Abstract
Extracellular superoxide dismutase (EC-SOD) is the major antioxidant enzyme present in the vascular wall, and is responsible for both the protection of vessels from oxidative stress and for the modulation of vascular tone. Concentrations of EC-SOD in human pulmonary arteries are very high relative to other tissues, and the expression of EC-SOD appears highly restricted to smooth muscle. The molecular basis for this smooth muscle–specific expression of EC-SOD is not known. Here we assessed the role of epigenetic factors in regulating the cell-specific and IFN-γ–inducible expression of EC-SOD in human pulmonary artery cells. The analysis of CpG site methylation within the promoter and coding regions of the EC-SOD gene demonstrated higher levels of DNA methylation within the distal promoter region in endothelial cells compared with smooth muscle cells. Exposure of both cell types to DNA demethylation agents reactivated the transcription of EC-SOD in endothelial cells alone. However, exposure to the histone deacetylase inhibitor trichostatin A (TSA) significantly induced EC-SOD gene expression in both endothelial cells and smooth muscle cells. Concentrations of EC-SOD mRNA were also induced up to 45-fold by IFN-γ in smooth muscle cells, but not in endothelial cells. The IFN-γ–dependent expression of EC-SOD was regulated by the Janus tyrosine kinase/signal transducers and activators of transcription proteins signaling pathway. Simultaneous exposure to TSA and IFN-γ produced a synergistic effect on the induction of EC-SOD gene expression, but only in endothelial cells. These findings provide strong evidence that EC-SOD cell-specific and IFN-γ–inducible expression in pulmonary artery cells is regulated, to a major degree, by epigenetic mechanisms that include histone acetylation and DNA methylation.
Keywords: extracellular superoxide dismutase, promoter, epigenetic, transcription, pulmonary arteries, endothelial cells, smooth muscle cells
Clinical Relevance
We identified the critical role of epigenetic factors in regulating the expression of human extracellular superoxide dismutase (EC-SOD) in pulmonary arteries. Because oxidative stress contributes to the development of pulmonary hypertension and other pulmonary diseases, understanding factors that regulate the expression and activity of EC-SOD in the lung may provide novel therapeutic strategies.
The extracellular superoxide dismutase (EC-SOD) enzyme is the main antioxidant enzyme in the lung and in the vascular wall. In the lung, its expression is restricted primarily to Type II pneumonocytes and fibroblasts, whereas in healthy vessels, EC-SOD is expressed almost exclusively by smooth muscle cells (1), with little to no detectable expression in endothelial cells (2). Vascular EC-SOD is mostly localized in the space between the endothelium and smooth muscle cells, where its concentration may be 3,000 times higher than in the surrounding spaces (3). The very high concentrations of EC-SOD in the vascular wall suggest that this enzyme plays a critical role in the regulation of vascular tone by modulating the extracellular redox state near the juncture of the endothelium and smooth muscle cells. On a physiological level, EC-SOD was shown to protect arteries from the detrimental effects of superoxide radicals in endothelium-dependent relaxation (4), to reduce angiotensin-induced blood pressure (5), and to attenuate the size of infarcts (6). The most recent findings regarding the role of EC-SOD indicate that it plays an important role in the regulation of vascular tone during the oxidative stress produced by angiotensin II in cerebral arteries (7), protects renal arteries against oxidative stress by attenuating the expression of p22phox (8), and protects against the vascular dysfunction associated with aging (9, 10). Furthermore, the overexpression of EC-SOD attenuated the development of pulmonary hypertension and vascular remodeling in mice exposed to chronic hypoxia and in mice exposed to bleomycin (11, 12). In addition, many cardiovascular pathophysiological conditions are known to be associated with an increased production of superoxide radicals, which, in turn, can undergo an extremely rapid reaction with nitric oxide, resulting in the production of the peroxynitrite anion (ONOO−) (13). Thus, in vessel walls, EC-SOD not only regulates the pool of bioavailable NO, but also prevents the formation of highly toxic peroxynitrite.
Despite the obviously important role EC-SOD plays in vascular protection and the regulation of its functions, remarkably little is known about the molecular mechanisms involved with EC-SOD's cell-specific and inducible expression in lung tissues. The basal and inducible expression of murine EC-SOD was shown to be regulated by the transcription factors specificity protein (Sp) 1, Sp3, E-twenty six, Kruppel-like, and myeloid zinc finger–1, which bind to the corresponding cis-element in the promoter region (14, 15). A new promoter for human EC-SOD was more recently identified, and was shown to interact with the Sp1 and Sp3 transcription factors, further contributing to its basal expression (16). Previously, we showed that a lack of EC-SOD gene expression in a pulmonary adenocarcinoma cell line (A549) was mostly attributable to the hypermethylation of its promoter that prevents the binding of the Sp1/Sp3 transcription factor to its putative cis-elements (17). Similar epigenetic silencing mechanisms were described for the expression of manganese superoxide dismutase in breast cancer cells (18) In addition, the expression of EC-SOD can be modulated by copper through the activation of the copper chaperone antioxidant–1 (Atox1). Atox1 binds directly to the GAAAGA sequence in the EC-SOD promoter in a copper-dependent manner (19). Nevertheless, the molecular mechanism for the cell-specific and inducible expression of EC-SOD in pulmonary artery cells remains unresolved.
IFN-γ is a major proinflammatory and antiviral cytokine that produces multiple effects in vascular cells through an orchestration of expression of a wide range of genes. In the arteries, IFN-γ is predominantly produced by CD4+ and CD8+ T cells, as well as by natural killer cells, dendritic cells, macrophages, and smooth muscle cells (20). After vascular responses to injury, IFN-γ is secreted into the arterial intima, where it can modulate the proliferation of many cell types, including vascular smooth muscle cells. Earlier studies sought to elucidate the role of IFN-γ in the regulation of smooth muscle proliferation, and reported on its antiproliferative effects (21). This antiproliferative effect of IFN-γ was successfully used to reduce arterial stenosis in a rat model of vascular injury (22). More recent studies using in vivo models indicated that IFN-γ can induce a robust proliferation of vascular cells (23, 24). Despite its controversial effects on the proliferation of vascular cells, IFN-γ is a key proinflammatory mediator that is expressed at high concentrations in atherosclerotic lesions. IFN-γ profoundly contributes to changes in levels of oxidative stress in the vascular wall, mostly through increasing the production of endothelial-derived NO by inducing the expression of iNOS (25). It also stimulates the secretion of reactive oxygen species (ROS) in the vascular wall by up-regulating the expression of NADPH oxidase and xanthine oxidase (26, 27). Thus, although the role of IFN-γ in regulating the proliferation and inflammation of vascular cells remains controversial, it has become more obvious that IFN-γ can significantly alter concentrations of reactive oxygen and nitrogen species in the tunica intima and tunica media regions of the vascular wall. Because EC-SOD is a major antioxidant enzyme in the pulmonary arteries, we investigated the molecular mechanisms that govern its cell-specific and IFN-γ–dependent expression.
