Abstract
The physical interaction between a lipid vesicle and a silver nanoparticle (AgNP)-human serum albumin (HSA) protein “corona” has been examined. Specifically, the binding of AgNPs and HSA was analyzed by spectrophotometry, and the induced conformational changes of the HSA were inferred from circular dichroism spectroscopy. The fluidity of the vesicle, a model system for mimicking cell membrane, was found to increase with the increased exposure to AgNP-HSA corona, though less pronounced compared to that induced by AgNPs alone. This study offers additional information for understanding the role of physical forces in nanoparticle-cell interaction and has implications for nanomedicine and nanotoxicology.
Understanding biological response to engineered nanomaterials is essential to the continued development of nanomedicines, whose expanding repertoire includes design of novel assemblies for gene and drug delivery and of highly specific and localized bioimaging and disease and tumor detection. On the other hand, the mass production of nanomaterials and rapid commercialization of nanotechnologies further justify research addressing occupational and environmental exposure to administered or accidentally released nanoparticles.1, 2, 3, 4 Central to these crucial research needs is a mechanistic description of the unique interplay between biological systems and engineered nanomaterials,1, 3, 4 especially at the cellular level which manifests the unit of life.
It has been realized that nanoparticles, upon their entry into the bloodstream or—more generically—when dispersed in a biological fluid, interact readily with proteins, peptides, amino acids, fatty acids, lipids, and other soft and organic matters.5, 6, 7 Consequently, the nanoparticles acquire an enhanced mobility as well as biocompatibility and may elicit their impact on the host system through a collective entity of nanoparticle-protein “corona,” rather than the physicochemistry of the nanoparticle “core” alone.5, 6, 7, 8, 9, 10 Such nanoparticle-protein corona may further initiate its contact with the cell through physical adsorption or recognition by the membrane receptors specific for the proteins that constitute the corona. Uptake of nanoparticles is thought—as agreed upon by a majority of the research community—to be realized via the energy-dependent biological process of endocytosis, in addition to passive diffusion and mechanical or biochemical damage in the lipid membrane induced by the trespassing nanoparticle.1 However, despite intensive research efforts, both experimentally11, 12, 13, 14, 15, 16, 17, 18, 19, 20 and through atomistic and coarse-grained computer simulations,21, 22, 23, 24, 25 it remains unclear and often controversial as to what extent the thermodynamic and endocytotic pathways may individually contribute to the convoluted process of nanoparticle cell uptake.
In this paper, we show a facile method of examining the physical interaction between a lipid vesicle—a model cell membrane—and a nanoparticle-protein corona. The lipid vesicle consists of zwitterionic dimyristoyl phosphatidylcholine (DMPC) doped with 10% anionic dimyristoyl phosphatidylglycerol (DMPG). The equal chain length of the fatty acyl tails and comparable head group sizes of DMPC and DMPG minimized phase separation in the vesicle.26 Physically, a dipole moment existed in DMPC that pointed from the O− to the N+ within the lipid head, while only a negatively charged O− was present in the lipid head of DMPG (Fig. 1a). Such net negative charge of the vesicle, afforded by the 10% DMPG lipids, conformed to the natural composition of weakly negative charge of cell membranes. In addition, the consideration of protein corona, instead of bare nanoparticle “core” alone, provided a more realistic system for examining the physical interactions between nanoparticles and the cell.
Figure 1.
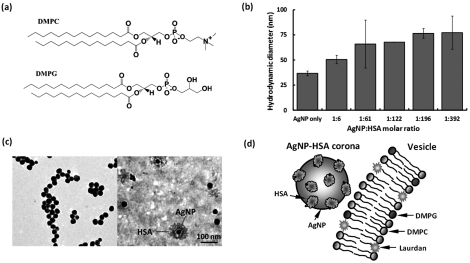
(Color online) (a) Chemical structures of DMPC and DMPG. (b) Hydrodynamic sizes of AgNP-HSA at molar ratios of 1:6–1:392. Incubation: 1 h. (c) TEM images of bare AgNPs (left panel) and AgNP-HSA corona (right panel). (d) Schematic (not to scale) of AgNP-HSA corona interacting with a Laurdan-labeled, DMPG-doped DMPC vesicle.
