Abstract
Pneumocystis pneumonia (PCP) is a life-threatening condition in immunosuppressed patients. Current treatments are inadequate, and new drug leads are needed. This fungus depends on its host for S-adenosylmethionine (AdoMet), a critical metabolic intermediate ordinarily synthesized by individual cells as needed. Pneumocystis contains a gene coding for the AdoMet-synthesizing enzyme methionine ATP transferase (MAT), and the protein is expressed. However, the fungus lacks MAT activity, and infection causes the depletion of host plasma AdoMet. The uptake of Pneumocystis AdoMet was shown to be exquisitely specific, which suggests the transport of AdoMet as a potential drug target. Here we report on the discovery of PcPET8, a Pneumocystis gene with homology to mitochondrial AdoMet transporters. When expressed by Saccharomyces cerevisiae, it locates properly to the mitochondrion and complements a strain of S. cerevisiae lacking its native mitochondrial AdoMet transporter. The importance of AdoMet transport is demonstrated by the ability of the AdoMet analogue sinefungin to block the uptake of Pneumocystis AdoMet and inhibit growth in culture. Because PcPET8 is likely critical for Pneumocystis, the yeast construct has potential as a surrogate for testing compounds against Pneumocystis.
Keywords: Pneumocystis, S-adenosylmethionine, mitochondrial transporter, PET8, PcPET8, PCP pneumonia
Clinical Relevance
The Pneumocystis mitochondrial S-adenosylmethionine (AdoMet) transporter may be of assistance in discovering new drugs by serving as a surrogate for testing the ability of compounds to block the acquisition and use of AdoMet by Pneumocystis.
The genus Pneumocystis contains genetically distinct but morphologically similar species, each limited to a single mammalian host species (1, 2). A species of Pneumocystis has been found in every mammalian species where it has been sought. Although the prevalence of colonization is high in humans and other mammals, Pneumocystis pneumonia (PCP) occurs only when the host is immunosuppressed (3). PCP in humans is associated with advanced HIV disease, severe malnourishment in children, and treatments for cancer, rheumatic disease, and the prevention of organ transplant rejection (3). It is frequently fatal if untreated (4). The antifolate combination of trimethoprim with sulfamethoxazole has been the mainstay of treatment, but is unsatisfactory because of significant adverse drug effects (5), especially for patients with advanced HIV, and treatment failures exceed 20%. Genetic data suggest increasing drug resistance (6). Alternative drugs exist, but all have drawbacks that include lower efficacy or greater toxicity. The mortality rate for severe infections has not changed appreciably for 30 years (1, 7). New drugs and new drug leads are urgently needed.
Among the unusual properties of Pneumocystis is the dependence on its host for S-adenosylmethionine (AdoMet), a key metabolic intermediate with a wide range of functions, including protein and nucleic acid methylation, phospholipid synthesis, polyamine synthesis, folate metabolism, and the production of glutathione, and it also exists as a bound prosthetic group for a class of enzymes (8). The condensation of methionine and ATP is catalyzed by methionine ATP transferase (MAT), yielding AdoMet, phosphate, and pyrophosphate. With the exceptions of Pneumocystis, some species of Rickettsia, and to some extent Leishmania, all cells examined were found capable of producing the AdoMet they require (9). Whereas AdoMet-requiring Rickettsia lacks a gene for MAT, Pneumocystis and Leishmania contain genes that code for functional MAT enzymes, and both are expressed (10, 11). Cultured Leishmania expresses MAT activity when in the log phase of growth, but activity ceases during the stationary phase, despite the continued expression of MAT, and AdoMet is scavenged from the culture medium. Leishmania and Pneumocystis both possess high-affinity, highly selective AdoMet transporters. Although Pneumocystis may express MAT activity at some stage of its life cycle, this activity has been undetectable in Pneumocystis isolated from infected animals, even when sensitive assays and procedures to eliminate possible interference are used (9). The dependence of Pneumocystis on host AdoMet is supported by evidence from experimental animals: PCP reduces pulmonary AdoMet and depletes plasma AdoMet, the selective reduction of pulmonary AdoMet attenuates the infection, and an infusion of AdoMet exacerbates the disease (9, 12, 13). For humans with PCP, plasma AdoMet is depleted and recovers rapidly upon the initiation of treatment (12, 14, 15).
