Abstract
Altered T cell function in systemic lupus erythematosus (SLE) is determined by various molecular and cellular abnormalities including increased IL-17 production. Recent evidence suggests a crucial role for signaling lymphocyte activation molecules (SLAMs) in the expression of autoimmunity. In this report, we demonstrate that SLAMF3 and SLAMF6 expression is increased on the surface of SLE T cells compared to normal cells. SLAM co-engagement with CD3 under Th17 polarizing conditions results in increased IL-17 production. SLAMF3 and SLAMF6 T cell surface expression and IL-17 levels significantly correlate with disease activity in SLE patients. Both naïve and memory CD4+ T cells produce more IL-17 in response to SLAM co-stimulation as compared to CD28 co-stimulation. In naïve CD4+ cells, IL-17 production after CD28 co-stimulation peaks on day 3, whereas co-stimulation with anti-SLAMF3 and anti-SLAMF6 antibodies results in a prolonged and yet increasing production over 6 days. Unlike co-stimulation with anti-CD28, SLAM co-stimulation requires the presence of the adaptor molecule SLAM-associated protein (SAP). Thus, engagement of SLAMF3 and SLAMF6 along with antigen-mediated CD3/TCR stimulation represents an important source of IL-17 production and disruption of this interaction with decoy receptors or blocking antibodies should mitigate disease expression in SLE and other autoimmune conditions.
Introduction
For optimal T cell activation, recognition of the Ag/MHC complex by the TCR is accompanied by signals mediated through co-stimulatory pathways (1, 2). CD28 co-stimulation is best characterized for T cell activation (3), however there is evidence for other co-stimulatory molecules including signaling lymphocyte activation molecule (SLAM) receptor family members (4, 5). Recently, the SLAM family of type I transmembrane receptors has been reported to mediate important regulatory signals between immune cells through their homophilic or heterophilic interactions. SLAM receptors are expressed on hematopoietic cells, including cells of the innate immune system, T and B cells. By virtue of their ability to transduce tyrosine phosphorylation signals through immunoreceptor tyrosine-based switch motif (ITSM) sequences, SLAM receptors play an important role in regulating both innate and adaptive immune responses. Upon activation, SLAM molecules associate with intracellular adaptor proteins, e.g. those of the SLAM-associated protein family (6-11). SLAM-associated proteins (SAPs) contribute to SLAM receptor activation as they mediate dimerization of SLAM receptors and compete with SLAM-induced signals. SAP deficiency is associated with severe natural killer (NK), T and B cell abnormalities and reduced antibody production (12).
Recent evidence indicates that SLAM signaling is also involved in the pathogenesis of autoimmune diseases including systemic lupus erythematosus (SLE) (13, 14). SLE is a chronic autoimmune inflammatory disease that is characterized by improper regulation of B and T cell function (15). The SLAM gene cluster encodes several co-stimulatory receptors, including SLAMF3 and SLAMF6. It is located within a genomic region, which entails genes with key immunological functions, including the Fc receptor cluster, the SLAM cluster, CD58, IL23R, TLR5 and complement receptor CR1 (16, 17). Polymorphisms in the SLAM cluster have been associated with autoimmune diseases in mice and humans for which this region was designated Sle1b locus and is considered as a genetic susceptibility region for the development of SLE (16, 18, 19). Polymorphisms in the Ly108 gene, one of the members of the SLAM family receptors (corresponding to SLAMF6 in humans), result in the generation of a Ly108 splice variant in lupus-prone mice that is involved in the pathogenesis of SLE (16, 20).
Differentiation of CD4+ T helper (Th) cells into distinct effector populations is one of the hallmarks of adaptive immune responses. Previous reports suggest that co-stimulation of Th cells through SLAMF6 promotes a Th1 phenotype under polarizing and non-polarizing conditions and furthermore, SLAMF6 appears to have superior co-stimulatory capacities when compared to CD28, especially on CD8+ and CD4/CD8 double-negative T cells (14, 21). Another member of the SLAM family, SLAMF3 (also known as CD229/Ly9 in mice), which is expressed on T and B cells, has been reported to promote Th2 differentiation (22). The most recently discovered T helper cell subset, denoted Th17 cells, is characterized by abundant production of IL-17A (herein referred to as IL-17), IL-21 and IL-22 plays a major role in host responses against bacterial infections and the development of autoimmune diseases including SLE (23, 24). Indeed, higher serum concentrations of IL-17 have been reported in SLE patients (25, 26) and studies in lupus-prone mice provide evidence for IL-17 as a crucial mediator of disease pathology in SLE (27-29).
Since SLAMF6 and SLAMF3 have been shown to be engaged in Th cell differentiation, we asked whether SLAM receptors play a role in the pathogenesis of SLE, and whether they contribute to Th17 differentiation. We report that surface expression of SLAMF6 and SLAMF3 is increased in SLE T cells and mirrors disease activity. Our results indicate that co-activation of SLAMF6 and SLAMF3 receptors along with CD3/TCR stimulation potently induces IL-17 production and thus, accounts for Th17 generation in T cells from both healthy controls and SLE patients.
