Abstract
Exposure to aristolochic acid (AA) is associated with human nephropathy and urothelial cancer. Individual susceptibility to AA-induced disease likely reflects individual differences in enzymes that metabolize AA. Herein, we evaluated AAI metabolism by human cytochrome P450 (CYP) 1A1 and 1A2 in two CYP1A-humanized mouse lines that carry functional human CYP1A1 and CYP1A2 genes in the absence of the mouse Cyp1a1/1a2 orthologs. Human and mouse hepatic microsomes and human CYPs were also studied. Human CYP1A1 and 1A2 were found to be principally responsible for reductive activation of AAI to form AAI-DNA adducts and for oxidative detoxication to 8-hydroxyaristolochic acid (AAIa), both in the intact mouse and in microsomes. Overall, AAI-DNA adduct levels were higher in CYP1A-humanized mice relative to wild-type mice, indicating that expression of human CYP1A1 and 1A2 in mice leads to higher AAI bioactivation than in mice containing the mouse CYP1A1 and 1A2 orthologs. Furthermore, an exclusive role of human CYP1A1 and 1A2 in AAI oxidation to AAIa was observed in human liver microsomes under the aerobic (i.e., oxidative) conditions. Because CYP1A2 levels in human liver are at least 100-fold greater than those of CYP1A1 and there exists a > 60-fold genetic variation in CYP1A2 levels in human populations, the role of CYP1A2 in AAI metabolism is clinically relevant. The results suggest that, in addition to CYP1A1 and 1A2 expression levels, in vivo oxygen concentration in specific tissues might affect the balance between AAI nitroreduction and demethylation, which in turn would influence tissue-specific toxicity or carcinogenicity.
Keywords: aristolochic acid nephropathy, Balkan endemic nephropathy, cytochrome P450, metabolism, aristolochic acid Ia, DNA adducts, CYP1A-humanized mouse models
The herbal drug aristolochic acid (AA), derived from Aristolochia species, has been shown to be the cause of so-called Chinese herbs nephropathy, now termed aristolochic acid nephropathy (AAN) (Debelle et al., 2008; Schmeiser et al., 2009). The plant extract AA is a mixture of structurally related nitrophenanthrene carboxylic acids, the major components being aristolochic acid I (AAI; Fig. 1) and aristolochic acid II (AAII). AAN is a rapidly progressive renal fibrosis that was observed initially in a group of Belgian women who had ingested weight loss pills containing Aristolochia fangchi (Nortier et al., 2000; Vanherweghem et al., 1993). Within a few years of taking the pills, AAN patients also showed a high risk of upper urothelial tract carcinoma (about 50%) and, subsequently, bladder urothelial carcinoma (Lemy et al., 2008). In the meantime, similar cases have been reported elsewhere in Europe and Asia (Schmeiser et al., 2009). More recently, exposure to AA has been linked to Balkan endemic nephropathy (BEN) and its associated urothelial cancer (Arlt et al., 2007; Grollman et al., 2007). This nephropathy is endemic in certain rural areas of Serbia, Bosnia, Croatia, Bulgaria, and Romania.
FIG. 1.
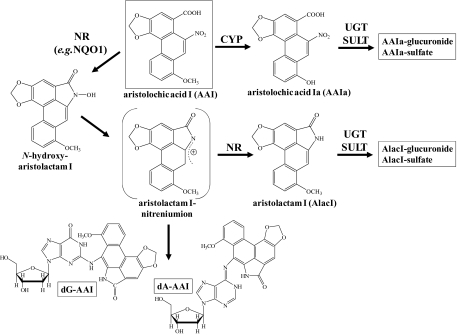
Pathways of AAI biotransformation and AAI-DNA adduct formation. dA-AAI, 7-(deoxyadenosin-N6-yl)aristolactam I; dG-AAI, 7-(deoxyguanosin-N2-yl)aristolactam I; NR, nitroreductase; UGT, UDP glucuronosyl transferase; and SULT, sulfotransferase.
Exposure to AA was demonstrated by the identification of specific AA-DNA adducts in urothelial tissue of AAN and BEN patients (Arlt et al., 2002; Grollman et al., 2007; Schmeiser et al., 1996). The most abundant DNA adduct detected in patients is 7-(deoxyadenosin-N6-yl)-aristolactam I (dA-AAI), which leads to characteristic AT→TA transversions. Such AT→TA mutations have been observed in the TP53 tumor suppressor gene in tumors from AAN and BEN patients (Arlt et al., 2007; Grollman et al., 2007), indicating a probable molecular mechanism associated with AA-induced carcinogenesis (Arlt et al., 2011b). AA was recently classified as carcinogenic to humans (group 1) by the International Agency for Research on Cancer (Grosse et al., 2009).
The activation pathway for AA is nitroreduction catalyzed by both cytosolic and microsomal enzymes; in this process NAD(P)H:quinone oxidoreductase (NQO1) is the most efficient cytosolic enzyme (Stiborová et al., 2003, 2008) (Fig. 1). In human hepatic microsomes, AAI is activated by cytochrome P450 (CYP) 1A2 and to a lesser extent by CYP1A1; P450 oxidoreductase (POR) also plays a minor role (Stiborová et al., 2001). Whereas the enzymes catalyzing the reductive activation of AAI leading to covalent DNA adducts have been widely investigated, those participating in its detoxication have not been extensively studied so far. Several studies have indicated that induction of CYP1A protects mice from AAI-induced acute renal injury (Xue et al., 2008). One detoxication metabolite identified is 8-hydroxyaristolochic acid I (aristolochic acid Ia, AAIa; Fig. 1), which is formed by AAI demethylation and which leads, in turn, to glucuronide or sulfate esters (Chan et al., 2006; Shibutani et al., 2010). Human and rodent CYP1A1 and 1A2 can oxidize AAI to AAIa in vitro (Levová et al., 2011; Rosenquist et al., 2010; Sistkova et al., 2008), and CYP1A1 and 1A2 in mice appear to mediate this reaction in vivo (Arlt et al., 2011a; Rosenquist et al., 2010). CYP1A1 and 1A2 also reductively activate AAI in human and rodent livers (Levová et al., 2011; Stiborová et al., 2001, 2005a,c, 2008). However, knowledge of the balance between oxidative detoxication and reductive activation of AAI by these human enzymes in vitro and in vivo is still lacking.
Much information about CYP regulation and function has been obtained by in vitro studies, but before extrapolation from in vitro data to in vivo pharmacokinetics can be made, additional factors need to be considered such as route-of-administration, absorption, renal clearance, and tissue-specific CYP expression (Nebert, 2006). Moreover, human/rodent CYP1A1 and 1A2 orthologs are known to exhibit species-specific differences in substrate preference and rates of metabolism (Aoyama et al., 1989; Turesky, 2005).
In the present study, we evaluated the oxidative detoxication and the reductive activation of AAI mediated by human CYP1A1 and 1A2 expressed in transgenic mouse models. We employed two humanized mouse lines, both carrying functional human CYP1A1 and CYP1A2 genes in place of the orthologous mouse genes; one line carries the high-affinity aryl hydrocarbon receptor (AHR) [hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1] (Dragin et al., 2007), whereas the other line carries the poor-affinity AHR [hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd] (Shi et al., 2008). The latter line is believed to be more relevant to human risk assessment vis-à-vis human CYP1A1 and CYP1A2 substrates because poor-affinity, rather than high-affinity, AHR is known to exist in human populations. DNA adduct formation in vivo and in vitro was investigated by the 32P-postlabeling method. Urinary AAIa and the CYP-mediated formation of AAIa in vitro in hepatic microsomes and by human recombinant CYPs were measured by high performance liquid chromatography (HPLC).
MATERIALS AND METHODS
Chemicals.
The natural mixture of AA consisting of 38% AAI and 58% AAII was purchased from Sigma Chemical Co (St Louis, MO). AAI (as sodium salt) was isolated from the mixture by preparative HPLC; its purity was 98% as estimated by HPLC (Schmeiser et al., 1996).
Animal treatment.
The hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 and hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd lines, both on a > 99.8% C57BL/6J background, were generated as reported (Dragin et al., 2007; Shi et al., 2008). Age-matched C57BL/6J mice wild-type (WT) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved by, and conducted in accordance with, the National Institute of Health standards for the care and use of experimental animals and the University of Cincinnati Medical Center Institutional Animal Care and Use Committee. Groups of female mice (3 months old; 25–30 g; n = 4 per group) were treated with a single dose of 50 mg/kg body weight AAI by oral gavage. AAI was dissolved in water at a concentration of 5 mg/ml. Control mice received gavage of solvent (water) only. Animals were killed 24 h after treatment. Several organs (liver, lung, kidney, bladder, spleen, and colon) were removed, snap frozen, and stored at −80°C until analysis. It is noteworthy that treatment of CYP1A-humanized mice with AAI was carried out in parallel with Cyp1a1(−/−), Cyp1a2(−/−), and Cyp1a1/1a2(−/−) mouse line treatments (Arlt et al., 2011a), whereas the WT mouse line was used as reference in both studies to allow direct comparison of the results.
