Abstract
The Cadm (cell adhesion molecule) family of cell adhesion molecules (also known as IGSF4, SynCAM, Necl and TSLC) has been implicated in a multitude of physiological and pathological processes, such as spermatogenesis, synapse formation and lung cancer. The precise mechanisms by which these adhesion molecules mediate these diverse functions remain unknown. To investigate mechanisms of action of these molecules during development, we have identified zebrafish orthologs of Cadm family members and have examined their expression patterns during development and in the adult. Zebrafish possess six cadm genes. Sequence comparisons and phylogenetic analysis suggest that four of the zebrafish cadm genes represent duplicates of two tetrapod Cadm genes, whereas the other two cadm genes are single orthologs of tetrapod Cadm genes. All six zebrafish cadms are expressed throughout the nervous system both during development and in the adult. The spatial and temporal patterns of expression suggest multiple roles for Cadms during nervous system development.
Keywords: Cadm, SynCAM, IGSF4, Necl, zebrafish, central nervous system
INTRODUCTION
The Cadms (cell adhesion molecules) are a family of type I transmembrane proteins that have been described in several pathological and physiological processes such as the progression of lung and other cancers (reviewed in Murakami, 2005), mast cell adhesion (Ito et al., 2003, 2007b; Furuno et al., 2005; Ito and Oonuma, 2006), spermatogenesis (Wakayama et al., 2001; Wakayama et al., 2003; Fujita et al., 2006; van der Weyden et al., 2006; Yamada et al., 2006), epithelium development and homeostasis (Ito et al., 2007b), and central nervous system development (Biederer et al., 2002; Sara et al., 2005; Spiegel et al., 2007).
Cadms have been independently identified in various model systems, thus these genes and the proteins they encode have acquired several different names, such as Necl (Nectin-like molecules), Igsf4 (Ig-like spermatogenic factor), TSLC (tumor suppressor in lung cancer), and SynCAM (synaptic cell adhesion molecule; Table 1). Recently, the Human Genome Organization (HUGO) Gene Nomenclature Committee renamed the genes CADM (Table 1). So far, four Cadm genes have been identified in tetrapod vertebrates, and the proteins they encode show a strict conservation of their structural organization. Cadm proteins are composed of three extracellular Ig-like loop domains, a trans-membrane domain, and a highly conserved short cytoplasmic tail containing two known protein–protein interaction domains, namely a juxtamembrane protein 4.1 binding motif and a C-terminus type II PDZ-binding domain (Fig. 1A; Biederer, 2006). Interestingly, genes with this protein structure are only found in vertebrates and appear to be an innovation of this phylum (Biederer, 2006).
TABLE 1.
Nomenclature of the Cadm Gene Family a
| Human gene name | CADM1 | CADM2 | CADM3 | CADM4 |
| Mouse gene name | Cadm1 | Cadm2 | Cadm3 | Cadm4 |
| Zebrafish gene name | cadm1 | cadm2 | cadm3 | cadm4 |
| Alternate gene and protein names | Necl2 | Necl3 | Necl1 | Necl4 |
| SynCAM1 | SynCAM2 | SynCAM3 | SynCAM4 | |
| IGSF4A | IGSF4D | IGSF4B | IGSF4C | |
| Tslc1 | Tsll1 | |||
| sgIGSF | ||||
| RA175 |
The human and mouse gene names were recently changed from IGSF4 to CADM by the HUGO gene nomenclature committee. We have adopted this nomenclature. Previous names given to these genes and proteins are shown at the bottom. GenBank ID numbers for the novel zebrafish cadm genes and isoforms are: cadm1a, EU182349, EU182350, EU182351; cadm1b, EU182352; cadm2b, EU182353; cadm3, EU182354.
Fig. 1.
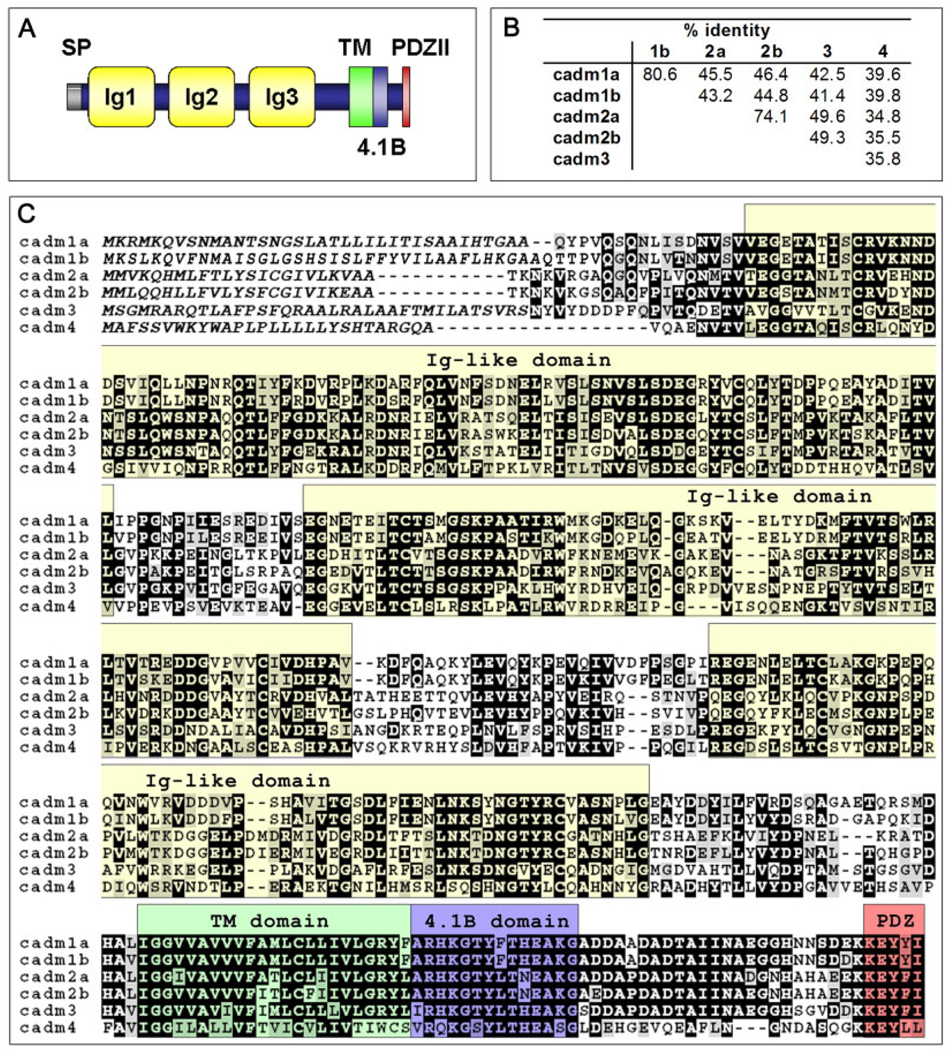
Structure and sequence of zebrafish Cadm proteins. A: Schematic representation of Cadm protein structure. All Cadms contain a signal peptide (SP), three Ig-like domains (Ig), a transmembrane (TM) domain, and a cytoplasmic tail with a 4.1B binding domain (4.1B) and the PDZ type II binding domain (PDZII). B: Amino acid identity as a percentage in pairwise alignments for the six Cadm protein sequences. C: Protein alignment of the zebrafish Cadm protein sequences. Conserved positions with an identical amino acid (black) and conserved subtitutions (gray) have been shadowed. Putative signal peptide for each cadm is in italic; each structural domain is indicated above the aligned sequences and color shaded as in A.
The extracellular Ig-like domains of Cadm proteins can mediate both hetero- and homophilic interactions that are Ca2+ and Mg2+-independent. To date, in addition to various combinations of Cadm to Cadm interactions (Takai et al., 2003; Maurel et al., 2007), two types of heterophilic binding partners have been identified, the nectins and class I-restricted T-cell-associated molecules (CRTAM). Furthermore, the intracellular tails of the Cadms have been shown to interact with several scaffolding molecules in various in vitro systems. These include CASK (Biederer et al., 2002), syntenin (Biederer et al., 2002; Meyer et al., 2004), GRIP (Meyer et al., 2004), MPP3 (ortholog of the Drosophila tumor suppressor gene Dlg; Fukuhara et al., 2003), and DAL-1 (Yageta et al., 2002). However, very little is known about how these interactions relate to the functions of the Cadms during development.
Extensive analyses have revealed that Cadm1 is expressed during early development in rodents (Fujita et al., 2005; Ohta et al., 2005). It has been found in most epithelia and neuroepithelia, such as hair follicles, lung, liver, gut, tongue, olfactory epithelium, dorsal root ganglia, various regions of the central nervous system, being particularly enriched in the marginal zone of the cortex, the external granule layer of the cerebellum, the habenular nucleus, and the thalamus (Fujita et al., 2005; Ohta et al., 2005). Additionally, Cadm1 has been shown to be highly expressed in the testis during germ cell development.