This requirement of exogenous AdoMet suggests transport as a drug target, a suggestion supported by our previous demonstration that rat-specific P. carinii possesses a high-affinity, high-specificity AdoMet transporter, and close AdoMet analogues such as S-adenosylhomocysteine and S-adenosylethionine exert no effect on the uptake of P. carinii AdoMet, despite inhibiting uptake by Saccharomyces cerevisiae (9, 16). To exploit this finding, we searched for potential Pneumocystis AdoMet transporter genes with the expectation of improving our understanding of Pneumocystis AdoMet metabolism and producing a surrogate test system for drug development. Using the limited Pneumocystis genomic data available, we identified a sequence predicting a protein with similarity to the S. cerevisiae miochondrial AdoMet transporter (PET8). Here we describe the identification of the P. carinii PET8 gene (PcPET8), demonstrate transcription by P. carinii, show the functionality of the PcPET8 gene product by the complementation of an S. cerevisiae PET8 knockout strain, and demonstrate that sinefungin, a PET8 inhibitor, blocks the uptake of Pneumocystis AdoMet in a dose-dependent fashion.
Materials and Methods
cDNA Synthesis
P. carinii was isolated from lungs that had been removed from infected rats and stored frozen, as previously described (12, 17). RNA was isolated from P. carinii cells, using the TRIZOL reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), and was then treated with RQ1 RNase-free DNase (Promega, San Luis Obispo, CA) to destroy any trace of genomic DNA. cDNA was synthesized using 1.0 μg of extracted RNA and the Superscript III One-Step RT-PCR system (Invitrogen) in a 50-μl reaction mix with the forward primer ATG GAT TTG AAA CTA ATT TAT G and the reverse primer AAT GCA CGT ATA CCT TCT TC. cDNA synthesis proceeded with the reverse transcription reaction at 56°C for 30 minutes, followed by 40 PCR amplification cycles (15 seconds at 94°C, 30 seconds at 56°C, and 60 seconds at 68°C), with a final extension for 5 minutes at 68°C. As a negative control to ensure a lack of contamination with genomic DNA, pseudo-cDNA synthesis was performed using Platinum Taq (Invitrogen) without reverse transcriptase and 1 μg of DNase-treated RNA.
Expression of Green Fluorescent Protein–Tagged PcPET8 in S. cerevisiae
To produce PcPET8 tagged at the C-terminus with green fluorescent protein (GFP) for cellular localization studies, we used the shuttle vector PKT128 (18), designed to produce such fusion proteins in yeast. Detailed methods are described in the online supplement.
S. cerevisiae PET8 Knockout (PET8∆) Complementation by PcPET8
The S. cerevisiae PET8∆ strain was obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). PcPET8 was cloned into pGREG506, to allow for expression in S. cerevisiae without extra amino acids. Detailed methods are described in the online supplement.
Measurement of AdoMet Transport
AdoMet transport was measured as previously described (9) and summarized here. P. carinii cells cultured for 4–8 days after isolation from rat lungs were washed three times with Eagle's minimum essential medium and suspended in PBS. Assays included 45 μl of P. carinii cell suspension and 5 μl of PBS buffer containing S-[methyl-14C]AdoMet (52.7 mCi/mmol; Moravek Biochemicals, Inc., Brea, CA) and sinefungin, to yield 33 μM of AdoMet and the indicated sinefungin concentrations. After mixing, assays were incubated for 30 seconds at 37°C, and then cells were washed by microfuging through 100 μl of dibutyl phthalate and mineral oil (1:1), to remove unincorporated AdoMet. The supernatants were discarded, and the pellets were counted by liquid scintillation.
