Abstract
Background
Leishmania major complex is the main causative agent of zoonotic cutaneous leishmaniasis (ZCL) in the Old World. Phlebotomus papatasi and Phlebotomus duboscqi are recognized vectors of L. major complex in Northern and Southern Sahara, respectively. In Mali, ZCL due to L. major is an emerging public health problem, with several cases reported from different parts of the country. The main objective of the present study was to identify the vectors of Leishmania major in the Bandiagara area, in Mali.
Methodology/Principal Findings
An entomological survey was carried out in the ZCL foci of Bandiagara area. Sandflies were collected using CDC miniature light traps and sticky papers. In the field, live female Phlebotomine sandflies were identified and examined for the presence of promastigotes. The remaining sandflies were identified morphologically and tested for Leishmania by PCR in the ITS2 gene. The source of blood meal of the engorged females was determined using the cyt-b sequence. Out of the 3,259 collected sandflies, 1,324 were identified morphologically, and consisted of 20 species, of which four belonged to the genus Phlebotomus and 16 to the genus Sergentomyia. Leishmania major DNA was detected by PCR in 7 of the 446 females (1.6%), specifically 2 out of 115 Phlebotomus duboscqi specimens, and 5 from 198 Sergentomyia darlingi specimens. Human DNA was detected in one blood-fed female S. darlingi positive for L. major DNA.
Conclusion
Our data suggest the possible involvement of P. duboscqi and potentially S. darlingi in the transmission of ZCL in Mali.
Introduction
Cutaneous leishmaniasis (CL) is a common infection and a major public health importance in 88 countries with approximately 1.5 million new cases each year [1]. Zoonotic cutaneous leishmaniasis (ZCL) caused by Leishmania major Yakimoff & Shokkor, 1914 is endemic in the Mediterranean basin, sub-Saharan Africa and Asia. In these areas, the proven vectors are sandflies belonging to the subgenus Phlebotomus: Phlebotomus (Phlebotomus) papatasi (Scopoli, 1786) in Northern Sahara areas [2]–[5], Phlebotomus (Phlebotomus) duboscqi Neveu-Lemaire, 1906, its vicariant species, in sub-Saharan regions, specifically in Ethiopia and Senegal [6]–[8], Phlebotomus (Phlebotomus) bergeroti Parrot, 1934 and Phlebotomus (Phlebotomus) salehi Mesghali, 1965 suspected in Egypt and Iran, respectively [9], [10].
In Mali, Leishmania major has been the main causative agent of ZCL for the last 25 years. The first human Leishmania strain was isolated from a skin lesion of a European woman [11]. The first unambiguous autochthonous case of ZCL was reported in Mali in 1989 [12]. Thereafter, several autochthonous cases were observed and the incidence of ZCL was estimated at 0.8% [13]. In Mali, four zymodemes were described: MON-25, MON-26, MON-74 and MON-117 [11], [12], [14] and P. duboscqi species was incriminated as the primary vector [15].
Here, we report the results of an entomological survey conducted between the 21st and the 27Th of January 2010 in the Bandiagara area, where ZCL is know to be endemic. The objectives of the study were (i) to inventory the Phlebotomine sandfly species, (ii) to search for promastigote forms in the mid-gut of live female sandflies and, (iii) to detect Leishmania DNA in female sandflies using the Internal Transcribed Spacer 2 (ITS2) gene system.
Materials and Methods
Study area
Sandflies were collected from four neighbouring villages located in the district of Bandiagara, in Mali, Nando (14°14′55N, 3°55′01W), Koundou (14°30′01N, 3°14′59W), Youga Na (14°32′19N, 3°13′14W) and Doucombo (14°11′32N, 3°35′39W). The population size in four villages investigated is approximately 3000 inhabitants. Houses are constructed essentially with clay bricks plastered with mud and straw, and they are adjoining. The climate is characterized by two distinct seasons: a dry season from October to May (temperature range 25–45°C; annually average rainfall 200 mm), a rainy season from June to September (temperature range 21–35°C; annually average rainfall 500 mm). Vegetation is sparse and characterized by the presence of baobab trees, small shrubs (shea, acacia and neem) and other savannah grasses. Domestic animals such as chickens, sheep, goats and cow are maintained in corrals around human habitations.