Materials and Methods
Study subjects and T cell culture
All SLE patients included in our studies were recruited from the Division of Rheumatology at Beth Israel Deaconess Medical Center (Boston, MA, USA) and diagnosed according to the revised SLE classification criteria of the American College of Rheumatology (30). SLE disease activity index (SLEDAI) scores ranged from 0 to 10. Written informed consent was obtained from all patients. Control blood samples were obtained from healthy platelet donors at the Kraft Family Blood Donor Center (Dana-Farber Cancer Institute, Boston, MA, USA). Primary total T cells were isolated from peripheral venous blood by negative selection as described previously (14). All primary T cells were kept in RPMI medium supplemented with 10% fetal bovine serum. Naïve (CD4+ CD45RA+) and memory (CD4+ CD45RO+) T cells were purified using T cell isolation kits from Miltenyi Biotec according to the manufacturer’s instructions.
T cell stimulation, Th17 differentiation assays and ELISAs
Cell culture plates were pre-coated overnight with 0.5 μg/ml monoclonal anti-CD3 (BioXcell, clone OKT3), 0.5 μg/ml anti-CD28 (Biolegend), 0.5 μg/ml anti-SLAMF6 (Genentech, clone 24D8.1H5.1F5), 0.5 μg/ml anti-SLAMF3 antibodies (Biolegend, clone HLy-9.1.25) or 0.5 μg/ml control IgG1 as indicated. Naïve and memory CD4+ T cells (1×106 per well) were differentiated into Th17 cells in serum-free X-VIVO10 medium (Biowhittaker) by the addition of IL-6 (25 ng/ml), TGF-β1 (5 ng/ml), IL-1β (12.5 ng/ml), IL-21 (25 ng/ml) and IL-23 (25 ng/ml) for the indicated time periods. IL-6, IL-1β, IL-23 and TGFβ1 were obtained from R&D Systems, IL-21 was purchased from Cell Sciences. Supernatants were collected at different time points and tested for IFNγ (Endogen) and IL-17 (eBioscience) by ELISA.
Flow cytometry
Intracellular T cell staining for IL-17 and IFNγ was performed at the indicated time points using the BD Cytofix/Cytoperm Kit and Alexa-647-labelled anti-IL-17 (BD Biosciences) and PE-labelled anti-IFNγ (Biolegend) antibodies. Cells were stimulated with phorbol-12-myristate 13-acetate (PMA; 50 ng/ml) and ionomycin (0.5 μg/ml) for a total period of 6 h. Golgistop (Monensin) was added 30 minutes after T cell stimulation was initiated. Surface staining was performed using a Pacific Blue-labelled anti-CD4 antibody (Biolegend) for 20 min on ice. Samples were analyzed on a LSRII flow cytometer (BD Biosciences) and analysis was performed with FlowJo software v. 8.3.3 (Tree Star).
siRNA experiments
Naïve (CD4+ CD45RA+) T cells were purified from healthy donors and transfected with different concentrations of siRNA oligonucleotides specific for SLAMF3, SLAMF6, SAP or a non-specific control siRNA (all from Applied Biosystems) using the Amaxa transfection system (Lonza). SAP siRNA transfection efficiency was confirmed by immunoblotting of cytosolic protein lysates using anti-SAP (Cell Signal Technology), anti-β-actin (Sigma) and suitable HRP-linked secondary antibodies. SLAMF3 and SLAMF6 siRNA transfection efficiency was confirmed by analyzing their surface expression by flow cytometry using anti-SLAMF3 and anti-SLAMF6 antibodies (both from Biolegend). All subsequent assays were performed 4 days after transfection at the point of maximal knock-down.
Statistical analyses
The paired two-tailed Student t-test and the Pearson product moment correlation coefficient r were used for statistical analyses.
Results
SLAMF3 and SLAMF6 are up-regulated on SLE T cells and serve as major co-stimuli for Th17 differentiation
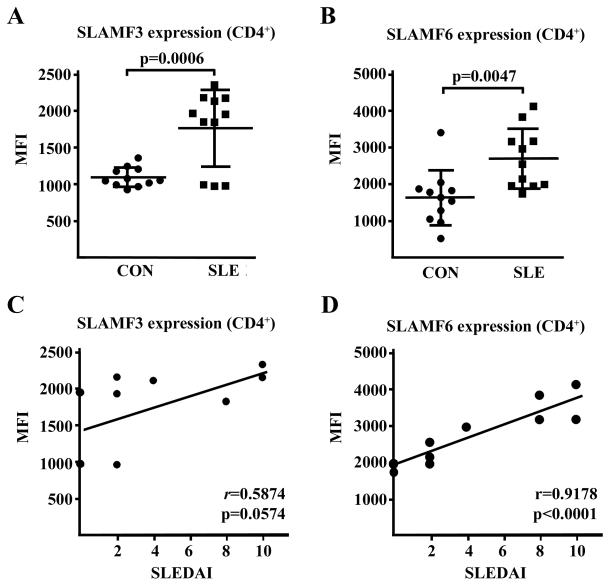
In order to assess the involvement of SLAM co-stimulation in SLE pathogenesis, we performed flow cytometric analysis of SLAM receptor surface expression on CD4+ T cells obtained from 11 SLE patients at different levels of disease activity as measured by SLE disease activity index (SLEDAI) and 11 healthy control subjects. We observed a significantly increased surface expression of both SLAMF3 and SLAMF6 on CD4+ T cells from SLE patients in comparison to the healthy individuals (Fig. 1A,B). Of note, there was a robust correlation between the individual cell surface expression of SLAMF6 on SLE CD4+ T cells and the corresponding SLEDAI indices (r=0.9178; Fig. 1D), however we did not find a significant correlation in case of SLAMF3 (Fig. 1C).