Results
Identification of PcPET8
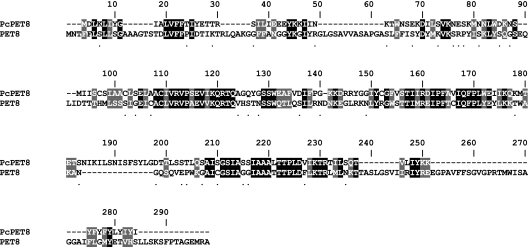
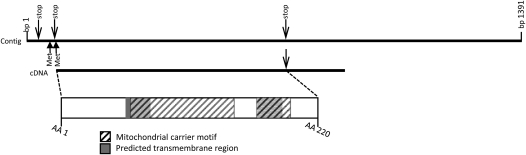
A basic local alignment search tool (BLAST) query of the Pneumocystis Genome Project (http://pgp.cchmc.org) based on PET8 identified a 1,391-base pair (bp) sequence in the contig database that contained an open reading frame (ORF) of 663 bp, coding for a 220 amino acid polypeptide of 24.99-kD molecular mass and a predicted isoelectric point of 8.9. Alignment of the theoretical protein with the amino acid sequence of S. cerevisiae PET8 (19) showed a 24% identity and 39% homology (Figure 1). Signal prediction did not reveal a signal peptide, but did identify two mitochondrial carrier domains between amino acids 61–150 and 170–198 (MotifScan; http://myhits.isb-sib.ch/cgi-bin/motif_scan), as well as two transmembrane regions between amino acids 59–79 and 172–192 (TopPred; available at http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) (20) (Figure 2). To confirm the sequence, we cloned the gene de novo. PCR amplification from P. carinii DNA, using primers based on the already mentioned 663-bp ORF, produced a single product of approximately 663 bp, which we cloned into the vector pET161/GW/D-TOPO. Sequencing demonstrated the new clone sequence to be identical to that of the Pneumocystis Genome Project contig sequence. Following the nomenclature guidelines for P. carinii (21), we named the sequence PcPet8 (GenBank accession number FJ769260).
Figure 1.
Alignment of the Pneumocystis carinii PET8 gene (PcPET8) and the S. cerevisiae miochondrial AdoMet transporter (PET8). The predicted PcPET8 protein sequence is aligned with PET8 of Saccharomyces cerevisiae (S. cerevisiae accession number NP_014395.1). Dark shading, identical amino acid; lighter shading, strongly similar amino acid; *weakly similar amino acid.
Figure 2.
Structural characteristics of PcPET8. Scale diagram shows structural features of PcPET8, including positions of the putative mitochondrial carrier motif and transmembrane regions, as indicated.
Transcription of PcPET8 by P. carinii
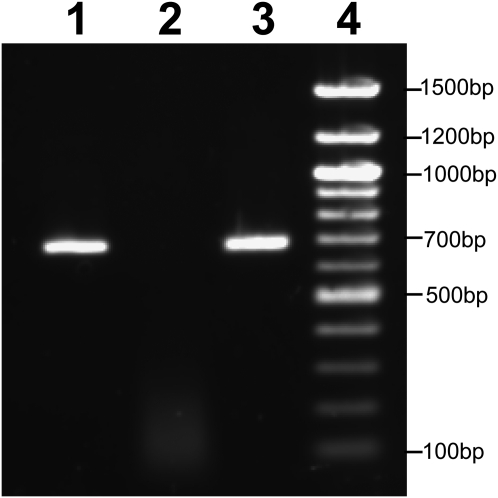
To demonstrate the presence of PcPET8-coded RNA in P. carinii, we cloned and sequenced the product of a one-step RT-PCR reaction, using primers based on PcPET8 and 1 μg of DNase-treated RNA extracted from P. carinii isolated from rat lungs (Figure 3). To ensure that the RT-PCR product did not result from genomic DNA contaminating the RNA preparation, a control reaction omitted the reverse transcriptase. The sequence of the RT-PCR product did not deviate from the genomic sequence, a result showing that the gene is transcribed and that it contains no introns.