Collection and identification of sandflies
Sandfly collection was performed using CDC miniature light and sticky paper traps according to published procedure [16]. Both types of traps were set during the late afternoon around human houses, animal shelters and entrance of the grottos. Sticky traps were also placed near rodent burrows and tree holes. Each morning, the traps were collected and the sandflies were organized according to trapping area. The female Phlebotomus sandflies captured alive were dissected immediately using sterile syringes (BD Microlance™3, France). The genitalia and sometime head were cut off and mounted under a cover slip in drop sterile saline water (0.9‰) for morphological identification using morphologic keys [17]–[19]. The mid-gut of engorged females was examined in a drop of sterile saline water (0. 9‰) with a light microscope to look for the presence of promastigotes. The remainder of the body was stored in sterile 1.5-ml microtubes in liquid nitrogen to research Leishmania DNA.
Sandflies dead at collection time were stored immediately in 70% alcohol and carried out to laboratory for later identification. Then, they were transferred into Marc André solution [17] and incubated at 37°C for 15 min. The head and genitalia were cut off using sterile and disposable syringes in one drop of Marc-André solution, and mounted under a cover slip in a drop of poly-vinylic-alcohol for morphological identification [17]–[19]. The remainder of the body was stored in sterile 1.5-ml microtubes at −20°C before DNA extraction.
DNA extraction, ITS2 Leishmania and cyt-b vertebrate genes amplifications
For each female sandfly, the remainder of the body was dry-ground in a 1.5-ml microtube using the Tissue Lyser apparatus (Qiagen, Germany). Total DNA was purified using the QIAamp DNA Mini Kit (Qiagen, Germany), and stored at −20°C until used. To confirm that viable DNA was present, PCR-amplify Internal Transcribed Spacer (ITS) 1-sandflies was performed (data not shown). To attempt detection of the ITS2 Leishmania rDNA gene in sandflies, a primer pair was designed 5.8S-AVZF 5′-GGAGGCGTGTGTTTGTGTTG-3′ and ITS2-AVZR 5′-GCGAAGTTGAATTCTCGTTT-3′. To identify the origin of the blood meal of engorged females, two primers (cyt-AVZF: 3′-CCTCAGAATGATATTTGTCCTC-5′and Cyt-AVZR 3′-ATCCAACATCTCAGCATTGATGAA-5′) were designed and used to PCR-amplify a region of cytochrome b (cyt-b) gene of vertebrates. Polymerase chain reaction (PCR) was performed in a 25-µl reaction volume containing 300 ng of DNA, 200 µM dNTPs, buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.3, and 1 mM MgCl2), 0.3-µM of each appropriate pair of primers: 5.8S-AVZF and ITS2-AVZR 5′ for Leishmania ITS2 gene, cyt-AVZF and Cyt-AVZR for cyt-b gene and 2.5 U of Thermus aquaticus DNA polymerase (AmpliTaq Gold; PerkinElmer Life and Analytical Sciences, Boston, MA). Cycling profile was 95°C for 10 min, then 40 repeats of 94°C-45 sec, 58°C-45 sec for ITS2 rDNA gene and 62°C- 50 sec for cyt-b partial gene, 72°C-45 sec, followed by 72°C-10 min. Non-inoculated PCR mix was used for each PCR run to detect contamination that could lead to false positive. DNA extracted from whole blood of dog, rabbit, chicken and human was used to validate the cyt-b PCR system.
Direct sequencing and analysing of PCR product
Four µl of each PCR product were subjected to electrophoresis in 1.5% agarose gel, stained with ethidium bromide and visualized under UV light. Bands of the expected size (350 bp) were purified using sephadex plate and sequenced in both directions using the same primers described above. Sequence correction was performed using System Navigator (Applied Biosystem). The ITS2 and cyt-b partial gene sequences obtained within sandfly specimens were blasted against the GenBank database (http://blast.ncbi.nlm.nih.gov). Neighbor-Joining tree based on Leishmania ITS2 gene was constructed using MEGA4 program [20], [21], after multiple alignments of our sequences with ITS2 reference sequences obtained from GenBank data base: L. chagasi (AJ000306.1), L. donovani (EU637919.1), L. infantum (AJ634354.1), L. major (AJ272383.1), L. mexicana (AB558238.1), L. guyanensis (DQ182537.1) and L. tropica (AJ300485.1).
