 ± 0.74 | 15.55 ± 0.61 | 17.88 ± 0.91 | |
|
| ||||
| τm | overall | 9.55 ± 0.21 | 10.48 ± 0.25 | 13.25 ± 0.34 |
|
| ||||
| NOE | α-helices | 0.81 ± 0.04 | 0.80 ± 0.04 | 0.83 ± 0.05 |
| β-strands | 0.88 ± 0.03 | 0.94 ± 0.04 | 0.80 ± 0.05 | |
| loops | 0.77 ± 0.03 | 0.76 ± 0.04 | 0.79 ± 0.06 | |
|
| ||||
| J(0) | α-helices | 5.79 ± 0.30 | 6.23 ± 0.14 | 6.65 ± 0.31 |
| β-strands | 4.57 ± 0.19 | 5.35 ± 0.10 | 6.32 ± 0.24 | |
| loops | 5.16 ± 0.24 | 5.83 ± 0.16 | 6.59 ± 0.30 | |
|
| ||||
| J(ωN) | α-helices | 0.33 ± 0.01 | 0.31 ± 0.01 | 0.26 ± 0.01 |
| β-strands | 0.33 ± 0.01 | 0.33 ± 0.01 | 0.25 ± 0.10 | |
| loops | 0.34 ± 0.01 | 0.32 ± 0.01 | 0.26 ± 0.01 | |
|
| ||||
| J(0.87ωH) | α-helices | 3.93 ± 0.80 | 4.46 ± 0.76 | 3.03 ± 0.83 |
| β-strands | 2.42 ± 0.65 | 2.56 ± 0.77 | 3.33 ± 0.77 | |
| loops | 5.02 ± 0.68 | 5.64 ± 0.72 | 4.40 ± 0.72 | |
In contrast, the R2 and {1H}-15N NOE values exhibit substantial variation along the protein backbone (panels B and C in Figure 1). The flexibility of the N-terminal loop L1 is also reflected in its R2 value of 8.06 s−1, much lower than the overall domain average R2 (Supplementary Tables S1). The R2 values of residues R20, Q47 (L3), and E60 are also significantly lower (≤ 8.88 s−1) than the overall R2, suggesting faster motions of these residues, as well (Figure 1). However, the 7 loop residues {C26 (L2), S57, L63 and T64 (L4), R87 (L6), L101 (L7), and E107 (L8)}, and 3 helical residues {R93 and R97 (α3), Q119 (α5)} exhibit significantly higher R2 (≥ 23.7 s−1) than the overall average, indicating comparatively restricted mobility of these residues. These higher R2 values may reflect individual contributions of these residues to the conformational exchange dynamics that occur on a slower (μsec-msec) time scale and/or the result of localized conformational effects resulting from the molecule’s non-isotropic tumbling behavior (see below).
{1H}-15N NOE is a measure of high frequency motions that decrease in value as the N-H bond vector mobility increases, and may actually be represented by negative values for those residues sampling high frequency motions. {1H}-15N NOE varied along the protein primary sequence (Figure 1C), with lowest values for N-terminal loop L1. β-strand residues exhibited higher NOE (0.88) than those in α-helices (0.81) or loops (0.77) (Table 1). Excluding those residues found in the highly flexible loop L1, loop residues D22 (L2), E60, V61 and D62 (L4), R87 and K89 (L6) and helix residues L76 (α2), L92 (α3) and Q119 (α5) showed the lowest NOE values (≤ 0.65) (Figure 1C). Among these, loop residues, D22, E60, V61, and D62 also exhibit lower than overall average R2 values (Supplementary Table S1), suggesting the increased mobility of these residues is due to internal motions occurring at faster (psec-nsec) time scale. The distribution of R2/R1 values tracked R2 values (Figure 1D), with an average value of < R2/R1 > = 10.89 ± 0.53.
Seventy-seven residues qualified for a valid R2/R1 data set by coarse-filter criteria (see Experimental Procedures), allowing the calculation of residue-specific local effective correlation times (τe) (Supplementary Figure 1) and overall rotational correlation time (τm). Non-qualified residues were largely from loop regions, whereas 51 of 65 residues (~79%) were localized to the non-loop regions stabilized by secondary structure. The majority of these qualified residues were from C-terminal secondary structured regions, with ~54% from theα3- α4 region. Only 5 residues found within the 15 residue unstructured N-terminal portion of the domain qualified. The overall rotational correlation time (τm) of 9.55 ± 0.21 ns is slightly higher than the 7.5 – 9.0 ns correlation time expected for a 15 kDa protein of 130 aa (including the 6 His tag) [24, 25]. This change may reflect non-isotropic molecular tumbling as indicated by the diffusion tensor analysis.
The principal components of the inertia tensor of the average Rv1739c STAS structure exhibit aratio of 1.00 : 0.92 : 0.77, suggesting that the STAS domain experiences conformational anisotropy. Diffusion tensor optimization yielded diffusion tensor parameters: D||/D⊥ = 0.827 ± 0.015, θ = 0.705 ± 0.055, and φ = 5.632 ± 0.057. These data suggested that the axially symmetric diffusion tensor better described the rotational behavior of Rv1739c STAS, which adopts an oblate ellipsoid shape in solution. Diffusion tensor parameters obtained from both actual and jackknife modes were in agreement (Supplementary Table S2).
(b) Model-free analysis
Using a trimmed data set of R1, R2, {1H}-15N NOE, structural coordinates, estimated τm, and diffusion tensor parameters, model-free formalism was applied to characterize the motional properties of 97 STAS residues. From these 97 residues, 86 were successfully fit to all five standard models. The models provided estimated parameters of the generalized order parameter (S2), fast S2 (S2f), slow S2 (S2s), internal effective correlation times (τe), and conformational exchange terms (Rex) (Supplementary Table S3). All appropriately fit models are plotted on a per residue basis in Supplementary Figure S2. 56 residues utilized model 1 (S2), 9 residues model 2 (S2, τe), 5 residues model 3 (S2, Rex), 8 residues model 4 (S2, τe, Rex), and 8 residues used model 5 (S2, S2f,τe) for appropriate data fit. The eleven residues Q9, Y19, R35, G46, W50, F51, R78, G80, A86, S100, and R124 did not fit well to any of the 5 models.
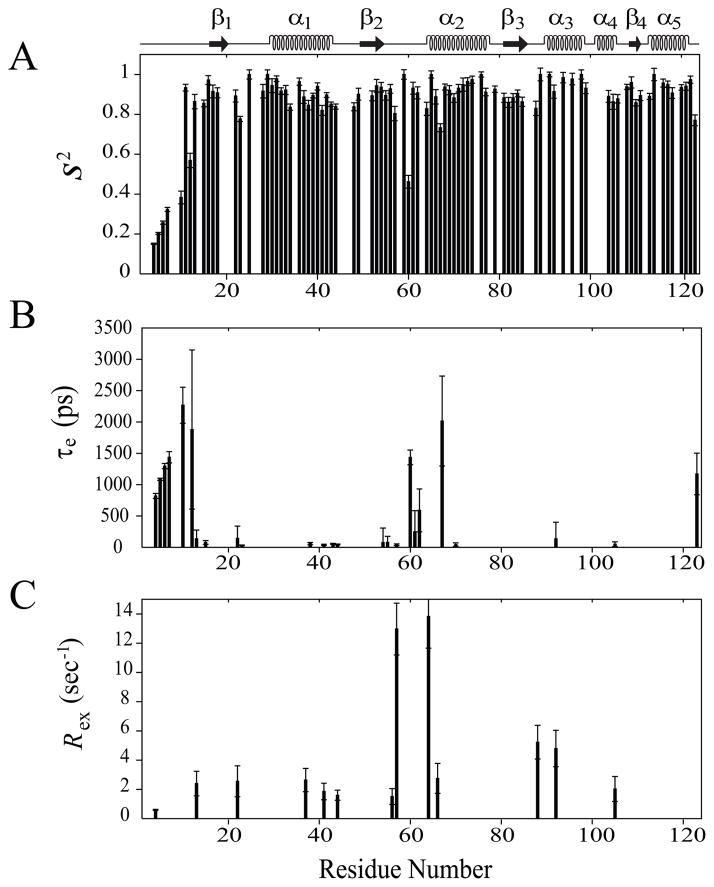
Residue-specific values of S2 (S2 f X S2s), τe, and Rex are plotted in Figure 2. S2 a measure of N-H bond vector amplitude of internal motions within the psec-nsec timescale, ranges between 0 (high amplitude, least ordered motion) and 1 (low amplitude, highly ordered motion) [25]. Overall average S2 of 0.89 ± 0.02 (Figure 2A) supports that Rv1739c STAS behaves as a rigid, structured molecule, as expected for a protein comprising well ordered secondary structured regions (S2 ~ 0.86) [26, 27]. Among secondary structured segments, α-helices and β-strands exhibit similar S2 values of 0.92 and 0.90, respectively, slightly higher than the average loop region S2 value of 0.85. Among individual secondary structured segments, the portion of the domain experiencing the greatest mobility is the N-terminus loop L1, that is characterized by an S2 of 0.50, a value consistent with its high R1, low R2 and {1H}-15N NOE. Importantly, these motional properties are highly congruent with our structural finding of Rv1739c STAS [15]. Exclusion of the loop L1 increased the overall average S2 value to 0.91. All other structured regions exhibit S2 values between 0.85 – 0.97 (Figures 2A and 3A, Supplementary Table S3).