Figure 1. Cell surface expression of SLAMF3 and SLAMF6 receptors is increased on SLE T cells.
(A) Total T cells from 11 healthy controls (CON) and 11 SLE patients were analyzed for surface expression of SLAMF3 by flow cytometry gating on CD4+ T cells. MFI: median fluorescence intensity. (B) Total T cells from the same individuals were analyzed for surface expression of SLAMF6 by flow cytometry gating on CD4+ T cells. (C) Surface expression of SLAMF3 on CD4+ T cells obtained from SLE patients was correlated to the individual SLE disease activity scores (SLEDAI). (D) Surface expression of SLAMF6 on CD4+ T cells obtained from SLE patients was correlated to the individual SLEDAI scores.
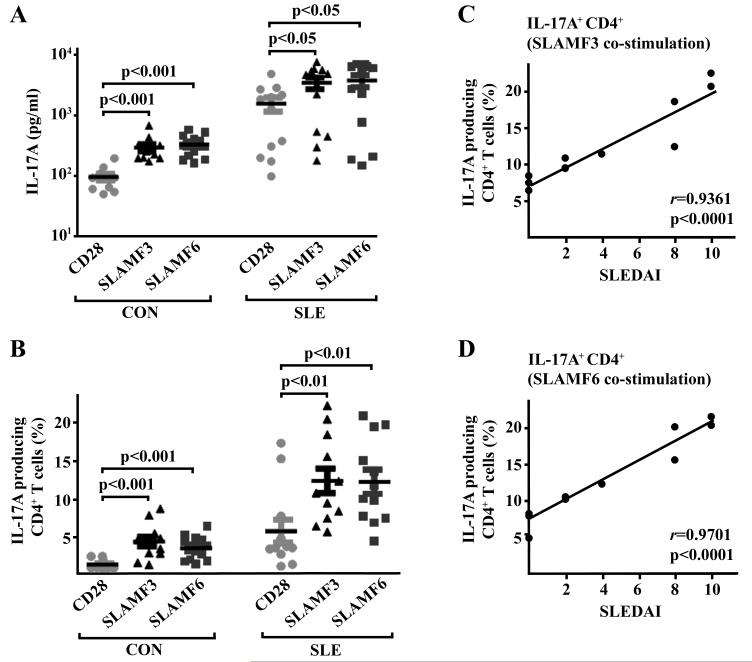
Since it has been reported that SLAM signaling is involved in T helper cell differentiation and that SLE patients display increased serum levels of IL-17, we hypothesized that the observed up-regulation of SLAMF3/SLAMF6 expression in SLE CD4+ T cells contributes to an augmented production of IL-17. Thus, we stimulated T cells from SLE patients and healthy controls with anti-CD3 and co-stimulatory anti-CD28, anti-SLAMF3 or anti-SLAMF6 antibodies in the presence of Th17 polarizing stimuli (i.e. IL-6, IL-1β, IL-21, IL-23 and TGFβ1). On day 6 after stimulation and differentiation had been initiated, we measured IL-17 levels in cell culture supernatants (Fig. 2A) and the percentage of IL-17-producing CD4+ T cells (Fig. 2B and Suppl. Fig. 1). Overall, SLE CD4+ T cells were found to produce more IL-17 under these conditions. Intriguingly, co-stimulation through both SLAM receptors was more potent in the induction of IL-17 production when compared to CD28 co-stimulation. We observed a strong correlation between the clinical disease activity and the corresponding percentage of IL-17-producing CD4+ T cells as induced through co-stimulation with either anti-SLAMF3 or anti-SLAMF6 antibodies (r=0.9361 and r=0.9701, respectively, Fig. 2C,D). To determine whether Th17 polarizing cytokines were necessary for the production of IL-17 following engagement of SLAMF3 and SLAMF6 molecules on the surface of SLE T cells, T cells from SLE patients and healthy controls were stimulated with anti-CD3 and anti-CD28, anti-SLAMF3 or anti-SLAMF6 antibodies in the absence of Th17 polarizing cytokines. We observed an increase in IL-17A production (Suppl. Fig. 2A,B) following SLAM co-stimulation compared to stimulation with CD28, suggesting that SLAMF3/6 co-stimulation can cause IL-17 production in SLE T cells in absence of Th17 polarizing cytokines.
Figure 2. SLAM receptor co-stimulation enhances IL-17 production and reflects disease activity in SLE patients.