Results
Morphological identification of collected sandflies
Of the 3259 sandflies trapped, 2416 were captured using 2 m2 of sticky papers (60 sandflies/night/m2), and 843 using 238 night-CDC light traps (4 sandflies/night/CDC light trap) (Table 1). All 843 sandflies captured using CDC traps were identified including, 314 live and 529 dead at collection time. Of the 2416 sandflies trapped using sticky papers, 481 were identified including all sandflies belonging to the Phlebotomus genus, sandflies morphologically similar (large size) to Phlebotomus and 222 other specimens randomly selected. Thus, a total of 1324 specimens were morphologically identified. Among them, 20 distinct species were identified, 4 within the Phlebotomus genus and 16 within the Sergentomyia genus (Table 2). S. (Spelaeomyia) darlingi was the most prevalent species with 505 specimens (38%), followed by P. (Phlebotomus) duboscqi with 177 specimens (13.4%) (Table 2).
Table 1. Distribution of sandflies in the Bandiagara area.
| CDC light miniatures | Sticky papers | |||
| Village | N° of traps set | N° of sandflies caught | N° of traps set | N° of sandflies caught |
| Nando | 16 | 615 | 385 | 1220 |
| Koundou | 5 | 35 | 227 | 416 |
| Doucombo | 6 | 54 | 196 | 333 |
| Youga Na | 6 | 139 | 37 | 447 |
| Total | 33 | 843 | 845 | 2416 |
Table 2. Distribution of sandfly species according to the capture method.
| CDC traps | Sticky traps | ||||
| Subgenus | Species | Live flies | Dead flies | Dead flies | Total |
| Phlebotomus | P. duboscqi | 96 | 41 | 40 | 177 |
| Paraphlebotomus | P. kazeruni | - | 22 | 11 | 33 |
| P. sergenti | 2 | 37 | 28 | 67 | |
| Anaphlebotomus | P. rodhaini | - | 2 | 5 | 7 |
| Spelaeomyia | S. darlingi | 31 | 298 | 176 | 505 |
| Sergentomyia | S. buxtoni | - | 3 | 14 | 17 |
| S. dubia | - | 12 | 33 | 45 | |
| S. fallax | - | 6 | 18 | 24 | |
| S. schwetzi | - | 1 | - | 1 | |
| S. davidsoni | - | - | 2 | 2 | |
| S. antennata | - | 29 | 51 | 80 | |
| Sintonius | S. affinis affinis | - | 2 | 3 | 5 |
| S. affinis vorax | - | 43 | 23 | 66 | |
| S. balmicola | - | 2 | 2 | 4 | |
| S. christophersi | - | 24 | 42 | 66 | |
| S. clydei | - | 2 | 24 | 26 | |
| S. wansoni | - | 3 | 4 | 7 | |
| Parrotomyia | S. africana africana | - | - | 1 | 1 |
| S. magna | - | - | 2 | 2 | |
| Grassomyia | S. squamipleuris | - | - | 1 | 1 |
| Sergentomyia sp. | 179 | 2 | 1 | 182 | |
| Phlebotomus sp. | 6 | - | 6 | ||
| Total | 314 | 529 | 481 | 1324 | |
Microscopic examination
None of the 35 female sandflies (29 P. duboscqi and 6 S. darlingi) which were dissected alive and examined was found to contain promastigotes.