Of interest, inactivation of the Cadm1 gene in mice produces infertile males (oligo-astheno-teratozoospermia; Fujita et al., 2006). Infertility is probably caused by a delay in the maturation of sperm cells, leading to an increase in apoptosis. The deficit in sperm cell maturation is due to an alteration of the adhesion between spermatogenic cells and the Sertoli cells (Wakayama et al., 2003; Fujita et al., 2006; van der Weyden et al., 2006; Yamada et al., 2006).
Given the widespread expression of Cadm1, it is interesting to note that no other deficits have been detected in Cadm1 knockout mice (Fujita et al., 2006), revealing probable compensation mechanisms in other tissues. Nonetheless, in vitro studies have determined several functions for Cadm1 in the nervous system. Cadm1 is expressed during the period of synaptogenesis and localizes to pre- and postsynaptic sites in the rodent brain. Its overexpression in cultured neurons increases spontaneous synaptic activity (Biederer et al., 2002; Sara et al., 2005). Moreover, the expression of recombinant Cadm1 in non-neuronal cells cocultured with neurons induces the formation of functional presynaptic terminals onto the non-neuronal cells. Cadm1 may therefore act as a synaptogenic molecule during nervous system development (Biederer et al., 2002; Sara et al., 2005).
In addition to promoting neuron to neuron interactions, Cadm1 also promotes a homophilic adhesion link between neurons and mast cells. This interaction grants Cadm1 the ability to modulate the immune system by enhancing mast cell response to nerve activation (Ito et al., 2003; Furuno et al., 2005; Ito and Oonuma, 2006; Ito et al., 2007a). Another function of Cadm1 in the immune system includes immunosurveillance. Loss of Cadm1 may provide an escape mechanism from detection by natural killer (NK) cells and cytotoxic T cells, which express CRTAM (Murakami, 2005). Also, cell adhesion through Cadms appears necessary to promote tumor suppression. For instance, absence of CADM1 expression and mutated forms of CADM1 in carcinoma have been found to enhance malignancy, through destabilization of cell–cell contacts (Fuchs and Colonna, 2006).
In contrast to Cadm1, Cadm3 and Cadm4 expression seems to be mostly restricted to neurons and glial cells (astrocytes, oligodendrocytes, and Schwann cells; Maurel et al., 2007; Spiegel et al., 2007). Recently, it has been proposed that a heterophilic interaction between Cadm3 and Cadm4 mediates Schwann cell adhesion to peripheral axons. Perturbation with dominant-negative forms of either Cadm3 or Cadm4 or knockdown of their expression blocks the myelination process (Maurel et al., 2007; Spiegel et al., 2007). Expression patterns and function of Cadm2 have not been reported to date.
As a prerequisite to better understanding the function and mechanism of all the Cadm genes during development, we have isolated orthologs of the tetrapod Cadm genes from the zebrafish (Danio rerio). We have characterized their pattern of expression during development and in the adult.
RESULTS AND DISCUSSION
Isolation and Characterization of the cadm Genes From Zebrafish
In tetrapods, four distinct Cadm gene family members have been identified to date (Biederer, 2006). By searching the zebrafish genome database (http://www.ensemble.org/Danio_rerio) using mammalian family members as a template, we have identified six orthologs in zebrafish. In accordance with the HUGO Gene Nomenclature Committee and the nomenclature guidelines proposed by the Zebrafish Model Organism Database (www.zfin.org), we will call these genes cadm (Table 1).
By using a combination of reverse transcriptase-polymerase chain reaction (RT-PCR) and 5′–rapid amplification of cDNA ends (RACE) -PCR, we have identified the coding sequences for the six cadm genes. This finding indicates that all six genes are expressed in zebrafish. The coding sequences show the same protein domain organization that characterizes the tetrapod Cadm family: a signal peptide followed by three immunoglobulin domains in the extracellular portions of the proteins, a transmembrane domain and a short cytoplasmic tail that encompass a juxtamembrane 4.1B binding domain and a C-terminal PDZ type II binding domain (Fig. 1A,C).
Alignment of the protein sequences of the six cadm genes (Fig. 1B,C) and comparison with tetrapod species (Fig. 2B; Supplementary Figure S1A–D, which can be viewed at http://www.interscience.wiley.com/jpages/1058-8388/suppmat) revealed that two genes are very similar to mouse Cadm1, whereas two other genes are very similar to mouse Cadm2. Given that the ancestor of teleost fish underwent a genome duplication before branching off from the tetrapod lineage during evolution (Postlethwait et al., 2004), we hypothesized that the genes similar to mouse Cadm1 and 2 are co-orthologs. We have named these cadm1a and cadm1b, and cadm2a and cadm2b. The respective co-ortholog protein sequences share higher homology between each other (80.6% and 74.1% identity for cadm1 and 2 genes, respectively, Fig. 1B) than with any other cadm genes (42.2% average identity in pairwise comparison).
Fig. 2.
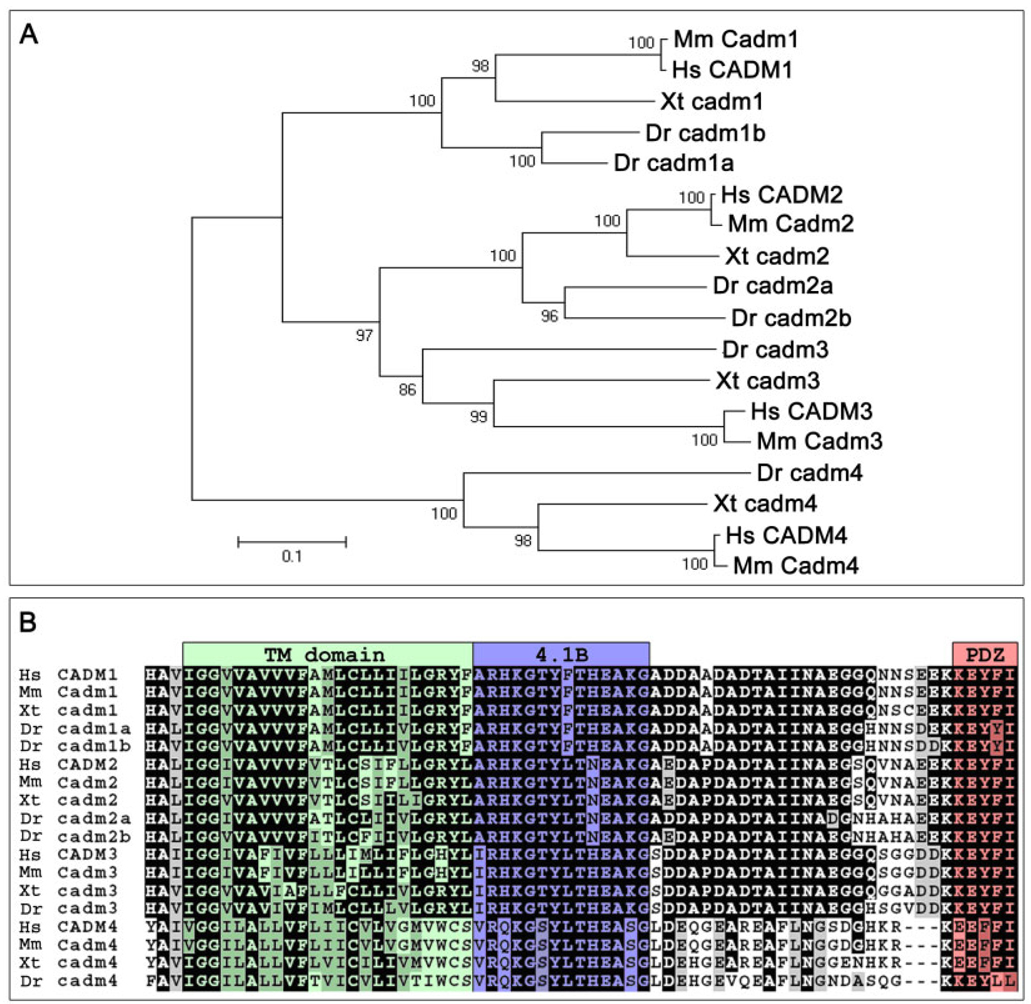
Phylogeny of the Cadm proteins. A: Relationships between the Cadm protein sequences of various vertebrates (Hs, Homo sapiens; Mm, Mus musculus; Xt, Xenopus tropicalis; Dr, Danio rerio) are shown in a phylogenetic tree. Amino acid sequences were aligned by Clustal-X. Sequences were trimmed to include unambiguously aligned regions, and phylogenic analysis used the Poisson-corrected neighbor-joining method. The branch lengths (numbers) are the percentage of bootstrap values for 1,000 replicas. Scale bar = 0.1 substitutions per site. B: Alignment of the transmembrane domain and cytoplasmic tail of the Cadm proteins highlights the high level of conservation of the structural domains among vertebrates. Conserved positions with an identical amino acid (black) and conserved subtitutions (gray) have been shadowed; the color shading of domains is as in Figure 1A.