DNA adduct analysis by 32P-postlabeling.
DNA from tissues was isolated by standard phenol/chloroform extraction. 32P-postlabeling analysis (Phillips and Arlt, 2007) using the nuclease P1 enrichment version and thin layer chromatography (TLC), and HPLC was performed as described (Schmeiser et al., 1996). Chromatographic conditions for TLC on polyethylenimine-cellulose plates (10 × 20 cm; Macherey-Nagel, Düren, Germany) were: D1, 1.0M sodium phosphate, pH 6.8; D3: 3.5 lithium-formate, 8.5M urea, pH 4; D4, 0.8M lithium chloride, 0.5M Tris-HCl, 8.5M urea, pH 9; and D5, 1.7M sodium phosphate, pH 6. After chromatography, TLC sheets were scanned using a Packard Instant Imager (Downers Grove, IL) and DNA adduct levels (RAL, relative adduct labeling) were calculated as described (Schmeiser et al., 1996). Results were expressed as DNA adducts per 108 nucleotides. AA-DNA adducts were identified using reference compounds as described (Schmeiser et al., 1996). Urothelial DNA samples from AAN patients were included in the analysis for comparison (Nortier et al., 2000).
Determination of AAIa by HPLC in mouse urine.
Urine samples (0.4–1.7 ml) collected from all mice treated with AAI were mixed with 4 vol of methanol, centrifuged (1000 revolutions per minute) for 4 min, and the supernatants then evaporated to dryness. The residues were dissolved in 100 μl of methanol and analyzed by HPLC. HPLC was performed with a reverse-phase column (Nucleosil 100-5 C18, 25 × 4.0 mm, 5 μm; Macherey-Nagel) proceeded by a C-18 guard column, using a linear gradient of 20–60% acetonitrile in 100mM triethylammonium acetate in 55 min with a flow rate of 0.6 ml/min. HPLC was carried out with a Dionex HPLC pump P580 with UV/VIS UVD 170S/340S spectrophotometer detector set at 254 nm, and peaks were integrated with CHROMELEON 6.01 integrator. A peak eluting at retention time (r.t.) 23.1 min was identified as AAIa using mass-spectroscopy analysis (compare Fig. 3A in Arlt et al., (2011a)). Mass spectra were measured on matrix assisted laser desorption/ionization-time of flight/time of flight (MALDI-TOF/TOF) ultraFLEX III mass spectrometers (Bruker-Daltonics, Bremen, Germany). Positive spectra were calibrated externally using the monoisotopic [M + H]+ ions of PepMixII calibrant (Bruker-Daltonics) or matrix peaks. A 10 mg/ml solution of α-cyano-4-hydroxy-cinnamic acid, or 50 mg/ml solution of 2,5-dihydrobenzoic acid in 50% MeCN/0.1% TFA, was used as a MALDI matrix. A 0.5-μl sample dissolved in MeCN was directly mixed with 0.5 μl of matrix solution and allowed to dry at ambient temperature on the target. The MALDI-TOF positive spectra were collected in reflector mode (Arlt et al., 2011a; Levová et al., 2011).
FIG. 3.
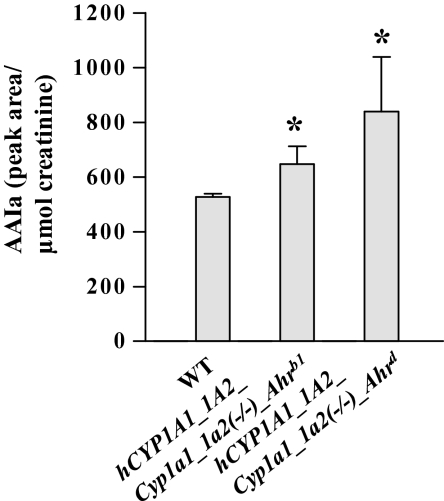
Urinary AAIa levels in CYP1A-humanized and WT mice (Arlt et al., 2011a) treated orally with 50 mg/kg body weight AAI for 24 h. All values are given as means ± SD (n = 4). Comparison was performed by t-test analysis; *p < 0.01, different from WT.
Determination of creatinine in urine samples.
Creatinine concentrations in urine samples were determined spectrophotometrically using a HELIOS Alpha spectrophotometer (Thermo Spectronics, U.K.) by the procedure described in the Creatinine Kit (BioSystems, Spain).
Preparation of microsomes and cytosols.
Hepatic, renal and pulmonary microsomes, and hepatic and renal cytosols from CYP1A-humanized and WT mice were isolated as described (Stiborová et al., 2001, 2005a). Because treating mice with AAI might influence levels and activities of biotransformation enzymes, microsomes and cytosols were isolated both from organs of control (untreated) mice and from those of mice treated with a single dose of 50 mg/kg body weight AAI by oral gavage (see above) and used for further analysis. Pooled microsomal and cytosolic fractions (n = 4 mice per group) were used for further analysis. Each mouse microsomal sample was analyzed for specific CYP1A1 and 1A2 activities by monitoring the following reactions: ethoxyresorufin O-deethylation (EROD) (CYP1A1/2) and methoxyresorufin O-deethylation (MROD) (CYP1A2). CYP1A1 activity was also determined using Sudan I oxidation to its C-hydroxylated metabolites (4′-hydroxy- and 6-hydroxy-Sudan I) in the presence of ketoconazole, a CYP3A4 inhibitor (Stiborová et al., 2002, 2005b). POR activity in hepatic microsomes was measured using cytochrome c as substrate (Stiborová et al., 2001).
Male human (pooled sample; catalog number 452172) and female human hepatic microsomes (pooled sample; cat. no. 452183) were purchased from Gentest Corp. (Woburn, MI). Human hepatic microsomes from individual donors (HG93, HG03, HG74, HG06, HK27, HG42, HG112, HG56, HG43, HG89, HG32, HK31, HK23, and HK34; Gentest) were also included. Donors ranged in age from 2 to 71 years and included seven men and seven women; drug and/or alcohol abuse history of the samples is described in Gentest protocols. Each human microsomal sample has been characterized for CYP and protein contents and specific CYP activities by Gentest Corp. We reanalyzed each microsomal preparation for specific CYP and POR activities by assays described in the protocols of Gentest Corp. Our data were similar to those reported by Gentest Corp. in their specification sheets. Moreover, we characterized these samples for the CYP1A1 marker activity using Sudan I, as described above. Data are shown in Table 1. Protein concentrations in microsomal and cytosolic fractions were assessed using the bicinchoninic acid protein assay with bovine serum albumin as a standard.
TABLE 1.
CYP-Mediated Formation of AAIa Form AAI in Human Hepatic Microsomes
| CYP-dependent catalytic activitiesa | |||||||||||||
| No. | pmol CYP per mg protein |
Sudan I oxidation in presence of ketoconazole (CYP1A1)b |
Phenacetin O-deethy-lation (CYP1A2) |
Coumarin 7-hydroxylation (CYP2A6) |
(S)-Mephenytoin N-demthy-lation (CYP2B6) |
Paclitaxel 6α-hydroxy-lation (CYP2C8) |
Diclofenac 4′-hydroxy-lation (CYP2C9) |
(S)-Mephenytoin 4′-hydroxy-lation (CYP2C19) |
Burfuralol 1′-hydroxy-lation (CYP2D6) |
Chloroxazone 6-hydroxy-lation (CYP2E1) |
Testosterone 6β-hydroxy-lation (CYP3A4) |
Lauric acid 12-hydroxy-lation (CYP4A) |
AAIs Peak area |
| HG93 | 290 | 1.1 | 750 | 340 | 25 | 180 | 2100 | 41 | 56 | 1800 | 2400 | 1600 | 6.0 |
| HG03 | 290 | 0.9 | 170 | 2000 | 51 | 200 | 1700 | 44 | 110 | 1800 | 6100 | 1600 | 2.0 |
| HG74 | 220 | 1.2 | 520 | 360 | 13 | 130 | 2100 | 55 | 120 | 1400 | 2700 | 1300 | 7.8 |
| HG06 | 300 | 0.9 | 770 | 580 | 3.0 | 130 | 2650 | 36 | ND | 1200 | 2990 | 740 | 5.8 |
| HK27 | 300 | 1.6 | 1320 | 1320 | 31 | 180 | 480 | 460 | 130 | 3000 | 4910 | 1110 | 10 |
| HG42 | 670 | 0.9 | 700 | 2200 | 150 | 480 | 1600 | 7.0 | 95 | 1600 | 15,000 | 1400 | 4.0 |
| HG112 | 560 | 0.5 | 320 | 490 | 150 | 180 | 4000 | 380 | 32 | 2400 | 25,000 | 1800 | 2.7 |
| HG56 | 550 | 3.0 | 2400 | 1500 | 48 | 180 | 3100 | 480 | 120 | 1900 | 5300 | 3000 | 18 |
| HG43 | 270 | 1.2 | 630 | 850 | 34 | 60 | 1400 | 700 | 20 | 1100 | 5600 | 2100 | 3.8 |
| HG89 | 400 | 1.6 | 1500 | 650 | 60 | 160 | 2100 | 190 | 13 | 1700 | 12,000 | 2200 | 16 |
| HG32 | 170 | 1.3 | 730 | 520 | 1.0 | 20 | 450 | 5.0 | 46 | 1200 | 2000 | 680 | 4.8 |
| HK31 | 580 | 2.5 | 1220 | 2160 | 8.0 | 130 | 1690 | 170 | 3.0 | 1660 | 8210 | 2010 | 17.5 |
| HK23 | 380 | 1.6 | 960 | 1100 | 24 | 160 | 2100 | 110 | 140 | 2100 | 6800 | 780 | 7.2 |
| HK34 | 500 | 1.6 | 1000 | 1500 | 39 | 220 | 1900 | 45 | 100 | 6000 | 5200 | 1100 | 6.15 |
| Meansc | 390 (150) | 1.4 (0.6) | 930 (540) | 1110 (650) | 46 (46) | 170 (100) | 1960 (890) | 200 (210) | 76 (47) | 2060 (1200) | 7440 (6010) | 1530 (630) | 8.0 (5.2) |
CYP activities in picomoles per minute per milligram protein. CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, and CYP4A determined by Gentest.