(A) Total T cells obtained from 11 healthy controls (CON) and 11 SLE patients were cultured under Th17 differentiation conditions along with anti-CD3 antibodies and co-stimulatory anti-CD28, anti-SLAMF3 or anti-SLAMF6 antibodies as indicated for a total period of 6 days. Subsequently, supernatants were subjected to IL-17 measurement (ELISA). (B) Total T cells were cultured as outlined under (A). Intracellular IL-17 staining was performed gating on CD4+ T cells. (C) Total T cells were isolated from SLE patients and cultured under Th17 conditions along with anti-CD3 and anti-SLAMF3 antibodies for 6 days. Percentages of IL-17-producing CD4+ T cells were correlated to the individual SLE disease activity scores (SLEDAI). (D) Percentages of IL-17-producing CD4+ T cells isolated from SLE patients (under anti-CD3/anti-SLAMF6 co-stimulation) were correlated to the individual SLEDAI scores.
Taken together, our data suggest that T cells from active SLE patients are characterized by an increased SLAM receptor expression, which is linked to increased IL-17 production.
SLAM-mediated co-stimulation promotes Th17 differentiation in human T cells
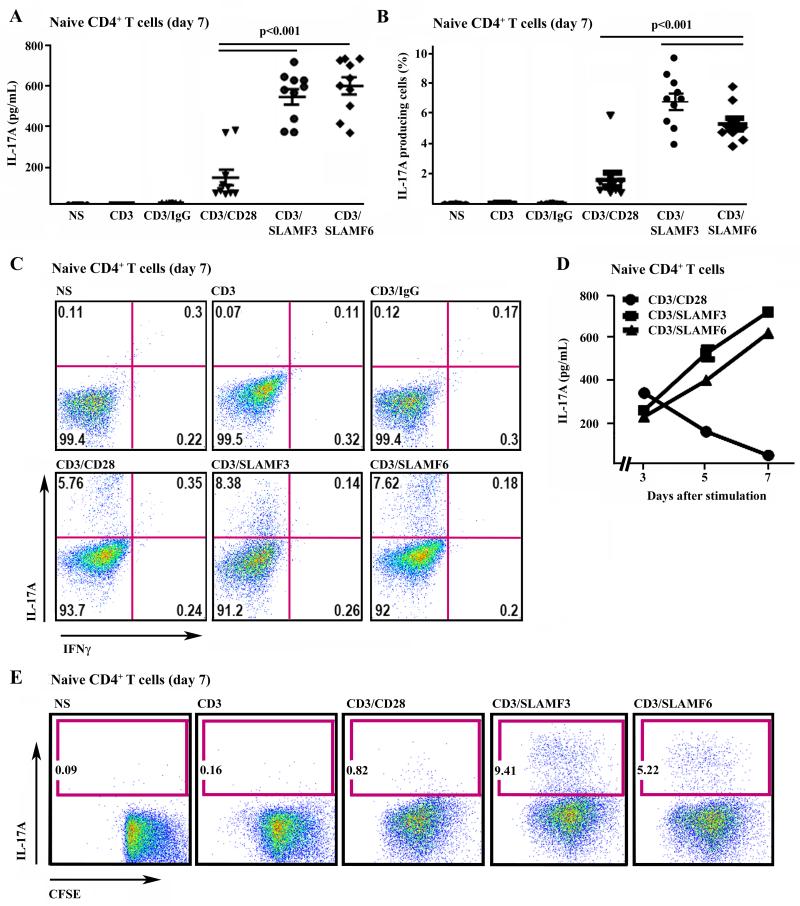
Next, we studied the involvement of SLAM receptors in human naïve (CD45RA+) and memory (CD45RO+) CD4+ T cells. Thus, we stimulated naïve CD4+ T cells with plate-bound anti-CD3 and co-stimulatory anti-CD28, anti-SLAMF3 or anti-SLAMF6 antibodies under Th17 differentiation conditions for 7 days and subsequently analyzed IL-17 production by flow cytometry and ELISA. In line with our observations in total CD4+ T cells, Th17-differentiated naïve CD4+ T cells produced significantly more IL-17 in response to SLAM co-stimulation when compared to CD28 co-stimulation (Fig. 3A-C). Interestingly, time kinetics of CD28 and SLAM co-stimulation were profoundly different. While IL-17 protein expression peaked on day 3 following T cell activation through CD3/CD28 co-stimulation and gradually decreased over time until day 7, it further increased in response to either SLAMF3 or SLAMF6 co-stimulation throughout the observation period (Fig. 3D). To exclude the possibility that IL-17 production is an effect of increased proliferation vs. Th17 differentiation in response to SLAM co-stimulation, we performed comparative proliferation analysis of SLAM and CD28 co-stimulated naïve CD4+ T cells. Naïve CD4+ T lymphocytes exhibited comparable rates of cell-division in response to CD28 and SLAM co-stimulation as assessed by CSFE staining, while IL-17 production was increased upon SLAM co-stimulation (Fig. 3E). This suggests that activated SLAM signaling transfers enhanced differentiation capacities to naïve CD4+ T cells towards a Th17 phenotype under polarizing conditions, rather than solely exerting proliferative effects on IL-17 producing cells.
Figure 3. SLAM co-stimulation promotes Th17 differentiation in naïve CD4+ T cells.