Materials and Methods
Reagents
Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Human IFN-γ and human TNF-α were purchased from R&D Systems (Minneapolis, MN). Janus tyrosine kinase (JAK) inhibitor I, AG 490 (a JAK2 inhibitor), and the inhibitor of signal transducers and activators of transcription proteins–3 (STAT3) were from Calbiochem (Gibbstown, NJ). All other chemicals and enzymes were from Boehringer Mannheim (Indianapolis, IN), Sigma Chemical Co. (St. Louis, MO), or Invitrogen (Carlsbad, CA).
Quantitative RT-PCR
Total RNA was prepared from cultured cells, using an RNAqueous-Micro Kit (Applied Biosystems, Foster City, CA). The synthesis of single-stranded DNA from RNA was performed using the SuperScript First-Strand Synthesis System for RT-PCR and random hexamers (Invitrogen), according to the manufacturer's protocols. To quantitate the abundance of gene-specific mRNAs, quantitative PCR was undertaken using the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and an iQ SYBR Green Master Mix. The PCR cycles involved 95°C for 3 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. The EC-SOD primers included forward (5′-TGC CCC GCG TCT TCA G -3′) and reverse (5′-CCA AAC ATT CCC CCA AAG G -3′). The human gp91phox primers included forward (5′- GTC ACA CCC TTC GCA TCC ATT CTC AAG TCA GT-3′) and reverse (5′- CTG AGA CTC ATC CCA GCC AGT GAG GTA G-3′). The human interferon responsive factor–1 (IRF-1) primers included forward (5′-GAT GAT CTT CCA GAT CCC AT-3′) and reverse (5′-TCT TTC ACC TCC TCG ATA TC-3′). PCR assays were run in triplicate, and concentrations of EC-SOD, gp91phox, and IRF-1 mRNA were normalized to concentrations of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The GAPDH primers included forward (5′-CAT GGA CTG TGG TCA TGA GT-3′) and reverse (5′-CCA TGT TCG TCA TGG GTG TGA-3′).
Bisulfite Genomic Sequencing to Analyze the Methylation Patterns of CpG Sites within the EC-SOD Promoter and Coding Regions
Genomic DNA was isolated from human pulmonary artery endothelial cells (HPAECs) and smooth muscle cells (HPASMCs), using a DNeasy Kit (Qiagen, Chatsworth, CA). The bisulfite modification of genomic DNA was performed with an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA), according to the manufacturer's protocol. An aliquot of the bisulfite-treated DNA (25–50 ng) was subjected to 35 cycles of PCR amplification. PCR primers for the amplification of the EC-SOD promoter were designed for the sodium bisulfite–modified template, using MethPrimer software (28). The PCR primer sets used in sodium bisulfite genomic sequencing to amplify the coding and 5′-flanking regions are presented in Table 1. The PCR fragments were subcloned using a TA cloning kit (Invitrogen), and at least 10 individual plasmid clones were sequenced.
TABLE 1.
PRIMERS USED FOR AMPLIFICATION OF SODIUM BISULFITE–TREATED GENOMIC DNA
| Primer Name | Sequence | Amplicon Location | Amplicon Size |
| Forward CpG (−848) | 5′-AGTTTTTATTTTTGGAAAGGGTTGT-3′ | 5′-flanking region | 367 bp |
| Reverse CpG (−481) | 5′-ACTCAAACTACAAAATCCCAACCT-3′ | ||
| Forward CpG (−506) | 5′-TAAGGTTGGGATTTTGTAGTTTGAG-3′ | 5′-flanking region | 472 bp |
| Reverse-CpG (−183) | 5′-CTACCCCTCCCATTTTTAAATTTT-3′ | ||
| Forward CpG (−204) | 5′-AATTTAAAAATGGGAGGGGTAGAG-3′ | 5′-flanking region | 246 bp |
| Reverse CpG (+42) | 5′-AAAAAACTTTCTCTCCTAAAAAAAA-3′ | ||
| Forward CpG (+3937) | 5′-TGTGTTTTTGTTTGTTTTTGGTAGT-3′ | Coding region | 139 bp |
| Reverse CpG (+4076) | 5′-CTACATAACCTCCTACCAAATCTCC-3′ | ||
| Forward CpG (+4054) | 5′-AGATTTGGTAGGAGGTTATGTAG-3′ | Coding region | 243 bp |
| Reverse CpG (+4297) | 5′-CAACCCTAACTCAAATCCCC-3′ | ||
| Forward CpG (+4278) | 5′-GGGGATTTGAGTTAGGGTTG-3′ | Coding region | 238 bp |
| Reverse CpG (+4516) | 5′-TTCTCCACGCTAACCTAATTAC-3′ | ||
| Forward CpG (+4378) | 5′-GCGACGGTAGTTTTTGGAGGTAT-3′ | Coding region | 176 bp |
| Reverse CpG (+4554) | 5′-CCACCACGCAGCAAACCAAC-3′ | ||
| Forward CpG (+4497) | 5′-AATTAGGTTAGCGTGGAGAA-3′ | Coding region | 209 bp |
| Reverse CpG (+4706) | 5′-AAACTCAAAAACAAAAAAAAAC-3′ |
Definition of abbreviations: bp, base pairs.
Results
Analysis of EC-SOD mRNA Expression in Different Tissues and Cells
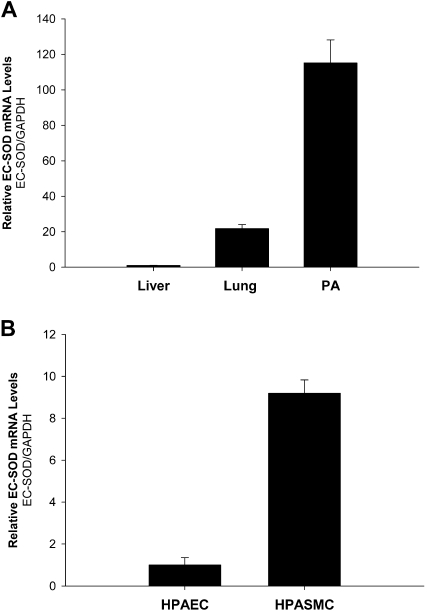
To evaluate baseline levels of EC-SOD expression, we purified total RNA from several murine tissues and performed quantitative RT-PCR. We found that the expression of EC-SOD was significantly higher in lung tissue versus liver tissue. Furthermore, concentrations of EC-SOD–specific mRNA in the pulmonary artery were six times higher than in lung tissue (Figure 1A). Next, we analyzed the expression of EC-SOD in primary pulmonary artery endothelial and smooth muscle cells. The concentrations of EC-SOD mRNA were ninefold higher in HPASMCs compared with endothelial cells (Figure 1B).