Picomole total C-hydroxylated metabolites per minute per milligram protein.
Arithmetic means for hepatic microsomal samples of 14 human donors and (SD), representing interindividual variability.
Microsomal incubations to study AAI demethylation.
Incubation mixtures, in a final volume of 250 μl, consisted of 100mM potassium phosphate buffer (pH 7.4), 1mM nicotinamide adenine dinucleotide phosphate reduced (NADPH), 1 mg human or mouse hepatic or mouse renal and pulmonary microsomal protein, and 10μM AAI. Incubations with microsomes were carried out at 37°C for 20 min; AAI oxidation (demethylation) to AAIa was determined to be linear up to 25 min. Control incubations were carried out (1) without microsomes, (2) without NADPH, or (3) without AAI. Supersomes isolated from insect cells transfected with baculovirus constructs containing complementary DNA of single human CYPs (CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A5), and expressing POR and/or cytochrome b5 were obtained from Gentest and tested for their efficiencies to oxidize AAI. Incubation mixtures, in a final volume of 250 μl, consisted of 100mM potassium phosphate buffer (pH 7.4), 1mM NADP+, 10mM MgCl2, 10mM D-glucose 6-phosphate, 1 U/ml D-glucose 6-phosphate dehydrogenase (NADPH-generating system), 50nM CYPs in Supersomes, and 10μM AAI. For controls, Supersomes containing POR alone were used. AAI and its metabolites (i.e., AAIa) were extracted from incubation mixtures twice with ethyl acetate (2 × 1 ml) and evaporated to dryness; residues were dissolved in 30 μl of methanol and subjected to reverse-phase HPLC as described above.
Microsomal AAI-DNA adduct formation.
Deaerated and argon-purged incubation mixtures, in a final volume of 750 μl, consisted of 50mM potassium phosphate buffer (pH 7.4), 1mM NADPH, 1 mg of hepatic or renal microsomal protein, 0.5 mg of calf thymus DNA (2mM dNp), and 0.5mM AAI. The reaction was initiated by adding NADPH. Microsomal incubations were carried out at 37°C for 60 min; microsomal-mediated AAI-derived DNA adduct formation was found to be linear up to 2 h (Stiborová et al., 2005a). Control incubations were carried out (1) without microsomes, (2) without NADPH, (3) without DNA, or (4) without AAI. After extraction with ethyl acetate, DNA was isolated from the residual. Water phase by the phenol/chloroform extraction method as described (Arlt et al., 2011a; Stiborová et al., 2005a).
Cytosolic AAI-DNA adduct formation.
The deaerated and argon-purged incubation mixtures, in a final volume of 750 μl, consisted of 50mM Tris-HCl buffer (pH 7.4), containing 0.2% Tween 20, 1mM NADPH, 1 mg mouse hepatic or renal cytosolic protein, 0.5 mg calf thymus DNA (2mM dNp), and 0.5mM AAI as described previously (Stiborová et al., 2003). Incubations with human cytosolic fractions were carried out at 37°C for 60 min; AAI-derived DNA adduct formation was found to be linear up to 2 h (Stiborová et al., 2003). Control incubations were carried out (1) without cytosol, (2) without NADPH, (3) without DNA, or (4) without AAI. DNA was then isolated as described above.
Inhibition studies.
The following chemicals were used to inhibit AAI demethylation by human hepatic microsomes and human recombinant CYP enzymes to AAIa: α-naphthoflavone (α-NF), which inhibits CYP1A1 and 1A2 (Stiborová et al., 2001, 2005a); furafylline, which inhibits CYP1A2 (Stiborová et al., 2001); diamantane, which inhibits CYP2B6 (Stiborová et al., 2002); sulfaphenazole, which inhibits CYP2C; quinidine, which inhibits CYP2D6; diethyldithiocarbamate (DDTC), which inhibits CYP2E1 and CYP2A6; and ketoconazole (KC), which inhibits CYP3A4 (Rendic and DiCarlo, 1997). Inhibitors were dissolved in 2.5 μl methanol (except for DDTC that was dissolved in distilled water) to yield final concentrations of 0.001–1mM in the incubation mixtures containing microsomes or human recombinant CYPs. These were incubated at 37°C for 10 min with the NADPH-generating system prior to adding AAI and then incubated for an additional 20 min at 37°C. AAI and its metabolite AAIa were extracted from incubation mixtures twice with ethyl acetate (2 × 1 ml) and analyzed by HPLC as described above.
In experiments investigating the inhibition of reductive AAI bioactivation (i.e., DNA adduct formation) in mouse hepatic microsomes, the following chemicals were used: α-NF; ellipticine (E), which competes with CYP1A1 substrates, thus inhibiting efficiently CYP1A1-mediated oxidation of other substrates (Stiborová et al., 2004); furafylline; and α-lipoic acid (α-LA), which inhibits POR (Stiborová et al., 2005a). Inhibitors were dissolved in 7.5 μl of methanol, to yield a final concentration of 0.1mM in the incubation mixtures. Mixtures were incubated at 37°C for 10 min with NADPH prior to adding AAI and then incubated further for 1 h at 37°C. After the incubation, DNA was isolated as described above.
RESULTS
AAI-DNA Adduct Formation in WT versus CYP1A-Humanized Mice
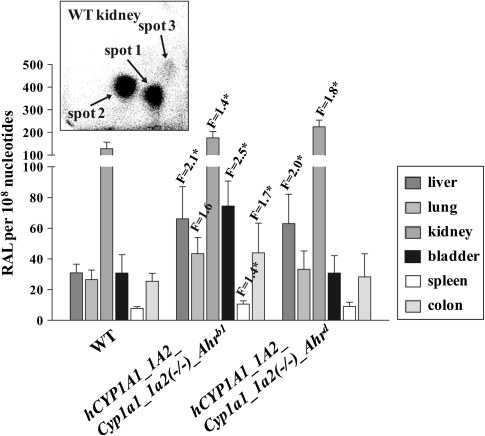
AAI-DNA adduct formation was determined by 32P-postlabeling in the liver, lung, kidney, bladder, spleen, and colon of WT, hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 and hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice treated with a single oral dose of 50 mg/kg body weight AAI for 24 h. Adduct patterns induced by AAI in all organs tested were qualitatively similar to those found in vivo in humans, consisting of two major adducts (spot 1 and 2) and one minor adduct (spot 3) (see Fig. 2, insert) (Nortier et al., 2000; Schmeiser et al., 1996). These adducts have previously been identified (Schmeiser et al., 1996) as 7-(deoxyadenosin-N6-yl)aristolactam I (spot 1; dA-AAI), 7-(deoxyguanosin-N2-yl)aristolactam I (spot 2; dG-AAI), and 7-(deoxyadenosin-N6-yl)aristolactam II (spot 3; dA-AAII). No adducts were observed in DNA of untreated animals.
FIG. 2.
Quantitative TLC 32P-postlabeling analysis of AAI-DNA adduct formation in organs of CYP1A-humanized and WT mice (Arlt et al., 2011a) treated orally with 50 mg/kg body weight AAI for 24 h. F, fold increase in DNA adduct formation in CYP1A-humanized mice compared with WT mice. -Ahrb1 = high-affinity -AHR. Ahrd = poor-affinity AHR. Values are given as means ± SD (n = 4); each DNA sample was determined by two postlabeled analyses. RAL, relative adduct labeling. Comparison was performed by t-test analysis; *p < 0.01, different from WT. Insert: Autoradiographic profile of AAI-DNA adducts in kidney from WT mice by using the nuclease P1 enrichment version of the assay. The adduct profile shown is representative of those obtained in CYP1A-humanized mice and other organs (i.e., liver, lung, bladder, spleen, and colon). The origin, in the bottom left-hand corner, was cut off before exposure. Spot 1, dA-AAI; spot 2, dG-AAI; and spot 3, dA-AAII.