(A)-(C) Naïve (CD45RA+ CD62Lhi) CD4+ T cells were stimulated with anti-CD3 and co-stimulatory anti-CD28, anti-SLAMF3, SLAMF6 and control IgG under Th17 conditions for 7 days. IL-17 and IFNγ production was analyzed by intracellular staining and ELISA. (A) Secretion of IL-17 by naïve CD4+ T cells on day 7 was measured by ELISA. (B) shows percentages of IL-17 producing cells. The relative number of IL-17 producing cells is given as a percentage of naïve CD4+ T cells. (C) One representative experiment is displayed (from day 7). (D) Kinetics of IL-17 secretion by naïve CD4+ T cells in response to co-stimulation through CD28, SLAMF3 and SLAMF6. On days 3, 5 and 7 the IL-17 levels were monitored by ELISA. (E) Naïve CD4+ T cells were labeled with CFSE, then stimulated and differentiated until day 7. Staining on day 7 was as above. No differences in CFSE staining were detected between the different groups, indicating comparable proliferation rates.
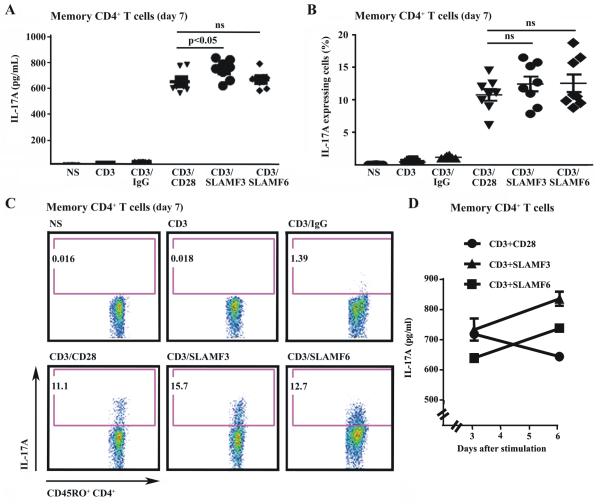
Next, we performed similar studies in memory CD4+ T cells. As expected, CD28 co-stimulation yielded higher IL-17 concentrations when compared to naïve CD4+ T cells. CD3/SLAM-mediated co-stimulation of memory T cells induced levels of IL-17 comparable to CD3/CD28 co-stimulation (Fig. 4A-C), however time kinetics revealed an increase in IL-17 production by memory CD4+ T cells from day 3 to 6 in response to SLAM co-stimulation, whereas CD28 co-stimulation led to a mild decrease during the observed time period (Fig. 4D).
Figure 4. SLAM co-stimulation promotes Th17 differentiation in memory CD4+ T cells.
(A)-(C) Memory (CD45RO+) CD4+ T cells were stimulated with anti-CD3 and co-stimulatory anti-CD28, anti-SLAMF3, SLAMF6 and control IgG under Th17 conditions for 7 days. IL-17 production was analyzed by intracellular staining. (A) Secretion of IL-17 by memory CD4+ T cells on day 7 was measured by ELISA. (B) shows percentages of IL-17 producing cells. The relative number of IL-17 producing cells is given as a percentage of memory CD4+ T cells. (C) One representative experiment is displayed (from day 7). (D) Kinetics of IL-17 secretion by memory CD4+ T cells in response to co-stimulation through SLAMF3, SLAMF6 and CD28. IL-17 levels were monitored on days 3 and 6 by ELISA.
SLAM depletion reduces IL-17 production in response to T cell stimulation
In order to investigate the specificity of SLAM-promoted Th17 differentiation, we silenced SLAMF3 and SLAMF6 expression in naïve CD4+ T cells with siRNA. Efficiency of SLAM knock-down was assessed by surface staining (Suppl. Fig. 3A,C). 4 days after siRNA transfection, cells were stimulated with anti-CD3 and anti-SLAM antibodies under Th17 polarizing conditions. Naïve CD4+ T cells that had been transfected with SLAM siRNA were co-stimulated with anti-CD3/anti-SLAM antibodies which resulted in a severely impaired IL-17 production as determined by ELISA analyses (Suppl. Fig. 3B,D).
SLAM-promoted Th17 differentiation in human T cell involves SAP-dependent pathways
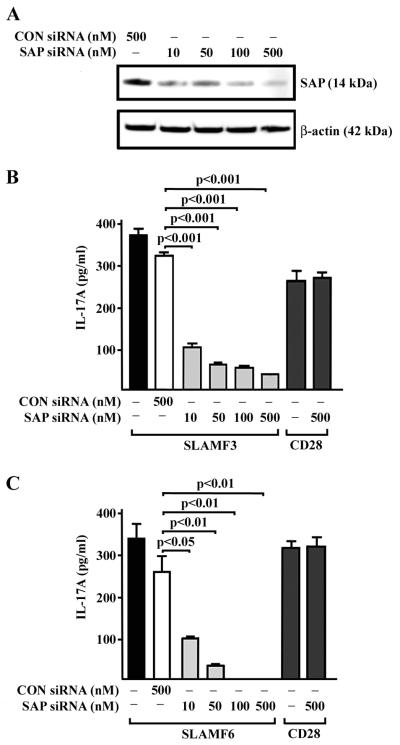
Next, we sought to analyze whether the adaptor protein SAP is involved in SLAM-mediated Th17 differentiation. Therefore, we knocked down endogenous SAP protein, using siRNA transfections in naïve CD4+ T cells (Fig. 5A). After SAP depletion, we observed a severely impaired IL-17 production following anti-CD3/anti-SLAM but not following anti-CD3/anti-CD28 stimulation under Th17 polarizing conditions (Fig. 5B,C). This suggests that SAP is engaged in SLAM-mediated Th17 differentiation of human naïve CD4+ T cells.