Figure 1.
Expression of extracellular superoxide dismutase (EC-SOD) in different tissues and cells. (A) Expression of EC-SOD in liver, lung, and pulmonary arteries (PA). (B) Analysis of EC-SOD expression in human pulmonary artery endothelial cells (HPAECs) and in human pulmonary artery smooth muscle cells (HPASMCs). Total RNA from liver, lung, and pulmonary artery cells was isolated and purified. Real-time PCR was used to determine relative concentrations of EC-SOD mRNA in those tissues and cells. Concentrations of EC-SOD mRNA were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). At least four independent amplifications were performed for each analysis. The results shown represent the mean ± SD.
Methylation Pattern of the EC-SOD Promoter in HPASMCs and HPAECs
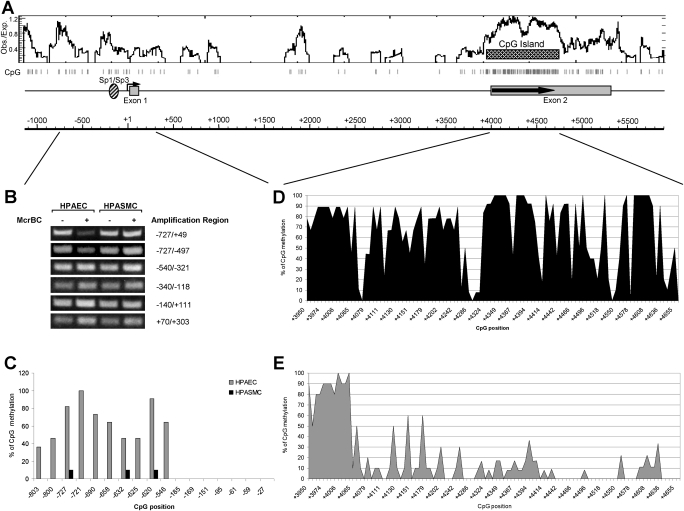
We suggested, based on previous studies, that differences in the cell-specific expression of EC-SOD may be regulated by epigenetic factors. Thus, we investigated whether the methylation of DNA in the promoter or coding regions could be responsible for the differential expression of EC-SOD in pulmonary artery cells. For this purpose, we purified genomic DNA from HPAECs and HPASMCs, and digested it with methylation-requiring nuclease McrBC. Subsequent PCR amplification of the EC-SOD promoter region from −727 to +49 indicated that HPASMC genomic DNA is more resistant to McrBC cleavage compared with HPAECs, implying that the 5′-flanking region of the human EC-SOD gene is methylated to a higher degree in HPAECs versus HPASMCs (Figure 2B). To delineate further the methylated regions in HPAECs, we amplified genomic DNA with primer pairs that spanned shorter DNA sequences. Only the primer pair that amplified the distal part of the EC-SOD promoter (from −727 to −497) produced less prominent bands from McrBC-treated, HPAEC-derived genomic DNA (Figure 2B). These data indicate that only the distal promoter region of the EC-SOD gene is methylated at a higher rate in endothelial cells compared with smooth muscle cells.
Figure 2.
Methylation status of CpG sites within 5′-flanking and coding regions of the EC-SOD gene. (A) Schematic representation of EC-SOD gene organization. The distribution of CpG sites within EC-SOD genomic DNA are shown as vertical gray bars. The ratio of observed versus expected CpG frequency is plotted, and the CpG island is marked as a pattern bar. (B) McrBC-based assay. The genomic DNA from HPAECs and HPASMCs was purified and digested with McrBC methylation–sensitive nuclease. The integrity of DNA was analyzed using primer pairs that amplify different region of the EC-SOD promoter. Amplified products were separated on 1.2% agarose gel and visualized using ethidium bromide under ultraviolet light. (C) Methylation status of CpG sites within the EC-SOD promoter, using bisulfite sequencing. At least 10 individual clones were sequenced for each cell type. (D and E) Methylation status of CpG sites within the EC-SOD coding region in HPASMCs (D) and HPAECs (E). From 9–12 clones were sequenced for each CpG site.
To analyze the methylation status of individual CpG sites in regions from −848 to +42, we used a genomic bisulfite sequencing method. CpG sites located in the region from −185 to −27 were not methylated in either cell line (Figure 2C). Analysis of the 5′-flanking region from −803 to −546 indicated a significantly higher percentage of methylated cytosines in endothelial cells compared with smooth muscle cells. For example, only three CpG sites at positions −727, −632, and −620 were methylated at a frequency of 10% in smooth muscle cells, whereas all these CpG sites were methylated at frequencies between 40–100% in endothelial cells. These data are in good agreement with our McrBC results. The importance of this differential DNA methylation is not clear, because no transcriptionally important elements were detected in this region of the EC-SOD gene.
Next, we analyzed the methylation status of CpG dinucleotides in the coding region. The EC-SOD gene exhibits a very high density of CpG sites within its coding region, forming a “CpG island.” CpG islands are usually associated with transcriptionally active regions of chromatin. The high frequency of methylated CpG sites (from 80–100%) in both cells was evident only in the region from +3950 to +4068 base pairs (bp). The rest of the downstream coding region was methylated at a higher frequency in HPASMCs compared with HPAECs. The importance of this differentially methylated region for regulating EC-SOD gene expression in human pulmonary artery cells is unclear at present.
Modulation of EC-SOD Gene Expression by the Demethylation Agent, 5-Azacytidine, and by the Histone Deacetylase Inhibitor, Trichostatin A
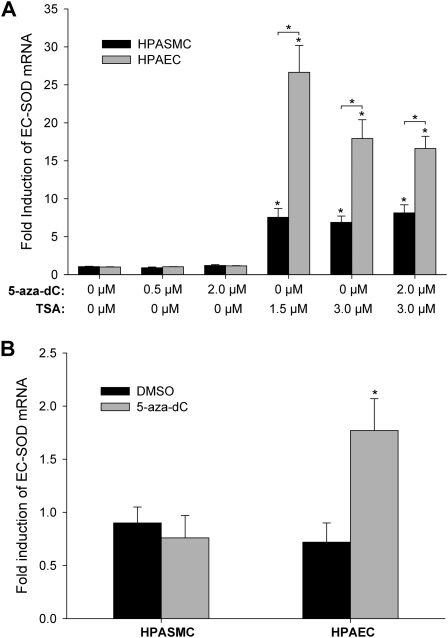
To investigate further the significance of methylation in regulating the cell-specific expression of EC-SOD in the pulmonary artery, HPAECs and HPASMCs were treated with 0.5 μM and 2 μM of the demethylating agent 5-azacytidine (5-aza-dC) for 48 hours. Concentrations of EC-SOD mRNA were unaffected by this treatment in both cell lines (Figure 3A). However, when cells were exposed to 2 μM of 5-aza-dC for a prolonged time (96 hours), concentrations of EC-SOD–specific mRNA increased up to 2.5-fold in endothelial cells but not in smooth muscle cells (Figure 3B). During this extended timeframe, cells grew from a confluence of approximately 30–35% to almost 100%, which would correspond to an approximately 1.5- to 2-fold population doubling. The differences in fold of EC-SOD gene induction between the 48-hour and 96-hour exposures can be explained by the mechanism of 5-aza-dC action. This inhibitor does not cause active DNA demethylation, but rather leads to passive demethylation through multiple rounds of cell division in the absence of DNA methyl transferase activity. Thus, only prolonged exposure to 5-aza-dC, during which cells were able to go through several division cycles, resulted in an up-regulation of EC-SOD expression.