In all mouse lines, the adduct levels were much higher in kidney than in other organs (Fig. 2). Compared with WT mice, up to 2.5-fold higher levels of AAI-DNA adducts were found in liver, lung, kidney, urinary bladder, spleen, and colon of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 and in liver and kidney of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice (p < 0.01). These findings indicate that expression of human CYP1A1 and 1A2 in mice lacking the mouse orthologous enzymes, increases the activation of AAI to reactive intermediates capable of forming DNA adducts in several organs.
Determination of AAIa in Mouse Urine
CYP1A-humanized mice also exhibited AAI oxidation, as measured by HPLC analysis of urine (compare peak with r.t. 23.1 min in Fig. 3A in Arlt et al. [2011a]), representing AAI detoxication. using positive MALDI-TOF/TOF peaks at m/z 327.029 and 328.043 were recorded, representing the molecular ions [M-H]+ and [M]+ of AAIa, respectively, and peaks at m/z 283.021 and 311.031, representing ions of AAIa fragments; these data confirmed that the metabolite is AAIa (Arlt et al., 2011a; Levová et al., 2011). AAI was not detected in urine from any mouse line. Compared with WT mice, 1.2- and 1.6-fold higher concentrations of AAIa were found in urine of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 and hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice, respectively (Fig. 3). Because the high-affinity AHR mouse would be more easily induced, in turn, it would also be more likely to exhibit enhanced clearance (detoxication, degradation) of any inducer (e.g., AAI) that can be metabolized by CYP1A.
Microsomal versus Cytosolic Activation of AAI
CYP-mediated AAI-DNA adduct formation was also investigated in hepatic microsomes isolated from WT and CYP1A-humanized mice either untreated (i.e., control) or pretreated with 50 mg/kg body weight AAI by oral gavage (see “Materials and Methods” section). These incubations were performed under hypoxic conditions in which each incubation mixture was purged with a stream of argon for 3 min before the addition of AAI. Although most of the oxygen was removed, we cannot exclude the negligible presence of O2 in the microsomal membranes and lumen.
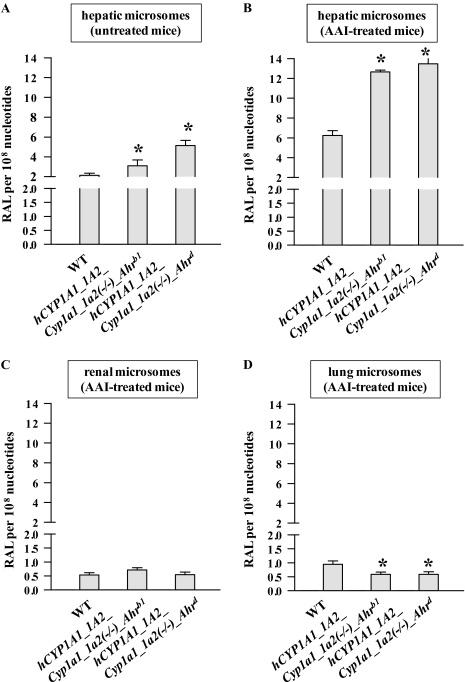
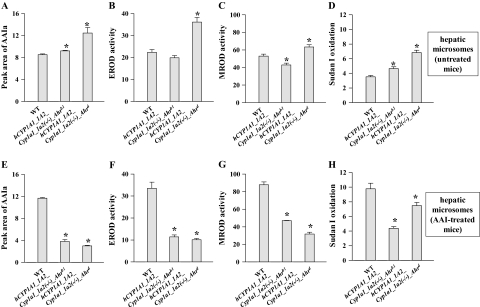
AAI was reductively activated by hepatic microsomes from all mouse lines, as evidenced by AAI-DNA adduct formation (Fig. 4), generating the same pattern of DNA adducts as that obtained in vivo in mice (compare Fig. 2, insert). No adducts were observed in control incubations carried out in parallel without microsomes, or without DNA, or without AAI. Higher levels (1.5- to 2.4-fold) of AAI-DNA adducts were detected in incubations with hepatic microsomes from untreated CYP1A-humanized mice relative to microsomal incubations from WT mice (Fig. 4A). The increase in total AAI-DNA adduct levels correlated with an increase in Sudan I oxidation (marker for human CYP1A1) in hepatic microsomes of both CYP1A-humanized mice and with EROD (marker for CYP1A1/2) or MROD (marker for CYP1A2) activity in hepatic microsomes of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice (Figs. 5B–D). With hepatic microsomes of WT mice pretreated with AAI, more than threefold higher levels of AAI-DNA adducts were found relative to those of untreated WT mice (p < 0.01) (Figs. 4A and B). Again, these higher levels of AAI-DNA adducts corresponded to increased activities of CYP1A1 and 1A2 (Sudan I oxidation, EROD, and MROD) in these microsomes (compare Fig. 5). Collectively, these results clearly show that human CYP1A1 and 1A2 activate AAI to intermediates capable of forming DNA adducts and that hepatic microsomes containing the human enzymes are more effective in AAI bioactivation, compared with mouse microsomal CYP1A1 and 1A2.
FIG. 4.
DNA adduct formation by AAI in microsomes isolated from CYP1A-humanized and WT mice (Arlt et al., 2011a), untreated (A) or pretreated with AAI (B–D). (A) Hepatic microsomes from untreated mice were used. Hepatic (B), renal (C), and pulmonary (D) microsomes from mice pretreated with AAI were used. Values are given as means ± SD of three parallel incubations (n = 3). RAL, relative adduct labeling. Comparison was performed by t-test analysis; *p < 0.01, different from WT.
FIG. 5.
Oxidation of AAI to AAIa by mouse hepatic microsomes isolated from CYP1A-humanized and WT mice (Arlt et al., 2011a), untreated (A) or pretreated with AAI (E). CYP1A enzymatic activity in hepatic microsomes as measured by EROD activity (picomoles resorufin per minute per milligram protein) (B, F) MROD activity (C, G) (picomoles resorufin per minute per milligram protein) or Sudan I oxidation (picomole total C-hydroxylated metabolites per minute per milligram protein) (D, H). All values are given as means ± SD of three parallel incubations (n = 3). Comparison was performed by t-test analysis; *p < 0.01, different from WT.
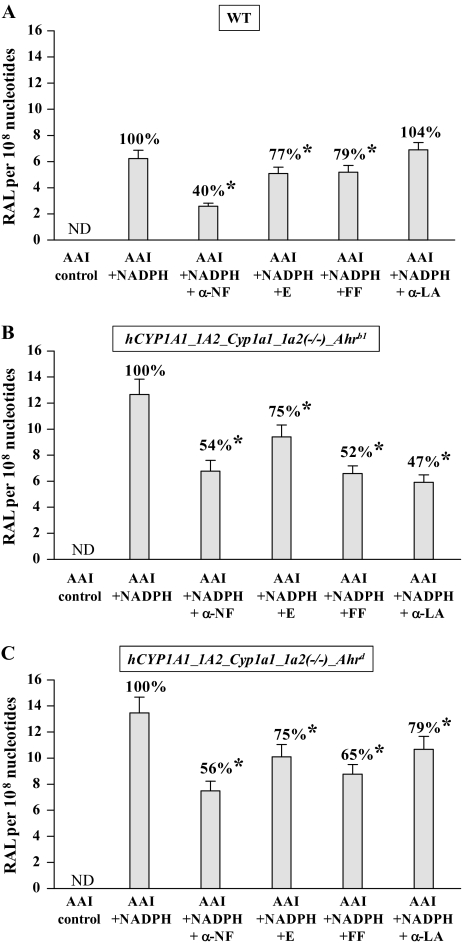
However, the increase in AAI-DNA adduct formation found in in vitro incubations with hepatic microsomes isolated from CYP1A-humanized mice treated with AAI (Fig. 4B) did not correspond to CYP1A activities in these microsomes (compare Figs. 5F–H). Lower activities of CYP1A1 and 1A2 were found in these microsomes with their marker substrates relative to those of WT mice (compare Figs. 5F–H). Results of experiments using inhibitors of CYP1A1, CYP1A2, and POR might explain these discrepancies (Fig. 6). With all samples of hepatic microsomes isolated from mice pretreated with AAI, AAI-DNA adduct formation was found to be inhibited by α-NF, most effectively in WT microsomes (down to 40%), followed by microsomes from both CYP1A-humanized mice (down to 55%). Ellipticine decreased AAI-DNA adduct formation to ∼75% in all hepatic microsomes. Furafylline decreased DNA adduct levels by AAI to ∼50–80% in all hepatic microsomal samples. Interestingly, even though POR activities in all hepatic microsomes did not differ significantly (Supplementary table 1), the POR inhibitor α-LA prevented AAI-DNA adduct formation, but only in hepatic microsomes isolated from CYP1A-humanized mice. α-LA was most effective as an inhibitor in the hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 line, whereas no effect was observed in microsomes isolated from WT mice (Fig. 6).
