Abstract
Aberrant cellular responses to pro-inflammatory cytokines, such as TNF-α, are pathogenic features in most chronic inflammatory diseases. A variety of extracellular and intracellular feedback pathways have evolved to prevent an inappropriate cellular reaction to these pro-inflammatory cytokines. Here, we report that TNF-α treatment of human and mouse cholangiocytes and hepatocytes downregulated expression of p300/CBP-associated factor (PCAF), a co-activator and an acetyltransferase that promotes histone acetylation and gene transcription. Of these upregulated microRNAs (miRNAs) in TNF-α-treated cells, miR-181a/b (miR-181a and miR-181b) suppressed translation of PCAF mRNA. Functional manipulation of miR-181a/b caused reciprocal alterations in PCAF protein expression in cultured cholangiocytes and hepatocytes. Inhibition of miR-181a/b function with anti-miRs blocked TNF-α-induced suppression of PCAF expression. Promoter recruitment of PCAF was shown to be associated with TNF-α-induced transcription of inflammatory genes. Intriguingly, pretreatment of cells with TNF-α inhibited transcription of inflammatory genes in response to subsequent TNF-α stimulation. Overexpression of PCAF or inhibition of miR-181a/b function with anti-miRs attenuated the inhibitory effects of TNF-α pretreatment on epithelial inflammatory response to subsequent TNF-α stimulation. Downregulation of PCAF and the inhibitory effects of TNF-α pretreatment on liver epithelial inflammatory response were further confirmed in a mouse model of TNF-α intraperitoneal injection. These data suggest that PCAF is a target for miR-181a/b, and downregulation of PCAF by TNF-α provides negative feedback regulation to inflammatory reactions in liver epithelial cells, a process that may be relevant to the epigenetic fine-tuning of epithelial inflammatory processes in general.
Keywords: miRNAs, PCAF, Negative feedback, Epithelial cells, Inflammation, TNF-α
INTRODUCTION
Chronic inflammation and cellular damage are common pathogenic features of most chronic inflammatory diseases. Recent advances in the understanding of immunity and inflammation have suggested that aberrant host responses, rather than the pathogen-specific toxins or gene products, may be the real pathogenetic factors that account for the chronicity of inflammatory reactions (1). Considerable clinical data suggest that pro-inflammatory cytokines, such as TNF-α, are key factors driving persistent inflammation and cellular injury in chronic hepatobiliary diseases (2). Liver epithelial cells (hepatocytes and cholangiocytes) play a role in the initiation, regulation, and resolution of inflammatory reactions in the liver. Both cell types express receptors for TNF-α, and activation of downstream signaling cascades of TNF-α receptors initiates a series of epithelial inflammatory reactions (3,4). Such epithelial cell responses are finely controlled at physiological conditions and reflect a delicate balance between effector functions and their potential to cause subsequent damage to liver tissues (3). To carry out a fine-tuning of inflammatory responses, epithelial cells have developed multiple strategies for the feedback regulation of intracellular signaling pathways. Several endogenous proteins have been identified to counter-regulate TNF-α-associated signaling cascades and promote resolution of inflammation (5).
Remodeling of chromatin within the nucleus, controlled by the degree of acetylation/deacetylation of histone residues on the histone core around which DNA is coiled, is important in allowing access for transcription factor DNA binding and gene transcription. Nuclear histone acetylation is a reversible process and is regulated by a group of histone acetyltransferases (HATs)4 that promote acetylation and histone deacetylases (HDACs) that promote deacetylation (6). TNF-α induces histone acetylation and promotes NF-κB- and AP-1-regulated pro-inflammatory IL-8 release in epithelial cells (7,8). NF-κB- and AP-1-mediated transcriptional activation of pro-inflammatory genes, such as IL-8, involves gene-specific promoter recruitment of co-activators, including p300/CBP-associated factor (PCAF) (9,10). Like other co-activators, PCAF can bridge the transcriptional factors to the basal transcriptional complex to maintain the appropriate level of gene activities during various physiological processes, such as the cell cycle, apoptosis, and carcinogenesis (11). PCAF also acts as an acetyltransferase that acetylates specific lysine residues in H3 and H4, resulting in remodeling of chromatin structure (6). Nevertheless, the role of PCAF-mediated transcriptional gene regulation in the control of epithelial inflammatory reactions in response to TNF-α stimulation is still unclear.