Analysis of conserved synteny between phyla can provide evidence of orthology and is necessary to determine whether genes are duplicates arising from a genome duplication (Postlethwait et al., 2004). Radiation hybrid mapping determined that cadm1a, 1b, 2a, 2b, 3, and 4 map to linkage groups 21, 16, 2, 15, 10, and 15, respectively. Because some of these locations differ from the assigned chromosomal locations in the latest zebrafish genome assembly (Zv7), we have compared the scaffolds containing the zebrafish cadm genes with the chromosomal region surrounding the murine Cadm genes. This analysis demonstrates that the neighboring genes of the zebrafish co-orthologs of Cadm1 are also found flanking the murine gene. In fact, three orthologous genes are found in all three chromosomal regions: jam3, igsf9b, and grit (Supplementary Figure S2). This confirms that cadm1a and 1b are derived from a duplication of the ancestral chromosomal region (Postlethwait et al., 2004).
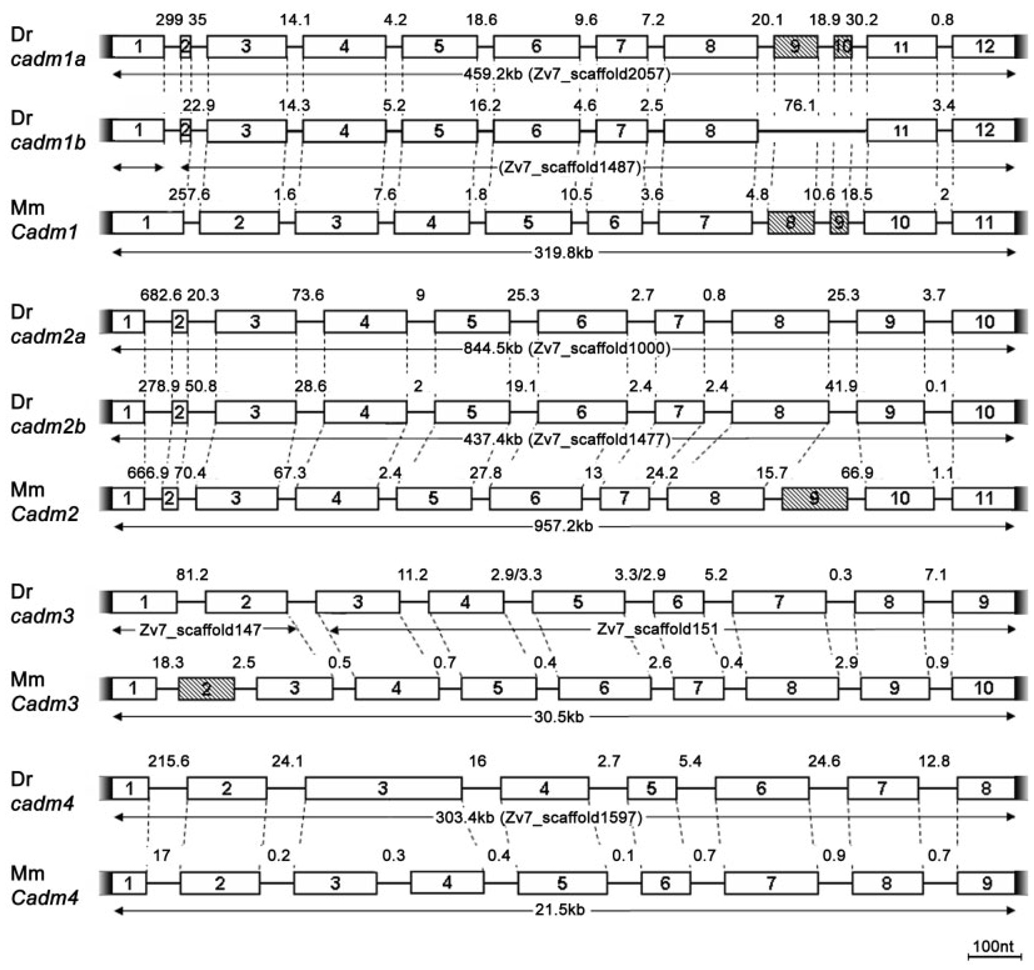
Similar analyses revealed conserved syntenies between zebrafish and murine Cadm3 and 4 genes (not shown). However, comparison of syntenic regions for cadm2a and 2b was not possible due to the paucity of neighboring genes. Further supporting the conclusion that cadm1a, 1b, 2a, and 2b arose from the teleost genome duplication, all cadm genes share the same gene structures as the respective mouse ortholog (Fig. 3). No duplicates have been found to date for cadm3 and 4. Because no duplicates have been found for these genes in other bony fish, we conclude that, subsequent to the genome duplication within the teleost lineage, these gene duplicates were probably lost.
Fig. 3.
Genomic analysis of zebrafish cadm genes. Comparison of the zebrafish (Dr) and mouse (Mm) loci. Exons are represented by numbered boxes. Shaded boxes show exons subjected to alternative splicing. Exon–intron boundaries that correspond between zebrafish and mouse genes are represented by dashed lines. All corresponding exons between the two species are of identical length, unless indicated otherwise in the text. The scale bar (100 nt) is for exons; the sizes of introns are indicated in kilobases. Notice that zebrafish cadm1b and 3 are both aligned on two different scaffolds in Zv7 assembled zebrafish genome. The first exon of cadm1b is found on Zv7_scaffold1472, whereas the other exons on Zv7_scaffold1487, suggesting a large first intron. Similarly, the first two exons of cadm3 are found in Zv7_scaffold147, while the rest of the gene is on Zv7_scaffold151.
Alignment of the six zebrafish Cadm protein sequences (Fig. 1B,C) reveals that the Cadm2 protein sequences are more closely related to Cadm3 (49.5% average identity) than to Cadm1 and 4 (45.9% and 35.2% average identity, respectively). Cadm4 is a more distant member of the family, with an average amino acid identity of 37.1%. The phylogenetic comparison between, human, mouse, toad, and zebrafish sequences confirms this relationship of zebrafish Cadm proteins (Fig. 2A).
When compared with their murine orthologs, the zebrafish Cadm proteins show on average 55% identity (Supplementary Fig. 1A–D). The strongest divergence is seen in the N-terminus of the proteins, encoding the signal peptides. In contrast, the C-terminal portions of the Cadms, including the intracellular protein–protein interaction domains are highly conserved across phyla (Fig. 2B). This striking conservation reflects constraints upon evolutionary mechanisms, suggesting an important function for both the 4.1B and PDZ type II binding domains. The Cadm1 PDZ binding domain is relevant to synaptogenesis as demonstrated by in vitro experiments (Biederer et al., 2002), while the 4.1B binding domain has been shown to-mediate interactions with proteins important for actin cytoskeleton stabilization (Yageta et al., 2002).
We have also compared the genomic organization of mouse and zebrafish Cadm genes. We focused our attention on their coding sequences, excluding 5′- and 3′-untranslated regions of the genes. Thus, the number of exons might be under-represented. Nevertheless, there are striking similarities between the mouse and zebrafish gene structures (Fig. 3). Almost all of the exons are of the same size and encode the same protein region, suggesting that, despite the large evolutionary distance that separates these species, the genes are remarkably well conserved. Here, we highlight the few differences.
When comparing mouse and zebrafish genes, we see three cases of missing introns, resulting in extended exons. Loss of introns appears to have happened both in mouse and in zebrafish, such that zebrafish cadm1a and 1b exons 1 and 2 correspond exactly to mouse Cadm1 exon 1, and mouse Cadm4 exons 3 and 4 appear to have been contracted into zebrafish exon 3. This loss of an intron from zebrafish cadm4 gene is substantiated by the medaka cadm4 gene (EnsEmbl ENSORGL00000004868), which possesses the same structure as its zebrafish ortholog. Unusually, the lengths of the first two exons of the cadm3 gene do not correspond to the lengths of the murine counterparts. We have confirmed the sequence of zebrafish cadm3 by identifying expressed sequence tags that cover at least the first three exons of the gene (for example, EST CN507252).
Another interesting point is that the murine Cadm genes show an increased number of alternatively spliced exons when compared with the zebrafish genes (hatched boxes in Fig. 3). Both Cadm2 and Cadm3 show additional exons 9 and 2, respectively, which are alternatively spliced for exons 3 and 8. The zebrafish cadm1 genes contain two exons (9 and 10) that can be alternatively spliced, and potentially produce four different isoforms, just as in mouse and human (Biederer, 2006). Using zebrafish adult brain cDNA, we were able to recover three of these isoforms for cadm1a, revealing an identical splice pattern between mouse and zebrafish. However, we have identified only one isoform for the cadm1b locus. Additionally, a third alternatively spliced exon in Cadm1 has been identified in the mouse and human genomes (Biederer, 2006). It is probable that additional splice variants also exist in zebrafish and that they are differentially regulated during development and in various tissues. This finding would explain the different number of isoforms we recovered for the cadm1 loci by screening adult central nervous system cDNA.