Fig. 2.
Model-free parameters of Rv1739c STAS as determined considering axially symmetric tumbling of the molecule. The values of (A) generalized order parameter (S2), (B) effective correlation time for internal motions (τe), and (C) exchange terms (Rex) with their error values were deduced by fitting the relaxation data to the five models of the model-free formalism, and plotted as a function of amino acid sequence. STAS secondary structure is shown at top.
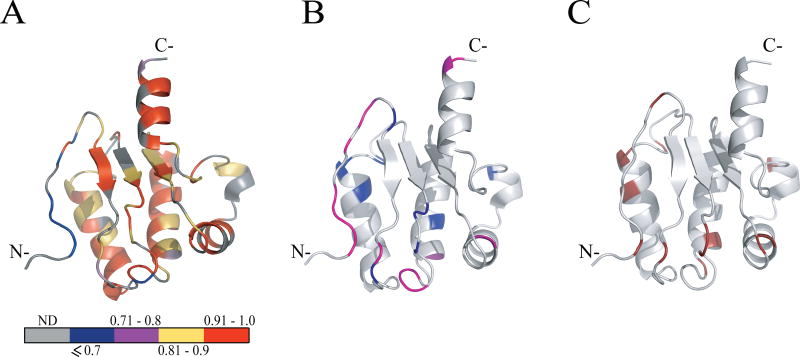
Fig. 3.
Model-free parameters mapped onto the ribbon representation of Rv1739c STAS. (A) Values of generalized order parameters (S2) are highlighted for each residue as follows: blue, S2 ≤ 0.7; magenta, 0.71 ≤ S2 > 0.8; yellow, 0.81 ≤ S2 > 0.9; red, 0.91 ≤ S2 > 1.0; gray, data unavailable (ND). (B) Values of effective correlation times (τe) for each residue: light magenta, residues experiencing faster motions (τe ≤ 100 ps); blue, residues experiencing slower motions (τe > 100 ps). (C) Residues with exchange terms (Rex) that account for conformational and/or chemical exchange are dark red. In (B) and (C), STAS ribbon structure is light gray (“hydrogen” in the PyMOL carbon pseudocolor scheme).
Twenty five residues contribute to Rv1739c STAS internal motions (τe), of which the 11 residues G15 (L1), 23 (L2), L38 and V41 (α1), Q43 and D44 (L3), 54 (β2), A55 (L4), S57 (L4), D70 (α2), and I105 (α4) show faster internal motions (τe ≤ 100 psec), while the 14 residues including I4, D5, D6, Y7, A10, R12, and V13 (L1), D22 (L2), E60, V61, and D62 (L4), D67 (α2), L92 (α3), and R123 (L10) account for slower internal motions (τe > 100 psec). Among these latter, D5, D6, Y7, A10, R12, E60, D67, and R123 show the slowest motions, on the sub-nanosecond time scale (Figures 2B and 3B).
The 13 Rv1739c STAS residues accounting for the chemical and/or conformational exchange contribution (Rex) to the relaxation data [I4 and V13 (L1), D22 (L2), A37 and V41 (α1), D44 (L3), E56, S57, and T64 (L4), L66 (α2), V88 (L6), L92 (α3), and I105 (α4)] reflected slowed internal motions on the μsec-msec time scale, and were fit with model 3 or 4. This residue pool comprises 5 and 8 residues from helical and loop segments, respectively; no β-strand residue required a Rex term to fit the data. Five residues (A37, E56, T64, L66, and V88) were fit with model 3. The remaining eight residues (I4, V13, D22, V41, D44, S57, L92, and I105) required the additional parameter, τe, to fit the data (Figures 2C and 3C).
15N backbone dynamics of liganded vs. unliganded STAS
(a)relaxation NMR data
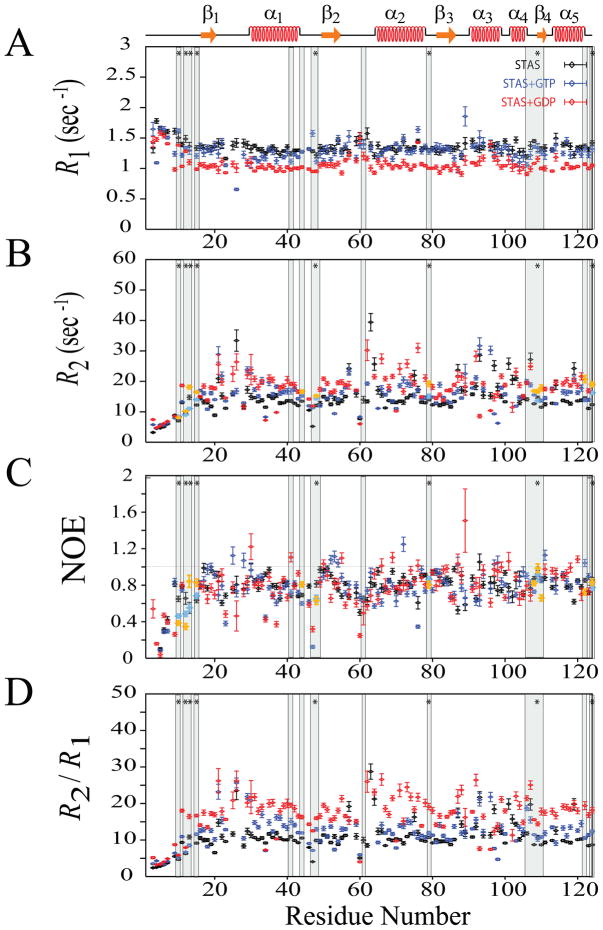
The effect of guanine nucleotide binding on Rv1739c STAS domain dynamics was studied by performing backbone 1H-15N relaxation measurements; T1, T2, and {1H}-15N NOE in the absence and presence of GTP or GDP. R1, R2, and {1H}-15N NOE data for STAS in the presence of GTP were obtained for 108, 107, and 107 residues, respectively (Supplementary Table S1 and Figure 4). Spectral overlap and poor resolution prevented measurement of R1 for the four residues L25, D62, T64, and Q119, of R2 for the five residues V61, L63, T64, L101, and L102, and {1H}-15N NOE for the three residues L102, Q119, and F121, respectively. {1H}-15N NOE values for residues Q47 and G106 were resolved in GTP-bound Rv1739c STAS but not in the unliganded protein. Average overall values of these 15N-relaxation parameters for 104 residues of Rv1739c STAS in presence of GTP were: < R1 > = 1.29 ± 0.05 s−1; < R2 > = 15.45 ± 0.60 s−1; < NOE > = 0.78 ± 0.04; and < R2/R1 > = 12.14 ± 0.57. GTP decreased R1 by 5.15 %, increased R2 by 5.68 %, decreased {1H}-15N NOE by 1.27%, and increased overall R2/ R1 by 11.48 % (Figure 4). Excluding residues in highly flexible loop L1, the loop residues A23 (L2), D44, G46, Q47, and V48 (L3), V61 and T64 (L4), and helix residues R34 and A37 (α1), L66, D70, and L76 (α2), L92 and R93 (α3) showed the lowest NOE values (≤ 0.65) (Figure 4C). Among these, loop residues A23, D44, G46, Q47, and V48 and helix residues R34 and D70 exhibit lower than overall average R2 values (Supplementary Table S1), suggesting the increased mobility of these residues is due to internal motions occurring on a faster (psec-nsec) time scale.
Fig. 4.
15N-relaxation data for unliganded Rv1739c STAS, (~0.5 mM in 50 mM Na phosphate, pH 7.2, 275 mM NaCl) and for Rv1739c STAS in the presence of 20 mM GTP and 20 mM GDP recorded at 600.13 MHz, 300 K. (A) Superposed sequence-specific longitudinal relaxation rates (R1). (B) Superposed sequence-specific transverse relaxation rates (R2). (C) Superposed sequence-specific {1H}-15N heteronuclear NOEs. (D) Superposed sequence-specific R2/R1 ratios. Shaded bars highlight residues experiencing chemical shift perturbation (CSP) upon nucleotide binding; bars marked by ‘*’ indicate residues perturbed by both GTP and GDP {15}. CSP residues that exhibit increased R2 and J(0) and decreased NOE and J(ωN) values are aquamarine for GTP and yellow for GDP; see text for details. STAS secondary structure is shown at top.