Figure 5. SAP knockdown abrogates IL-17 production.
Naïve CD4+ T lymphocytes were transfected with SAP siRNA or control siRNA as indicated. Efficacy of siRNA transfection was determined by SAP protein levels using western blot analysis (A). Three days after transfection cells were co-stimulated with anti-SLAMF3 (B) or anti-SLAMF6 antibodies (C) in addition to anti-CD3 under Th17 differentiation conditions for another 4 days. Levels of secreted IL-17 were measured by ELISA.
Discussion
In the present study, we investigated classical/canonical and non-canonical TCR co-stimulatory molecules and their contribution to the differentiation of total, naïve and memory CD4+ T cells towards a Th17 phenotype. We document that the non-canonical co-stimulatory molecules SLAMF3 and SLAMF6 exhibit more potent co-stimulatory capacities in Th17 generation in both, control and SLE T cells. This is of special interest, since SLE T cells display increased surface expression of SLAM receptor proteins. The degree of SLAMF3 and SLAMF6 expression correlated with disease severity as assessed by individual SLEDAI scores.
In the presence of Th17-polarizing cytokines, co-stimulation through SLAMF3 or SLAM6 results in increased production of IL-17 in naïve CD4+ T cells and follows different time kinetics than the one elicited by CD28. IL-17 production following CD28 co-stimulation peaks at day 3 and may relate to a normal immune response elicited by the antigen involving the CD3/TCR complex. In contrast, engagement of SLAMF3 and SLAMF6 after exposure to antigen may relate to a prolonged inflammatory response, which may cause organ damage. In SLE patients the CD3/TCR complex may be engaged by autoantigens or circulating anti-CD3/TCR autoantibodies (37).
It is well-documented that effector memory CD4+ T cells are principal producers of IL-17 in vivo (31, 32). Our observations provide evidence that under Th17 polarizing conditions IL-17 production by memory CD4+ T cells is significantly increased upon TCR co-stimulation through SLAMF3 when compared to canonical TCR co-stimulation by CD28. This is of special interest since pathogenic memory CD4+ T cells are expanded and naïve CD4+ T cells are decreased in SLE patients (33-36). This suggests that in addition to increased Th17 differentiation from naïve CD4+ T cells, SLAM signaling contributes to increased IL-17 expression from pathologically expanded memory CD4+ T cells in SLE patients.
SLAM molecules associate with intracellular adaptor proteins, such as SAP family members. SAP proteins contribute to SLAM receptor activation as they mediate dimerization of SLAM receptors. SAP proteins have been reported to promote Th1 and Th2 differentiation (38-42). Our observation that SAP blockade with SAP siRNA results in significantly reduced IL-17 expression from T cells suggests a key role for SAP as a common proximal signal in SLAM-promoted IL-17 production. As the aforementioned SLAM receptors, SAP proteins may be promising targets for pharmacological blockade. Unlike further downstream molecules, such as MAPK, SAP proteins are limited to SLAM-associated pathways, which contribute to (pathological) Th17 generation and IL-17 expression in SLE and presumably other autoimmune diseases (43). However, further downstream molecules through which SLAM molecules promote Th17 differentiation remain to be elucidated and future studies are warranted to unravel mechanisms how SLAM receptors promote and sustain an increased IL-17 production.
Our observation that TCR co-stimulation with SLAMF3 and SLAMF6 promotes Th17 differentiation, and that SLAMF3 and SLAMF6 surface expression on SLE T cells is significantly increased when compared to healthy controls and reflects disease activity may also link the genomic location of SLAM family genes within the lupus susceptibility locus Sle1b to reported SLAM polymorphisms in lupus-prone mice and SLE patients (13, 16, 18, 20). Previous studies indicated that SLAM polymorphisms contribute to SLE pathogenesis through increased gene expression (16). Polymorphisms in the Ly108 (SLAMF6) gene result in overexpression of a Ly108 splice variant in lupus-prone mice. This is in agreement with our previous report that SLAMF6 co-stimulation of human SLE T cells results in reduced secretion of IL-2 and Th1 cytokines resembling a SLE phenotype (14).
This is the first report to demonstrate that the non-canonical co-stimulatory molecules SLAMF3 and SLAMF6 promote Th17 differentiation. Both these molecules are more potent inducers of Th17 differentiation when compared to the canonical co-stimulatory molecule CD28. Furthermore, SLAMF3 and SLAMF6 surface expression is increased on SLE T cells in a disease activity-dependent manner. The superior capacities of SLAM molecules in Th17 generation may be attributed to yet unidentified downstream signal transducers. We believe that our studies hold the promise to create a new platform to study Th17 cells from different perspective and SLAM family of co-stimulators could be chosen as therapeutic targets to control SLE and autoimmune disorders. Future studies are needed in order to (i) investigate SLAM receptor expression in different autoimmune diseases, (ii) to dissect SLAM propensities in T helper cell differentiation under polarizing and non-polarizing conditions and (iii) to analyze the involvement of downstream factors that account for SLAM-promoted IL-17 gene expression.