FIG. 6.
Effect of CYP and POR inhibitors on AAI-DNA adduct formation catalyzed by hepatic microsomes isolated from CYP1A-humanized and WT mice (Arlt et al., 2011a) pretreated with AAI. Values are given as means ± SD of three parallel incubations (n = 3). RAL, relative adduct labeling. Comparison was performed by t-test analysis; *p < 0.01, different from incubation with NADPH alone. Inhibition is compared with percentage DNA adduct formation in incubations with NADPH as cofactor only. Control, without cofactor. α-NF, α-naphthoflavone. E, ellipticine. FF, furafylline. α-LA, α-lipoic acid. ND, not detected.
Our experiments with CYP1A inhibitors support the above finding that most AAI activation in hepatic microsomes from WT mice pretreated with AAI is attributed to CYP1A1 and 1A2. In contrast, the inhibitory results with α-LA in hepatic microsomes from CYP1A-humanized mice pretreated with AAI (principally seen in hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 mice) suggest that POR, in addition to CYP1A1 and 1A2, might also contribute to AAI-DNA adduct formation (Figs. 6B and C).
Renal and pulmonary microsomes isolated from mice pretreated with AAI were much less effective in activating AAI than hepatic microsomes (compare Figs. 4C and D). Pulmonary microsomes from CYP1A-humanized mice were slightly less effective in activating AAI than microsomes from WT mice (Fig. 4D).
Because NQO1 is an important enzyme catalyzing the reductive activation of AAI, we compared renal and hepatic cytosols of WT and CYP-humanized mice isolated from AAI-treated mice. No differences were observed in cytosols from either organ in the three mouse lines. As expected, hepatic cytosols were more active than renal cytosols (Supplementary figs. 1A and B).
Human and Mouse Hepatic Microsomes Oxidize AAI to AAIa
To examine the role of human CYP1A1 and 1A2 in oxidative detoxication of AAI, we performed in vitro incubations using the same hepatic mouse microsomes as those used in the experiments above, except that incubations were carried out under aerobic conditions. In addition, human hepatic microsomes, both pooled microsomes as well as microsomes from fourteen individual human donors, were employed in this study.
All mouse and human hepatic microsomes were able to metabolize AAI to AAIa. A 1.1- to 1.5-fold increase in AAIa formation was found in hepatic microsomes from untreated CYP1A-humanized mice, compared with that in WT mice (Fig. 5A). This increase correlated with enhanced Sudan I oxidation (marker for CYP1A1; Fig. 5D) in hepatic microsomes of both CYP1A-humanized mouse lines and with EROD (marker for CYP1A1/2; Fig. 5B) and with MROD (marker for CYP1A2; Fig. 5C) activity in hepatic microsomes of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice. Collectively, these data show that CYP1A1 and 1A2 are responsible for AAI demethylation to form AAIa and that microsomes containing human CYP1A1 and 1A2 seem to be more effective than microsomes containing the mouse orthologous enzymes.
The amounts of AAIa formed in hepatic microsomes of mice pretreated with AAI were greater than those of untreated mice. In microsomes isolated from WT mice pretreated with AAI, 1.4-fold greater amounts of AAIa were formed than from untreated WT mice (p < 0.01) (compare Figs. 5A and E). In contrast, AAI pretreatment of mice led to ∼70% decrease in AAIa formation catalyzed by hepatic microsomes from CYP1A-humanized mice. Again, the decrease in AAI demethylation to AAIa correlated with marker activities of CYP1A enzymes (compare Figs. 5E–H). These results indicate that AAI induces CYP1A activities in WT mice, whereas AAI does not induce human CYP1A in the humanized mouse lines.
In contrast to mouse hepatic microsomes, mouse renal and pulmonary microsomes did not oxidize AAI to AAIa under the same experimental conditions (data not shown).
To resolve further the importance of human CYP1A1- and 1A2-mediated demethylation of AAI to AAIa, three more experimental approaches were employed: (1) use of Supersomes, (2) correlation of CYP enzyme activities in human hepatic microsomes with AAIa formation by the same microsomes, and (3) selective enzyme inhibition in human microsomes.
Oxidation of AAI to AAIa in Supersomes
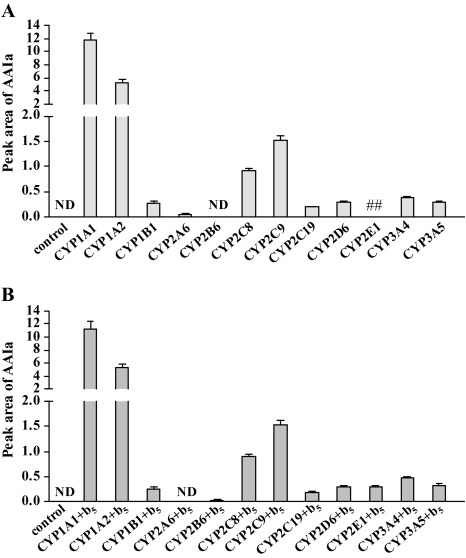
Experiments were conducted using Supersomes containing human CYPs, POR, and/or cytochrome b5 (Fig. 7). Recombinant human CYPs used in these experiments efficiently oxidized their typical substrates (data not shown). Human CYP1A1 was the most efficient at demethylating AAI to AAIa, followed by CYP1A2. Other human CYPs, including CYP1B1, 2C, 2D6, and 3A, were also capable of AAI demethylation, but to a much lesser extent. Cytochrome b5 had essentially no influence on product yield (Fig. 7). No AAIa formation occurred in control incubations with Supersomes containing POR alone.
FIG. 7.
AAIa formation after incubation of AAI with Supersomes, each containing a different human recombinant CYP (50nM) (A) and containing these enzymes in combination with cytochrome b5 (b5) (B). Values are given as means ± SD of three parallel incubations (n = 3). ND, not detected. ##, not determined.
Correlation of CYP Enzyme Activities in Human Hepatic Microsomes with AAI Demethylation to AAIa
All human hepatic microsomes used in the present study were able to catalyze reactions known to be associated with specific CYP enzymes (Table 1). Large interindividual variations in the catalytic activities were evident among different hepatic microsomal samples (Table 1). As shown in Table 1, AAIa formation in human hepatic microsomes also displayed wide interindividual variation. The levels of AAIa formed in these microsomes were highly significantly correlated with Sudan I oxidation, a marker for CYP1A1, and with phenacetin O-deethylase activity, a marker for CYP1A2 (Table 2). Likewise, using bilinear regression between activities of CYP1A1 (Sudan I oxidation) plus CYP1A2 (phenacetin O-deethylation) and AAIa formation, a highly significant correlation was found (R = 0.899). No significant correlation was seen with any of the other nine CYPs examined, or with total levels of CYP content, vis-à-vis AAIa formation. These findings indicate that human CYP1A1 and 1A2 are the major hepatic enzymes catalyzing AAI demethylation.
TABLE 2.
Linear Regression Correlation Coefficients among CYP Enzyme Activities and AAIa Formation in Human Hepatic Microsomesa
| pmol CYP per mg protein | Sudan I oxidation in presence of ketoconazole (CYP1A1)b | Phenacetin O-deethy-lation (CYP1A2) | Coumarin 7-hydroxylation (CYP2A6) | (S)-Mephenytoin N-demthy-lation (CYP2B6) | Paclitaxel 6α-hydroxy-lation (CYP2C8) | Diclofenac 4′-hydroxy-lation (CYP2C9) | (S)-Mephenytoin 4′-hydroxy-lation (CYP2C19) | Burfuralol 1′-hydroxy-lation (CYP2D6) | Chloroxazone 6-hydroxy-lation (CYP2E1) | Testosterone 6β-hydroxy-lation (CYP3A4) | Lauric acid 12-hydroxy-lation (CYP4A) | |
| Correlation coefficient | 0.307 | 0.884 | 0.932 | 0.268 | −0.265 | −0.119 | 0.067 | 0.215 | −0.123 | −0.030 | −0.110 | 0.559 |
| Significance | 0.285 | <0.0001 | <0.0001 | 0.355 | 0.360 | 0.685 | 0.820 | 0.460 | 0.688 | 0.919 | 0.707 | 0.058 |
Data calculated from Table 1.
CYP enzymes are shown in brackets.
Effect of Inhibitors of CYP Enzymes on AAI Demethylation Catalyzed by Human Hepatic Microsomes and Human CYPs
Inhibition experiments supported the role of CYP1A1 and 1A2 in AAI demethylation in human hepatic microsomes. As shown in Table 3, α-NF and furafylline were effective in inhibiting AAIa formation. Moreover, most inhibitors of other CYP enzymes (diamantane, sulfaphenazole, quinidine, and DDTC) were ineffective.