R1, R2, and {1H}-15N NOE data for STAS in the presence of GDP were obtained for 110, 108, and 110 residues, respectively (Supplementary Table S1 and Figure 4). Spectral overlap and poor resolution prevented measurement of R1 for the two residues A28 and F121, of R2 for the four residues V61, L63, A65, and L101, and of {1H}-15N NOE for residues L102 and Q119. {1H}-15N NOE values for residues D3, I4, Q47 and G106 were resolved in GDP-bound Rv1739c STAS but not in the unliganded protein. Average overall values of these 15N-relaxation parameters for Rv1739c STAS in presence of GDP were: < R1 > = 1.08 ± 0.04 s−1; < R2 > = 17.66 ± 0.86 s−1; < NOE > = 0.77 ± 0.05; and < R2/R1 > = 16.86 ± 0.84. GDP decreased R1 by 20.59 %, increased R2 by 20.79%, decreased {1H}-15N NOE by 2.53%, and increased overall R2/ R1 by 54.82 % (Figure 4). Excluding residues in highly flexible loop L1, the loop residues A23 and C26 (L2), G46, Q47, and V48 (L3), E56, V59, E60, V61, and D62 (L4), helix residues R34 and A37 (α1), A65 (α2), L96 (α3), and K104 (α4) and strand residues V18 (β1) and A86 (β3) showed lowest NOE values (≤ 0.65) (Figure 4C). Among these, loop residues A23, G46, Q47, V48, V59, and E60, helix residues R34, A37, and L96, and β-strand residue V18 exhibit lower than overall average R2 values (Supplementary Table S1), suggesting the increased mobility of these residues is due to internal motions occurring at faster (psec-nsec) time scales. Determination of R2 values for STAS residues V61 and L63 in the ligand-bound conformational state was precluded by line-broadening of these resonances. These spectral features likely reflect additional slow/intermediate exchange processes that are sampled by these residues. Interestingly, the relaxation parameters of residues immediately adjacent to these residues suggest that these residues do experience high R2 values.
Despite the overall increase in R2 elicited by GTP binding, individual residue and local segmental R2 values varied along the protein sequence, whereas the increase in R2 by GDP binding was uniform (Figure 4 and Supplementary Table S1). Both ligands decreased R1 across most of the structured segments of the protein. However, the nucleotide-induced changes in R1 and R2 values differed in the L6-α3-L7 region, perhaps reflecting the enhanced inherent mobility experienced locally by this region. These ligand-induced changes in the measured relaxation parameters suggest distinct effects of GTP and GDP on internal dynamics. The increased R2/R1 ratios of ligand-bound STAS suggest slower tumbling of the liganded protein, and may reflect increased ligand-induced anisotropy of the STAS domain. Further implications of the increased R2/R1 ratio with respect to estimated relaxation parameters are discussed below.
These R1 and R2 results were used to further estimate the overall rotational correlation times (τm) of Rv1739c STAS alone and in the presence of these nucleotides. Coarse-filter criteria applied to the R1, R2, and {1H}-15N NOE data sets of liganded STAS were as described above for unliganded STAS. In the presence of GTP, 71 filter-qualified STAS residues exhibited a τm of 10.48 ± 0.25 ns, an increase of ~10% over that of unliganded STAS. In the presence of GDP, 73 filter-qualified STAS residues permitted calculation of τm = 13.25 ± 0.34 ns, a value ~39% longer than that of unliganded STAS (Supplementary Figure S1). These estimated (τm) values for free, GTP-bound, and GDP-bound STAS agree with the values determined from spectral density functions within ~6% [28]. Thus, nucleotide binding substantially altered 15N backbone relaxation parameters of Rv1739c STAS. GDP-induced changes in STAS dynamic parameters exceeded those induced by GTP, consistent with the higher STAS binding affinity for GDP [15].
(b) reduced spectral density function analysis
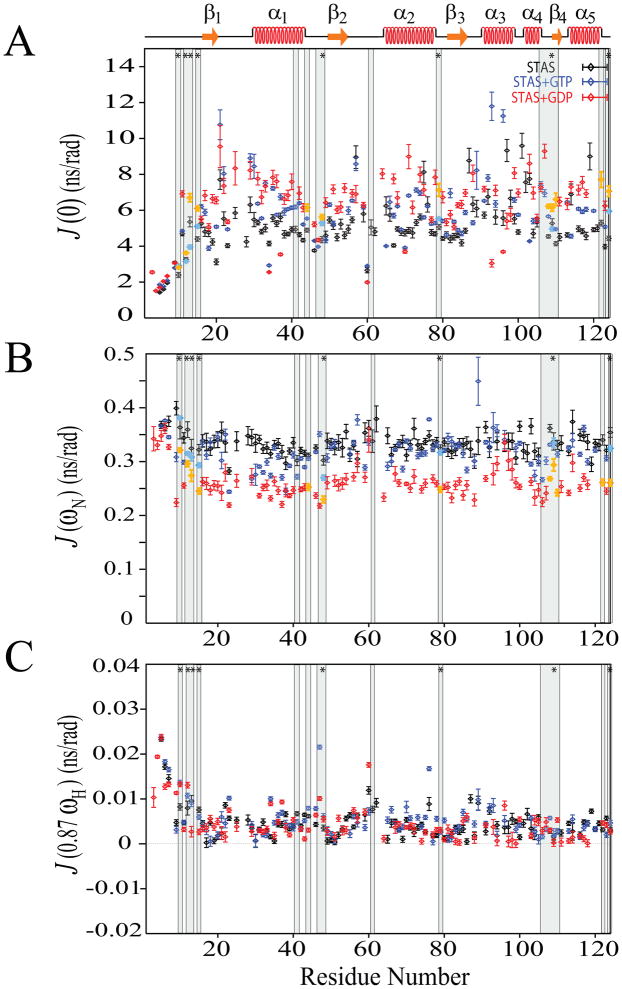
The above guanine nucleotide binding effects on Rv1739c STAS relaxation parameters encouraged us to estimate additional dynamic parameters. However, model-free formalism requirements for rotational diffusion tensor information unavailable for nucleotide-Rv1739c STAS supported our reduced spectral density mapping studies of STAS backbone dynamics [22, 23, 28, 29]. Reduced spectral density mapping analysis yielded motional information of N-H bond vectors at three different frequencies; J(0), J(ωN), and J(0.87ωH) (Figure 5). For 103 residues of unliganded STAS these values were: < J(0) > = 5.13 (nsec); < J(ωN) > = 0.33 (nsec); and < J(0.87ωH) > = 4.57 (psec). For 89 residues of GTP-liganded STAS the values were: < J(0) > = 5.76 (nsec); < J(ωN) > = 0.32 (nsec); and < J(0.87ωH) > = 5.43 (psec). For 90 residues of GDP-liganded STAS, the values were: < J(0) > = 6.19 (nsec); < J(ωN) > = 0.26 (nsec); and < J(0.87ωH) > = 4.59 (psec) (Supplementary Table S4). Figure 6 shows reduced spectral density functions plotted as the difference between liganded and unliganded Rv1739c STAS.
Fig. 5.
Reduced spectral density functions deduced from 15N relaxation data for free, GTP-bound, and GDP-bound STAS. (A) Superposed sequence-specific J(0) functions. (B) Superposed sequence-specific J(ωN) functions. (C) Superposed sequence-specific J(0.87ωH) functions. Shaded bars highlight residues experiencing chemical shift perturbation (CSP) upon nucleotide binding; bars marked by ‘*’ indicate residues perturbed by both GTP and GDP { 15}. CSP residues that exhibit increased R2 and J(0) and decreased NOE and J(ωN) values are aquamarine for GTP and yellow for GDP; see text for details. STAS secondary structure is shown at top.
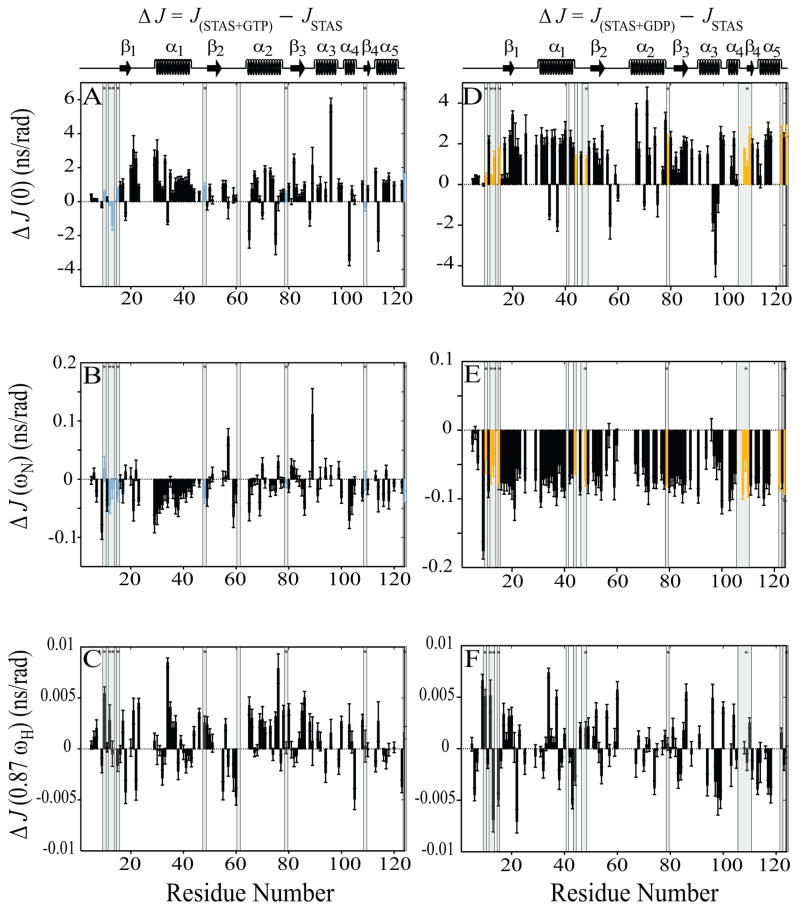
Fig. 6.