Silver nanoparticles (AgNPs, coated with citrate) were purchased from NanoComposix and used in our experiments. Human serum albumin (HSA, MW: 66 478) proteins were obtained from Sigma. Silver nanoparticles were selected due to their increasing mass production and domestic use,27 while the selection of HSA was based on its high abundance among plasma proteins. The zeta-potentials of AgNPs and HSA in Milli-Q water (pH = 6.5) were determined to be −31 mV and −17 mV, respectively (ZetaSizer Nano, Malvern). The stronger surface charge provided a stable suspension for AgNPs, while HSA molecules could be multimeric due to their weaker charge. AgNPs and HSA of molar ratios from 1:6 to 1:392 were incubated at room temperature for 1 h, and the hydrodynamic sizes of their mixtures were determined by dynamic light scattering (DLS) (Nanosizer S90, Malvern). Figure 1b shows the increased size with the increased ratio of HSA to AgNPs. Multi-layer coating of HSA onto AgNPs was evident at the molar ratio of 1:122 and above, considering the size of an HSA monomer is ∼8 nm.28 The formation of AgNP-HSA corona was confirmed by transmission electron microscopy (TEM, Hitachi H7600), where AgNPs (4.92 × 10−4 μM) were incubated with HSA (7.16 μM) at 4 °C overnight and negatively stained with phosphotungstic acid for 45 min prior to imaging. The average diameter of bare AgNPs was ∼30 nm, in agreement with that provided by the vendor (Fig. 1c, left panel). With incubation, a thick layer of optically less dense material, believed to be HSA, was clearly visible on the AgNP surfaces (Fig. 1c, right panel). The size of the AgNP-HSA corona determined by TEM was ∼80 nm, in agreement with the DLS measurement. Since both the AgNPs and the HSA were net negatively charged, the formation of AgNP-HSA corona could result from the hydrogen bonding between the hydroxyls/oxygens of the citrate coating on the AgNPs and the nitrogen/sulfur electron acceptors/donors on the HSA, and from the hydrophobic interaction between exposed silver atoms (due to incomplete citrate coating29) of the AgNPs and the hydrophobic domains of the HSA.
The formation of AgNP-HSA corona was further confirmed by measuring the absorption spectra of AgNPs (9.8 × 10−5 μM), HSA (1.42 μM), and their mixture AgNP-HSA using a UV-vis spectrophotometer (Cary 300 BIO, Varian). As shown in Fig. 2 inset, a characteristic peak of surface plasmon resonance (SPR) was identified for AgNPs at 404 nm. After incubation with HSA, the absorption peak was red-shifted to 412 nm. This phenomenon can be understood by the following analysis. Assume the dielectric constant of a AgNP relative to its surrounding medium is , where ɛs and ɛm are the dielectric constants of the AgNP and the medium, respectively. Here, ɛ′ is negative and decreases with the increasing wavelength of light, while ɛ″ is approximately constant for wavelength longer than 300 nm. According to the Clausius-Mossotti relation,30 the extinction cross-section of the AgNP can be expressed as , and its extinction peak occurs at ɛ′ = −2 or . Thus, when ɛm was increased due to the binding of (dielectric) HSA molecules onto the AgNP, a red-shift in wavelength occurred for the extinction to reach its new peak value.
Figure 2.
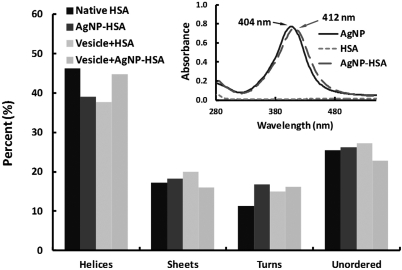
(Color online) Percent of secondary HSA structures inferred from the CD spectra for native HSA, HSA pre-incubated with AgNPs, HSA in the presence of vesicles, and HSA pre-incubated with AgNPs in the presence of vesicles. Inset: UV-vis spectra showing a red-shift of the extinction peak of AgNPs pre-incubated with HSA. Pre-incubation time: 8 h.
To examine the physical interaction between AgNP-HSA and cell membranes, artificial vesicles were generated by lipid extrusion. Specifically, 10 mg of DMPC lipids, doped with 10% DMPG, were first dissolved in 1 mL of chloroform in a flask, and then dried under airflow to form thin lipid sheets on the flask bottom. After that 1 mL of Milli-Q water (at 30 °C) was added to the flask to hydrate the lipid sheets, and the mixture of lipids and water was agitated for ∼2 min to form large multilamellar vesicles (LMVs). This process was performed in a warm water bath to avoid gel-liquid crystal transition. A water-bath sonication was then applied to the mixture for 5–10 min to form large unilamellar vesicles (LUVs). After sonication, the vesicle suspension was extruded through a porous polycarbonate membrane (pore size: 100 nm) to yield uniformly sized LUVs (100 nm). In particular, for the detection of vesicle phase transition, Laurdan (6-dodecanoyl-2-dimethylaminonaphthalene, AnaSpec) was added to the chloroform solution to partition into the vesicle bilayers.