Figure 3.
Acetylation status of histones is important for cell-specific expression of EC-SOD. (A) HPAECs and HPASMCs were exposed to 5-aza-2′-deoxycytidine (5-aza-dC; 0.5 μM and 2 μM) or trichostatin A (TSA; 1.5 μM and 3 μM), or a combination of both substances for 48 hours. (B) Cells were exposed to DMSO or 2 μM of 5-aza-dC for 96 hours. Total RNA was extracted, and EC-SOD mRNA was measured by quantitative real-time RT-PCR. The data were normalized to concentrations of GAPDH mRNA. The results shown represent the mean ± SD from at least two independent experiments. *P < 0.01, significant difference, according to ANOVA/Bonferroni post hoc test.
In addition, we analyzed the role of histone modifications in the regulation of EC-SOD expression. The exposure of HPAECs and HPASMCs to the histone deacetylase inhibitor significantly induced the expression of EC-SOD in both cell lines. In HPAECs, concentrations of EC-SOD mRNA were up-regulated by 26.6 ± 3.6-fold and 17.9 ± 2.5-fold after exposure to 1.5 μM and 3 μM trichostatin A (TSA), respectively. The magnitude of induction in HPASMCs was significantly lower, reaching only 7.5 ± 1.2-fold and 6.9 ± 0.8-fold induction, respectively (Figure 3). The simultaneous treatment of cells with TSA and 5-aza-dC did not change the fold induction of EC-SOD mRNA. These data suggest that the expression of EC-SOD positively correlates with the acetylation status of histones alone. In HPAECs, which express relatively low concentrations of EC-SOD, the induction of EC-SOD by TSA was considerably higher compared with HPASMCs. A strong possibility exists, therefore, that histone modifications within the 5′-flanking or coding regions may regulate the cell-specific expression of EC-SOD in pulmonary arteries.
Induction of EC-SOD Expression by IFN-γ in Pulmonary Vascular Cells
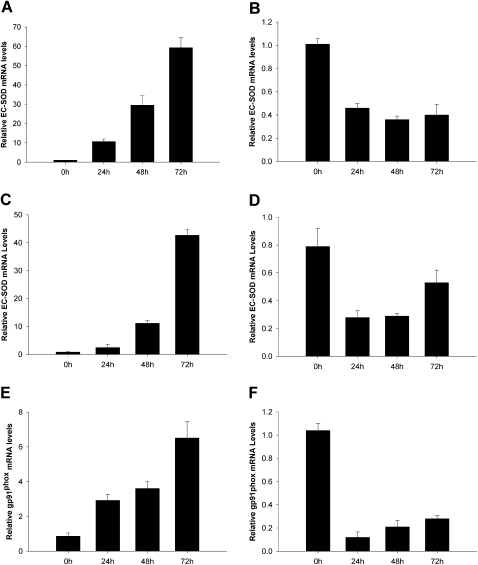
We were interested in the effect of cytokines (in particular, IFN-γ and TNF-α) on the modulation of EC-SOD concentrations in pulmonary cells. To study the effects of these cytokines on EC-SOD gene expression, HPASMCs and MRC5 cells were exposed to 5,000 U/ml of IFN-γ or 30 ng/ml of TNF-α for 24, 48, and 72 hours. As shown in Figure 4A, exposure of the cells to IFN-γ for 24 hours resulted in modest increases of EC-SOD mRNA concentrations, whereas prolonged exposure for up to 72 hours showed significantly increased EC-SOD gene expression (as high as 45-fold). The slow activation course of EC-SOD by IFN-γ suggests that epigenetic factors may be responsible. The exposure of cells to TNF-α attenuated the expression of EC-SOD in both cells (Figures 4B–4D). Interestingly, the expression of the gp91phox subunit of the NADPH oxidase complex, responsible for the generation of ROS in the vascular wall, closely follows the pattern of EC-SOD expression (Figures 4E and 4F). This observation implies that the expression of EC-SOD in vascular smooth muscle cells is coordinated with the expression of gp91phox to maintain concentrations of superoxide radicals in the intima of the vascular wall under nontoxic, physiologic levels.
Figure 4.
Regulation of EC-SOD and gp91phox transcription by cytokines. MRC5 cells (A and B) and HPASMCs (C–F) were exposed to 5,000 U/ml of IFN-γ (A, C, and E) or 30 ng/ml of TNF-α (B, D, and F) for the times indicated. Total RNA was extracted, and EC-SOD and gp91phox mRNA was measured by quantitative real-time RT-PCR. The data were normalized to concentrations of GAPDH mRNA. The results shown represent the mean ± SD from at least two independent experiments.