TABLE 3.
Effects of CYP Inhibitors on AAI Demethylation to AAIa by Human Hepatic Microsomes versus Human Recombinant CYP Enzymes
| Inhibitora | IC50 (μM) for AAIa formationb |
||
| Human microsomes | Human recombinant CYPs | ||
| α-Napththoflavone (CYP1A1/2) | 80.9 ± 6.5 | CYP1A1 | 0.2 ± 0.02 |
| Furafylline (CYP1A2) | 26.8 ± 2.2 | CYP1A2 | 1.5 ± 0.1 |
| Diamantane (CYP2B6) | NIc | — | — |
| Sulfaphenzazole (CYP2C) | NI | — | — |
| Quinidine (CYP2D6) | NI | — | — |
| DDTC (CYP2A, CYP2E1) | NI | — | — |
| Ketoconazole (CYP3A4) | 23.5 ± 2.1 | CYP3A4 | — |
| Ketoconazole | — | CYP1A1 | 0.16 ± 0.02 |
| Ketoconazole | — | CYP1A2 | 14.3 ± 0.95 |
—, not determined.
CYP enzymes that are known to be inhibited by selective inhibitors are shown in brackets.
Estimated from concentration-dependent inhibition of AAI metabolite formation by interpolation (inhibitors were 1−1000μM depending on the chemical). AAI (10μM) and 0.1 nmol of CYP were present in the incubation mixture. All values represent the mean ± SD of three parallel determinations (n = 3).
NI, no inhibition IC50 > 100μM.
However, although human liver is enriched for many CYPs, some of these enzymes are expressed in liver at very low levels (e.g., CYP1A1, 1B1, and 2B6); this feature can therefore influence the degree of their inhibition. In addition, it should be noted that results with inhibitors are sometimes difficult to interpret because one inhibitor may be more effective with one substrate than another. Therefore, we also studied the effects of these specific CYP inhibitors on AAIa formation by the recombinant human CYPs.
As shown in Table 3, α-NF and furafylline strongly inhibited AAI oxidation to AAIa catalyzed by human recombinant CYP1A1 and 1A2. Therefore, inhibition of AAI demethylation by these compounds in human microsomes must reflect inhibition of these enzymes. Because of the low efficiency of human recombinant CYP3A4 to catalyze AAIa formation (compare Fig. 7), CYP3A4 inhibition by ketoconazole could not be determined in Supersomes. However, we did find that ketoconazole inhibits oxidation of AAI to AAIa catalyzed by human recombinant CYP1A1 and 1A2 (Table 3). Therefore, the effect of ketoconazole on AAI demethylation in human hepatic microsomes is probably not due to inhibition of CYP3A4 but rather to inhibition of CYP1A1 and 1A2.
Contribution of Individual Human Liver Microsomal CYP Enzymes to AAI Demethylation
Based on our data showing AAI oxidation to AAIa by individual human recombinant CYPs (see Fig. 7) and inhibition of individual CYPs in human hepatic microsomes (see Table 3), as well as the typical expression levels of CYP enzymes in human hepatic microsomes (Rendic and DiCarlo, 1997), we were able to estimate the contribution of individual CYPs to the formation of AAIa in human hepatic microsomes. Even though human CYP1A1 is the most effective enzyme in AAI demethylation (Fig. 7), its contribution to AAIa formation in human hepatic microsomes is lower than that of CYP1A2 due to its extremely low expression in human livers (i.e., < 1% of total hepatic CYP) (Ikeya et al., 1989; Stiborová et al., 2002, 2005a). CYP1A1 contributes ∼13% to AAIa formation in human liver, whereas CYP1A2 contributes ∼87%. No other human hepatic CYP appears to participate significantly in AAI demethylation to AAIa in human liver microsomes. These findings correspond well with results showing high correlation coefficients that were seen only between activities of CYP1A1 and 1A2 and AAIa formation (compare Table 2).
DISCUSSION
A large body of evidence indicates that the nephrotoxic and carcinogenic effects of AAI are reflected largely by metabolism. Whereas AAI reduction leads to bioactivation to a cyclic acylnitrenium ion that generates DNA adducts, oxidation of AAI to AAIa is a detoxication reaction because AAIa is much less toxic than AAI (Shibutani et al., 2010). In addition, AAIa and its conjugates, the glucuronide, the acetate, and the sulfate esters, are readily excreted in urine (Chan et al., 2006).
AAIa formation was recently found to be catalyzed mainly by human and rat CYP1A1 and 1A2 in vitro (Levová et al., 2011; Rosenquist et al., 2010; Sistkova et al., 2008) and by mouse CYP1A1 and 1A2 in vivo (Arlt et al., 2011a; Rosenquist et al., 2010). However, depending on experimental conditions (i.e., oxygen concentrations), human CYP1A1 and 1A2 also reductively activate AAI to form DNA adducts in vitro (Levová et al., 2011; Stiborová et al., 2001, 2005a,c). In the present report, we therefore investigated the balance between oxidative detoxication and reductive activation of AAI in vitro and in vivo in detail.
Herein, we employed the CYP1A-humanized hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 and hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mouse lines that carry functional human CYP1A1 and CYP1A2 genes and lack the mouse orthologous genes; the former line harbors the high-affinity AHR, the latter has the poor-affinity AHR (Dragin et al., 2007; Shi et al., 2008). We have demonstrated for the first time that human CYP1A1 and 1A2 enzymes are responsible for both AAI activation (to a cyclic nitrenium ion that forms DNA adducts) and AAI demethylation to AAIa (a detoxication pathway), both in vitro and in the intact animal. Both reductive activation and oxidative detoxification of AAI in the humanized lines are more extensive than in the WT mouse line, without a clear difference between the contribution of CYP1A1 and 1A2. Also, we have proven the participation of mouse CYP1A1 and 1A2 in reductive activation by using selective inhibitors of these enzymes in in vitro experiments.
Our results are consistent with a recent study employing the HRN (Hepatic Cytochrome P450 Reductase Null) mouse model in which POR-mediated CYP activity is deleted specifically in liver (Levová et al., 2011). Moreover, the essential role of mouse CYP1A1 and 1A2 in AAI metabolism in vivo has also been confirmed in a study using Cyp1a knock out mouse models from which the lines used in the present study were created (Arlt et al., 2011a).
These results indicate that AAI is a ligand substrate for the CYP1A1 and 1A2 enzymes at low oxygen concentrations and is reduced instead of oxidized during the CYP-mediated reaction cycle. In contrast, under aerobic conditions, AAI is a classical substrate of CYP1A1 and 1A2 in which one atom of atmospheric oxygen is used to O-demethylate the methoxy group of AAI to generate AAIa.
Our present results also demonstrate that the CYP1A-humanized mouse lines used in this study are excellent models to further investigate the toxic effects of AAI, as well as the effects of other toxic agents (Cheung et al., 2005).
Curiously, whereas AAI pretreatment of WT mice led to increased CYP1A enzyme activities in liver, decreases in these hepatic enzyme activities were observed in both CYP1A-humanized mouse lines. This is in contrast to the enhanced CYP1A1 expression and also of CYP1A2 by 2,3,7,8-tetrachlorodibenzodioxin used by Shi et al. (2008) to distinguish the AHR receptors in the two lines. There a clear dependence of the induction levels on the AHR receptor affinity was observed. The conclusion of these observations is that AAI does not induce human CYP1A via the AHR. Because AHR is the only promoter known for CYP1A, AAI is probably not an inducer of CYP1A in humans.
The two CYP1A-humanized mouse lines employed in the study differ in their efficiencies to metabolize AAI in different organs. Higher levels of AAI-DNA adducts were formed in kidney and liver of both lines, compared with that in WT mice; however, only the high-affinity AHR line showed higher AAI-DNA adduct levels also in lung, bladder, spleen, and colon. This finding might be explained by the high-affinity AHR being more “sensitive” to local concentrations of the inducer AAI in these tissues (than the poor-affinity AHR), thereby leading to enhanced reductive metabolism to form the AAI-DNA adducts. Collectively, these results indicate that human CYP1A enzymes expressed in these mice favor AAI reduction. The overall enhanced DNA adduct formation in high-affinity AHR mice, compared with low-affinity AHR mice, is inversely correlated with the levels of AAIa urinary excretion; higher amounts of urinary AAIa were seen in hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice than in hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 mice (see Fig. 3). Nevertheless, it should be noted that the levels of free AAIa in urine might not reflect the total detoxication of AAI (Arlt et al., 2011a). Previous studies (Chan et al., 2006; Shibutani et al., 2010) showed that AAIa, which is a major metabolite in urine, can form conjugates with glucuronide, sulfate, and acetate, which could also be excreted in urine and feces. However, the measurement of these conjugates was beyond the scope of the present study.