To investigate the effect of AgNP binding on the conformation of HSA, circular dichroism (CD) spectroscopy was performed. AgNPs and HSA were incubated for 8 h prior to the measurement, using the same molar ratio but diluted 16× as in the UV-vis measurement to comply with the sensitivity of the spectropolarimeter (Jasco J-810). The vesicle-HSA sample was prepared by mixing the DMPC vesicles (10% DMPG-doped) and HSA immediately before the CD measurement. The vesicle-AgNP-HSA sample was obtained from the mixture of DMPC vesicles (10% DMPG-doped) and pre-formed AgNP-HSA corona. The CD spectra were acquired at room temperature over a wavelength range of 200-300 nm using quartz cuvettes and were averaged over three scans taken at a speed of 50 nm/min. The backgrounds of the AgNPs and vesicles were subtracted accordingly.
The readout values of the HSA ellipticity (θ, in mdeg) were converted to a standard unit of deg·cm2/dmol ([θ]) using equation , where M0 is the mean residue molecular weight (118 g/mol), Csoln is the protein concentration in solution (in g/mL), and L is the path length through the buffer (1 cm). As shown in Fig. 2, the α-helix content in HSA decreased by 15.7% after incubation with AgNPs, compared to that for the native state of HSA, and decreased by 18.4% after incubation with the vesicles. However, incubating vesicles with the pre-formed AgNP-HSA corona reversed the conformational change of HSA induced by AgNPs, causing a decrease of only 3.2% in the α-helix content of the HSA. For β-sheets, increases of 5.8% and 15.9% were observed for the samples of AgNP-HSA and vesicle-HSA, respectively, and a decrease of 7.3% was measured for the sample of vesicle-AgNP-HSA, compared to that for the sample of HSA alone. These results suggest that, in the presence of either vesicles or AgNPs, HSA could undergo significant conformational changes to alter its α-helices into β-sheets and other secondary structures. Such changes can be attributed to the interaction between the hydrophobic domains in the HSA and the hydrophobic surface areas of the AgNPs, and electrostatic attraction between the negatively charged surface domains of HSA and the positively charged DMPC lipid head groups. In contrast, the net negative charge of the vesicles further compromised the relatively weak affinity of the negatively charged AgNPs for the HSA, leading to the partial recovery of protein conformation.
The effect of AgNP-HSA protein corona on the fluidity of DMPG-doped DMPC vesicles was evaluated based on the fluorescence emission of the Laurdan dyes partitioned within the vesicle bilayers (Fig. 1d). First, the samples of AgNP-HSA mixtures were prepared at different concentrations (6.15 × 10−7–492 × 10−7 μM for AgNPs and, accordingly, 8.95 × 10−3–716 × 10−3 μM for HSA) and incubated at 4 °C overnight to ensure the equilibrium of their binding. Then the samples were separately added to the vesicle suspensions of 0.05 mg/mL. A spectrofluorometer (Cary Eclipse, Varian) was used to excite the Laurdan at 340 nm, and the fluorescence intensities of the dyes were collected at both 416 nm and 473 nm to derive the generalized polarization (GP) values for the vesicles:13, 19. An increasing GP value indicates a phase transition toward gelatin, while a decreasing GP value represents fluidization. The spectrofluorometer chamber was operated at 25 °C, above the lipid phase transition temperature. Measurements were repeated for AgNPs and HSA, respectively.
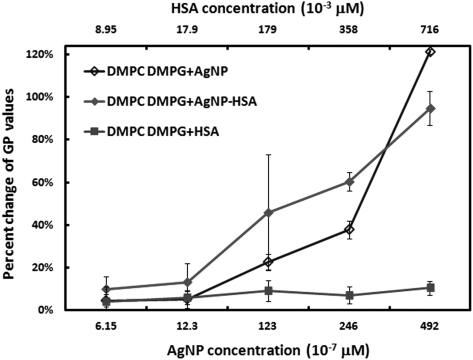
Compared with the control vesicles, HSA showed little impact while both AgNPs and AgNP-HSA gave rise to decreased GP values in the vesicles (Fig. 3). This indicates an enhanced fluidization of the vesicle bilayers, which may be attributed to the structural reorganizations of the vesicles in response to the nanoparticle adsorption. In addition to the prevalent weak forces of hydrogen bonding and van der Waals interaction, strong and long range electrostatic interactions between the charged domains of the lipids (N+ in the DMPC and O− in the DMPG, Fig. 1a) and that of the AgNPs (e.g., citrate coating) or AgNP-HSA also took place to alter the vesicle fluidity. As shown in Fig. 3, at low nanoparticle/protein concentrations, the effect of AgNPs on vesicle fluidity was similar to that induced by AgNP-HSA, implying that both AgNPs and HSA in the corona interacted with the lipid vesicles. In contrast, at high nanoparticle/protein concentrations, due to the presence of excess unbound HSA molecules, the vesicles experienced less perturbation from the AgNPs in the corona than from bare AgNPs.