Leishmania DNA detection from sandflies species
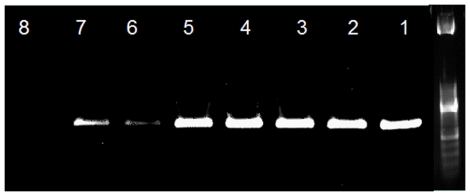
A total of 446 female sandflies were analyzed: 115 P. (Phlebotomus) duboscqi, 27 P. (Paraphlebotomus) kazeruni, 6 P. (Paraphlebotomus) sergenti, 198 S. (Spelaeomyia) darlingi, 20 S. (Sintonius) affinis vorax, 20 S. (Sergentomyia) antennata, 20 S. (Sintonius) clydei, 20 S. (Sergentomyia) fallax, 10 S. (Sintonius) christophersi and 10 S. (Sergentomyia) dubia. Leishmania DNA was detected by PCR in 7 dead specimens (1.6%), of which 2 P. duboscqi and 5 S. darlingi (Figure 1). No amplification was observed in the negative controls tested with Leishmania PCR system.
Figure 1. Leishmania ITS2 PCR products from collected female sandflies.
Leishmania DNA amplified within S darlingi (1 to 5) and P. duboscqi (6–7). N = negative control, band size of the PCR product is 400 bp.
Leishmania DNA sequencing and Neighbour-Joining analysis
The seven ITS2 PCR positive products obtained within 2 P. duboscqi and 5 S. darlingi were sequenced and aligned. Comparative analysis of the two consensus sequences identified a polymorphic microsatellite region consisting of 8 AT repeats in 2 identical P. duboscqi sequences versus 9 AT repeats in 5 identical S. darlingi sequences. The two ITS2 sequence types obtained were submitted to GenBank and the access numbers are HQ591339 for S. darlingi and HQ591340 for P. duboscqi.
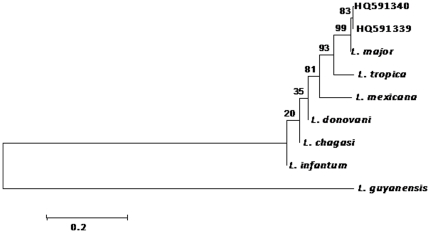
In the NJ tree, the 7 Leishmania major sequences determined in this study were grouped with L. major reference sequence (Figure 2) in a unique cluster supported with 99% bootstrap value. The identification of L. major DNA was blindly confirmed by the French National Reference Center on Leishmaniasis (Montpellier Academic Hospital, University of Montpellier, France) using Multilocus sequence typing (MLST) approach on seven genomic loci (4677 bp in all). Sequences are deposited in GenBank under the following accession numbers: JF732921-JF732927.
Figure 2. Dendrogram constructed using sequences of Leishmania major from S. darling (HQ591339) and P. duboscqi (HQ591340).
ITS2 polymerase chain reaction (PCR) fragment of the sequences amplified were aligned with the reference strains (L. major, L. tropica, L. infantum, L. donovani, L. mexicana, L. chagasi and L. guyanensis) and tree constructed using NJ. Bootstrap analysis was performed with 1000 replicates and P values reported at nodes.
Vertebrate DNA detection in female sandflies
A total of 20 engorged female sandflies (15 S. darlingi including one contained Leishmania DNA and 5 P. duboscqi) were tested to determine the vertebrate source of the blood meal using cyt-b PCR. A positive PCR was obtained in 11 of the 15 S. darlingi and in none of the 5 P. duboscqi. Direct sequencing of the 11 PCR products was performed and sequence analysis demonstrated that the 11 sequences were 100% identical to human sequences. One of these 11 engorged S. darlingi that contained human blood DNA contained also Leishmania major DNA.
Discussion
In Mali, Leishmania major is the main agent of ZCL. P. duboscqi was incriminated as the primary vector of ZCL very recently [15]. The Bandiagara area, where ZCL is endemic, was chosen to organize an entomologic study to inventory sandfly species present in the region, and to address whether sandfly species other than P. duboscqi may play a role in the epidemiology of ZCL.
In this study, a total of 20 different sandfly species were trapped, of which five were identified for the first time in Mali. Thus, with the 17 species previously described [15], [17], [22], [23], the new inventory counts 22 species of sandflies in Mali. The new species, previously unrecognized in Mali, were P. (Paraphlebotomus) kazeruni, S. (Sintonius) affinis affinis, S. (Sintonius) balmicola, S. (Parrotomyia) magna and S. (Sergentomyia) davidsoni.