Signal Transduction Pathways Responsible for the Induction of EC-SOD by IFN-γ
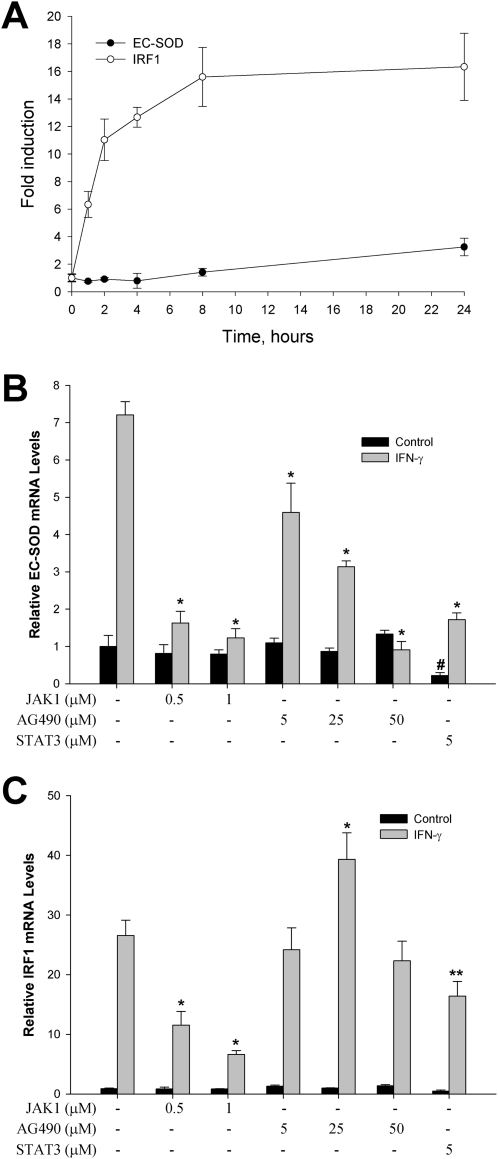
To analyze the signal transduction pathways responsible for the IFN-γ–mediated induction of EC-SOD in HPASMCs, we compared its time-dependent mRNA steady-state concentrations with those of IRF-1. IRF-1 is one of the immediate early genes induced by IFN-γ, predominantly through the JAK/STAT signaling pathway. Figure 5A illustrates that the accumulation of IRF-1 mRNA was detected after 1 hour of exposure to IFN-γ, whereas statistically significant increases in EC-SOD steady-state mRNA concentrations were evident only after a 24-hour exposure. Because IFN-γ is known to activate the JAK/STAT pathway, we analyzed the effects of JAK1 and JAK2 inhibitors on the expression of EC-SOD in HPASMCs. As shown in Figure 5B, the JAK1 inhibitor attenuated IFN-γ–induced EC-SOD gene expression by 96.6% in a dose-dependent manner. The exposure of cells to AG490 (an inhibitor of JAK2 autophosphorylation) also significantly reduced the magnitude of EC-SOD gene induction, starting at a concentration of 5 μM. The basal concentrations of EC-SOD mRNA were unchanged after exposure to JAK1 and JAK2 inhibitors. Interestingly, IRF-1 gene induction was attenuated only by the JAK1 inhibitor, and not by the inhibitor of JAK2 autophosphorylation, AG490 (Figure 5C). A similar dependence on JAK1 activation alone was evident for the induction of IRF-1 by respiratory syncytial virus in A549 cells. Neither IRF-1 promoter activity nor the binding of STAT to the IRF-1 promoter was inhibited by the exposure of cells to AG490 (29).
Figure 5.
Role of Janus tyrosine kinase (JAK)/signal transducers and activators of transcription proteins (STAT) signaling pathway in IFN-γ–dependent induction of EC-SOD. (A) Time course of inducible expression of interferon-responsive factor–1 (IRF-1) and EC-SOD in HPASMCs. Cells were incubated with 5,000 U/ml of IFN-γ for the indicated times. (B and C) Effects of JAK1, JAK2 (AG490), and STAT3 inhibitors on IFN-γ–induced EC-SOD (B) and IRF-1 (C) mRNA concentrations in HPASMCs. Cells were incubated with vehicle (Control) or IFN-γ (5,000 U/ml) in the presence or absence of the tested compound for 48 hours. Total RNA was isolated, and gene-specific mRNA concentrations were analyzed, using quantitative RT-PCR. Concentrations of EC-SOD and IRF-1 mRNA were normalized to the expression of GAPHD. The results shown represent the mean ± SD. *P < 0.001 and **P < 0.01, compared with cells treated with IFN-γ, and #P < 0.001, compared with cells treated with vehicle, according to ANOVA/Bonferroni post hoc test.
Our data indicate that the induction of EC-SOD by IFN-γ in smooth muscle cells required the activation of JAK1 and JAK2 protein kinases. The next step after the activation of JAK kinases involves the phosphorylation and nuclear translocation of the STAT family of transcription factors. We became interested in the role of STAT3 in the activation of EC-SOD transcription because IFN-γ was shown to activate STAT3 only in human vascular smooth muscle cells, and weakly or not at all in human endothelial cells (30). The exposure of HPASMCs to 5 μM of the STAT3 inhibitor attenuated the induction of EC-SOD by 88.4%, from 7.21 ± 0.35-fold to 1.72 ± 0.18-fold (Figure 5B). However, the basal concentration of EC-SOD mRNA was also affected. Based on these results, the STAT3 signaling pathway seems to contribute, in part, to EC-SOD gene induction by IFN-γ in HPASMCs.
Effect of IFN-γ on the Stability of EC-SOD mRNA
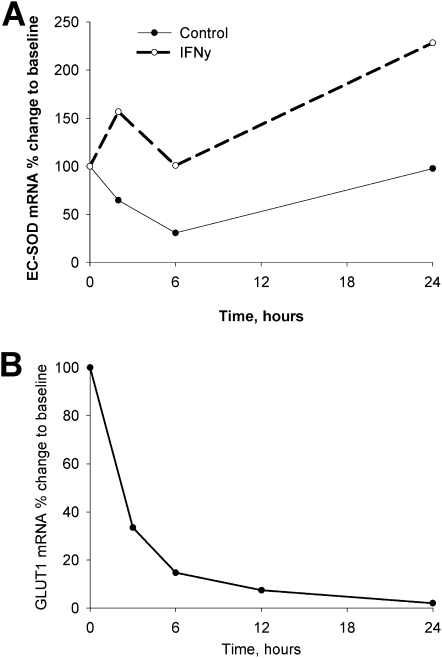
Experiments were performed to determine whether the increased expression after exposure to IFN-γ was likely attributable to transcriptional or posttranscriptional mechanisms. In these experiments, smooth muscle cells were exposed to IFN-γ for 72 hours, followed by the addition of actinomycin D. We found that IFN-γ increased the stability of EC-SOD mRNA only approximately twofold, whereas concentrations of EC-SOD–specific mRNA increased up to 45-fold (Figure 6A). To verify that the high stability of EC-SOD mRNA detected by this assay was not an artifact, we analyzed the half-life of glucose transporter–1 mRNA, and found that it corresponds to the previously published half-life of 2–2.5 hours (Figure 6B) (31). Based on these data, we conclude that the induction of EC-SOD gene expression by IFN-γ is most likely regulated at the transcriptional level.
Figure 6.
Stability of EC-SOD mRNA after exposure to IFN-γ. (A) HPASMCs were exposed to IFN-γ (5,000 U/ml) or PBS for 72 hours, followed by treatment with actinomycin D (10 μg/ml). Total RNA was collected at the times indicated, and concentrations of EC-SOD mRNA were determined by quantitative RT-PCR. (B) Stability of glucose transporter–1 (GLUT1) mRNA under control conditions is shown for comparison with mRNA stability. The data shown represent at least two independent experiments.