In conclusion, despite their relatively large evolutionary distance, the cadm genes are remarkably conserved between zebrafish and mammals, suggesting that their functions are probably also conserved. In addition, the maintenance of the duplicated cadm1 and cadm2 loci in zebrafish indicates that multiple functions of the mammalian orthologs may have been partitioned to the co-orthologs. On the other hand, it is possible that the co-orthologs may carry out the same function but in different tissues or at different times during development, as has been seen for other co-orthologs in teleost fish (Postlethwait et al., 2004).
Expression Patterns of cadm Genes
We assayed the temporal and spatial distribution of cadm gene expression during zebrafish development by whole-mount RNA in situ hybridization (ISH), from approximately 10 hours postfertilization (hpf) through 72 hpf. We are confident that the probes to all six genes are specific and do not cross-react, given their distinct expression patterns and the fact that ISH using sense probes did not show any staining.
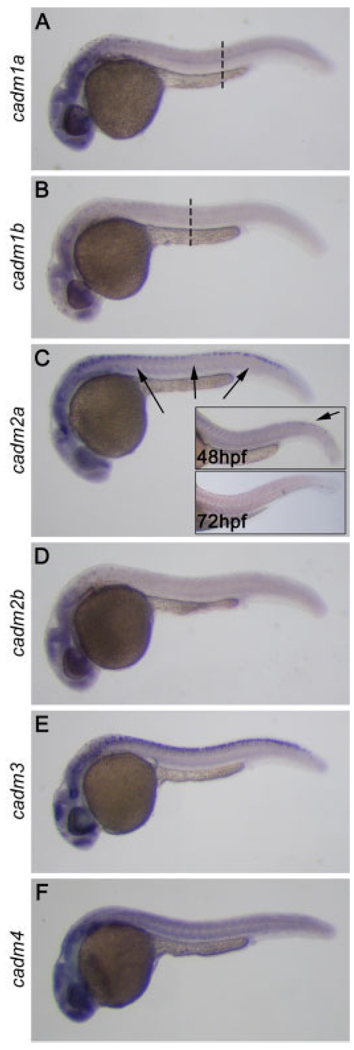
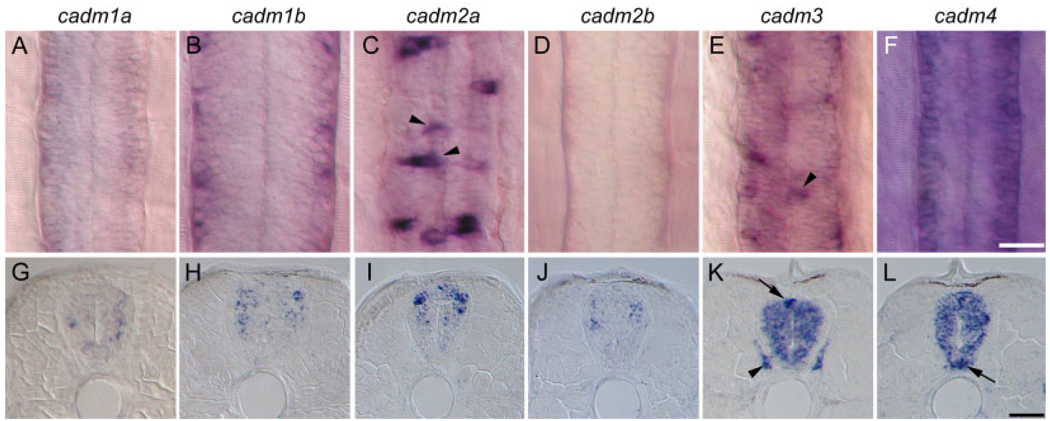
No expression was seen at 10 hpf, but by 15 hpf, all of the cadm genes showed evidence of expression, in particular in the developing head (not shown). At 24 hpf, development has progressed to the point that most major organ systems have begun to form, and expression of all six cadms was seen throughout the central nervous system, including the eye and spinal cord (Fig. 4). While expression for all cadm genes was consistently stronger in the head than in the trunk and tail, their expression patterns in the spinal cord were dynamic and divergent between the cadm family members. Expression was evident in different domains of the spinal cord at 24 hpf (Fig. 4, discussed below, see Fig. 7). Furthermore, the spinal cord expression of cadm genes was evident as a wave of expression during development. For example, the expression of cadm2a was decreased in the rostral spinal cord by 48 hpf, becoming undetectable throughout the spinal cord by 72 hpf (Fig. 4C, insets). Also, the cadm1 genes were present in a rostrocaudal gradient at 24 hpf, with no expression seen above the yolk tube and caudal to this (Fig. 4A,B, dashed lines). These patterns are confirmed for all cadm genes by ISH on sections at 24 and 48 hpf (not shown). Because many neurons in the spinal cord develop in a rostrocaudal gradient (Lewis and Eisen, 2003), it is probable that the cadm genes are important for a specific event during neuronal maturation.
Fig. 4.
Expression of cadms at 24 hours post-fertilization (hpf). In situ hybridization performed on 24 hpf whole-mount zebrafish embryos reveals that the six cadm genes are expressed throughout the central nervous system, in particular in the developing brain, the visual system and the spinal cord. A–C: Expression for cadm1a and 1b is in a rostrocaudal gradient (A,B), being undetectable caudal to the dashed lines. cadm2a expression decreases rostrally at 48 hpf and is lost by 72 hpf (insets in C).
Fig. 7.
Expression of cadms in the developing spinal cord. A–L: The cadm expression revealed by in situ hybridization in whole-mount (dorsal view, A–F) and cross-sections (G–L) of 48 hpf embryos. Due to the strong rostrocaudal gradient of cadm1b expression, panels B and H show an anterior localized region of the spinal cord (somites 3–5), immediately behind the hindbrain, whereas all other images are from a region dorsal to the anus (somites 12–15). Staining for cadm2a and 3 is in dorsal cells at the midline of the spinal cord, suggestive of sensory Rohon-Beard neurons (arrowheads in C and E, arrow in K). Expression for cadm3 is also seen in the dorsal root ganglion (arrowhead in K). cadm4 is strongly expressed in the ventral domain of the spinal cord indicative of floor plate or motoneurons. Scale bars = 20 µm in A–F, 20 µm in G–L.
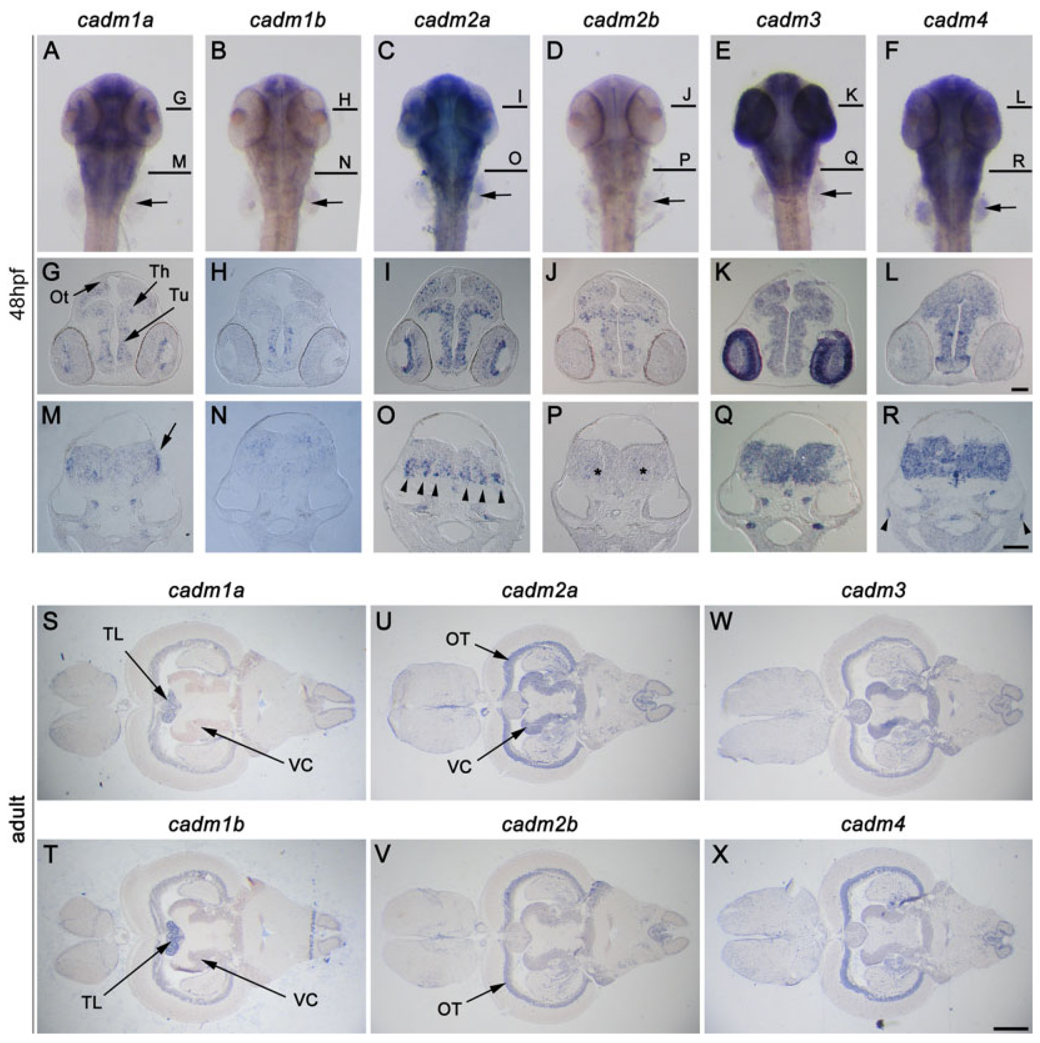
The general pattern of strong expression throughout the brain persisted to 72 hpf. However, some expression was also seen outside of the nervous system; all 6 cadms were expressed in the precartilage of the pectoral fin buds at 48 hpf (arrows in Fig. 5A–F), the pancreas, gut, and developing swim bladder (not shown). All cadm genes were expressed at low levels in the adult testis, with cadm3 showing the strongest expression (not shown), suggesting that, as in mammals (Fujita et al., 2006), these cell adhesion molecules may play a role in the maturation of sperm. Due to the predominant expression of the cadm genes in the nervous system during development, we have focused our attention on the brain, visual system and spinal cord of the zebrafish.