MicroRNAs (miRNAs) mediate gene suppression at the posttranscription level through mRNA cleavage and/or translational repression (12). Studies have revealed key roles for miRNAs in diverse regulatory pathways, including timing control in development, cell differentiation, apoptosis, and cell proliferation and, more recently, in regulation of inflammatory responses (13). We previously demonstrated that transcription of miRNA genes in cholangiocytes can be elaborately controlled by nuclear transcription factors associated with inflammation, such as the NF-κB pathway (14,15). Functionally, miRNAs may modulate epithelial inflammatory responses, including production and release of cytokines/chemokines, expression of adhesion and co-stimulatory molecules, and feedback regulation of epithelial homeostasis (16,17). Therefore, miRNAs act as critical regulators to the fine-tuning of epithelial inflammatory responses.
In this study, we investigated the expression of PCAF in liver epithelial cells in response to TNF-α, its relationship to miRNA-mediated posttranscriptional gene regulation, and finally its role in controlling the expression of inflammatory genes in response to secondary TNF-α stimulation. The data we report here show that TNF-α downregulates PCAF expression in liver epithelial cells with the involvement of miR-181a/b (i.e., miR-181a and miR-181b). Such downregulation of PCAF may provide negative feedback control of transcription of inflammatory genes, relevant to the epigenetic fine-tuning of epithelial inflammatory processes.
MATERIALS AND METHODS
Reagents, antibodies, and plasmid constructs
Human TNF-α was from Biomax Technologies (San Diego) and mouse TNF-α was from R&D Systems (Minneapolis). Anti-PCAF was from Santa Cruz Biotechnology (Santa Cruz) and anti-β-actin from Sigma-Aldrich (St. Louis). PCAF expression vector pCX-PCAF (Flag-tagged) was a gift from Dr. Tony Kouzarides (University of Cambridge). Chromatin immunoprecipitation (ChIP) assay kit was from Upstate Biotechnology (Charlottesville).
Cell culture
H69 cells are SV40-transformed normal human cholangiocytes originally derived from a liver harvested for transplant (18). 603B cells are immortalized normal mouse cholangiocytes (a gift of Dr. Y. Ueno, Sendai, Japan). These cholangiocytes continue to express biliary epithelial cell markers, including cytokeratin 19, gamma glutamyl transpeptidase, and ion transporters consistent with biliary function, and have been extensively characterized (18,19). AML12 is a hepatocyte cell line obtained from mouse (CD1 strain, line MT42) and was from the American Type Culture Collection.
Western blot
Whole cell lysates were obtained with the M-PER Mammalian Protein Extraction Reagent (ThermoScientific, Rockford) plus several protease inhibitors (1 mM PMSF; 10 µg/mL leupeptin, 2 µg/mL pepstatin). Antibodies to PCAF and β-actin were used. Densitometric levels of PCAF were quantified and expressed as their ratio to β-actin.
Real-time PCR
Total RNAs were prepared and comparative real-time PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) (14–17). The PCR primers were as follows: human PCAF (forward, 5’-CTGGAGGCACCATCTCAACGAA-3’, and reverse, 5’-ACAGTGAAGACCGAGCGAAGCA-3’); mouse PCAF (forward, 5’-CCGTGTCATTGGTGGTATCTGTT-3’, and reverse, 5-AGGAAGTTGAGGATCTCGTGCTT-3’); human IL-8 (forward, 5’-ATGACTTCCAAGCTGGCCGT-3’, and reverse, 5’-CCTCTTCAAAAACTTCTCCACACC-3’); human COX-2 (forward, 5'-CCAGCACTTCACGCATCAGTT-3’, and reverse, 5'-AAAGGCGCAGTTTACGCTGT-3'); mouse MIP-2 (forward, 5’-TGTCAATGCCTGAAGACCCTGCC-3’, and reverse, 5’-AACTTTTTGACCGCCCTTGAGA-3’); human GAPDH (forward, 5’-TGCACCACCAACTGCTTAGC-3’, and reverse, 5’-GGCATGGACTGTGGTCATGAG-3’); and mouse β-actin (forward, 5’-TGGTGGGAATGGGTCAGAA-3’, and reverse, 5’-TCTCCATGTCGTCCCAGTTG-3’). For real-time PCR analysis of mature miRNAs, LNA™ primer sets for miR-181a/b and snRNA RNU6B for human and mice were obtained from Exiqon (Vedbaek, Denmark). Total RNAs were reverse-transcribed using the Universal cDNA Synthesis Kit (Exiqon), and real-time PCR was performed in triplicate. The Ct values were analyzed using the comparative Ct (ΔΔCt) method, and the amount of target was obtained by normalizing to snRNA RNU6B and relative to the control (non-treated cells) (14–17).