Sequence-specific reduced spectral density function value differences (ΔJ) between nucleotide-bound and free STAS. Left panels, (A–C),ΔJ between GTP-bound STAS and free STAS; Right panels, (D-F), ΔJ between GDP-bound STAS and free STAS. (A & D) ΔJ(0), (B & E) ΔJ(ωN), and (C & F) ΔJ (0.87ωH). Shaded bars highlight residues experiencing chemical shift perturbation (CSP) upon nucleotide binding; bars marked by ‘*’ indicate residues perturbed by both GTP and GDP {15}. CSP residues that exhibit increased R2 and J(0) and decreased NOE and J(ωN) values are aquamarine for GTP and yellow for GDP. STAS secondary structure is at top of each column of panels.
Overall J(0) was increased ~12 % in the presence of GTP and ~21 % in the presence of GDP as compared to values for unliganded STAS. In contrast, overall J(ωN) was modestly decreased by ~3 % following GTP binding, but significantly decreased by ~21 % in the presence of GDP. Overall J(0.87ωH) was increased ~19% in the presence of GTP, but decreased by ~0.5 % by GDP. These changes likely reflect localized individual residues within secondary structured segments (Supplementary Table 4 and Figure 5).
J(0) is sensitive to internal motions of the N-H bond vectors on fast (psec-nsec) as well as slow (μsec-msec) time scales; A value of J(0) < 2/5τm reflects the presence of faster internal motions, whereas, high J(0) values indicate slower internal motions often observed in the presence of chemical or conformational exchange. J(ωN) and J(0.87ωH) are sensitive to faster (sub-nanosecond) internal motions but insensitive to the slower (μsec-msec) internal motions of N-H bond vectors [30].
Filter criteria for higher and lower residue-specific J(0) were chosen as one standard deviation (S.D.) above and below the average J(0) value, as determined by residues within structured regions of the core characterized by uniform J(0) distribution along the sequence [31]. Average J(0) of core region residues was 5.30 ns/rad for STAS, 6.03 for STAS+GTP, and 6.46 for STAS+GDP. Higher J(0) values were those satisfying the inequality J(0) > J(0)cutoff ; where J(0)cutoff = {J(0)avg + one S.D.}. The {2/5τm} values of STAS, STAS+GTP, and STAS+GDP were 8% below those characterized by lower values {J(0)avg - one S.D.}. Therefore, residues with lower J(0) were identified by the criterion of J(0) < 2/5τm [29, 30].
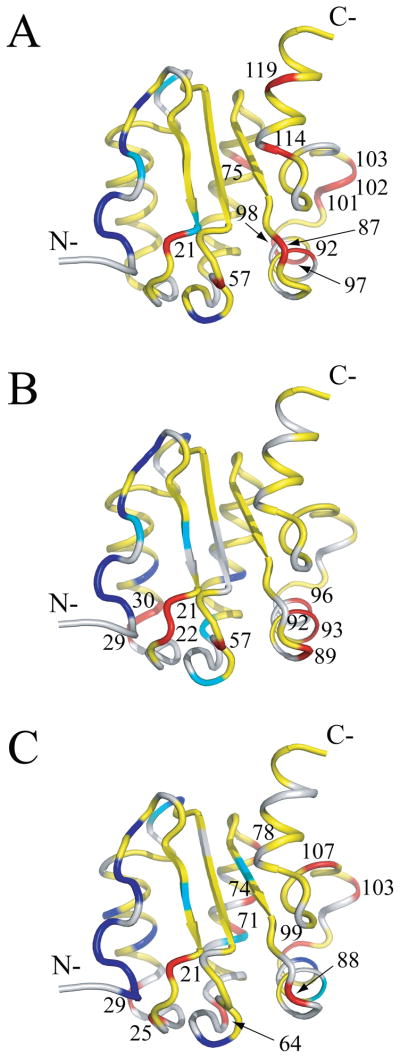
Residue-specific J(0) values determined by the above criteria in the absence and presence of nucleotides are mapped onto the Rv1739c STAS solution structure in Figure 7. Residues with J(0) < 2/5τm and J(0.87ωH) > 7.5 ps/rad exhibited internal flexibility on a faster (sub-nanosecond) time scale. They include (in blue) for STAS, D5, D6, Y7, A10, R12, and E60; for STAS+GTP, D5, D6, Y7, A10, R12, V13, R34, Q47, and D70; and for STAS+GDP, D3, I4, D5, D6, Y7, Q9, A10, R12, R34, A37, Q47, E60, and L96. Residues with J(0) < 2/5τm and J(0.87ωH) < 7.5 ps/rad also demonstrated internal flexibility on a fast (sub-nanosecond) time scale, including (in cyan); Q9, R20, and G46 for STAS alone; Q9, V18, E60, and A65 for STAS+GTP; and V18, G46, D70, F83, and R93 for STAS+GDP. Residues with J(0) > J(0)cutoff revealed internal flexibility on a slow time scale (μsec-msec) that may represent chemical or conformational exchange. These residues (in red) include Y21, S57, E75, R87, L92, R97, A98, L101, L102, D103, L114, and Q119 for STAS alone; Y21, D22, N29, A30, S57, K89, L92, R93, and L96 for STAS+GTP; and Y21, L25, N29, T64, Q71, T74, R78, V88, A99, D103, and E107 for STAS+GDP. These residues also exhibited low J(0.87ωH) values supporting the absence of faster time scale internal motions. Residues with 2/5τm < J(0) < J(0)cutoff (in yellow) exhibit nsec time scale internal motions which may be relatively slower than those identified with J(0) < 2/5τm. Among these residues, V13, G15, D22,V61, D62, L76, and K89 of STAS, and A23, D44, G46, and L76 of STAS+GTP exhibit J(0.87ωH) > 7.5 ps/rad (Supplementary Table S4) that may reflect their relatively increased motion as compared to residues with J(0.87ωH) < 7.5 ps/rad.
Fig. 7.
J(0) values mapped onto the Rv1739c STAS average structure. (A) J(0) values of STAS domain; (B) J(0) values of STAS+GTP; (C) J(0) values of STAS+GDP. On the light gray ribbon structure, blue residues have J(0) < 2/5τm and J(0.87ωH) > 7.5 ps/rad; cyan residues have J(0) < 2/5τm and J(0.87ωH) < 7.5 ps/rad; yellow residues have 2/5τm < J(0) > J(0)cutoff rendered; red residues have J(0) > J(0)cutoff rendered red and are labeled by residue number. J(0)cutoff = J(0) + one RMSD.
In earlier studies, our 1H-15N HSQC data identified chemical shift (CSP) perturbations for 9 and 16 Rv1739c STAS residues following the binding of GTP and GDP [15]. These residues are denoted by shading under the bars in Figures 4 through 6 and are mapped onto the STAS structure (Supplementary Figure S3). Several of the GTP-perturbed residues V48, R79, and R124 and A10, as well as the GDP-perturbed residues R12, V48, R79, D108, I110, R122, and R124 exhibit increased R2 and J(0) values and decreased NOE and J(ωN) values (in red, Supplementary Figure S3). GDP-perturbed residues V13, G15, D44, and H109 exhibited increased R2 and J(0) values and decreased J(ωN) values (in yellow, Supplementary Figure S3). GTP- perturbed residues A10, R12, V13, G15, and H109 (in yellow, Supplementary Figure S3) were characterized by different patterns of R2, J(0), NOE, and J(ωN) values. GTP-perturbed residue V61 and GDP-perturbed residues V41, Q47, G106, and E107 (in blue, Supplementary Figure S3) lack data for at least two parameters among R2, J(0), NOE, and J(ωN), and so could not be further analyzed.
Nucleotide binding increased overall J(0) value uniformly across the STAS domain, suggesting both slowed internal motions and a possible contribution from increased molecular anisotropy (Supplementary Tables S4-S7). The low J(0) values of structured regions L1-β1, L3, L5-β3, L7-α4, and β4 in the GTP-STAS complex (Supplementary Table S6), and L1-β1, L3, L4, β3, and α3 in the GDP-STAS complex (Supplementary Table S7) support increased flexibility on faster time scale and lower τm than for other structured regions. J(0.87ωH), was non-uniform across the protein sequence, consistent with {1H}-15N NOE. Nucleotide-specific responses of J(0.87ωH) and {1H}-15N NOE parameters for secondary structured segments are summarized in Supplementary Tables S5–S7. The combined effect of these two parameters on individual secondary-structured segments is discussed below.
C-terminal aa 114–124 are significantly ordered by presence of the 8 aa α-helix, α5. The ligand-independent lack of internal flexibility in this region is reflected in the above-average NOE values (Supplementary Table S1), 2/5τm < J(0) ≪ J(0)cutoff, and low values of J(0.87ωH), with exceptions of residues L114 and Q119 in the absence of ligand. In contrast, many residues of N-terminal loop region L1 (aa 1–15) are characterized by NOE values below average and by lowest J(0) values and highest values of J(0.87ωH) (Table S1 and Supplementary Tables S5–S7). These properties indicate both internal flexibility on a faster time scale and ligand-independent lack of motional rigidity of this highly flexible segment.