Fig. 5.
Expression of cadms in the brain. A–R: In situ hybridization (ISH) staining in 48 hours postfertilization (hpf) zebrafish embryos; a dorsal view of whole-mount zebrafish (A–F), and cross-sections at the midbrain (G–L), and hindbrain (M–R) are presented for each cadm gene. Lines in A–F represent levels of the sections in G–L and M–R. S–X: cadm expression visualized in horizontal sections of adult brain. The cadm genes are expressed broadly in the developing and adult brain and show partially overlapping domains of expression. cadm2a is strongly expressed in the ventral medulla oblongata (MO) (arrowheads in O), while cadm2b is more diffuse in this region (asterisks in P). The posterior part of the MO shows strong staining for cadm1a (arrow in M). Ot, developing optic tectum; OT, optic tectum granular layer L3; Th, thalamus; TL, torus longitudinalis; Tu, tuberculum; VC, valvula cerebellis. Orientations are anterior at the top in A–F; dorsal at the top in G–R; anterior to the left in S–X. Scale bars = 50 µm in G–L, 50 µm in M–R, 0.5 mm in S–X.
cadm Expression in Developing and Adult Brain
The cadms were broadly expressed throughout the brain both during development and in the adult (Fig. 4, Fig 5). We have summarized the brain expression patterns in Table 2 and Table 3, and detailed descriptions of the expression patterns are in the Supplementary Materials. The tables and descriptions are based on analyses of both horizontal and coronal sections of the whole brain, the majority of which are not shown here.
TABLE 2.
Summary of cadm Expression in Brain at 48 Hours Postfertilization a
| 48hpf Forebrain |
||||||
|---|---|---|---|---|---|---|
| Pallium | ||||||
| Olfactory bulb | ||||||
| Epiphysis | ||||||
| Habenula | ||||||
| Subpallium | ||||||
| Olfactory epithelium | ||||||
| Dorsal thalamus | ||||||
| Ventral thalamus | ||||||
| Griseum tectale | ||||||
| Pretectum | ||||||
| Posterior tuberculum (dorsal part) | ||||||
| Posterior tuberculum (ventral part) | ||||||
| Midbrain | ||||||
| Tectum opticum | ||||||
| Medial longitudinal fasicle nucleus | ||||||
| nucleus | ||||||
| Midbrain tegmentum | ||||||
| Rostral hypothalamus | ||||||
| Intermediate hypothalamus | ||||||
| Hindbrain | ||||||
| Valvula cerebelli | ||||||
| Cerebellar plate | ||||||
| Torus semicircularis | ||||||
| Trigeminal ganglion | ||||||
| Medulla oblongata | ||||||
| Caudal hypothalamus | ||||||
| Hypophysis | ||||||
| Rhombic lip | ||||||
| Anterior lateral line ganglion | ||||||
| Posterior lateral line ganglion | ||||||
| Octaval ganglion | ||||||
| Facial ganglion | ||||||
| Glossopharyngeal ganglion | ||||||
| Vagal ganglion | ||||||
Expression for brain structures are summarized for the six cadm genes. A black box denotes the presence of cells expressing the cadm gene listed at the top of the columns.
TABLE 3.
Summary of cadm Expression in Adult Brain
| adult Telenchephalon |
||||||
|---|---|---|---|---|---|---|
| Glomerular layer of olfactory bulb | ||||||
| External cellular layer of | ||||||
| olfactory bulb | ||||||
| Internal cellular layer of olfactory | ||||||
| bulb | ||||||
| Lateral dorsal telencephalic cell | ||||||
| mass | ||||||
| Medial dorsal telencephalic cell | ||||||
| mass | ||||||
| Ventral dorsal telecephalic cell | ||||||
| mass | ||||||
| Ventral telencephalon nuclei | ||||||
| Entopedoncular nucleus | ||||||
| Diencephalon | ||||||
| Parvocellular preoptic nuclei | ||||||
| (anterior) | ||||||
| Parvocellular preoptic nuclei | ||||||
| (posterior) | ||||||
| Magnocellular preoptic nucleus | ||||||
| Habenula | ||||||
| Suprachiasmatic nucleus | ||||||
| Preoptic nuclei | ||||||
| Thalamic nuclei (ventromedial) | ||||||
| Thalamic nuclei (central | ||||||
| posterior) | ||||||
| Thalamic nuclei (anterior) | ||||||
| Preglomerular nucleus (lateral) | ||||||
| Preglomerular nucleus (anterior) | ||||||
| Corpus mamillare | ||||||
| Torus lateralis | ||||||
| Periventricular hypothalamus | ||||||
| Inferior lobe (diffuse nucleus) | ||||||
| Inferior lobe (central nucleus) | ||||||
| Magnocellular pretectal nucleus | ||||||
| Periventricular pretectal nucleus | ||||||
| Mesencephalon | ||||||
| Tectum opticum (white matter) | ||||||
| Tectum opticum (granular zone) | ||||||
| Tectum opticum (periventricular | ||||||
| zone) | ||||||
| Torus semicircularis (central | ||||||
| nucleus) | ||||||
| Torus semicircularis | ||||||
| (ventrolateral) | ||||||
| Torus longitudinalis | ||||||
| Nucleus isthmi | ||||||
| Rhombencephalon | ||||||
| Eminentia granularis | ||||||
| Corpus cerrebelli | ||||||
| Valvula cerebelli (lateral division) | ||||||
| Valvulae (nucleus lateralis) | ||||||
| Lobus caudalis cerebelli | ||||||
| Facial lobe | ||||||
| Vagal lobe | ||||||
| Descending octaval nucleus | ||||||
| Anterior octaval nucleus | ||||||
| Medial octavolateralis nucleus | ||||||
| Caudal octavolateralis nucleus | ||||||
Expression for brain structures are summarized for the six cadm genes. A black box denotes the presence of cells expressing the cadm gene listed at the top of the columns.
In general, the cadm1 and 2 genes showed weaker and more punctate expression than cadm3 and 4 (Fig. 5). This may suggest that the four cadm1 and 2 genes are expressed in subsets of cells within a structure. This expression may be in distinct cells within each structure, however, more detailed analyses will be required to determine whether this is true. Conversely, cadm3 and 4 appeared to be expressed in more cell types and more brain regions throughout the nervous system than cadm1a, 1b, 2a, and 2b (Table 2, Table 3). Whereas as the co-orthologs were sparsely expressed in cells at 48 hpf, the same structures solidly expressed cadm3 and 4 (for example Fig. 5K,L). This finding may reflect the expression of cadm3 and 4 in both neurons and glial cells, as has been demonstrated in the mouse (Kakunaga et al., 2005; Gruber-Olipitz et al., 2006; Maurel et al., 2007; Spiegel et al., 2007).
One of the areas with the most striking expression at 48 hpf was the medulla oblongata (MO). Expression varied considerably for each cadm gene through the rostrocaudal length of the MO. While cadm1a showed weak punctate expression medially in the anterior portion of the MO, it was expressed predominantly in the periphery of the posterior MO (Fig. 5M, arrow). In contrast, cadm1b was hardly expressed in the anterior MO, showing a scattered punctate staining toward the middle, becoming concentrated dorsally toward the posterior end of the structure (Fig. 5N). Cadm2a showed strong staining mostly in the ventral region of the whole MO in a series of stripes (arrowheads in Fig. 5O), whereas cadm2b was only weakly expressed in the ventral region of the MO (Fig. 5P, asterisks). Both cadm3 and 4 were expressed strongly throughout the MO (Fig. 5Q,R).