The significance of our findings is manifold: The strong correlation between SLE disease activity and SLAMF3 and SLAM6 expression on the surface of SLE T cells suggests that their expression levels may gain biomarker value. The fact that SLAM engagement leads to increased and prolonged production of IL-17 strongly suggests that blockade of their engagement either with blocking antibodies or decoy receptors may suppress the inflammatory response without affecting IL-17 production through the canonical engagement of CD28.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant PO1 AI065687 and Deutsche Forschungsgemeinschaft (grant RA1927/1-1, to T.R.).
Abbreviations
- SLAM
signaling lymphocyte activation molecule
- SLE
systemic lupus erythematosus
- SAP
SLAM-associated protein
References
- 1.Carreno BM, Collins M. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annu Rev Immunol. 2002;20:29–53. doi: 10.1146/annurev.immunol.20.091101.091806. [DOI] [PubMed] [Google Scholar]
- 2.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 3.Veillette A, Latour S, Davidson D. Negative regulation of immunoreceptor signaling. Annu Rev Immunol. 2002;20:669–707. doi: 10.1146/annurev.immunol.20.081501.130710. [DOI] [PubMed] [Google Scholar]
- 4.Lucas PJ, Negishi I, Nakayama K, Fields LE, Loh DY. Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response. J Immunol. 1995;154:5757–5768. [PubMed] [Google Scholar]
- 5.Shahinian A, Pfeffer K, Lee KP, Kundig TM, Kishihara K, Wakeham A, Kawai K, Ohashi PS, Thompson CB, Mak TW. Differential T cell costimulatory requirements in CD28-deficient mice. Science. 1993;261:609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 6.Howie D, Okamoto S, Rietdijk S, Clarke K, Wang N, Gullo C, Bruggeman JP, Manning S, Coyle AJ, Greenfield E, Kuchroo V, Terhorst C. The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood. 2002;100:2899–2907. doi: 10.1182/blood-2002-02-0445. [DOI] [PubMed] [Google Scholar]
- 7.Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C. Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions. J Immunol. 2005;174:5931–5935. doi: 10.4049/jimmunol.174.10.5931. [DOI] [PubMed] [Google Scholar]
- 8.Henning G, Kraft MS, Derfuss T, Pirzer R, de Saint-Basile G, Aversa G, Fleckenstein B, Meinl E. Signaling lymphocytic activation molecule (SLAM) regulates T cellular cytotoxicity. Eur J Immunol. 2001;31:2741–2750. doi: 10.1002/1521-4141(200109)31:9<2741::aid-immu2741>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Mehrle S, Schmidt J, Buchler MW, Watzl C, Marten A. Enhancement of anti-tumor activity in vitro and in vivo by CD150 and SAP. Mol Immunol. 2008;45:796–804. doi: 10.1016/j.molimm.2007.06.361. [DOI] [PubMed] [Google Scholar]
- 10.Rethi B, Gogolak P, Szatmari I, Veres A, Erdos E, Nagy L, Rajnavolgyi E, Terhorst C, Lanyi A. SLAM/SLAM interactions inhibit CD40-induced production of inflammatory cytokines in monocyte-derived dendritic cells. Blood. 2006;107:2821–2829. doi: 10.1182/blood-2005-06-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27:698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Cunninghame Graham DS, Vyse TJ, Fortin PR, Montpetit A, Cai YC, Lim S, McKenzie T, Farwell L, Rhodes B, Chad L, Hudson TJ, Sharpe A, Terhorst C, Greenwood CM, Wither J, Rioux JD. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9:93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee M, Kis-Toth K, Thai TH, Terhorst C, Tsokos GC. SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity. 2011;44:211–218. doi: 10.3109/08916934.2010.530627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsokos GC. Systemic lupus erythematosus. New England Journal of Medicine. 2011 doi: 10.1056/NEJMra1100359. in press. [DOI] [PubMed] [Google Scholar]
- 16.Wang A, Batteux F, Wakeland EK. The role of SLAM/CD2 polymorphisms in systemic autoimmunity. Curr Opin Immunol. 2010;22:706–714. doi: 10.1016/j.coi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Flajnik MF, Kasahara M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 18.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr., Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci U S A. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–1669. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 21.Valdez PA, Wang H, Seshasayee D, van Lookeren Campagne M, Gurney A, Lee WP, Grewal IS. NTB-A, a new activating receptor in T cells that regulates autoimmune disease. J Biol Chem. 2004;279:18662–18669. doi: 10.1074/jbc.M312313200. [DOI] [PubMed] [Google Scholar]
- 22.Graham DB, Bell MP, McCausland MM, Huntoon CJ, van Deursen J, Faubion WA, Crotty S, McKean DJ. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J Immunol. 2006;176:291–300. doi: 10.4049/jimmunol.176.1.291. [DOI] [PubMed] [Google Scholar]