J(ωN) was nearly invariant along the polypeptide sequence in the absence or presence of ligand (Figure 5 and Supplementary Table S4), consistent with the unchanging R1 values (Supplementary Table S1). However, J(ωN) values were reduced by nucleotide binding, suggesting increased psec-nsec internal motions. Ligand binding decreased J(0.87ωH) in structured C-terminal segments suggesting limited faster motions in this region.
Taken together, these data show that Rv1739c STAS dynamic properties are dominated by ligand binding-induced increases of τm. Modulation by GDP was greater than by GTP, again reflecting the higher binding affinity of GDP [15].
Discussion
STAS domain structures reported to date include those of internally deleted rat prestin [14], M. tuberculosis Rv1739c [15], and E. coli ychM in complex with acyl carrier protein [9]. Backbone dynamics were previously reported for the unliganded state of structurally related bacterial response regulators, Spo0F and SpoIIAA. While the SpoIIAA backbone dynamics were nearly invariant throughout the protein [32], significant local variations in the backbone dynamics were observed within Spo0F regions involved in protein-protein interactions. These regions were characterized by slow mobility on the msec time scale even in the absence of ligand [33]. However, investigation of backbone dynamics for STAS domains of SulP/SLC26 anion transporter proteins has not been reported. In this report we have explored the 1H-15N backbone dynamics of Rv1739c STAS in the absence and presence of the two nucleotide ligands GTP and GDP.
Protein backbone flexibility is normally greater at the N- and C-termini of a protein than within the protein core. The measured relaxation parameters R1, R2, and {1H}-15N NOE), and the estimated reduced spectral density parameters J(0), J(ωN), and J(0.87ωH) revealed that the Rv1739c STAS N-terminal L1 loop is more flexible than the protein core and, unlike the rest of the protein, was unaltered following nucleotide binding. In contrast, the Rv1739c STAS C-terminal helix α5 is well-ordered and rigid, in agreement with our previous structural studies [15]. Beyond the L1 loop, relaxation parameter values were homogeneously distributed among secondary structured elements of helices and strands. Heterogeneity of measured and estimated relaxation parameters was largely restricted to loop regions L2, L4, and L6. R2 and J(0) values were heterogeneously distributed additionally in the L7-L8 regions near helix α4.
The slight increase in overall rotational correlation time of Rv1739c suggests a degree of anisotropic molecular tumbling. This anisotropy is consistent with modest (< 8 %) aggregation that was occasionally observed in low ionic strength buffers for STAS concentrations of 0.7–1.0 mM [15], necessitating increased salt concentrations [275 mM] used during extended periods of protein NMR data acquisition. The increased R2/R1 ratio paralleled the increased τm and the slower tumbling of GDP-STAS and (to a lesser degree) of GTP-STAS (see above). This slowed tumbling behavior dominated the observed and calculated relaxation properties.
For ωHτm ≫ 1, and with consideration of 15N CSA and 1H-15N dipolar interaction, overall NOE for a rigidly spherical tumbling molecule should have a value of 0.82 [34]. Thus, the overall NOE value of 0.79 suggested Rv1739c STAS to be a relatively rigid protein. The marginally reduced NOE values of the guanine nucleotide-bound states (Table 1) may reflect slight decreases in overall STAS rigidity. SpoIIAA backbone dynamics data suggested an average NOE of 0.77; these data were not analyzed further for estimation of either model-free or spectral density parameters. Based on limited conformational fluctuation and flexibility of terminal residues, the SpoIIAA structure was interpreted as a rigid isotropic fold, despite the anisotropy (and possible prolate ellipsoid shape) reflected in the inertia tensor components of 1:1.2:1.4 [32]. The S2 parameter measures faster time scale motions, and the distribution of (1-S2) values pattern is proportional to J(0.87ωH) values when ωHτm ≫ 1 [28]. Since this condition holds for Rv1739c STAS in both unliganded and liganded states, J(0.87ωH) of the liganded complexes may correlate with similar patterns of (1- S2) and provide an estimate of S2 of the complexes as discussed below.
Rv1739c STAS exhibited differences between its binding of GTP and GDP. GTP binding increased R1 value of residue S57 and immediately adjacent residues, as well as of K89. GDP binding increased R1 values of E60 and its adjacent region, as well as of L76, E94, L96, and R97. GTP binding lowered R2 values of L53, R97, and A98, and increased R2 values of R93 and L96. In contrast, GDP binding lowered R2 values only for R93 and L96. However, Rv1739c STAS binding of either GTP or GDP lowered R2 values of Q47 and E60 and increased R2 of Y21. The NOE mirrors the J(0.87ωH) distribution. In the absence or presence of nucleotide, all secondary-structured segments exhibited NOE values > 0.7 and J(0.87ωH) values < 7.5 ps/rad, except for the loop regions L1, L3 and L4, for which values of these parameters exhibited specific nucleotide-sensitivity (Table 1 and Supplementary Tables S5–S7). The majority of these segments exhibited similar responses of NOE and J(0.87ωH) values to both nucleotide ligands. However, segments β1, L2, β2, L4, and α4 exhibited nucleotide-specific changes in motional rigidity, which was marginally increased by GTP and marginally lowered by GDP, whereas helix α2 exhibited opposite effects. These nucleotide-specific responses likely reflect differences in packing and orientation of the bound ligands.
In both unliganded and liganded forms of Rv1739c STAS the spectral density term J(0.87ωH) is lower than J(ωN), and both, in turn, are lower than J(0) globally and for individual residues. These relationships, characteristic of structured, well-folded proteins, support the predominant contribution of faster internal motions (psec-nsec time scale) to the high frequency terms J(0.87ωH) and J(ωN), and the additional contribution to J(0) of slower internal motions (μsec-msec time scale) reflecting Rex. This key feature of different time scale motions in J(0.87ωH) and J(ωN) vs. J(0) increases the utility of reduced spectral density analysis in the study of motions across a wide range of time scales encompassing those of enzymatic catalysis and protein-ligand interactions. As described by Lefevre et al., Rv1739c STAS spectral density parameters in both the absence and presence of nucleotide ligands exhibit linear dependence of J(0) with J(ωN) and with J(0.87ωH) across most of the protein [35] except for residues of the N-terminal region including loop L1 (Supplementary Figure S4). The core region plot of J(0.87ωH) vs. J(0) shows select off-set residues (red in Supplementary Figure S4) which are those with J(0) > J(0)cutoff (red in Figure 7). The high J(0) values of these residues indicate slower internal motions of μsec-msec time scale, probably reflecting chemical and/or conformational exchange processes.
Most N-terminal L1 region residues and some core residues of J(0) < 2/5τm follow a linear fit deviating from the protein core fit (Supplementary Figure S4). These residues (blue and cyan in Supplementary Figure S4) follow J(0.87ωH) value criteria set for similar residues in Figure 7, and experienced high amplitude motions on a faster NMR time scale. Within the L1 region, in both free and bound states, motion increases towards the N-terminus, as reflected in decreasing J(0) and J(ωN) and increasing J(0.87ωH). Loop regions experienced fast internal motions compared to helical and strand regions, as reflected by J(0.87ωH) > 7.5 psec/rad [29, 36] for nearly all L1 residues of unliganded and liganded STAS. In loop L3, J(0.87ωH) exceeded 7.5 psec/rad for D44, G46, and Q47 in the GTP-bound state, but for Q47 only in the GDP-bound state. In loop L4, adjacent to the apparently unphosphorylated residue S58 [15] corresponding to the SpoIIAA phosphorylation site [37], J(0.87ωH) exceeded 7.5 psec/rad for V61 and D62 of both unliganded and liganded STAS, in contrast to nearby E60, for which J(0.87ωH) exceeded 7.5 psec/rad only for unliganded and GDP-bound STAS, but not for GTP-bound STAS. J(0.87ωH) also exceeded 7.5 psec/rad for most residues of region L6-α3, but for R93 exceeded 7.5 psec/rad in the presence of GTP and fell to 3.77 psec/rad with GDP. The locally restricted variation in backbone motions evident in Rv1739c might indicate interaction surfaces, as has been observed for SpoII0F [33] and Cdc42Hs [38].
Relaxation data and spectral density parameters for some residues of Rv1739c STAS could not be obtained in the presence of nucleotide, possibly due to spectral overlap. Most of these residues also failed filter criteria and exhibited very high R2 values, perhaps reflecting increased linewidths correlating with increased τm. Among residues that exhibited nucleotide-induced CSP [15], (Supplementary Figure S3) some showed increased R2 and J(0) and decreased NOE and J(ωN) values, in agreement with most core residues of the STAS domain, suggesting decreased rigidity, slowed internal motions, and larger tumbling times (τm). The 9 GTP-perturbed residues exhibited average R2/R1 ratio of 12.14 and average τm of 9.42 ns, a tumbling time lower than the 10.48 ns overall average of GTP-bound STAS. The 16 GDP-perturbed residues exhibited average R2/R1 ratio of 16.84 and average τm of 12.65 ns, a tumbling time lower than the 13.25 ns overall average of GDP-bound STAS. These analyses suggest that the observed increases in R2 and J(0) values for residues that experience CSP partially contribute to the overall increase in τm as well as to the ligand-induced increase in molecular anisotropy of the STAS domain. However, we cannot rule out additional factors governing intermediate-to-slow time scale motions associated with these residues.