It was interesting to note that the co-orthologs for the cadm genes 1 and 2 demonstrated overlapping, yet more restricted expression patterns both at 48 hpf and in adult. For example, cadm1a was expressed throughout the dorsal thalamus and the posterior tuberculum at 48 hpf (arrows in Fig. 5G), while cadm1b was only expressed in the ventral part of the posterior tuberculum (arrow in Fig. 5N). Analogously, cadm2a was expressed throughout both the granular zone of the optic tectum (OT) and lateral division of the valvula cerebelli (VC) in adult zebrafish (arrows in Fig. 5U), whereas cadm2b was only present in the OT (arrows in Fig. 5V). The restriction of expression of one of the co-orthologs, rather than a distinct expression pattern, may suggest a partitioning of function between the two co-orthologs.
On close examination, it was apparent that, in many areas, the cadm1 and 2 genes showed inverse expression patterns in the developing and adult brain. This is especially clear in Table 2 and Table 3, which reveal that inverse staining was seen for the olfactory epithelium, griseum tectale, VC, cerebellar plate, and the trigeminal ganglion at 48 hpf. In the adult, the olfactory bulb, the anterior thalamic nuclei, both the magnocellular and periventricular pretectal nuclei, the torus longitudinalis (TL), the valvula, the caudal lobe of the cerebellum and the octaval nucleus all showed inverse staining for cadm1 and 2. Clear examples are the TL and VC. Both cadm1 genes were expressed in the TL (arrows in Fig. 5S,T), while cadm2a and 2b were absent (Fig. 5U,V). In contrast, both cadm1a and 1b were not detected in the VC, whereas cadm2a and 2b were present (arrows in Fig. 6S–V). This inverse expression pattern suggests that transcription for these genes may be coregulated in an exclusionary manner.
Fig. 6.
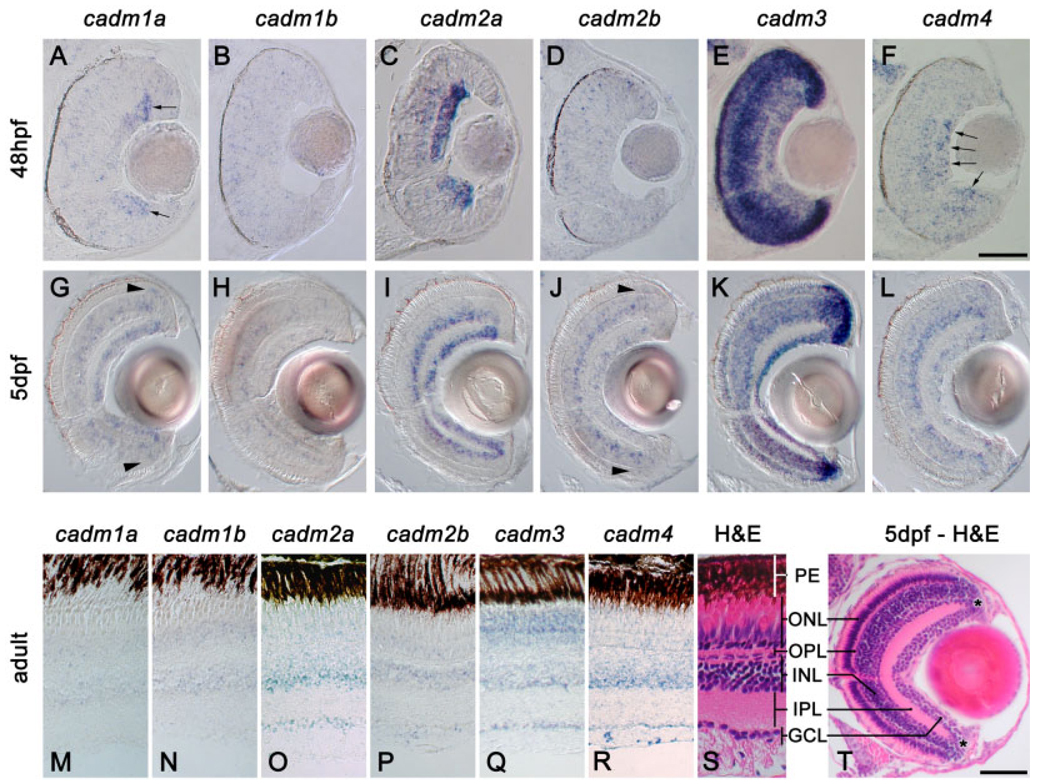
Expression of cadms in the visual system. In situ hybridization staining in sections of retina at 48 hpf (A–F), 5 days postfertilization (dpf; G–L), and adult zebrafish (M–R). S and T show hematoxylin and eosin staining (H&E) of sections of adult and 5 dpf, respectively; the different layers of the retina are indicated: PE, pigment epithelium; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Asterisk in T denotes the location of the marginal zone. cadm1a is expressed in the dividing ganglion cells (arrows in A), while cadm4 is more strongly expressed in the mature cells of the GCL (arrows in F). Arrowheads in G and J reveal precursors at the margin of the outer nuclear layer (ONL). Scale bars = 50 µm in A–F, 50 µm in G–L,T.
cadm Expression in the Developing and Adult Visual System
We examined in detail the cadm gene expression patterns in retina at 48 hpf, 5 days postfertilization (dpf) and adult eyes. Lamination of the retina occurs with a central to peripheral and inner to outer gradient of maturation and is generally complete by 48 hpf (Schmitt and Dowling, 1994). At this age of development, most cadm genes were expressed in a punctate, scattered manner. For instance, cadm1b and 2b were expressed throughout the retina, lens, and the cornea in this manner (Fig. 6B and D, respectively). A similar scattered expression was seen for cadm1a, 2a, and 4 in the inner nuclear layer (INL), outer nuclear layer (ONL), and the marginal zone (Fig. 6A, C, and F, respectively). Because the neuroepithelial cells within these regions are still actively dividing during this period of rapid growth, these expression patterns might represent mitotically active cells. Expression in mitotically active cells would then suggest a novel function for these cadms during early neuronal differentiation.
The ganglion cell layer (GCL) displayed the most varied expression pattern of all the layers of the retina at 48 hpf. cadm1a expression appeared to follow the wave of differentiation of this layer: high expression of cadm1a was seen in the periphery of the layer where the neuroblasts are still mitotically active at this stage, while being down-regulated in the more mature GCL (Fig. 6A, arrows). Conversely, cadm4 expression in the GCL was higher in more mature neurons (Fig. 6F, arrows), while cadm2a was expressed in the entire GCL independently of the level of maturation of the ganglion cells (Fig. 6C). In contrast, and as seen in the brain, cadm3 was expressed in the vast majority of cells in the retina (Fig. 6E).
Visual function is observed as early as 3 dpf and is fully functional by 5 dpf (Schmitt and Dowling, 1994). At this time, cadm genes were expressed in partially overlapping domains within most retinal layers. In the GCL, cadm1a, 2a, and 3 were expressed in most of the neurons (Fig. 6G,I,K), while cadm1b, 2b, and 4 (Fig. 6H, J and L) were seen in subsets of cells. In the INL, cadm1a, 1b, 2a, 2b, and 4 were expressed in restricted populations of cells (Fig. 6G–J,L). These expression patterns suggest that all cadm genes are expressed in amacrine cells, bipolar cells, and Müller glia. In addition, it appears that cadm1a, 2b, 3, and 4 are expressed in horizontal cells at the outer limit of the INL (Fig. 6G,J–L) and cadm3 and 4 in the ONL (Fig. 6K,L). Notably, the periphery of the ONL also expressed cadm1a, 2b, and 4, and might represent dividing retinal precursor cells (Fig. 6G,J,L, arrowheads).
All cadm genes remain expressed in the adult eyes with partially overlapping domains of expression, generally mirroring the expression at 5 dpf. However, cadm1a and 1b were down-regulated in the GCL in adult eyes (Fig. 6M,N). Cadm2a, 2b, 3, and 4 were detected in the GCL, albeit at different levels (Fig. 6O–R). Cadm2a and 2b were expressed in a small subset of the GCL cells (Fig. 6O,P), whereas cadm3 and 4 were present in most of the ganglion cells (Fig. 6Q,R). All six cadm genes were expressed in the INL: cadm1a, 1b, 2b, and 4 were restricted to the medial domain of the INL, suggesting they are expressed in bipolar, but not amacrine or horizontal cells. In contrast, cadm2a and 3 presented a broader expression in the INL, including the amacrine cell domain; cadm3 was expressed at a higher level in the amacrine cells than in the other regions of the INL. The ONL presented a sparse expression of cadm1a, 1b, 2a, and 4 in both inner and outer segments, suggesting they are present in a small subpopulation of rods and cones; cadm3 was detected in virtually all cells of the ONL. In contrast, cadm2b was restricted to the outer segment and, therefore, expressed in the rod population of the ONL.