Figure 3.
Transcription of PcPET8 by P. carinii. A “one-step” RT-PCR reaction was performed using total RNA isolated from P. carinii and primers, based on the PcPET8 sequence. Lane 1, DNase-treated total RNA template. The product demonstrates the presence of PcPET8 transcript in RNA. Lane 2, DNase-treated total RNA template, with omission of reverse transcriptase. The lack of product demonstrates a lack of genomic DNA contamination in the DNase-treated RNA template. Lane 3, Genomic DNA template, with omission of reverse transcriptase. The correct size of the product shows primer specificity, and confirms the lack of introns.
Expression of PcPET8 in S. cerevisiae and Intracellular Localization
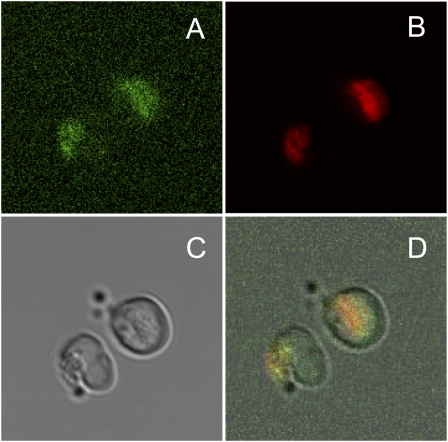
PcPET8 was inserted into the yeast expression vector PKT128 (18) to produce a fusion protein with GFP, linked to the C-terminus of PcPET8. To allow for the simultaneous detection of yeast mitochondria and this fusion protein, transformed yeast cells were grown in liquid medium with MitoTracker Red 580 (Invitrogen), a fluorescent stain specific for mitochondria. Confocal images (Figure 4) show the colocalization of GFP and MitoTracker Red 580, indicating a mitochondrial localization for the recombinant protein.
Figure 4.
Cellular localization of PcPET8 expressed by S. cerevisiae. A wild-type S. cerevisiae cell expresses PcPET8 tagged with green fluorescent protein (GFP) at the C-terminus, and grown in the presence of a mitochondrial marker. (A) PcPET8–GFP. (B) MitoTracker Red 580. (C) Phase contrast. (D) Merged images show the colocalization of GFP and MitoTracker Red 580.
PcPET8 Complementation of an S. cerevisiae PET8 Knockout
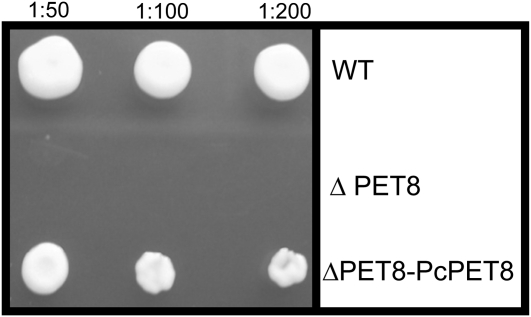
We tested the function of PcPET8 according to its expression in a PET8 knockout strain of S. cerevisiae (PET8∆). Because the conversion of desthiobiotin to biotin occurs in mitochondria, which requires that AdoMet be transported from the cytosol, PET8∆ is a biotin auxotroph (22). Therefore, if recombinant PcPET8 can function as a mitochondrial AdoMet transporter in S. cerevisiae, the expression of PcPET8 by PET8∆ should relieve the requirement for biotin. PET8∆ was transformed using pGREG506 with a PcPET8 insert (PET8∆–PcPET8). For positive and negative controls, respectively, the PET8-competent strain BY4741 and PET8∆ were transformed with an empty vector. After selection with uracil dropout medium, all three transformed types grew well when supplemented with biotin, but only BY4741 and PET8∆–PcPET8 grew on plates containing desthiobiotin but lacking biotin (Figure 5). This successful complementation demonstrates that the product of PcPet8 functions as a mitochondrial AdoMet transporter in S. cerevisiae.