miRCURY™ LNA array analysis of miRNAs
The Exiqon (Vedbaek, Denmark) miRCURY LNA microRNA arrays and service to process the samples were used (14,15). Briefly, H69 cells were grown to 80% confluence and exposed to TNF-α (10 ng/ml) for 8h. Total RNAs were prepared and the quality of isolated RNAs was verified. A mixture of equal amounts of total RNAs from the control and TNF-α treated cells were used as the reference pool. A total of 2 µg RNA from each sample was then labeled with the Hy5™ fluorescent label, and the reference pool labeled with Hy3™ using the miRCURY™ LNA Array labeling kit (Exiqon). The labeled samples and reference pool were then mixed pair-wise and hybridized to the miRCURY™ LNA array containing capture probes targeting all human miRNAs listed in the miRBASE version 8.1 (Exiqon). After hybridization, the slides were scanned and quantified signals were normalized by Exiqon using the global Lowess regression algorithm. Normalized Hy5/Hy3 ratios were used for further analysis, as previously reported (14–15).
Anti-miRs and miRNA precursors
Specific antisense oligonucleotides to miRNAs (anti-miRs) were used to inhibit miRNA function and specific miRNA precursors to increase miRNA expression (20). Anti-miR-181a/b and precursors to miR-181a/b were obtained from Ambion. For experiments, cells were grown to 90% confluent and treated with anti-miRs or precursors to miR-181a/b (0–30 nM) using the lipofectamineTM 2000 reagent (Invitrogen). Nonspecific anti-miR (anti-miR-Ctrl) and precursor (precursor-Ctrl) (Ambion) were used as controls.
Luciferase reporter constructs and luciferase assay
Complementary 38 bp DNA oligonucleotides containing the putative target site for miR-181a/b within the 3’ untranslated region (3′UTR) of human PCAF were synthesized with flanking SpeI and HindIII restriction enzyme digestion sites (Sense, 5’-ctagATAACATTTTCAGACCATGAATGAATGTTTCCAT-3’; antisense, 5’-agctATGGAAACATTCATTCATGGTCTGAAAATGTTAT-3’) and cloned into the multiple cloning site of the pMIR-REPORT Luciferase vector (Ambion). 3′UTR of mouse PCAF were the following (Sense, 5’-ctagTCTAGAGAAAAGTTTGACCATGAATGTCTCCAT-3’; antisense, 5’-AGCTATGGAGACATTCATGGTCAAACTTTTCTCTAGA-3’). pMIR-REPORT Luciferase constructs containing mutant 3′UTR (human: AATGAATGT to TTACTTACA; mouse: ATGAATGT to TACTTACA) were also generated. We then transfected cells with each reporter construct and β-gal (as the internal control), as well as anti-miRs or precursors to miR-181a/b. Luciferase activity was measured and normalized to the control β-gal level, as previously reported (16,17).
ChIP assay
ChIP analysis was performed with a commercially available ChIP Assay Kit (Upstate Biotechnologies) in accordance with the manufacturer’s instructions. In brief, 1×106 603B cells were cultured in 10-cm culture dishes and treated with TNF-α (10 ng/ml) in 2% FBS media for 1h. The chromatin fraction was immunoprecipitated overnight at 4°C using anti-PCAF antibody. PCR amplification was performed in a total volume of 20 µl with specific primers. PCR primers for ChIP analysis for NF-κB binding to the promoter of MIP-2 were forward (5’-GCCTATCGCCAATGAGC-3’) and reverse (5’-CAATTTTCTGAACCAAGGG-3’), and to IL-8 promoter were forward (5’-GGGCCATCAGTTGCAAATC-3’) and reverse (5’-TTCCTTCCGGTGGTTTCTTC-3’).
Injection of TNF-α in mice and immunohistochemistry
The C57BL/6J mice (The Jacksons Laboratory) were used and approved by the Creighton University Biosafety and Institutional Animal Care and Use Committees. Animals received treatment of TNF-α (2.5 µg in 200 µl of saline) by intraperitoneal (i.p.) injection, as previously reported (21). For TNF-α pretreatment animals, mice were injected i.p. with TNF-α for 24h and then received the same dose of TNF-α by i.p. injection. A portion of the liver was fixed with formaldehyde solution for H&E staining and immunohistochemisty, and the rest was used to obtain extracts for RNA and protein extraction for PCR or Western blot.