In summary, cadm genes are highly regulated in the course of the development of the retina. They show partially overlapping domains of expression within the retina, but can be restricted to distinct cell populations composing each layer. These dynamic expression patterns suggest that the various cadms may perform distinct functions at discrete times during differentiation of retinal cells.
cadm Expression in the Developing Spinal Cord
We examined the expression of the six cadm genes in the spinal cord at 48 hpf, because the wave of expression of these genes was in the widest variety of cell types at this time. We present both dorsal views of RNA ISH in whole-mount embryos (Fig. 7A–F) and in sections (Fig. 7G–L). This strategy allows appreciation of the spatial distribution of expression along both rostrocaudal and dorsoventral axes. We used the following spatial subdivisions along these axes to assign expression to particular cell types. The dorsal domain is defined by the sensory Rohon-Beard neuron, whereas the intermediate domain is composed of various types of interneurons. Finally, the ventral domain comprises floor plate cells and motoneurons. The morphology, localization, and axon projections of zebrafish spinal cord neurons have been well described (Hale et al., 2001; Lewis and Eisen, 2003).
Diffuse expression was seen for all cadms in the various domains of the spinal cord; however, several expression patterns were suggestive of strong expression in distinct populations of spinal neurons (Fig. 4, Fig. 7). cadm1a was mostly detected in the intermediate domain of the spinal cord, probably in a subpopulation of interneurons. It was more strongly detected in the ventral part of this domain in the caudal spinal cord, suggesting that it is expressed first in a ventral population of interneurons, before expanding to a more dorsal domain. cadm1b was expressed in the intermediate domain of the spinal cord and, in particular, in its dorsal aspect, suggesting that cadm1b is expressed in dorsal interneurons.
cadm2a was expressed throughout the dorsal and intermediate region of the spinal cord and was particularly strong in cells at the midline in the dorsal spinal cord, suggesting expression in sensory Rohon-Beard neurons (Fig. 7C, arrowheads). In contrast, cadm2b was weakly expressed and excluded from the dorsal and ventral domain, suggesting that it may be exclusively present in interneurons.
cadm3 and 4 were widely expressed in the spinal cord, including the proliferating ventricular zone. cadm3 showed strong staining in dorsal neurons, including Rohon-Beard neurons (Fig. 7, arrowhead in E and arrow in K). In contrast, cadm4 was more highly expressed in the ventral domain of the spinal cord, suggestive of floor plate cells and motoneurons (arrow Fig. 7L). Additionally, cadm3 was present in other neuronal tissues in the trunk and tail, the localization suggesting expression in dorsal root and sympathetic ganglia (Fig. 7K, arrowhead). cadm4 was also detected in the developing neuromasts of the lateral line system (Fig. 5R).
As Cadm1 is a potential mediator of synaptogenesis (Biederer et al., 2002) and the expression of all six cadm genes is found in discrete neuronal subtypes of the developing spinal cord at a time when neuronal circuits are becoming functional (16–48 hpf; Drapeau et al., 2002), it is reasonable to hypothesize that these molecules could mediate synaptogenesis or determine synaptic specificity for the early circuits in spinal cord during development. With this expression data in hand, it may now be possible to perform a functional analysis of this family of cell adhesion molecules in vivo.
Concluding Remarks
The zebrafish cadm genes show strong similarities with their tetrapod orthologs in terms of genetic structure and protein organization. Their expression is highly regulated during development of the central nervous system, and they show partially overlapping domains of expression within each structure. The maintenance of two duplicated cadm loci in zebrafish with similar expression patterns indicates the probable acquisition of novel functions or the partition of functions during the course of evolution. The dynamic expression patterns during development suggest that these cell adhesion molecules may play multiple roles during neuronal differentiation, including steps in neuronal cell fate decisions and synaptogenesis.
EXPERIMENTAL PROCEDURES
Cloning the cadm Genes
To clone the zebrafish cadm genes, we searched the zebrafish genome assembly Zv6 from the Sanger Institute (http://www.ensemble.org/Danio_rerio) by tBLASTn (http://www.ncbi.nlm.nih.gov/BLAST) comparison with the mouse Cadm1 to Cadm4 protein sequences (NP_997558.2, NP_848836, NP_444429 and NP_694752 respectively). This search revealed six cadm genes in zebrafish, with a duplication of the cadm1 and cadm2 loci. The best hits on the zebrafish genome were then compared with the GenBank database to recover published sequences. Full-length cDNA for one cadm2 (Igsf4d, renamed in this study cadm2a) and cadm4 were found in the Genbank references NM_200664.1 (IMAGE clone 5603809) and BC085419 (IMAGE clone 7227860), respectively. Partial cloning of cadm1a (third Ig domain, transmembrane domain and cytoplasmic tail of the protein), based on a partial sequence of RA175 (AB183400) was performed by RT-PCR of zebrafish adult brain cDNA (forward primer: ATGCTAGCAAGGAGAGATAT reverse primer TCAGGACTCAGATATAGTAT). The database search with the mouse Cadm1 did not allow recovery of the highly divergent 5′ fragment of zebrafish cadm1a. Comparison of the medaka cadm1 sequence (EnsEmbl ENSORLG00000005159) by BLASTn with the zebrafish genome revealed a 5′ fragment (without the signal peptide sequence), which was cloned by RT-PCR (forward primer, CAGAATCTCATATCGGACAACGTC and reverse primer, GCCTGAAAGTCCTTGACTGC). We used the FirstChoice RLM-RACE Kit (Ambion) to recover the signal peptide of cadm1a (outer primer: TCCTCGCGGGACTCTATGAT and inner primer: CGAGCATCTTTCAGGGGTCT). Zebrafish cadm1b is derived from partial zebrafish cDNA sequences (XM_ 001337106, XM_685581 and BU710444) and cloned by RT-PCR of 72 hpf whole zebrafish embryo cDNA. These published sequences did not contain the signal peptide of cadm1b, which was cloned by 5′-RACE PCR (outer primer, CTCGCGGGACTCTAAGATTG and inner primer, CGGTGGATCCGTGTAGAGTT). Cadm2b and cadm3 were deduced from XM_001342480 and XM_695311 (cadm2b) and NM_001045246 (cadm3) and cloned by RT-PCR from zebrafish adult brain cDNA (forward primer, TGCACGCAACAAATATCCTC and reverse primer, CTAAATGAAGTACTCTTTCTTTTCC for cadm2b and forward primer, CTATTGGCTGTAGCGTGCTG and reverse primer, GAAGCGTGTGAAGGAAGAGG for cadm3).
Protein Alignment and Phylogenetic Analysis
Deduced protein sequences for the zebrafish cadm genes based on the cloned cDNA sequences have been aligned with ClustalW. This alignment, using only the shortest isoforms, was used to generate a phylogenetic tree with the MEGA package, based on Poisson-corrected neighbor-joining amino-acid distances and considering gap pairwise deletion. Protein accession number (GenBank for human and mouse and EnsEmbl for Xenopus) used for the analysis were, NP_055148.3 (Homo-sapiens; Hs_CADM1), NP_694854.2 (Hs_CADM2), NP_067012.1 (Hs_CADM3), NP_660339.1 (Hs_CADM3), NP_997559.1 (Mus musculus, Mm_Cadm1), NP_848836.1 (Mm_Cadm2), NP_444429.1 (Mm_Cadm3), NP_694752.1 (Mm_Cadm4), ENSXETP00000033613 (Xenopus tropicalis, Xt_cadm1), ENSXETG00000026489 (Xt_cadm2), ENSXETP00000041942 (Xt_cadm3), and ENSXETP00000033087 (Xt_cadm4).
Mapping and Conserved Synteny
The zebrafish cadm genes were mapped on the LN54 radiation hybrid mapping panel (Hukriede et al., 1999) as described (Postlethwait, 2000). The chromosomal position of each gene was determined through the use of the mapping program of Dr. I. Dawid (http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi). Two sets of primers were used to confirm the localization on the LN54 panel (primer sequences available on demand). For comparative mapping with murine Cadm genes, zebrafish cadm genes were considered orthologs when the human and zebrafish genes were best hits on the reciprocal BLASTp searches on the other genome. The mouse Cadm sequences used for this analysis are as indicated in Protein Alignment and Phylogenetic Analysis.
In Situ Hybridization
Zebrafish embryos, larvae and adults (AB/Tübingen strain) were raised at 28.5°C according to standard protocols (Westerfield, 2000). Sense and anti-sense probes were in vitro transcribed and digoxigenin tagged according to the manufacturer’s protocol (Roche) from linearized pCRII-TOPO (Invitrogen) cadm1a plasmid or from PCR fragments of cadm1b, 2a, 2b, 3, and 4 linked to a T7 or T3 promoter. cadm1a probes encompass the cadm1a cDNA from nt 623 to 1384 of the CDS; cadm1b, from nt 381 to 856, cadm2a, from nt 331 to 1154; cadm2b, from nt 351 to 1048; cadm3, from nt 239 to 992; cadm4, from nt 278 to 992. ISH on frozen sections was performed according to the protocol described by Jensen et al. (2001) with the following changes. Labeled probes were diluted 1:200 in hybridization buffer and heated to 68°C for 30 min, 200 µl of diluted probe was used per slide. Hybridization was carried out at 68°C. Posthybridization washes were increased to 3 washes in 50% form-amide, 1× standard saline citrate (SSC), 0.1% Tween-20 at 68°C for 30 min and three washes in MABT (100 mM maleic acid, 150 mM NaCl, pH 7.5, 0.1% Tween-20) for 30 min at room temperature. Anti-digoxigenin conjugated to AP antibody (Roche) was used at a 1:1500 dilution. Whole-mount ISH was carried out according to the protocol as seen at http://courses.mbl.edu/zebrafish/faculty/houart_wilson/pdf/in_situ_hybridization.pdf. BM purple AP Substrate (Roche) was used for coloration. Images of sections and whole-mount embryos were taken on a Zeiss Axioplan microscope or a Leica MZ6 stereomicroscope using a Nikon Coolpix 990 or 4500 digital camera. The images were arranged for presentation purposes using Adobe Photoshop. Annotations of the anatomy of the embryonic and adult brain are done according to Wulliman et al. (1996) and Muller and Wulliman (2005).