The results found in our in vitro inhibition experiments show that, besides human CYP1A1 and 1A2, POR also contributes to AAI-DNA adduct formation, mainly in hepatic microsomes of hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 mice. Thus, the spectrum of enzymes involved in AAI reductive bioactivation appear to be different in WT (predominantly CYP1A-mediated) and CYP1A-humanized lines (CYP1A- and POR-mediated), but the contribution of CYP1A1 and 1A2 appear not to be very different. These results are consistent with our earlier findings that purified POR alone is able to catalyze AAI-DNA adduct formation in vitro (Stiborová et al., 2001). However, POR seems to have a rather low, but detectable, capacity to activate AAI in hepatic microsomes, compared with CYP1A1 or 1A2 (Stiborová et al., 2001; present work). The contribution of POR or CYP1A1 and 1A2 to AAI reduction is thus dictated by a competition between the active sites of these enzymes and their quantitative amounts. As demonstrated by the effective inhibition of AAI-DNA adduct formation by α-LA in hepatic microsomes from hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1, the contribution of POR to AAI activation in hepatic microsomes of this mouse line is higher than that in hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice. In addition, we found lower efficiencies of AAI demethylation to AAIa by human CYP1A1 and 1A2 in in vitro experiments using hepatic microsomes from hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrb1 than from hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mice. Moreover, these findings were in good agreement with the CYP1A1 and 1A2 marker activities (i.e., Sudan I oxidation, EROD, and MROD). The observations confirm the lower basal CYP1A1 messenger RNA levels in the low-affinity AHR line (Shi et al., 2008).
AAN patients rapidly progress to end-stage renal disease despite cessation of AA-containing products (Debelle et al., 2008), and tubulointerstitial hypoxia in chronic kidney disease plays a major role in the progression to end-stage renal disease (Mimura and Nangaku, 2010). One of the fundamental physiological differences between tumor cells and normal cells is the cellular ability to survive under hypoxic conditions, indicating that hypoxia is a key regulatory factor in tumor growth (Harris, 2002). Our recent studies in the HRN and Cyp1a knock out mouse models (Arlt et al., 2011a; Levová et al., 2011) as well as our findings of the present study with CYP1A-humanized mice suggest that, in addition to the influence of hepatic CYP1A expression, the in vivo oxygen concentrations in tissues may affect the balance between AAI nitroreduction and demethylation, thereby influencing the level of (geno)toxicity induced. Therefore, a low in vivo oxygen level in the kidney could be a critical factor not only in AAI metabolism but also in the potential development of tubulointerstitial hypoxia.
It has been suggested that the hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd line is more relevant to human risk assessment vis-à-vis human CYP1A1 and CYP1A2 substrates because humans have the low-affinity AHR (Shi et al., 2008). Based on this and the present data showing a higher potency of the low-affinity AHR CYP1A-humanized line to detoxify AAI, we suggest that a higher proportion of human CYP1A1- and 1A2-catalyzed reactions might be attributed to AAI detoxication. In contrast, the role of human CYP1A in AAI activation appears to be important only in some organs, predominantly those in which conditions that are more hypoxic occur. Therefore, we evaluated in detail the contribution of human CYP1A1 and 1A2 as the key enzymes for AAI demethylating to AAIa in human liver, which is the major organ that demethylates AAI.
Studying hepatic microsomes from fourteen human donors and a panel of human recombinant CYPs, we identified the impact of human CYP1A1 and 1A2 in demethylation of AAI. High correlation coefficients were found between the activity of CYP1A1 or CYP1A2 and the AAIa formation. This finding, and the results showing inhibition of only these enzymes in human hepatic microsomes, indicates an exclusive role of CYP1A1 and 1A2 in AAIa formation in human liver. A major contribution of CYP1A2 (∼87%) to this reaction was also confirmed by strong inhibition of CYP1A2-mediated AAI demethylation in human hepatic microsomes by furafylline. CYP1A1-mediated oxidation of AAI to AAIa is also important (∼13% contribution), even considering the very low levels of CYP1A1 expression in human liver (< 1% of total liver CYP content) (Ikeya et al., 1989; Stiborová et al., 2002, 2005b). This can be explained by the fact that human recombinant CYP1A1 is the most effective CYP enzyme catalyzing AAI demethylation. Although other human recombinant CYPs (including CYP2C8, 2C9, 3A4, 1B1, 2D6, and 2E1) were also found to oxidize AAI, their contributions to this reaction in human liver appear to be negligible.
Taking into account previous data showing a major role of human NQO1 in AAI activation (Stiborová et al., 2003, 2008, 2011) and results of the present work indicating also a role for CYP1A1 and 1A2 in this process, we propose that the pathways of AAI metabolism in humans are mainly dictated by the Km of AAI for human NQO1 versus CYP1A1 and 1A2, and their enzymatic turnover, as well as the balance between CYP1A efficiency of AAI oxidation versus reduction. Because human CYP3A5 also activates AAI (Levová et al., 2011) and is expressed in kidney, the target organ for AA toxicity, its participation in AAI-DNA adduct formation in the human urothelium may be of considerable importance. Each of these enzymes exhibits polymorphisms (http//cypalleles.ki.se), which are likely to influence further each individual’s susceptibility to AA. Indeed, it was reported that human NQO1 genetic polymorphisms are important in AA-induced BEN (Toncheva et al., 2004). One of the NQO1 polymorphisms, the genotype NQO1*2/*2, has already been shown to predispose BEN patients to urothelial cancer (odds ratio = 13.75, 95% confidence interval 1.17–166.21) (Toncheva et al., 2004). In addition, a pilot study has shown a weak association between the human CYP3A5*1 allele and BEN (Atanasova et al., 2005).
Even though CYP1A1 and 1A2 exhibit genetic polymorphisms, which subsequently could modulate cancer development (Rendic and DiCarlo, 1997), there are still no epidemiological studies available investigating the involvement of CYP1A1 and 1A2 gene polymorphisms in AA-induced kidney injury and cancer development in BEN or AAN patients. Therefore, the evaluation of variations in activities and genetic polymorphisms of these human enzymes remains a major challenge to explain human interindividual susceptibility to AA and to predict individual risk of cancer among AAN and BEN patients.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Grant Agency of Czech Republic (303/09/0472 and 305/09/H008); Ministry of Education of Czech Republic (MSM0021620808 and 1M0505). Work at the Institute of Cancer Research is supported by Cancer Research UK. Portions of this work were funded by National Institutes of Health (R01 ES014403 and P30 ES006096 to D.W.N. and Z.S.).
Supplementary Material
Acknowledgments
The authors would like to thank the Grant Agency of the Czech Republic and the Ministry of Education of the Czech Republic.