Figure 3.
(Color online) Percent change of GP values for vesicles incubated with different concentrations of AgNPs, HSA, or AgNP-HSA. The concentration of the (DMPC + 10% DMPG) lipids was 0.05 mg/mL for all cases. The percent changes were calculated by comparing the actual GP values of the samples with that of the vesicle suspension (control). Since the GP values of the control were negative, a positive percent change corresponds to a decreasing GP value.
Taken together the results from the UV-vis, TEM, and CD measurements, it is evident that the binding of AgNP-HSA was mediated by the physical forces of hydrogen bonding, electrostatic interaction, and hydrophobic interaction. Upon the formation of AgNP-HSA corona, the percent of α-helices was reduced while that of β-sheets was increased in the HSA secondary structures, possibly resulting from breakage of the hydrogen bonds between neighboring α-helices and configuration of new, sterically less ordered hydrogen bonds between the α-helices and the citrate coating of the AgNPs, similarly to that observed for tubulins exposed to a fullerene derivative.31 As shown in the CD measurement, the presence of lipid vesicles alleviated the conformational changes of the proteins induced by the nanoparticles, likely due to the electrostatic repulsion between the vesicles and the nanoparticles. Conversely, the GP measurement demonstrated that both nanoparticles and protein corona interacted with lipid vesicles to enhance fluidity of the latter, although free proteins did not exert much effect on the vesicle conformation. Overall, our study suggests that the formation of nanoparticle-protein corona may negate, to certain extent, the physical interactions between the nanoparticle core and cell membranes. Such physical perspective, when combined with the biological and biochemical mechanisms of endocytosis, lipid peroxidation, and enzymatic activity,32 may prove essential for our understanding and prediction of the behavior of nanomaterials in biological systems for the advancement of nanomedicine and nanotoxicology.
Acknowledgments
This research was sponsored by NSF CAREER Award No. CBET-0744040 and U.S. EPA Grant No. R834092 to Ke and NIH RO1 No. ES019311 grant to Brown. The authors thank Pengyu Chen for insightful discussions.
References
- Nel A. E., Madler L., Velegol D., Xia T., Hoek E. M. V., Somasundaran P., Klaessig F., Castranova V., and Thompson M., Nature Mater. 8, 543 (2009). 10.1038/nmat2442 [DOI] [PubMed] [Google Scholar]
- Lewinski N., Colvin V., and Drezek R., Small 4, 26 (2008). 10.1002/smll.v4:1 [DOI] [PubMed] [Google Scholar]
- Ke P. C. and Lamm M. H., Phys. Chem. Chem. Phys. 13, 7273 (2011). 10.1039/c0cp02891f [DOI] [PubMed] [Google Scholar]
- Verma A. and Stellacci F., Small 6, 12 (2010). 10.1002/smll.200901158 [DOI] [PubMed] [Google Scholar]
- Klein J., Proc. Natl. Acad. Sci. U.S.A. 104, 2029 (2007). 10.1073/pnas.0611610104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch I., Salvati A., and Dawson K. A., Nat. Nanotechnol. 4, 546 (2009). 10.1038/nnano.2009.248 [DOI] [PubMed] [Google Scholar]
- Lynch I. and Dawson K. A., Nanotoday 3, 40 (2011). [Google Scholar]
- Cedervall T., Lynch I., Lindman S., Berggård T., Thulin E., Nilsson H., Dawson K. A., and Linse S., Proc. Natl. Acad. Sci. U.S.A. 104, 2050 (2007). 10.1073/pnas.0608582104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laera S., Ceccone G., Rossi F., Gilliland D., Hussain R., Siligardi G., and Calzolai L., Nano Lett. 11, 4480 (2011). 10.1021/nl202909s [DOI] [PubMed] [Google Scholar]