In our traps, S. darlingi was the predominant identified species (38%), followed by P. duboscqi (13.4%). In contrast with our data, S. darlingi was previously seldom identified during a three years investigation in two villages in central Mali [15]. S. dubia was reported as the predominant species in the Bamako area, the Capital City of Mali [22]. However, our inventory remains limited to the Bandiagara area and investigations were done during a short period of time. Other captures in other regions at other periods could change these results.
Usually, the vector role of a sandfly species is epidemiologically suspected in Leishmaniasis focus when the species is predominant and proved anthropophilic behavior. This suspicion is strengthened when the same sandfly species is found infected with metacyclic promastigotes of the same Leishmania species isolated within human and potential animal reservoir host of parasites. The vector role is confirmed when the transmission of Leishmania to human is experimentally demonstrated by the bite of the sandfly [24]. More recently, molecular techniques allowed detecting the Leishmania DNA within human and sandflies. The technical approach developed in our study based on ITS2-PCR proved to be easy to implement in field conditions.
P. duboscqi is a vector of Leishmania major in Ethiopia and Senegal, where the evidence was brought by isolation of parasite strains [6]–[8,]. More recently, females of P. duboscqi were found infected with L. major in two villages in central Mali [15]. In our study, the detection of Leishmania major DNA in two female specimens of P. duboscqi caught in the ZCL Bandiagara foci provide additional evidence in favour of the role of P. duboscqi as vector of Leishmania major in Mali.
Perhaps the most intriguing and interesting finding of our study is the detection of L. major DNA in five females of S. darlingi for the first time. To date, there is no data in the literature about the possible role of S. darlingi as vector of L. major or any other species of Leishmania. No data is available on the trophic preferences of S. darling. Whether S. darlingi might be a vector for Leishmaniasis remains to be investigated. The dogma is that Sergentomyia species do not bite humans, and as a consequence cannot transmit either Leishmania or any other pathogen. However, recent reports are questioning this dogma. Sergentomyia schwetzi, Sergentomyia garnhami and Sintonius clydei were reported to bite humans [17]. Studies conducted in endemic foci in India, Iran and Kenya showed that Sergentomyia babu, Sergentomyia sintoni and Sergentomyia garnhami, respectively, can be naturally infected by mammalian Leishmania, incriminating them as potential vectors [25]–[27]. Although Lawyer et al [28] reported in 1990 that L. major cannot develop in S. schwetzi. More recently, a study conducted in Senegal showed that S. schwetzi and S. dubia might be capable of transmitting canine leishmaniasis [29]. In our case, S. darlingi was the predominant species and the most frequently infected (5 females) with Leishmania major DNA. The molecular identification was blindly confirmed by the French Reference Center using a different detection methodology i.e. the MLST approach. Human blood was amplified from infected and blood-fed female specimens on the one hand and from blood-fed but not infected on the other. Taken together, our data suggest that S. darlingi might be a vector for L. major in the Bandiagara region of Mali. This hypothesis needs to be more thoroughly evaluated in further studies aiming to isolate metacyclic promastigotes of L. major in the digestive tubes of S. darlingi, and to demonstrate experimentally its capacity to transmit the parasite by biting to mammals.
Together, these observations challenge the dogma that both visceral and cutaneous leishmaniasis are exclusively transmitted by the species belonging to the Phlebotomus genus in Old World.
To confirm our hypothesis that ZCL can be transmitted by S darlingi in Mali, it is necessary to pursue field investigations and to isolate L major from S darlingi, and to initiate experimental studies aiming at the demonstration of L major transmission by the bite of S darlingi.