Role of Histone Acetylation in the Regulation of IFN-γ–Inducible EC-SOD Expression in HPASMCs
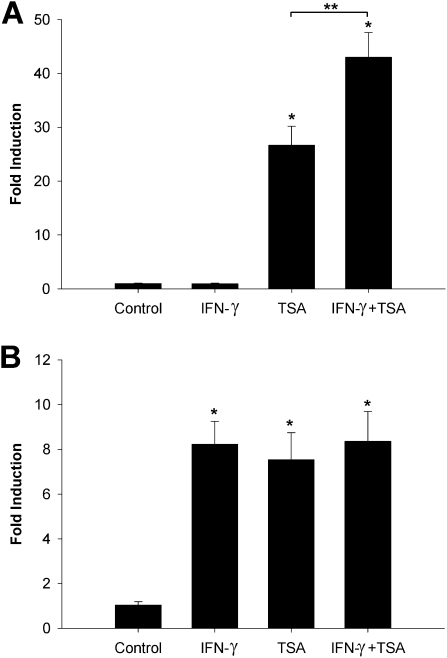
Next, we analyzed whether the status of histone acetylation will have any effect on IFN-γ–dependent expression in pulmonary artery cells. To increase histone acetylation in cells, we used the histone deacetylase inhibitor TSA. The treatment of both cell lines with 1.5 μM TSA for 48 hours increased concentrations of EC-SOD mRNA up to 26-fold in HPAECs, and eightfold in HPASMCs (Figures 7A and 7B). IFN-γ alone was able to activate the transcription of EC-SOD only in HPASMCs. The exposure of smooth muscle cells to both agents did not produce any synergistic effects (Figure 7B). However, the simultaneous exposure of HPAECs to both agents produced an additive activating effect, enhancing the expression of EC-SOD from 26.64 ± 3.54-fold to 42.96 ± 4.61-fold, suggesting that histone modifications and chromatin organization play an important role in the regulation of IFN-γ–inducible EC-SOD expression in vascular endothelial cells.
Figure 7.
Role of histone acetylation in IFN-γ–inducible EC-SOD expression. HPAECs (A) and HPASMCs (B) were exposed to either IFN-γ (5,000 U/ml) or TSA (1.5 μM) or a combination of both substances for 48 hours. Total RNA was extracted, and EC-SOD mRNA was measured by quantitative real-time RT-PCR. The data were normalized to concentrations of GAPDH mRNA. The results shown represent the mean ± SD from at least two independent experiments. *P < 0.001, compared with nontreated cells, according to ANOVA/Bonferroni post hoc test. **P < 0.01, according to t test.
Discussion
Our results demonstrate that the expression of EC-SOD can be induced by IFN-γ in both a dose-dependent and time-dependent manner via the activation of JAK/STAT signaling pathways. This induction is, at least in part, selective for HPASMCs, where the basal level of EC-SOD expression is already higher compared with that in endothelial cells. In addition, we show, for the first time to the best of our knowledge, that histone acetylation and DNA methylation are involved in the cell-specific expression of EC-SOD in pulmonary vascular cells. EC-SOD is well-documented to regulate vascular tone by controlling the concentrations of superoxide radicals within the vascular wall. Superoxide radicals interact with nitric oxide at very rapid, diffusion-limited rates to produce peroxynitrate. This reaction not only inactivates the vasodilating activity of nitric oxide, but can also modify vascular proteins, lipids, and nucleic acids. The nitration of vascular proteins in tyrosine residues and the oxidation of methionine and cysteine residues can produce profound effects on signal transduction pathways and vascular responsiveness (32, 33). In this regard, we decided to investigate the molecular mechanisms that govern the expression of EC-SOD in vascular cells, and to elucidate potential epigenetic approaches to modify the expression of EC-SOD, using pharmacologic agents.
Our previously published data indicate that DNA methylation is important in the repression of EC-SOD expression in pulmonary adenocarcinoma A549 cells (17). We and others determined that EC-SOD is expressed at very high concentrations in pulmonary arteries, specifically in pulmonary artery smooth muscle cells. Therefore, we analyzed the role of DNA methylation in the suppression of EC-SOD production in endothelial cells. DNA methylation is an important regulator of gene expression. In mammals, the primary targets for methylation are cytosines, followed by guanosine bases. The majority of CpG dinucleotides are methylated and distributed throughout all the genome. However, the distribution pattern of CpG sites is not random throughout different genetic elements, including genes, noncoding sequences, and transposones. The promoters and 5′-flanking regions of many genes exhibit a high frequency of CpG sites (∼1 CpG per 10 bp), compared with 1 CpG site per 100 bp on average for the rest of the genome. These high-density CpG regions are called “CpG islands” (CGIs), and tend to be hypomethylated. The EC-SOD promoter and 5′-flanking region do not contain any CGIs, but the coding region is characterized by very high frequencies of CpG sites. This feature of EC-SOD cDNA attracted our attention as a potential epigenetically driven regulatory element. An analysis of DNA methylation within the coding region of the EC-SOD gene in smooth muscle and endothelial cells showed substantial differences. In pulmonary smooth muscle cells, the percentage of methylated cytosines was considerably lower compared with that in endothelial cells. The functional significance of these differences is supported by the finding that cytosine demethylation agents were able to reactivate EC-SOD gene transcription only in endothelial cells. Thus, this differential pattern of DNA methylation may regulate the cell-specific expression of EC-SOD in pulmonary arteries. Recently, Maunakea and colleagues demonstrated that the majority of methylated CGIs are associated with intragenic and intergenic regions, but not with promoters (34). They concluded that DNA methylation does not seem to play a major role in gene regulation by 5′ CGI promoters. These intergenic methylated CGIs can regulate the levels of alternative transcripts expressed in a cell-specific or tissue-specific fashion. The analysis of CpG sites within the EC-SOD promoter region found no differences in their methylation frequencies, and a very low percentage of them were methylated. The most striking difference in the methylation of cytosines was observed in the distal promoter, where CpG sites were mostly methylated in endothelial cells and completely unmethylated in smooth muscle cells. This pattern of methylation is in good agreement with current hypotheses stating that hypermethylated promoter regions are associated with repressive chromatin structures. It would be interesting to analyze the presence of cis-regulatory elements in this region of the distal promoter.