- 23.Garrett-Sinha LA, John S, Gaffen SL. IL-17 and the Th17 lineage in systemic lupus erythematosus. Curr Opin Rheumatol. 2008;20:519–525. doi: 10.1097/BOR.0b013e328304b6b5. [DOI] [PubMed] [Google Scholar]
- 24.Crispin JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, Kyttaris VC, Juang YT, Tsokos GC. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol. 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CK, Lit LC, Tam LS, Li EK, Wong PT, Lam CW. Hyperproduction of IL-23 and IL-17 in patients with systemic lupus erythematosus: implications for Th17-mediated inflammation in auto-immunity. Clin Immunol. 2008;127:385–393. doi: 10.1016/j.clim.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Wong CK, Ho CY, Li EK, Lam CW. Elevation of proinflammatory cytokine (IL-18, IL-17, IL-12) and Th2 cytokine (IL-4) concentrations in patients with systemic lupus erythematosus. Lupus. 2000;9:589–593. doi: 10.1191/096120300678828703. [DOI] [PubMed] [Google Scholar]
- 27.Hsu HC, Yang P, Wang J, Wu Q, Myers R, Chen J, Yi J, Guentert T, Tousson A, Stanus AL, Le TV, Lorenz RG, Xu H, Kolls JK, Carter RH, Chaplin DD, Williams RW, Mountz JD. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 28.Jacob N, Yang H, Pricop L, Liu Y, Gao X, Zheng SG, Wang J, Gao HX, Putterman C, Koss MN, Stohl W, Jacob CO. Accelerated pathological and clinical nephritis in systemic lupus erythematosus-prone New Zealand Mixed 2328 mice doubly deficient in TNF receptor 1 and TNF receptor 2 via a Th17-associated pathway. J Immunol. 2009;182:2532–2541. doi: 10.4049/jimmunol.0802948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HK, Liu M, Datta SK. Low-dose peptide tolerance therapy of lupus generates plasmacytoid dendritic cells that cause expansion of autoantigen-specific regulatory T cells and contraction of inflammatory Th17 cells. J Immunol. 2007;178:7849–7858. doi: 10.4049/jimmunol.178.12.7849. [DOI] [PubMed] [Google Scholar]
- 30.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 31.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Rohowsky-Kochan C. Regulation of IL-17 in human CCR6+ effector memory T cells. J Immunol. 2008;180:7948–7957. doi: 10.4049/jimmunol.180.12.7948. [DOI] [PubMed] [Google Scholar]
- 33.Gordon C, Matthews N, Schlesinger BC, Akbar AN, Bacon PA, Emery P, Salmon M. Active systemic lupus erythematosus is associated with the recruitment of naive/resting T cells. Br J Rheumatol. 1996;35:226–230. doi: 10.1093/rheumatology/35.3.226. [DOI] [PubMed] [Google Scholar]
- 34.Neidhart M, Pataki F, Michel BA, Fehr K. CD45 isoforms expression on CD4+ and CD8+ peripheral blood T-lymphocytes is related to auto-immune processes and hematological manifestations in systemic lupus erythematosus. Schweiz Med Wochenschr. 1996;126:1922–1925. [PubMed] [Google Scholar]
- 35.Watanabe T, Suzuki J, Mitsuo A, Nakano S, Tamayama Y, Katagiri A, Amano H, Morimoto S, Tokano Y, Takasaki Y. Striking alteration of some populations of T/B cells in systemic lupus erythematosus: relationship to expression of CD62L or some chemokine receptors. Lupus. 2008;17:26–33. doi: 10.1177/0961203307085246. [DOI] [PubMed] [Google Scholar]
- 36.Han BK, White AM, Dao KH, Karp DR, Wakeland EK, Davis LS. Increased prevalence of activated CD70+CD4+ T cells in the periphery of patients with systemic lupus erythematosus. Lupus. 2005;14:598–606. doi: 10.1191/0961203305lu2171oa. [DOI] [PubMed] [Google Scholar]
- 37.Juang YT, Wang Y, Solomou EE, Li Y, Mawrin C, Tenbrock K, Kyttaris VC, Tsokos GC. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J Clin Invest. 2005;115:996–1005. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nat Immunol. 2001;2:681–690. doi: 10.1038/90615. [DOI] [PubMed] [Google Scholar]
- 39.Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, Engel P, Terhorst C. X-linked lymphoproliferative disease: a progressive immunodeficiency. Annu Rev Immunol. 2001;19:657–682. doi: 10.1146/annurev.immunol.19.1.657. [DOI] [PubMed] [Google Scholar]
- 40.Nichols KE, Koretzky GA, June CH. SAP: natural inhibitor or grand SLAM of T cell activation? Nat Immunol. 2001;2:665–666. doi: 10.1038/90595. [DOI] [PubMed] [Google Scholar]
- 41.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 42.Vilar ML, Frutuoso MS, Arruda SM, Lima DM, Bezerra CS, Pompeu MM. The role of the SLAM-SAP signaling pathway in the modulation of CD4+ T cell responses. Braz J Med Biol Res. 2011;44:276–82. doi: 10.1590/s0100-879x2011007500038. [DOI] [PubMed] [Google Scholar]
- 43.Komori H, Furukawa H, Mori S, Ito MR, Terada M, Zhang MC, Ishii N, Sakuma N, Nose M, Ono M. A signal adaptor SLAM-associated protein regulates spontaneous autoimmunity and Fas-dependent lymphoproliferation in MRL-Faslpr lupus mice. J Immunol. 2006;176:395–400. doi: 10.4049/jimmunol.176.1.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.