In conclusion, the above relaxation parameters and the higher J(0) and R2 values of both unliganded and liganded Rv1739c STAS suggest that certain residues, in addition to contributing to increased molecular anisotropy of the STAS domain, may undergo slow internal motions accompanied by chemical/conformational exchange. These residues include, for STAS, Y21 and C26 (L2), S57 and L63 (L4), E75 (α2), R87 (L6), L92, R93, R97, and A98 (α3), L101 (L7), L102 and D103 (α4), E107 (L8), L114 and Q119 (α5); for STAS+GTP, Y21, D22, and N29 (L2), A30 (α1), S57 (L4), K89 (L6), L92, R93, and L96 (α3); and for STAS+GDP, Y21, L25, and N29 (L2), D62 and T64 (L4), Q71, T74, and L76 (α2), R78 (L5), V88 (L6), R92 and A99 (α3), D103 (α4), and E107 (L8). Several of these residues belong to segments L2, L4, and L6-α3, which constitute a single surface in the J(0) map of Rv1739c STAS; in L2 both GTP and GDP increased from 1 to 3 the number of residues with J(0) > J(0)cutoff (Figure 7). Additional regions of elevated J(0) in the absence of GTP include α2, α4 and L8; in α2 GDP increased from 1 to 3 the number of residues with J(0) > J(0)cutoff. α5 residues L114 and Q119 exhibited J(0) > J(0)cutoff only in the absence of nucleotide. With the single exceptions of V61 in GTP-bound STAS and E107 in GDP-bound STAS, these residues do not include those that underwent nucleotide-induced CSP.
Analysis of these data suggested that STAS binding by either GTP or GDP increased the number of residues undergoing slower motions in the region of loop L2. The residues of STAS domain helix α5 exhibiting slower motions were undetectable in the ligand-bound states. Residues undergoing slower motions in and near region of helix α4 were detected in ligand-free and in GDP-bound STAS, but undetected in GTP-bound STAS. GDP binding increased the number of residues undergoing slower motions in the region of helix α2, but these residues were undetected in GTP-bound STAS. The data together suggest that ligand-induced slower internal motions are confined to a molecular surface including loops L2, L4, and L6 and helix α3. Ligand binding also resulted in faster internal motions of residues within the same molecular surface, as well as in select aa residues from the region of helix α1. Binding of GDP (but not GTP) promoted slower internal motions in the region of helices α2 and α4, including the region near α4 identified by CSP and docking calculations to contribute to GDP binding [15]. The nucleotide-specific differences in internal motions may reflect the absence, presence, or hydrolysis of the γ-phosphate.
The Rv1739c STAS dynamics parameters studied here have identified residues that exhibited guanine nucleotide-sensitive internal motions on psec-nsec and μsec-msec time scales. We speculate that these nucleotide binding-induced internal motions are linked to localized conformational changes in a contiguous molecular surface of Rv1739c STAS. Although a few STAS residues exhibit both CSP and motional parameter changes in response to nucleotide, additional residues may be influenced allosterically by nucleotide binding. Allosteric regulation is a common phenomena in both enzymatic catalysis and signal transduction [39–41], as shown for the guanine nucleotide binding G proteins and transglutaminase 2 [42, 43].
Taken together, the elevated J(0) and R2 values of ligand-bound Rv1739c STAS suggest that nucleotide binding increased slow (μsec-msec) internal motions contributing to Rex terms, and decreased faster (psec-nsec) internal motions resulting in slower tumbling (increased τm) of the liganded protein. The increased R2/R1 ratio of liganded STAS is consistent with the increased τm, and these effects are more prominent for GDP than for GTP. The ligand-induced increase in overall rotational correlation time of Rv1739c STAS could reflect a modestly increased molecular anisotropy of liganded vs. unliganded STAS domain, and/or a modest shift in equilibrium between monomer and still hypothetical dimer.
Rat and zebrafish prestin, human SLC26A3, and P. aeruginosa putative SulP anion transporter PA1647 have been characterized as holoprotein dimers [44], Prestin dimers may be disulfide-linked [45] (but see [46]). Evidence for higher order oligomers has been presented for gerbil and mouse prestin holoprotein [45]. However, isolated STAS domain from SLC26A3 [47], isolated internal loop-deleted STAS domain from rat prestin [14], and isolated Rv1739c STAS [15] behaved as monomers, or at least lacked evident characteristics of oligomers. In contrast, the STAS domain of E. coli putative bicarbonate transporter ychM was isolated as a 1:1 heterodimer with acyl carrier protein [9]. The class of G proteins known as GADs are activated by (usually but not uniformly) ligand-induced homodimerization, proceeding through their exchange cycles independent of GAPs, GEFs, and GDIs [48]. GADs resemble Rv1739c STAS in their generally low guanine nucleotide affinities and low (as for hGBP1) or absent (as for some septins) GTPase activities. However, the canonical G protein switch regions present in GADs remain unidentified in Rv1739c or other STAS domains.
Future NMR experiments designed to detect slower motions on the μsec-sec time scale, including CPMG [49, 50], R1ρ [51, 52] relaxation dispersion, and ZZ exchange [53], may provide further insight into nucleotide-STAS interactions dominated by slower internal motions, and clues to their roles in SulP/SLC26 holoprotein functions. Comparison in these experiments of wildtype Rv1739c STAS with missense mutants in the conserved candidate phosphorylation site or in conformationally sensitive Trp and Tyr residues may address more directly the links between backbone motional properties and protein conformation and function.
Experimental Procedures
Protein sample preparation and NMR data acquisition
15N-labeled Rv1739c STAS domain (aa 437–560) with a C-terminal his6-tag was overexpressed and purified as previously reported [15, 16], including a final step of size exclusion chromatography [15] in 50 mM Na phosphate, 275 mM NaCl, pH 7.2 [15]. The NMR sample was prepared in a 92% H2O/8%D2O solvent system that contained 2–3 mM DTT-d10, 0.05% (w/v) NaN3, and 0.25 mM DSS (2,2-dimethyl-2-silapentanesulfonic acid) as an internal standard. STAS concentration was ~ 0.4 – 0.5 mM in both unliganded and liganded forms [16].
Longitudinal relaxation times (T1), transverse relaxation times (T2), and {1H}-15N heteronuclear NOE were measured from 2D 1H-15N HSQC spectra recorded at 298 K on a Bruker Avance 600.13 MHz spectrometer equipped with a 5 mm triple resonance PFG (z-axis) probe, using pulse programs as described [54]. All NMR experiments were executed in the gradient-selected sensitivity-enhanced mode using a matrix of 2048 (F2) x 128 (F1) data-points and spectral widths of 13 ppm and 36 ppm in the 1H (F2) and 15N (F1) dimensions, respectively. Frequency discrimination in the indirect dimensions of all multidimensional spectra was achieved using the States-TPPI mode of quadrature detection [55]. 1H and 15N carrier frequencies were set to 4.70 ppm and 119 ppm, respectively. Similar data sets of T1, T2, and {1H}-15N NOE were acquired for three different 15N labeled samples of nucleotide-free STAS, STAS in the presence of 20 mM GTP, and STAS in the presence of 20 mM GDP. T1 relaxation experiments were acquired with 5 s recycle delays and relaxation delays of 10, 50, 100, 200*, 500, 800, 1000, 1200*, and 1500 ms. T2 data were acquired with 5 s recycle delays and relaxation delays of 16, 32, 48, 64*, 96, 112, 144, 160*, 176, and 192 ms, where * indicates duplicate data set acquisition. Acquisition of relaxation delays was interleaved to avoid time-dependent sample degradation and/or field drift [56, 57]. {1H}-15N heteronuclear NOEs were determined as the ratio of spectral peak height recorded with 1H saturation (NOE, with 3 s recycle delay and 3 s proton saturation) to that recorded without 1H saturation (NONOE, using a 5 s recycle delay). After each NMR data set acquisition, protein stability was assessed by 2D 1H-15N HSQC spectrum.
NMR data processing and measurement of 15N backbone relaxation parameters
NMR data were processed and visualized by NMRPipe/NMRDraw [58] on an Intel PC workstation running RedHat Linux versions 7.1 or enterprise5. The directly and indirectly detected time domain data were processed by applying a 90° phase-shifted squared sinebell or a Gaussian filter with a line-broadening parameter of 10 Hz as weighting functions. Data sets were zero-filled once in each dimension prior to Fourier transformation. All chemical shifts were referenced to a DSS internal standard [59].
Relaxation times T1 and T2 were determined by fitting peak heights of respective experimental data sets to a single exponential decay using the nonlinear least-squares (NlinLS) routine in NMRPipe/NMRDraw distribution [58]. Errors in data fit were estimated from duplicate measurements of relaxation data points in both experiments.