IFN-γ is capable of activating a number of genes through the JAK and STAT pathways, and also of binding transcription factors to upstream regions or enhancers of inducible genes in specific DNA regulatory elements, known as the interferon-stimulated response element (ISRE) and the γ-activated sequence (GAS). IFN-γ was shown to induce the expression of EC-SOD in human vascular smooth muscle cells (35). A simultaneous instillation of IFN-γ and TNF-α into the lungs of rats induced the expression of EC-SOD through, at least in part, an NF-κB–dependent mechanism (36). Our data indicate a robust induction of EC-SOD in pulmonary fibroblasts and in smooth muscle cells of human pulmonary arteries. Thus, we analyzed the presence and location of ISRE with the consensus sequence 5′-A/GNGAANNGAAACT-3′, and of GAS with the consensus sequence 5′-TTCN(2–4)GAA-3′, in close proximity to the EC-SOD gene. We found several GAS sequences located within the 5′-flanking region and near transcription start sites. However, cloning 1,000 bp of the EC-SOD 5′-flanking region in front of the luciferase reporter gene did not produce an IFN-γ–responsive construct (data not shown). Multiple GAS sequences were found within the intronic region and the 3′-flanking region of the EC-SOD gene. In addition, we found sequence 5′-AGG AAA AGG AAt CT-3′, located in close proximity to transcription start sites, and with a very close resemblance to ISRE apart from only one mismatch (T instead of A). However, analysis of the functional importance of these cis-elements in the IFN-γ–dependent induction of EC-SOD requires further investigation.
How histone modifications and chromatin organization regulate the IFN-γ–inducible expression of EC-SOD remain unclear at present. In one putative mechanism, the exposure of cells to IFN-γ can change the nucleosomal organization within the EC-SOD promoter region. We showed that the Sp1 and Sp3 transcription factors bind to the proximal promoter and facilitate transcription of the EC-SOD gene in pulmonary cells. The relaxation of nucleosomes in this region may expose Sp1/Sp3 binding sites, and allow interactions with their trans-factors. A good example of nucleosomal reorganization within the Sp1 binding site of the IL-12 promoter after exposure to LPS and IFN-γ was described elsewhere (37). However, our data (not shown) indicate that the Sp1/Sp3–specific cis-element located in the proximal promoter is not likely to participate in the IFN-γ–mediated induction of the EC-SOD gene in vascular cells.
Another model suggests that exposure of endothelial cells to TSA increases the accessibility of some unidentified IFN–responsive elements to IFN-γ–inducible transcription factors, which can facilitate the transcription of EC-SOD. Exposure to cytokines was shown to induce a notable perturbation in the acetylation and methylation state of histones in specific regions of the genome. Similar synergistic effects of histone deacetylase inhibitors were described for a number of IFN-γ–inducible genes in murine trophoblast cells (38) and for the reactivation of the caspase-8 gene in cancer cells (39).
In our study, we observed a strikingly similar regulation of gp91phox expression in response to IFN-γ and TNF-α exposure. The main catalytic, membrane-bound gp91phox subunit (renamed Nox2) of the NADPH oxidase complex is mostly expressed in neutrophils, and is involved in the production of superoxide fluxes designed to kill invading pathogens. Recently, vascular smooth muscle cells were also shown to produce superoxide, mostly through NADPH oxidase activity. The production of superoxide radicals in the vascular wall at low concentrations is beneficial for vascular physiology because of their role in the regulation of multiple signaling pathways. However, very high concentrations of ROS can be detrimental for vascular cells. The IFN-γ–induced production of NO, combined with a burst in O2− concentrations, can lead to the formation of peroxynitrite, a strong oxidizing molecule that can induce an inflammatory response in surrounding cells and tissues (40). Hence, the orchestrated production of superoxide radicals and their dismutation may be a well-designed mechanism to maintain appropriate amounts of reactive oxygen and nitrogen species generated during an inflammatory response. Superoxide concentrations should be sufficiently high to keep pathogens from replication, but low enough not to cause any excessive damage to the vascular tissue of the host.
In conclusion, our results indicate that DNA methylation and histone deacetylation in human pulmonary endothelial cells are associated with the decreased expression of EC-SOD. In addition, we determined that the induction of EC-SOD by IFN-γ is regulated via JAK/STAT signaling pathways in smooth muscle cells and can be reactivated, at least in part, by exposure to the histone deacetylase inhibitor in endothelial cells.
Supplementary Material
Acknowledgments
The authors acknowledge the University of Louisville Sequencing Facility for help in sequencing DNA clones. The authors thank Dr. K. Ramos (University of Louisville) for providing quantitative RT-PCR equipment.
Footnotes
This work was supported by National Institute of Health grants HL64894 and HL074289 (R.J.F.), and by an internal grant from the University of Louisville.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0012OC on April 14, 2011
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Stralin P, Karlsson K, Johansson BO, Marklund SL. The interstitium of the human arterial wall contains very large amounts of extracellular superoxide dismutase. Arterioscler Thromb Vasc Biol 1995;15:2032–2036 [DOI] [PubMed] [Google Scholar]
- 2.Marklund SL. Expression of extracellular superoxide dismutase by human cell lines. Biochem J 1990;266:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlsson K, Marklund SL. Binding of human extracellular-superoxide dismutase C to cultured cell lines and to blood cells. Lab Invest 1989;60:659–666 [PubMed] [Google Scholar]
- 4.Abrahamsson T, Brandt U, Marklund SL, Sjoqvist PO. Vascular bound recombinant extracellular superoxide dismutase Type C protects against the detrimental effects of superoxide radicals on endothelium-dependent arterial relaxation. Circ Res 1992;70:264–271 [DOI] [PubMed] [Google Scholar]
- 5.Laursen JB, Rajagopalan S, Galis Z, Tarpey M, Freeman BA, Harrison DG. Role of superoxide in angiotensin II–induced but not catecholamine-induced hypertension. Circulation 1997;95:588–593 [DOI] [PubMed] [Google Scholar]
- 6.Hatori N, Sjoquist PO, Marklund SL, Ryden L. Effects of recombinant human extracellular-superoxide dismutase Type C on myocardial infarct size in pigs. Free Radic Biol Med 1992;13:221–230 [DOI] [PubMed] [Google Scholar]
- 7.Kitayama J, Yi C, Faraci FM, Heistad DD. Modulation of dilator responses of cerebral arterioles by extracellular superoxide dismutase. Stroke 2006;37:2802–2806 [DOI] [PubMed] [Google Scholar]