Figure 5.
Functional assay of PcPET8. Expression of PcPET8 allows the S. cerevisiae PET8 knockout strain (PET8∆) to grow in a medium lacking biotin but containing the biotin precursor desthiobiotin, a molecule that needs S-adenosylmethionine in the mitochondria to be transformed in biotin. Cells were plated in three serial dilutions. WT, wild type.
Sinefungin Inhibition of Pneumocystis AdoMet Transport
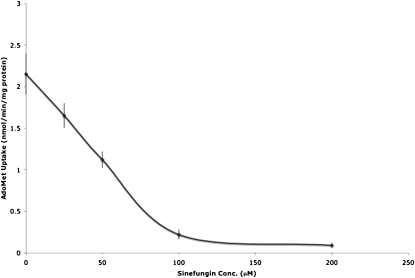
We measured the ability of the PET8 inhibitor sinefungin to block the uptake of AdoMet by isolated P. carinii. The data presented in Figure 6 show that 50 μM of sinefungin inhibited uptake by approximately 50%, and 100 μM inhibited uptake by approximately 90%.
Figure 6.
Dose-dependent inhibition of S-adenosylmethionine (AdoMet) transport by sinefungin. Uptake measurement used [14CH3]AdoMet at 33 μM. See Materials and Methods for details. Conc., concentration.
Discussion
We identified and confirmed by independent cloning the sequence of a contig in the Pneumocystis Genome Project database with similarity to the S. cerevisiae mitochondrial AdoMet transporter gene PET8. We named this gene PcPET8, in accordance with the suggested guidelines for nomenclature. The production of PcPET8 cDNA shows that the gene is transcribed, and the identification of cDNA and genomic sequences indicates a lack of introns. When expressed by S. cerevisiae, the gene product localizes to the mitochondrion, and complements a strain of S. cerevisiae lacking PET8.
PET8∆–PcPET8 does not grow quite as well as the wild type (Figure 5). Perhaps less PcPET8 protein is produced by the transfectant than the amount of PET8 produced by the wild type, or else S. cerevisiae and P. carinii are sufficiently different that the exotic mitochondrial AdoMet transporter functions less well than the native protein. Regardless, the ability to complement PET8∆ and allow growth without biotin clearly demonstrates that PcPET8 functions as a mitochondrial AdoMet transporter in S. cerevisiae. Despite the short 5′ untranslated region and an ORF coding for 220 amino acids versus the 298 of PET8, several points indicate that the coding sequence of PcPET8 is complete. Alignment of the predicted protein with PET8 (Figure 1) shows gaps interspersed throughout, yet PcPET8 codes for a functional protein. The N-terminus of PcPET8 is shorter by only three amino acids, suggesting the completeness of the 5′ end, an interpretation reinforced by the presence of two in-frame stop codons 5′ to the putative initiating met codon, with no intervening met codons. Although we did not find a canonical eukaryote ribosome–binding (Kozak) sequence surrounding the initiating met codon, the invariant characteristic of a G following that codon is present, despite the absence of the characteristic A/G, located three bases 5′ to the met codon. The lack of a recognizable pattern is of limited significance because Kozak sequences are variable, have been little studied in yeast, and have not been studied at all in Pneumocystis. The completeness of the 3′ end is supported by the sequence identity of cDNA, including the stop codon 660 bp after the initiating met codon.
Although PcPET8 is a mitochondrial AdoMet transporter, it could theoretically contribute to the cellular uptake of AdoMet. Our data show that PcPET8 localizes to S. cerevisiae mitochondria and substitutes for the native transporter, but we have no data for its localization in P. carinii, and therefore we cannot rule out the possibility that PcPET8 locates to the cell membrane, where it mediates the uptake of AdoMet. Another possibility is that the Pneumocystis cell membrane lacks an active AdoMet transporter, but has an AdoMet permease. If that were the case, then mitochondrial uptake could create an AdoMet concentration gradient across the cell membrane, thereby promoting cellular uptake. Regardless of whether PcPET8 is responsible for cellular uptake, mitochondrial uptake would be a viable drug target if it can be selectively blocked in Pneumocystis.