References
- Aoyama T, Gonzalez FJ, Gelboin HV. Human cDNA-expressed cytochrome P450 IA2: Mutagen activation and substrate specificity. Mol. Carcinog. 1989;2:192–198. doi: 10.1002/mc.2940020405. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Ferluga D, Stiborová M, Pfohl-Leszkowicz A, Vukelic M, Ceovic S, Schmeiser HH, Cosyns JP. Is aristolochic acid a risk factor for Balkan endemic nephropathy-associated urothelial cancer? Int. J. Cancer. 2002;101:500–502. doi: 10.1002/ijc.10602. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Levová K, Bárta F, Shi Z, Frei E, Schmeiser HH, Nebert DW, Phillips DH, Stiborová M. Role of CYP1A1 and CYP1A2 in bioactivation versus detoxication of the renal carcinogen aristolochic acid I: Studies in Cyp1a(−/−) knockout mice. Chem. Res. Toxicol. 2011a;24:1710–1719. doi: 10.1021/tx200259y. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Stiborová M, vom Brocke J, Simoes ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH. Aristolochic acid mutagenesis: Molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis. 2007;28:2253–2261. doi: 10.1093/carcin/bgm082. [DOI] [PubMed] [Google Scholar]
- Arlt VM, Zuo J, Trenz K, Roufosse CA, Lord GM, Nortier JL, Schmeiser HH, Hollstein M, Phillips DH. Gene expression changes induced by the human carcinogen aristolochic acid I in renal and hepatic tissue of mice. Int. J. Cancer. 2011b;128:21–32. doi: 10.1002/ijc.25324. [DOI] [PubMed] [Google Scholar]
- Atanasova S, von Ahsen N, Toncheva DI, Dimitrov TG, Oellerich M, Amstrong VM. Genetic polymorphism of cytochrome P450 among patients with Balkan endemic nephropathy (BEN) Clin. Biochem. 2005;38:223–228. doi: 10.1016/j.clinbiochem.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Chan W, Cu L, Xu G, Cai Z. Study of the phase I and phase II metabolism of nephrotoxin aristolochic acid by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2006;20:1755–1760. doi: 10.1002/rcm.2513. [DOI] [PubMed] [Google Scholar]
- Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem. Res. Toxicol. 2005;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: A worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- Dragin N, Uno S, Wang B, Dalton TP, Nebert DW. Generation of ‘humanized' hCYP1A1_1A2_Cyp1a1/1a2(−/−) mouse line. Biochem. Biophys. Res. Commun. 2007;359:635–642. doi: 10.1016/j.bbrc.2007.05.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse Y, Baan R, Straif K, Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Galichet L, Cogliano V. A review of human carcinogens-Part A: Pharmaceuticals. Lancet Oncol. 2009;10:13–14. doi: 10.1016/s1470-2045(08)70286-9. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat. Rev. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, Kimura S. Human CYP1A2: Sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expression. Mol. Endocrinol. 1989;3:1399–1408. doi: 10.1210/mend-3-9-1399. [DOI] [PubMed] [Google Scholar]
- Lemy A, Wissing KM, Rorive S, Zlotta A, Roumeguere T, Muniz Martinez MC, Decaestecker C, Salmon I, Abramowicz D, et al. Late onset of bladder urothelial carcinoma after kidney transplantation for end-stage aristolochic acid nephropathy: A case series with 15-year follow-up. Am. J. Kidney Dis. 2008;51:471–477. doi: 10.1053/j.ajkd.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Levová K, Moserova M, Kotrbova V, Sulc M, Henderson CJ, Wolf CR, Phillips DH, Frei E, Schmeiser HH, Mares J, et al. Role of cytochromes P450 1A1/2 in detoxication and activation of carcinogenic aristolochic acid I: Studies with the hepatic NADPH: Cytochrome P450 reductase null (HRN) mouse model. Toxicol. Sci. 2011;121:43–56. doi: 10.1093/toxsci/kfr050. [DOI] [PubMed] [Google Scholar]
- Mimura I, Nangaku M. The suffocating kidney: Tubulointerstitial hypoxia in end-stage renal disease. Nat. Rev. Nephrol. 2010;6:667–678. doi: 10.1038/nrneph.2010.124. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Comparison of gene expression in cell culture to that in the intact animal: Relevance to drugs and environmental toxicants. Focus on “development of a transactivator in hepatoma cells that allows expression of phase I, phase II, and chemical defense genes.“. Am. J. Physiol. 2006;290:C37–C41. doi: 10.1152/ajpcell.00444.2005. [DOI] [PubMed] [Google Scholar]
- Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De Pauw L, Abramowicz D, Vereerstraeten P, et al. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N. Engl. J. Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- Phillips DH, Arlt VM. The 32P-postlabeling assay for DNA adducts. Nat. Protoc. 2007;2:2772–2781. doi: 10.1038/nprot.2007.394. [DOI] [PubMed] [Google Scholar]
- Rendic S, DiCarlo FJ. Human cytochrome P450 enzymes: A status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab. Rev. 1997;29:413–480. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- Rosenquist TA, Einolf HJ, Dickman KG, Wang L, Smith A, Grollman AP. Cytochrome P450 1A2 detoxicates aristolochic acid in the mouse. Drug Metab. Dispos. 2010;38:761–768. doi: 10.1124/dmd.110.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeiser HH, Bieler CA, Wiessler M, van Ypersele de Strihou C, Cosyns JP. Detection of DNA adducts formed by aristolochic acid in renal tissue from patients with Chinese herbs nephropathy. Cancer Res. 1996;56:2025–2028. [PubMed] [Google Scholar]
- Schmeiser HH, Stiborová M, Arlt VM. Chemical and molecular basis of the carcinogenicity of Aristolochia plants. Curr. Opin. Drug Discov. Devel. 2009;12:141–148. [PubMed] [Google Scholar]
- Shi Z, Chen Y, Dong H, Amos-Kroohs RM, Nebert DW. Generation of a 'humanized' hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mouse line harboring the poor-affinity aryl hydrocarbon receptor. Biochem. Biophys. Res. Commun. 2008;376:775–780. doi: 10.1016/j.bbrc.2008.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S, Bonala RR, Rosenquist T, Rieger R, Suzuki N, Johnson F, Miller F, Grollman AP. Detoxification of aristolochic acid I by O-demethylation: Less nephrotoxicity and genotoxicity of aristolochic acid Ia in rodents. Int. J. Cancer. 2010;127:1021–1027. doi: 10.1002/ijc.25141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistkova J, Hudecek J, Hodek P, Frei E, Schmeiser HH, Stiborová M. Human cytochromes P450 1A1 and 1A2 participate in detoxication of carcinogenic aristolochic acid. Neuro. Endocrinol. Lett. 2008;29:733–737. [PubMed] [Google Scholar]
- Stiborová M, Frei E, Arlt VM, Schmeiser HH. Metabolic activation of carcinogenic aristolochic acid, a risk factor for Balkan endemic nephropathy. Mutat. Res. 2008;658:55–67. doi: 10.1016/j.mrrev.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Frei E, Hodek P, Wiessler M, Schmeiser HH. Human hepatic and renal microsomes, cytochromes P450 1A1/2, NADPH:Cytochrome P450 reductase and prostaglandin H synthase mediate the formation of aristolochic acid-DNA adducts found in patients with urothelial cancer. Int. J. Cancer. 2005a;113:189–197. doi: 10.1002/ijc.20564. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Frei E, Sopko B, Sopkova K, Markova V, Lankova M, Kumstyrova T, Wiessler M, Schmeiser HH. Human cytosolic enzymes involved in the metabolic activation of carcinogenic aristolochic acid: Evidence for reductive activation by human NAD(P)H:Quinone oxidoreductase. Carcinogenesis. 2003;24:1695–1703. doi: 10.1093/carcin/bgg119. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Frei E, Wiessler M, Schmeiser HH. Human enzymes involved in the metabolic activation of carcinogenic aristolochic acids: Evidence for reductive activation by cytochromes P450 1A1 and 1A2. Chem. Res. Toxicol. 2001;14:1128–1137. doi: 10.1021/tx010059z. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Mares J, Frei E, Arlt VM, Martínek V, Schmeiser HH. The human carcinogen aristolochic acid I is activated to form DNA adducts by human NAD(P)H:quinone oxidoreductase without the contribution of acetyltransferases or sulfotransferases. Environ. Mol. Mutagen. 2011;52:448–459. doi: 10.1002/em.20642. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Martínek V, Rýdlová H, Hodek P, Frei E. Sudan I is a potential carcinogen for humans: Evidence for its metabolic activation and detoxication by human recombinant cytochrome P450 1A1 and liver microsomes. Cancer Res. 2002;62:5678–5684. [PubMed] [Google Scholar]
- Stiborová M, Martínek V, Rýdlová H, Koblas T, Hodek P. Expression of cytochrome P450 1A1 and its contribution to oxidation of a potential human carcinogen 1-phenylazo-2-naphthol (Sudan I) in human livers. Cancer Lett. 2005b;220:145–154. doi: 10.1016/j.canlet.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Sejbal J, Borek-Dohalska L, Aimova D, Poljakova J, Forsterova K, Rupertova M, Wiesner J, Hudecek J, Wiessler M, et al. The anticancer drug ellipticine forms covalent DNA adducts, mediated by human cytochromes P450, through metabolism to 13-hydroxyellipticine and ellipticine N2-oxide. Cancer Res. 2004;64:8374–8380. doi: 10.1158/0008-5472.CAN-04-2202. [DOI] [PubMed] [Google Scholar]
- Stiborová M, Sopko B, Hodek P, Frei E, Schmeiser HH, Hudecek J. The binding of aristolochic acid I to the active site of human cytochromes P450 1A1 and 1A2 explains their potential to reductively activate this human carcinogen. Cancer Lett. 2005c;229:193–204. doi: 10.1016/j.canlet.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Toncheva DI, von Ahsen N, Atanasova SY, Dimitrov TG, Amstrong VM. Identification of NQO1 and GSTs genotype frequencies in Bulgarian patients with Balkan endemic nephropathy. J. Nephrol. 2004;17:384–389. [PubMed] [Google Scholar]
- Turesky RJ. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol. Nutr. Food Res. 2005;49:101–117. doi: 10.1002/mnfr.200400076. [DOI] [PubMed] [Google Scholar]
- Vanherweghem JL, Depierreux M, Tielemans C, Abramowicz D, Dratwa M, Jadoul M, Richard C, Vandervelde D, Verbeelen D, Vanhaelen-Fastre R, et al. Rapidly progressive interstitial renal fibrosis in young women: Association with slimming regimen including Chinese herbs. Lancet. 1993;341:387–391. doi: 10.1016/0140-6736(93)92984-2. [DOI] [PubMed] [Google Scholar]
- Xue X, Xiao Y, Zhu H, Wang H, Liu Y, Xie T, Ren J. Induction of P450 1A by 3-methylcholanthrene protects mice from aristolochic acid-I-induced acute renal injury. Nephrol. Dial. Transplant. 2008;23:3074–3081. doi: 10.1093/ndt/gfn262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.