- 8.Welch WJ, Chabrashvili T, Solis G, Chen Y, Gill PS, Aslam S, Wang X, Ji H, Sandberg K, Jose P, et al. Role of extracellular superoxide dismutase in the mouse angiotensin slow pressor response. Hypertension 2006;48:934–941 [DOI] [PubMed] [Google Scholar]
- 9.Brown KA, Chu Y, Lund DD, Heistad DD, Faraci FM. Gene transfer of extracellular superoxide dismutase protects against vascular dysfunction with aging. Am J Physiol Heart Circ Physiol 2006;290:H2600–H2605 [DOI] [PubMed] [Google Scholar]
- 10.Lund DD, Chu Y, Miller JD, Heistad DD. Protective effect of extracellular superoxide dismutase on endothelial function during aging. Am J Physiol Heart Circ Physiol 2009;296:H1920–H1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nozik-Grayck E, Suliman HB, Majka S, Albietz J, Van Rheen Z, Roush K, Stenmark KR. Lung EC-SOD overexpression attenuates hypoxic induction of EGR-1 and chronic hypoxic pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol 2008;295:L422–L430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rheen Z, Fattman C, Domarski S, Majka S, Klemm D, Stenmark KR, Nozik-Grayck E. Lung EC-SOD overexpression lessens bleomycin-induced pulmonary hypertension and vascular remodeling. Am J Respir Cell Mol Biol 2011;44:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA 1990;87:1620–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelko IN, Folz RJ. Myeloid zinc finger (MZF)-like, Kruppel-like and ETS families of transcription factors determine the cell-specific expression of mouse extracellular superoxide dismutase. Biochem J 2003;369:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelko IN, Folz RJ. Sp1 and Sp3 transcription factors mediate trichostatin A–induced and basal expression of extracellular superoxide dismutase. Free Radic Biol Med 2004;37:1256–1271 [DOI] [PubMed] [Google Scholar]
- 16.Zelko IN, Mueller MR, Folz RJ. Transcription factors Sp1 and Sp3 regulate expression of human extracellular superoxide dismutase in lung fibroblasts. Am J Respir Cell Mol Biol 2008;39:243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic Biol Med 2010;48:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitchler MJ, Oberley LW, Domann FE. Epigenetic silencing of SOD2 by histone modifications in human breast cancer cells. Free Radic Biol Med 2008;45:1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh S, Ozumi K, Kim HW, Nakagawa O, McKinney RD, Folz RJ, Zelko IN, Ushio-Fukai M, Fukai T. Novel mechanism for regulation of extracellular SOD transcription and activity by copper: role of antioxidant-1. Free Radic Biol Med 2009;46:95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogdan C, Schleicher U. Production of interferon-gamma by myeloid cells: fact or fancy? Trends Immunol 2006;27:282–290 [DOI] [PubMed] [Google Scholar]
- 21.Hansson GK, Jonasson L, Holm J, Clowes MM, Clowes AW. Gamma-interferon regulates vascular smooth muscle proliferation and IA antigen expression in vivo and in vitro. Circ Res 1988;63:712–719 [DOI] [PubMed] [Google Scholar]
- 22.Hansson GK, Holm J. Interferon-gamma inhibits arterial stenosis after injury. Circulation 1991;84:1266–1272 [DOI] [PubMed] [Google Scholar]
- 23.Gomez D, Reich NC. Stimulation of primary human endothelial cell proliferation by IFN. J Immunol 2003;170:5373–5381 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Bai Y, Qin L, Zhang P, Yi T, Teesdale SA, Zhao L, Pober JS, Tellides G. Interferon-gamma induces human vascular smooth muscle cell proliferation and intimal expansion by phosphatidylinositol 3–kinase dependent mammalian target of rapamycin raptor complex 1 activation. Circ Res 2007;101:560–569 [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama M, Hirata K, Kawashima S, Kawahara Y. Regulation of nitric oxide synthase gene expression by cytokines. J Card Fail 1996;2:S179–S185 [DOI] [PubMed] [Google Scholar]
- 26.Matsubara T, Ziff M. Increased superoxide anion release from human endothelial cells in response to cytokines. J Immunol 1986;137:3295–3298 [PubMed] [Google Scholar]
- 27.Pfefferkorn LC, Guyre PM, Fanger MW. Functional comparison of the inductions of NADPH oxidase activity and Fc gamma RI in IFN gamma-treated U937 cells. Mol Immunol 1990;27:263–272 [DOI] [PubMed] [Google Scholar]
- 28.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427–1431 [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Castro S, Brasier AR, Jamaluddin M, Garofalo RP, Casola A. Reactive oxygen species mediate virus-induced STAT activation: role of tyrosine phosphatases. J Biol Chem 2004;279:2461–2469 [DOI] [PubMed] [Google Scholar]
- 30.Bai Y, Ahmad U, Wang Y, Li JH, Choy JC, Kim RW, Kirkiles-Smith N, Maher SE, Karras JG, Bennett CF, et al. Interferon-gamma induces X-linked inhibitor of apoptosis-associated factor–1 and NOXA expression and potentiates human vascular smooth muscle cell apoptosis by STAT3 activation. J Biol Chem 2008;283:6832–6842 [DOI] [PubMed] [Google Scholar]
- 31.Maher F, Harrison LC. Stabilization of glucose transporter mRNA by insulin/IGF-1 and glucose deprivation. Biochem Biophys Res Commun 1990;171:210–215 [DOI] [PubMed] [Google Scholar]
- 32.Tien M, Berlett BS, Levine RL, Chock PB, Stadtman ER. Peroxynitrite-mediated modification of proteins at physiological carbon dioxide concentration: pH dependence of carbonyl formation, tyrosine nitration, and methionine oxidation. Proc Natl Acad Sci USA 1999;96:7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 2007;6:662–680 [DOI] [PubMed] [Google Scholar]
- 34.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D‘Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466:253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stralin P, Marklund SL. Multiple cytokines regulate the expression of extracellular superoxide dismutase in human vascular smooth muscle cells. Atherosclerosis 2000;151:433–441 [DOI] [PubMed] [Google Scholar]
- 36.Brady TC, Chang LY, Day BJ, Crapo JD. Extracellular superoxide dismutase is upregulated with inducible nitric oxide synthase after NF-kappa B activation. Am J Physiol 1997;273:L1002–L1006 [DOI] [PubMed] [Google Scholar]
- 37.Goriely S, Demonte D, Nizet S, De Wit D, Willems F, Goldman M. Van Lint C. Human IL-12(p35) gene activation involves selective remodeling of a single nucleosome within a region of the promoter containing critical Sp1-binding sites. Blood 2003;101:4894–4902 [DOI] [PubMed] [Google Scholar]
- 38.Choi JC, Holtz R, Murphy SP. Histone deacetylases inhibit IFN-gamma inducible gene expression in mouse trophoblast cells. J Immunol 2009;182:6307–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hacker S, Dittrich A, Mohr A, Schweitzer T, Rutkowski S, Krauss J, Debatin KM, Fulda S. Histone deacetylase inhibitors cooperate with IFN-gamma to restore caspase-8 expression and overcome trail resistance in cancers with silencing of caspase-8. Oncogene 2009;28:3097–3110 [DOI] [PubMed] [Google Scholar]
- 40.Li J, Baud O, Vartanian T, Volpe JJ, Rosenberg PA. Peroxynitrite generated by inducible nitric oxide synthase and NADPH oxidase mediates microglial toxicity to oligodendrocytes. Proc Natl Acad Sci USA 2005;102:9936–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.