15N backbone dynamics of unliganded and liganded STAS
Relaxation data were analyzed using the model-free formalism-based programs [19, 20] FastModelfree [60] interfaced with Modelfree v4.01 [27] and Tensor2 [61]. The overall rotational correlation time (τm) of the Rv1739c STAS domain polypeptide was estimated from a trimmed R2/R1 data set including values from residues obeying the filter criteria of heteronuclear NOE ≥ 0.65 and R1 and R2 values within one standard deviation (S. D.) from the average value (Ri < {Ri} − σRi), where i=1 and/or 2. Residues with R2 values greater than one S.D. (R2 ≥ {R2} + σR2) were considered only when their corresponding R1 values were lower by one S. D. (R1 ≤ {R1} −σR1) [39, 56, 62]. The coordinate system of the average Rv1739c STAS structure (PDB 2KLN) was moved to the molecule’s center of mass using PDBINERTIA [63], and principal components of moments of inertia were determined. The rotational diffusion tensor of the molecule was estimated for isotropic, axially symmetric, and fully anisotropic models by QUADRIC_DIFFUSION [63], and Tensor2 programs, using a filtered data set that excluded flexible residues according to described criteria [62]. Based on χ2 goodness-of-fit and F-test statistical parameters, an axially symmetric model was found appropriate to fit the relaxation data. This analysis yielded values for D||/D⊥, θ, and φ that, along with rNH = 1.02 Å, and a CSA tensor of −172 ppm, were used as input parameters in model-free analysis to obtain motional amplitudes and timescales of N-H bond vectors. The analysis yielded values for generalized order parameter (S2), internal correlation times of local internal motions (τe), and conformational exchange terms (Rex).
The relaxation input parameters of T1, T2, and {1H}-15N heteronuclear NOE for the Rv1739c STAS domain in the presence of GTP and GDP, for which crystal structures are not available, were analyzed using reduced spectral density mapping that does not require knowledge of the molecular rotational diffusion tensor [22, 23]. The reduced spectral density mapping assumes a single frequency term, J(0.87ωH) incorporating the three highest frequency terms J(ωH+ωN), J(ωH), and J(ωH-ωN) from the five spectral density terms [22, 35]. Reduced spectral density functions of J(0), J(ωN), and J(0.87ωH) were deduced for STAS, STAS+20 mM GTP, and STAS+20 mM GDP using the program “15N spectral density analysis” in Mathematica Notebook [64]. Estimation of parameters and the associated error in parameter measurement was from a series of 500 Monte Carlo simulations implemented to estimate reduced spectral density functions and the associated errors propagated from measured relaxation parameters T1, T2, and {1H}-15N heteronuclear NOE. To verify consistency between measured data and estimated spectral density functions, τm of Rv1739c STAS was also estimated in the absence and presence of guanine nucleotides from spectral density functions [28] by the following equation:
Figures were generated using GNUPLOT (linux version 3.5; http://www.gnuplot.info/) and PyMOL (Warren L. DeLano “The PyMOL Molecular Graphics System.” DeLano Scientific LLC, San Carlos, CA, USA. http://www.pymol.org).
Supplementary Material
Fig. S1. Sequence-specific rotational correlation times (τm) of Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Fig. S2. Residues with assigned models 1 to 5 obtained from deduction of model-free parameters from Rv1739c STAS 15N backbone relaxation data.
Fig. S3. Rv1739c STAS residues showing chemical shift perturbation (CSP) upon nucleotide binding, mapped onto the average STAS structure.
Fig. S4. Reduced spectral density correlation scatter plots of J(ωN) vs. J(0) and of J(0.87ωH) vs. J(0) for STAS, STAS +GTP, and STAS+GDP.
Table S1. Residue-specific 15N backbone relaxation data of R1 (sec−1), R2 (sec−1), and {1H}-15N heteronuclear NOE for Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Table S2. Inertia and diffusion tensors of Rv1739c STAS.
Table S3. Model-free 15N backbone relaxation parameters of Rv1739c STAS.
Table S4. Reduced spectral density function parameters of Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Table S5. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, generalized order parameters (S2), and reduced spectral density function parameters of each secondary-structured segment of Rv1739c STAS.
Table S6. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, and reduced spectral density function parameters of each secondary-structured segment of Rv1739c STAS in the presence of GTP.
Table S7. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, and reduced spectral density function parameters of each secondary structured segment of Rv1739c STAS in the presence of GDP.
Acknowledgments
This work was supported by National Institutes of Health grants R01DK43495 (to SLA) and P30DK34854 (to SLA from Harvard Digestive Diseases Center). NMR data were acquired at The Biological NMR Center, Tufts University, Boston. We thank Dr. Gillian Henry (Tufts University) for helpful discussions during NMR data acquisition.
Abbreviations
- STAS
sulfate transporter and anti-sigma factor antagonist
- HSQC
heteronuclear single quantum coherence
- R1
longitudinal relaxation rate
- R2
transverse relaxation rate
- τm
rotational correlation time
- S2
generalized order parameter
- τe
internal effective correlation times
- Rex
conformational exchange terms
- J(ω)
spectral density function at frequency ω
References
- 1.Shelden MC, Howitt SM, Price GD. Membrane topology of the cyanobacterial bicarbonate transporter, BicA, a member of the SulP (SLC26A) family. Mol Membr Biol. 2010;27:12–23. doi: 10.3109/09687680903400120. [DOI] [PubMed] [Google Scholar]
- 2.Dorwart MR, Shcheynikov N, Yang D, Muallem S. The solute carrier 26 family of proteins in epithelial ion transport. Physiology (Bethesda) 2008;23:104–114. doi: 10.1152/physiol.00037.2007. [DOI] [PubMed] [Google Scholar]
- 3.Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, Nimmo ER, Massey D, Blaszczyk K, Elliott T, Cotterill L, Dallal H, Lobo AJ, Mowat C, Sanderson JD, Jewell DP, Newman WG, Edwards C, Ahmad T, Mansfield JC, Satsangi J, Parkes M, Mathew CG, Donnelly P, Peltonen L, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, McCarthy MI, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Samani N, Trembath RC, Viswanathan AC, Wood N, Spencer CC, Barrett JC, Bellenguez C, Davison D, Freeman C, Strange A, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Perez ML, Potter SC, Ravindrarajah R, Ricketts M, Waller M, Weston P, Widaa S, Whittaker P, Deloukas P, Peltonen L, Mathew CG, Blackwell JM, Brown MA, Corvin A, McCarthy MI, Spencer CC, Attwood AP, Stephens J, Sambrook J, Ouwehand WH, McArdle WL, Ring SM, Strachan DP. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yusa A, Miyazaki K, Kimura N, Izawa M, Kannagi R. Epigenetic silencing of the sulfate transporter gene DTDST induces sialyl Lewisx expression and accelerates proliferation of colon cancer cells. Cancer Res. 2011;70:4064–4073. doi: 10.1158/0008-5472.CAN-09-2383. [DOI] [PubMed] [Google Scholar]
- 6.Ohana E, Yang D, Shcheynikov N, Muallem S. Diverse transport modes by the solute carrier 26 family of anion transporters. J Physiol. 2009;587:2179–2185. doi: 10.1113/jphysiol.2008.164863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felce J, Saier MH., Jr Carbonic anhydrases fused to anion transporters of the SulP family: evidence for a novel type of bicarbonate transporter. J Mol Microbiol Biotechnol. 2004;8:169–176. doi: 10.1159/000085789. [DOI] [PubMed] [Google Scholar]
- 8.Shibagaki N, Grossman AR. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem. 2006;281:22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- 9.Babu M, Greenblatt JF, Emili A, Strynadka NC, Reithmeier RA, Moraes TF. Structure of a SLC26 anion transporter STAS domain in complex with acyl carrier protein: implications for E. coli YchM in fatty acid metabolism. Structure. 2010;18:1450–1462. doi: 10.1016/j.str.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci U S A. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price GD, Howitt SM. The cyanobacterial bicarbonate transporter BicA: its physiological role and the implications of structural similarities with human SLC26 transporters. Biochem Cell Biol. 2011;89:178–188. doi: 10.1139/o10-136. [DOI] [PubMed] [Google Scholar]
- 12.Oliver D, He DZ, Klocker N, Ludwig J, Schulte U, Waldegger S, Ruppersberg JP, Dallos P, Fakler B. Intracellular anions as the voltage sensor of prestin, the outer hair cell motor protein. Science. 2001;292:2340–2343. doi: 10.1126/science.1060939. [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Long KB, Shen W, Madison LD, Dallos P. Prestin topology: localization of protein epitopes in relation to the plasma membrane. Neuroreport. 2001;12:1929–1935. doi: 10.1097/00001756-200107030-00032. [DOI] [PubMed] [Google Scholar]
- 14.Pasqualetto E, Aiello R, Gesiot L, Bonetto G, Bellanda M, Battistutta R. Structure of the cytosolic portion of the motor protein prestin and functional role of the STAS domain in SLC26/SulP anion transporters. J Mol Biol. 2010;400:448–462. doi: 10.1016/j.jmb.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Sharma AK, Ye L, Baer CE, Shanmugasundaram K, Alber T, Alper SL, Rigby AC. Solution Structure of the Guanine Nucleotide-binding STAS Domain of SLC26-related SulP Protein Rv1739c from Mycobacterium tuberculosis. J Biol Chem. 2011;286:8534–8544. doi: 10.1074/jbc.M110.165449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma AK, Ye L, Zolotarev AS, Alper SL, Rigby AC. NMR assignment and secondary structure of the STAS domain of Rv1739c, a putative sulfate transporter of Mycobacterium tuberculosis. Biomol NMR Assign. 2009;3:99–102. doi: 10.1007/s12104-009-9150-z. [DOI] [PubMed] [Google Scholar]