- Sund J., Alenius H., Vippola M., Savolainen K., and Puustinen A., ACS Nano 5, 4300 (2011). 10.1021/nn101492k [DOI] [PubMed] [Google Scholar]
- Ginzburg V. V. and Balijepalli S., Nano Lett. 7, 3716 (2007). 10.1021/nl072053l [DOI] [PubMed] [Google Scholar]
- Roiter Y., Ornatska M., Rammohan A. R., Balakrishnan J., Heine D. R., and Minko S., Nano Lett. 8, 941 (2008). 10.1021/nl080080l [DOI] [PubMed] [Google Scholar]
- Wang B., Zhang L., Bae S. C., and Granick S., Proc. Natl. Acad. Sci. U.S.A. 105, 18171 (2008). 10.1073/pnas.0807296105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen E., Lin S., Reid M. L., Allegood M. S., Wang X., Rao A. M., Vattulainen I., and Ke P. C., Small 4, 1986 (2008). 10.1002/smll.200701279 [DOI] [PubMed] [Google Scholar]
- Cho E. C., Xie J., Wurm P. A., and Xia Y., Nano Lett. 9, 1080 (2009). 10.1021/nl803487r [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yang M., Park J.-H., Singelyn J., Ma H., Sailor M. J., Ruoslahti E., Ozkan M., and Ozkan C., Small 5, 1990 (2009). 10.1002/smll.200900520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chompoosor A., Saha K., Ghosh P. S., Macarthy D. J., Miranda O. R., Zhu Z.-J., Arcaro K. F., and Rotello V. M., Small 6, 2246 (2010). 10.1002/smll.201000463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Ratnikova T. A., Stone M. B., Lin S., Lard M., Huang G., Hudson J. S., and Ke P. C., Small 6, 612 (2010). 10.1002/smll.200901911 [DOI] [PubMed] [Google Scholar]
- Chen R., Huang G., and Ke P. C., Appl. Phys. Lett. 97, 093706 (2010). 10.1063/1.3486681 [DOI] [Google Scholar]
- de Planque M. R. R., Aghdaei S., Roose T., and Morgan H., ACS Nano 5, 3599 (2011). 10.1021/nn103320j [DOI] [PubMed] [Google Scholar]
- Qiao R., Roberts A. P., Mount A. S., Klaine S. J., and Ke P. C., Nano Lett. 7, 614 (2007). 10.1021/nl062515f [DOI] [PubMed] [Google Scholar]
- Wong-ekkabut J., Baoukina S., Triampo W., Tang I. M., Tieleman D. P., and Monticelli L., Nat. Nanotechnol. 3, 363 (2008). 10.1038/nnano.2008.130 [DOI] [PubMed] [Google Scholar]
- Yuan H., Li J., Bao G., and Zhang S., Phys. Rev. Lett. 105, 138101 (2010). 10.1103/PhysRevLett.105.138101 [DOI] [PubMed] [Google Scholar]
- Yang K. and Ma Y.-Q., Nat. Nanotechnol. 5, 579 (2010). 10.1038/nnano.2010.141 [DOI] [PubMed] [Google Scholar]
- Shi X., von dem Bussche A., Hurt R. H., Kane A. B., and Gao H., Nat. Nanotechnol. 6, 714 (2011). 10.1038/nnano.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P. C. and Naumann C. A., Langmuir 17, 3727 (2001). 10.1021/la010116o [DOI] [Google Scholar]
- Hendren C. O., Mesnard X., Dröge J., and Wiesner M. R., Environ. Sci. Technol. 45, 2562 (2011). 10.1021/es103300g [DOI] [PubMed] [Google Scholar]
- Meierhofer T., van den Elsen J. M. H., Cameron P. J., Munoz-Berbel X., and Jenkins A. T. A., J. Fluoresc. 20, 371 (2010). 10.1007/s10895-009-0522-7 [DOI] [PubMed] [Google Scholar]
- Huynh K. A. and Chen K. L., Environ. Sci. Technol. 45, 5564 (2011). 10.1021/es200157h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohren C. F., Clothiaux E., and Huffman D. R., Absorption and Scattering of Light by Small Particles (Wiley VCH, Hoboken, NJ, 2010). [Google Scholar]
- Ratnikova T. A., Govindan P. N., Salonen E., and Ke P. C., ACS Nano 5, 6306 (2011). 10.1021/nn201331n [DOI] [PubMed] [Google Scholar]
- Mortimer M., Kasemets K., Vodovnik M., Marinšek-Logar R., and Kahru A., Environ. Sci. Technol. 45, 6617 (2011). 10.1021/es201524q [DOI] [PubMed] [Google Scholar]