Supplementary Material
ACKNOWLEDGMENTS
We thank Keith Beadle, Danielle Leavitt, and Elise Manalo for expert technical assistance and Poh Kheng Loi for assistance with histology. We thank the staff of the University of Oregon Zebrafish Facility for fish husbandry. Special thanks go to John Postlethwait, Judith Eisen, and Alexandra Tallafuss for input and comments on previous versions of this manuscript. P.W. received a Whitehall Foundation Fellowship and a Autism Speaks Research Award; T.P. received an Autism Speaks Postdoctoral Fellowship; and C.E.-N. received a NIH Developmental Biology Training Grant.
Grant sponsor: Whitehall Foundation; Grant sponsor: Autism Speaks; Grant sponsor: NIH.
Footnotes
The Supplementary Material referred to in this article can be found at http://www.interscience.wiley.com/jpages/1058-8388/suppmat
REFERENCES
- Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2006;87:139–150. doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC. SynCAM, a synaptic adhesion molecule thatdrivessynapseassembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Drapeau P, Saint-Amant L, Buss RR, Chong M, McDearmid JR, Brustein E. Development of the locomotor network in zebrafish. Prog Neurobiol. 2002;68:85–111. doi: 10.1016/s0301-0082(02)00075-8. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Colonna M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin Cancer Biol. 2006;16:359–366. doi: 10.1016/j.semcancer.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Fujita E, Urase K, Soyama A, Kouroku Y, Momoi T. Distribution of RA175/TSLC1/SynCAM, a member of the immunoglobulin superfamily, in the developing nervous system. Brain Res Dev Brain Res. 2005;154:199–209. doi: 10.1016/j.devbrainres.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Ozeki S, Tanabe Y, Toyama Y, Maekawa M, Kojima N, Senoo H, Toshimori K, Momoi T. Oligo-astheno-teratozoospermia in mice lacking RA175/TSLC1/SynCAM/IGSF4A, a cell adhesion molecule in the immunoglobulin superfamily. Mol Cell Biol. 2006;26:718–726. doi: 10.1128/MCB.26.2.718-726.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara H, Masuda M, Yageta M, Fukami T, Kuramochi M, Maruyama T, Kitamura T, Murakami Y. Association of a lung tumor suppressor TSLC1 with MPP3, a human homologue of Drosophila tumor suppressor Dlg. Oncogene. 2003;22:6160–6165. doi: 10.1038/sj.onc.1206744. [DOI] [PubMed] [Google Scholar]
- Furuno T, Ito A, Koma Y, Watabe K, Yokozaki H, Bienenstock J, Nakanishi M, Kitamura Y. The spermatogenic Ig superfamily/synaptic cell adhesion molecule mast-cell adhesion molecule promotes interaction with nerves. J Immunol. 2005;174:6934–6942. doi: 10.4049/jimmunol.174.11.6934. [DOI] [PubMed] [Google Scholar]
- Gruber-Olipitz M, Yang JW, Slavc I, Lubec G. Nectin-like molecule 1 is a high abundance protein in cerebellar neurons. Amino Acids. 2006;30:409–415. doi: 10.1007/s00726-006-0323-0. [DOI] [PubMed] [Google Scholar]
- Hale ME, Ritter DA, Fetcho JR. A confocal study of spinal interneurons in living larval zebrafish. J Comp Neurol. 2001;437:1–16. doi: 10.1002/cne.1266. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, Hudson TJ, Zon LI, McPherson JD, Chevrette M, Dawid IB, Johnson SL, Ekker M. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci U S A. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Oonuma J. Direct interaction between nerves and mast cells mediated by the SgIGSF/SynCAM adhesion molecule. J Pharmacol Sci. 2006;102:1–5. doi: 10.1254/jphs.cpj06014x. [DOI] [PubMed] [Google Scholar]
- Ito A, Jippo T, Wakayama T, Morii E, Koma Y, Onda H, Nojima H, Iseki S, Kitamura Y. SgIGSF: a new mast-cell adhesion molecule used for attachment to fibroblasts and transcriptionally regulated by MITF. Blood. 2003;101:2601–2608. doi: 10.1182/blood-2002-07-2265. [DOI] [PubMed] [Google Scholar]
- Ito A, Hagiyama M, Oonuma J, Murakami Y, Yokozaki H, Takaki M. Involvement of the SgIGSF/Necl-2 adhesion molecule in degranulation of mesenteric mast cells. J Neuroimmunol. 2007a;184:209–213. doi: 10.1016/j.jneuroim.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Ito A, Nishikawa Y, Ohnuma K, Ohnuma I, Koma Y, Sato A, Enomoto K, Tsujimura T, Yokozaki H. SgIGSF is a novel biliary-epithelial cell adhesion molecule mediating duct/ductule development. Hepatology. 2007b;45:684–694. doi: 10.1002/hep.21501. [DOI] [PubMed] [Google Scholar]
- Jensen AM, Walker C, Westerfield M. mosaic eyes: a zebrafish gene required in pigmented epithelium for apical localization of retinal cell division and lamination. Development. 2001;128:95–105. doi: 10.1242/dev.128.1.95. [DOI] [PubMed] [Google Scholar]
- Kakunaga S, Ikeda W, Itoh S, Deguchi-Tawarada M, Ohtsuka T, Mizoguchi A, Takai Y. Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci. 2005;118:1267–1277. doi: 10.1242/jcs.01656. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69:419–449. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Maurel P, Einheber S, Galinska J, Thaker P, Lam I, Rubin MB, Scherer SS, Murakami Y, Gutmann DH, Salzer JL. Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol. 2007;178:861–874. doi: 10.1083/jcb.200705132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Muller T, Wulliman MF. Atlas of early zebrafish brain development, a tool for molecular neurogenetics. Amsterdam: Elsevier; 2005. p. 183. [Google Scholar]
- Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Itoh K, Yaoi T, Tando S, Fukui K, Fushiki S. Spatiotemporal patterns of expression of IGSF4 in developing mouse nervous system. Brain Res Dev Brain Res. 2005;156:23–31. doi: 10.1016/j.devbrainres.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Postlehwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, Sudhof TC, Kavalali ET. Selective capability of Syn-CAM and neuroligin for functional synapse assembly. J Neurosci. 2005;25:260–270. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J Comp Neurol. 1994;344:532–542. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci. 2007;10:861–869. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655–667. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Arends MJ, Chausiaux OE, Ellis PJ, Lange UC, Surani MA, Affara N, Murakami Y, Adams DJ, Bradley A. Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis. Mol Cell Biol. 2006;26:3595–3609. doi: 10.1128/MCB.26.9.3595-3609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Ohashi K, Mizuno K, Iseki S. Cloning and characterization of a novel mouse immunoglobulin superfamily gene expressed in early spermatogenic cells. Mol Reprod Dev. 2001;60:158–164. doi: 10.1002/mrd.1072. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Koami H, Ariga H, Kobayashi D, Sai Y, Tsuji A, Yamamoto M, Iseki S. Expression and functional characterization of the adhesion molecule spermatogenic immunoglobulin superfamily in the mouse testis. Biol Reprod. 2003;68:1755–1763. doi: 10.1095/biolreprod.102.012344. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book; a guide for the laboratory use of zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Wulliman MF, Rupp B, Reichert H. Neuroanatomy of the zebrafish brain, a topological atlas. Basel: Birkhauser Verlag; 1996. [Google Scholar]
- Yageta M, Kuramochi M, Masuda M, Fukami T, Fukuhara H, Maruyama T, Shibuya M, Murakami Y. Direct association of TSLC1 and DAL-1, two distinct tumor suppressor proteins in lung cancer. Cancer Res. 2002;62:5129–5133. [PubMed] [Google Scholar]
- Yamada D, Yoshida M, Williams YN, Fukami T, Kikuchi S, Masuda M, Maruyama T, Ohta T, Nakae D, Maekawa A, Kitamura T, Murakami Y. Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule. Mol Cell Biol. 2006;26:3610–3624. doi: 10.1128/MCB.26.9.3610-3624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.