Acknowledgments
The work is dedicated to (feu) Professor Philippe Ranque, the founder of the DEAP at the Ecole Nationale de Medicine et de Pharmacie du Mali, who conducted the first field work on Leishmania and Phlebotomus in Mali.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The funding that supported this study was from the Foundation MERIEUX. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. The global burden of disease: 2004 update. Geneva: World Health Organization; 2008. [Google Scholar]
- 2.Sergent E, Parrot I, Donatien A, Beguet M. Transmission du clou de Biskra par le phlébotome Phlebotomus papatasi (Scop). C R Acad Sci. 1921:1030–1032. [Google Scholar]
- 3.Parrot L, Donatien A. Infection naturelle et infection expérimentale de Phlebotomus papatasi par le parasite du bouton d'orient. Bull Soc Path Ex. 1926;19:694–696. [Google Scholar]
- 4.Ben-Ismail R, Gramiccia M, Gradoni L, Helal H, Ben Rachid MS. Isolation of Leishmania major from Phlebotomus papatasi in Tunisia. Trans R Soc Trop Med Hyg. 1987;5:186–187. doi: 10.1016/0035-9203(87)90018-6. [DOI] [PubMed] [Google Scholar]
- 5.Izri A, Belazzoug S, Pratlong F, Rioux JA. Isolation of Leishmania major from Phlebotomus papatasi in Biskra (Algérie) completion of an epidemiological saga. Parasitol Hum Comp. 1992;1:31–32. doi: 10.1051/parasite/199267131. [DOI] [PubMed] [Google Scholar]
- 6.Gebre-Michael T, Pratlong F, Lane RP. Phlebotomus (Phlebotomus) duboscqi (Diptera: Phlebotominae), naturally infected with Leishmania major in southern Ethiopia. Trans R Soc Trop Med Hyg. 1993;1:118–125. doi: 10.1016/0035-9203(93)90399-b. [DOI] [PubMed] [Google Scholar]
- 7.Dedet JP, Desjeux P, Derouin F. Ecology of a focus of cutaneous leishmaniasis in the Thiès region (Senegal, West Africa). 4. Spontaneous infestation and biology of Phlebotomus duboscqi Neveu-Lemaire 1906. Bull Soc Pathol Ex. 1982;75:588–98. [PubMed] [Google Scholar]
- 8.Develoux M, Diallo S, Dieng Y, Mane I, Huerre M, et al. Diffuse Cutaneous leishmaniasis due to Leishmania major in Senegal. Trans R Soc Trop Med Hyg. 1996;4:396–7. doi: 10.1016/s0035-9203(96)90520-9. [DOI] [PubMed] [Google Scholar]
- 9.Hanafi HA, El Sawaf BM, Fryauff DJ, Modi GB. Experimental infection and transmission of Leishmania major by laboratory-reared Phlebotomus bergeroti Parrot. Am J Trop Med Hyg. 1996;6:644–646. doi: 10.4269/ajtmh.1996.54.644. [DOI] [PubMed] [Google Scholar]
- 10.Kasiri H, Javadian E, Seyedi-Rashti MA. List of Phlebotominae (Diptera:Psychodidae) of Iran. Bull Soc Path Ex. 2000;2:129–30. [PubMed] [Google Scholar]
- 11.Garin JP, Peyramond D, Piens MA, Rioux JA, Godfrey DG, et al. Presence of Leishmania major Yakimoff and Schokhor, 1914 in Mali. Enzymatic identification of a strain of human origin. Ann Parasitol Hum Comp. 1985;1:93–4. doi: 10.1051/parasite/198560193. [DOI] [PubMed] [Google Scholar]
- 12.Izri MA, Doumbo O, Belazzoug S, Pratlong F. Presence of Leishmania major MON-26 in Mali. Ann Parasitol Hum Comp. 1989;6:510–1. doi: 10.1051/parasite/1989646510. [DOI] [PubMed] [Google Scholar]
- 13.Keita SFO, Ndiaye Ht, Konare Hd. Epidémiologie et polymorphisme clinique de la leishmaniose cutanée observée au CNAM (ex-Institut Marchoux) Bamako (Mali). Mali Médical. 2003;XVIII:3. [Google Scholar]