The antibiotic sinefungin is active against P. carinii in culture (12), is known to compete with AdoMet for S. cerevisiae PET8 transport (23), and is the only compound we found able to block the uptake of AdoMet by Pneumocystis. Kinetic data for the inhibition of AdoMet transport by sinefungin are lacking, but because the observed inhibition occurred after 30 seconds and sinefungin is known to compete with the PET8-mediated transport of AdoMet (22), we are confident that sinefungin directly inhibits the uptake of Pneumocystis AdoMet by blocking transport rather than indirectly, though interference with AdoMet-dependent reactions. Our data showing that 100 μM of sinefungin block the uptake of AdoMet by 90% confirm our earlier result using that concentration (12). More importantly, this concentration also completely blocks growth in culture, a finding that supports AdoMet transport as a drug target. The fact that P. carinii AdoMet uptake is more sensitive to sinefungin than S. cerevisiae suggests that P. carinii AdoMet transport may be a sensitive drug target. Although adverse reactions make sinefungin an unlikely drug candidate, the ability of this compound to inhibit the growth of S. cerevisiae dependent on PcPET8 suggests that this construct may prove useful as a tool to identify other compounds that block Pneumocystis AdoMet transport, and may prove helpful in identifying viable drug candidates.
Furthermore, the ability of sinefungin to inhibit the growth of S. cerevisiae dependent on PcPET8 suggests that this construct may prove useful as surrogate when screening compounds for anti-Pneumocystis activity.
In conclusion, the newly discovered PcPET8 gene shows homology with the S. cerevisiae mitochondrial AdoMet transporter, is transcribed by Pneumocystis, and lacks introns. When expressed by S. cerevisiae, it locates to the mitochondrion and is able to complement a strain of S. cerevisiae lacking its native mitochondrial AdoMet transporter. Although this gene may or may not be involved in the uptake of AdoMet by Pneumocystis, it likely serves a critical function. Furthermore, sinefungin, which is known to inhibit S. cerevisiae PET8, also inhibited the uptake of Pneumocystis AdoMet in dose-dependent manner. PcPET8-dependent S. cerevisiae has potential as a surrogate when testing compounds for the ability of to block the acquisition or use of AdoMet by Pneumocystis. Compounds able to block PcPET8-dependent AdoMet transport may have further utility because BLAST searches of genomic databases of other human pathogens such as Plasmodium spp., Leishmania major, and Trypanosoma brucei suggest the presence of similar transporters in those parasites.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health grant RO1AI064017 (S.M.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0009OC on June 3, 2011
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Laakkonen J. Pneumocystis carinii in wildlife. Int J Parasitol 1998;28:241–252 [DOI] [PubMed] [Google Scholar]
- 2.Chabe M, Aliouat-Denis CM, Delhaes L, el Aliouat M, Viscogliosi E, Dei-Cas E. Pneumocystis: from a doubtful unique entity to a group of highly diversified fungal species. FEM Yeast Res 2011;11:2–17 [DOI] [PubMed] [Google Scholar]
- 3.Sepkowitz KA. Opportunistic infections in patients with and patients without acquired immunodeficiency syndrome. Clin Infect Dis 2002;34:1098–1107 [DOI] [PubMed] [Google Scholar]