- 17.Zolotarev AS, Unnikrishnan M, Shmukler BE, Clark JS, Vandorpe DH, Grigorieff N, Rubin EJ, Alper SL. Increased sulfate uptake by E. coli overexpressing the SLC26-related SulP protein Rv1739c from Mycobacterium tuberculosis. Comp Biochem Physiol A Mol Integr Physiol. 2008;149:255–266. doi: 10.1016/j.cbpa.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najafi SM, Harris DA, Yudkin MD. The SpoIIAA protein of Bacillus subtilis has GTP-binding properties. J Bacteriol. 1996;178:6632–6634. doi: 10.1128/jb.178.22.6632-6634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982a;104:4546–4559. [Google Scholar]
- 20.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982b;104:4559–4570. [Google Scholar]
- 21.Clore GMSA, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple 2-parameter model-free approach to the interpretation of N-15 nuclear magnetic-relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 22.Peng JW, Wagner G. Mapping of the spectral densities of N-H bond motions in eglin c using heteronuclear relaxation experiments. Biochemistry. 1992;31:8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- 23.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. Spectral density function mapping using 15N relaxation data exclusively. J Biomol NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 24.Maciejewski MW, Liu D, Prasad R, Wilson SH, Mullen GP. Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity. J Mol Biol. 2000;296:229–253. doi: 10.1006/jmbi.1999.3455. [DOI] [PubMed] [Google Scholar]
- 25.Reddy T, Rainey JK. Interpretation of biomolecular NMR spin relaxation parameters. Biochem Cell Biol. 2010;88:131–142. doi: 10.1139/o09-152. [DOI] [PubMed] [Google Scholar]
- 26.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 27.Mandel AM, Akke M, Palmer AG., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 28.Bracken C, Carr PA, Cavanagh J, Palmer AG., 3rd Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 29.Viles JH, Donne D, Kroon G, Prusiner SB, Cohen FE, Dyson HJ, Wright PE. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry. 2001;40:2743–2753. doi: 10.1021/bi002898a. [DOI] [PubMed] [Google Scholar]
- 30.Dyson HJ, Wright PE. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 2001;339:258–270. doi: 10.1016/s0076-6879(01)39317-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P, Dayie KT, Wagner G. Unusual lack of internal mobility and fast overall tumbling in oxidized flavodoxin from Anacystis nidulans. J Mol Biol. 1997;272:443–455. doi: 10.1006/jmbi.1997.1266. [DOI] [PubMed] [Google Scholar]
- 32.Kovacs H, Comfort D, Lord M, Yudkin M, Campbell ID, Nilges M. NMR studies of the sporulation protein SpoIIAA: implications for the regulation of the transcription factor sigmaF in Bacillus subtilis. J Biomol NMR. 2001;19:293–304. doi: 10.1023/a:1011247523112. [DOI] [PubMed] [Google Scholar]
- 33.Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–293. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- 34.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 35.Lefevre JF, Dayie KT, Peng JW, Wagner G. Internal mobility in the partially folded DNA binding and dimerization domains of GAL4: NMR analysis of the N-H spectral density functions. Biochemistry. 1996;35:2674–2686. doi: 10.1021/bi9526802. [DOI] [PubMed] [Google Scholar]
- 36.Kuang Z, Yao S, Keizer DW, Wang CC, Bach LA, Forbes BE, Wallace JC, Norton RS. Structure, dynamics and heparin binding of the C-terminal domain of insulin-like growth factor-binding protein-2 (IGFBP-2) J Mol Biol. 2006;364:690–704. doi: 10.1016/j.jmb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Masuda S, Murakami KS, Wang S, Anders Olson C, Donigian J, Leon F, Darst SA, Campbell EA. Crystal structures of the ADP and ATP bound forms of the Bacillus anti-sigma factor SpoIIAB in complex with the anti-anti-sigma SpoIIAA. J Mol Biol. 2004;340:941–956. doi: 10.1016/j.jmb.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 38.Loh AP, Pawley N, Nicholson LK, Oswald RE. An increase in side chain entropy facilitates effector binding: NMR characterization of the side chain methyl group dynamics in Cdc42Hs. Biochemistry. 2001;40:4590–4600. doi: 10.1021/bi002418f. [DOI] [PubMed] [Google Scholar]
- 39.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 40.Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol. 21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 42.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 43.Begg GE, Carrington L, Stokes PH, Matthews JM, Wouters MA, Husain A, Lorand L, Iismaa SE, Graham RM. Mechanism of allosteric regulation of transglutaminase 2 by GTP. Proc Natl Acad Sci U S A. 2006;103:19683–19688. doi: 10.1073/pnas.0609283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Detro-Dassen S, Schanzler M, Lauks H, Martin I, zu Berstenhorst SM, Nothmann D, Torres-Salazar D, Hidalgo P, Schmalzing G, Fahlke C. Conserved dimeric subunit stoichiometry of SLC26 multifunctional anion exchangers. J Biol Chem. 2008;283:4177–4188. doi: 10.1074/jbc.M704924200. [DOI] [PubMed] [Google Scholar]
- 45.Zheng J, Du GG, Anderson CT, Keller JP, Orem A, Dallos P, Cheatham M. Analysis of the oligomeric structure of the motor protein prestin. J Biol Chem. 2006;281:19916–19924. doi: 10.1074/jbc.M513854200. [DOI] [PubMed] [Google Scholar]
- 46.Bai JP, Surguchev A, Bian S, Song L, Santos-Sacchi J, Navaratnam D. Combinatorial cysteine mutagenesis reveals a critical intramonomer role for cysteines in prestin voltage sensing. Biophys J. 2010;99:85–94. doi: 10.1016/j.bpj.2010.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorwart MR, Shcheynikov N, Baker JM, Forman-Kay JD, Muallem S, Thomas PJ. Congenital chloride-losing diarrhea causing mutations in the STAS domain result in misfolding and mistrafficking of SLC26A3. J Biol Chem. 2008;283:8711–8722. doi: 10.1074/jbc.M704328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 49.Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J Biomol NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 50.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 51.Massi F, Johnson E, Wang C, Rance M, Palmer AG., 3rd NMR R1 rho rotating-frame relaxation with weak radio frequency fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 52.Palmer AG. Chemical Exchange Effects in Biological Macromolecules. Encyclopedia of Magnetic Resonance 2007 [Google Scholar]
- 53.Li Y, Palmer AG., 3rd TROSY-selected ZZ-exchange experiment for characterizing slow chemical exchange in large proteins. J Biomol NMR. 2009;45:357–360. doi: 10.1007/s10858-009-9385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 55.Marion D, Ikura M, Tschudin R, Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. Journal of Magnetic Resonance. 1989;85:393–399. [Google Scholar]
- 56.Savard PY, Gagne SM. Backbone dynamics of TEM-1 determined by NMR: evidence for a highly ordered protein. Biochemistry. 2006;45:11414–11424. doi: 10.1021/bi060414q. [DOI] [PubMed] [Google Scholar]
- 57.Tjandra N, Wingfield P, Stahl S, Bax A. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J Biomol NMR. 1996;8:273–284. doi: 10.1007/BF00410326. [DOI] [PubMed] [Google Scholar]
- 58.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 59.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J Mol Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 60.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 61.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 62.Pawley NH, Wang C, Koide S, Nicholson LK. An improved method for distinguishing between anisotropic tumbling and chemical exchange in analysis of 15N relaxation parameters. J Biomol NMR. 2001;20:149–165. doi: 10.1023/a:1011249816560. [DOI] [PubMed] [Google Scholar]
- 63.Palmer AG., III http://biochemistry.hs.columbia.edu/labs/palmer/software/diffusion.html.
- 64.Spyracopoulos L. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J Biomol NMR. 2006;36:215–224. doi: 10.1007/s10858-006-9083-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence-specific rotational correlation times (τm) of Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Fig. S2. Residues with assigned models 1 to 5 obtained from deduction of model-free parameters from Rv1739c STAS 15N backbone relaxation data.
Fig. S3. Rv1739c STAS residues showing chemical shift perturbation (CSP) upon nucleotide binding, mapped onto the average STAS structure.
Fig. S4. Reduced spectral density correlation scatter plots of J(ωN) vs. J(0) and of J(0.87ωH) vs. J(0) for STAS, STAS +GTP, and STAS+GDP.
Table S1. Residue-specific 15N backbone relaxation data of R1 (sec−1), R2 (sec−1), and {1H}-15N heteronuclear NOE for Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Table S2. Inertia and diffusion tensors of Rv1739c STAS.
Table S3. Model-free 15N backbone relaxation parameters of Rv1739c STAS.
Table S4. Reduced spectral density function parameters of Rv1739c STAS in free, GTP-bound, and GDP-bound states.
Table S5. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, generalized order parameters (S2), and reduced spectral density function parameters of each secondary-structured segment of Rv1739c STAS.
Table S6. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, and reduced spectral density function parameters of each secondary-structured segment of Rv1739c STAS in the presence of GTP.
Table S7. 15N relaxation rates R1 (sec−1), R2 (sec−1), {1H}-15N heteronuclear NOEs, and reduced spectral density function parameters of each secondary structured segment of Rv1739c STAS in the presence of GDP.