- 14.Pratlong F, Dereure J, Ravel C, Lami P, Balard Y, et al. Geographical distribution and epidemiological features of Old World cutaneous leishmaniasis foci, based on the isoenzyme analysis of 1048 strains. Trop Med Int Health. 2009;9:1071–1085. doi: 10.1111/j.1365-3156.2009.02336.x. [DOI] [PubMed] [Google Scholar]
- 15.Anderson JM, Samake S, Jaramillo-Gutierrez G, Sissoko I, et al. Seasonality and Prevalence of Leishmania major Infection in Phlebotomus duboscqi Neveu-Lemaire from Two Neighboring Villages in Central Mali. Plos Negl Trop Dis. 2011;5:e1139. doi: 10.1371/journal.pntd.0001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izri MA, Belazzoug S. Phlebotomus (Larroussius) perfiliewi naturally infected with dermotropic Leishmania infantum at Tenes, Algeria. Trans R Soc Trop Med Hyg. 1993;87:399. doi: 10.1016/0035-9203(93)90011-e. [DOI] [PubMed] [Google Scholar]
- 17.Abonnenc E. Les phlébotomes de la région éthiopienne (Diptera, Psychodidae). 1972. Mem. O.R.S.T.O.M. N° 55, 289. [PubMed]
- 18.Léger N, Pesson B, Madulo-Leblond G, Abonnenc E. Sur la différentiation des femelles du sous-genre Larroussius Nitzulescu, 1931 (Diptera, phlebotomidae) de la région méditerranéenne. Ann Parasitol Hum Comp. 1983;58:611–623. [PubMed] [Google Scholar]
- 19.Niang AA, Hervy JP, Depaquit J, Boussès P, Davidson I, et al. Sandflies of the Afro-tropical region. [CD] IRD Edition 2004 [Google Scholar]
- 20.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Nat Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Ranque P, Sangaré C, Abonnenc E, Léger N. Note préliminaire sur les Phlébotomes de la région de Bamako-Mali- Présence de Phlebotomus sergenti, Parrot 1917. Acta Trop. 1975;32:348. [PubMed] [Google Scholar]
- 23.Kervran P. Description de quelques espèces de phlébotomes du Soudan français. Ann Parasitol. 1946;21:153–163. [Google Scholar]
- 24.Killick-Kendrick R. Phlebotomine vectors of the leishmaniasis: a review. Med Vet Entomol. 1990;1:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee S, Mohammed Quamarul H, Ghosh A, Kashi Nath G, Bhattacharya A, et al. Leishmania DNA in Phlebotomus and Sergentomyia species during a Kala-azar epidemic. Am J Trop Med Hyg. 1997;4:423–5. doi: 10.4269/ajtmh.1997.57.423. [DOI] [PubMed] [Google Scholar]
- 26.Parvizi P, Amirkhani A. Mitochondrial DNA characterization of Sergentomyia sintoni populations and finding mammalian Leishmania infections in this sandfly by using ITS-rDNA gene. Iranian J Vet Res. 2008;22:9–18. [Google Scholar]
- 27.Mutinga MJ, Massamba NN, Basimike M, Kamau CC, Amimo FA, et al. Cutaneous leishmaniasis in Kenya: Sergentomyia garnhami (Diptera Psychodidae), a possible vector of Leishmania major in Kitui District: a new focus of the disease. East Afr Med J. 1994;7:424–8. [PubMed] [Google Scholar]
- 28.Lawyer PG, Ngumbi PM, Anjili CO, Odongo SO, Mebrahtu YB, et al. Development of Leishmania major in Phlebotomus duboscqi and Sergentomyia schwetzi (Diptera: Psychodidae. AM J Trop Med Hyg. 1990;1:31–43. doi: 10.4269/ajtmh.1990.43.31. [DOI] [PubMed] [Google Scholar]
- 29.Senghor MW, Faye MN, Faye B, Diarra K, Elguero E, et al. Ecology of phlebotomine sandflies in the rural community of Mont Rolland (Thiès region, Senegal): area of transmission of canine leishmaniasis. Plos one. 2011;3:e14773. doi: 10.1371/journal.pone.0014773. [DOI] [PMC free article] [PubMed] [Google Scholar]