- 4.McLennan G, Antic R, Seymour AE, Frith PA, Clarkson AR. Pneumocystis carinii pneumonitis successfully treated with trimethoprim–sulphamethoxazole. Aust N Z J Med 1977;7:299–301 [DOI] [PubMed] [Google Scholar]
- 5.Wilkin A, Feinberg J. Pneumocystis carinii pneumonia: a clinical review. Am Fam Phys 1999;60:1699–1708, 1713–1694 [PubMed] [Google Scholar]
- 6.Walker DJ, Meshnick SR. Drug resistance in Pneumocystis carinii: an emerging problem. Drug Resist Updat 1998;1:201–204 [DOI] [PubMed] [Google Scholar]
- 7.Walzer PD, Evans HE, Copas AJ, Edwards SG, Grant AD, Miller RF. Early predictors of mortality from Pneumocystis jirovecii pneumonia in HIV-infected patients: 1985–2006. Clin Infect Dis 2008;46:625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JA, Wingard JR. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011;52:e56–e93 [DOI] [PubMed] [Google Scholar]
- 9.Merali S, Vargas D, Franklin M, Clarkson AB., Jr S-adenosylmethionine and Pneumocystis carinii. J Biol Chem 2000;275:14958–14963 [DOI] [PubMed] [Google Scholar]
- 10.Kutty G, Hernandez-Novoa B, Czapiga M, Kovacs JA. Pneumocystis encodes a functional S-adenosylmethionine synthetase gene. Eukaryot Cell 2008;7:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laakkonen J, Sundell J, Soveri T. Lung parasites of least weasels in Finland. J Wildl Dis 1998;34:816–819 [DOI] [PubMed] [Google Scholar]
- 12.Merali S, Clarkson AB., Jr S-adenosylmethionine and Pneumocystis. FEMS Microbiol Lett 2004;237:179–186 [DOI] [PubMed] [Google Scholar]
- 13.Moncada CA, Clarkson A, Perez-Leal O, Merali S. Mechanism and tissue specificity of nicotine-mediated lung S-adenosylmethionine reduction. J Biol Chem 2008;283:7690–7696 [DOI] [PubMed] [Google Scholar]
- 14.Skelly M, Hoffman J, Fabbri M, Holzman RS, Clarkson AB, Merali S. S-adenosylmethionine concentrations in diagnosis of Pneumocystis carinii pneumonia. Lancet 2003;361:1267–1268 [DOI] [PubMed] [Google Scholar]
- 15.Skelly MJ, Holzman RS, Merali S. S-adenosylmethionine levels in the diagnosis of Pneumocystis carinii pneumonia in patients with HIV infection. Clin Infect Dis 2008;46:467–471 [DOI] [PubMed] [Google Scholar]
- 16.Murphy JT, Spence KD. Transport of S-adenosylmethionine in Saccharomyces cerevisiae. J Bacteriol 1972;109:499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merali S. Pneumocystis carinii polyamine catabolism. J Biol Chem 1999;274:21017–21022 [DOI] [PubMed] [Google Scholar]
- 18.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 2004;21:661–670 [DOI] [PubMed] [Google Scholar]
- 19.Sohn S, Eagan J, Sepkowitz KA. Safety-engineered device implementation: does it introduce bias in percutaneous injury reporting? Infect Control Hosp Epidemiol 2004;25:543–547 [DOI] [PubMed] [Google Scholar]
- 20.Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 1994;10:685–686 [DOI] [PubMed] [Google Scholar]
- 21.Stringer JR, Wakefield AE, Cushion MT, Dei-Cas E. Pneumocystis taxonomy and nomenclature: an update. J Eukaryot Microbiol 1997;44:5S–6S [DOI] [PubMed] [Google Scholar]
- 22.Walshe LJ, Malak SF, Eagan J, Sepkowitz KA. Complication rates among cancer patients with peripherally inserted central catheters. J Clin Oncol 2002;20:3276–3281 [DOI] [PubMed] [Google Scholar]
- 23.Zheng S, Shuman S, Schwer B. Sinefungin resistance of Saccharomyces cerevisiae arising from SAM3 mutations that inactivate the AdoMet transporter or from increased expression of AdoMet synthase plus mRNA CAP guanine-N7 methyltransferase. Nucleic Acids Res 2007;35:6895–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.