RESULTS
TNF-α decreases expression of PCAF protein in cholangiocytes without a change in PCAF mRNA level
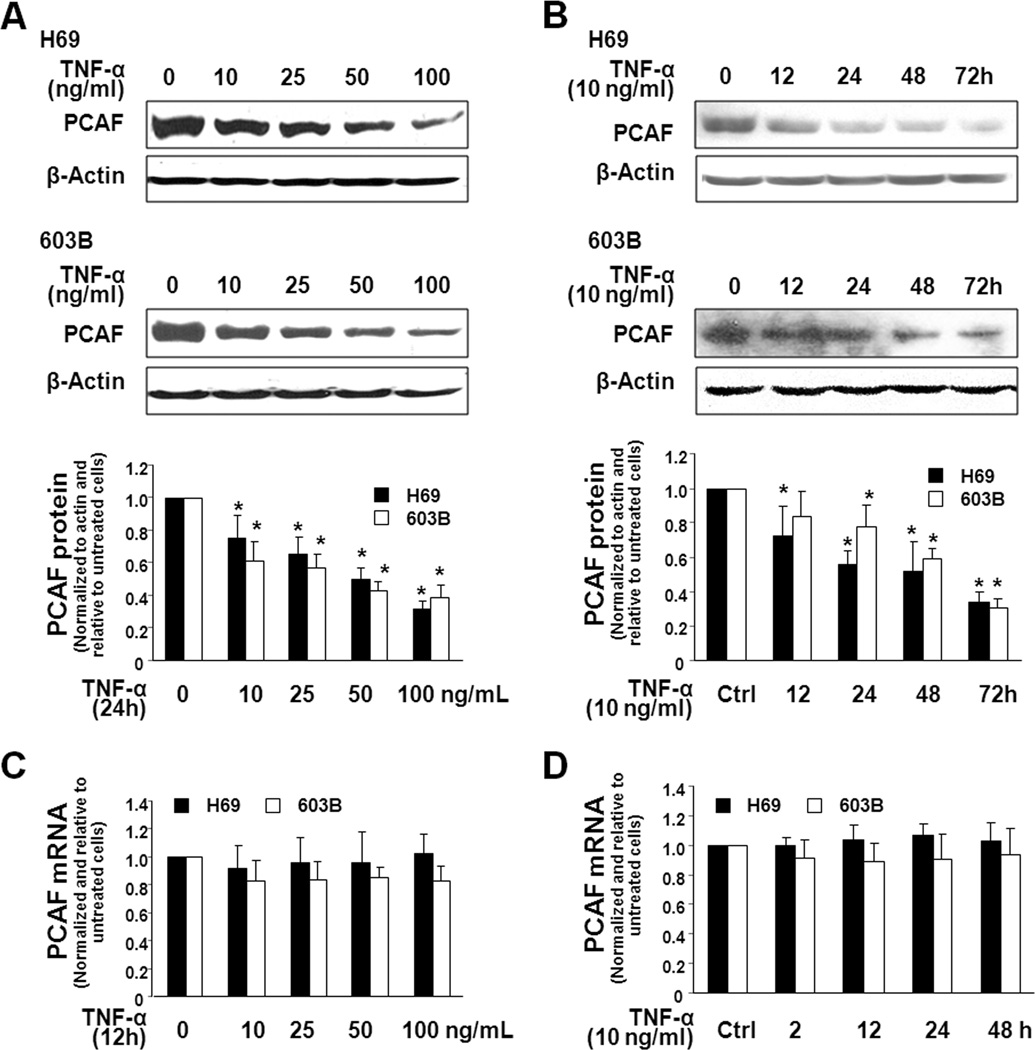
We assessed PCAF expression in H69 and 603B cells in response to TNF-α. A dose- and time-dependent decrease of PCAF protein expression was detected by Western blot in H69 and 603B cells following TNF-α stimulation (Fig. 1A and 1B). Interestingly, no significant change of PCAF mRNA levels was found in TNF-α-treated cells as measured by real-time PCR (Fig. 1C and 1D), suggesting posttranscriptional mechanisms.
FIGURE 1.
TNF-α decreases expression of PCAF protein in cholangiocytes without a change in PCAF mRNA level. H69 and 603B cells were exposed to TNF-α followed by Western blot for PCAF protein and real-time PCR for PCAF mRNA. A and B, A dose-dependent (A) and time-dependent (B) downregulation of PCAF protein was detected in H69 and 603B cells following TNF-α stimulation. Cells were exposed to TNF-α (10 ng/ml) for up to 72h or at various concentrations of TNF-α (10–100 ng/ml) for 24h. Representative Western blots were shown and densitometric ratio to β-actin was also presented. β-actin was blotted as the protein loading control. C and D, No changes in PCAF mRNA level were detected in cells after exposure to TNF-α. mRNAs of GAPDH (for H69) or β-actin (for 603B) were used to normalize the PCAF mRNA levels. Data in A–D are averages of three independent experiments. *, p < 0.05 ANOVA vs. the non-treated control.
TNF-α alters miRNA expression in cholangiocytes
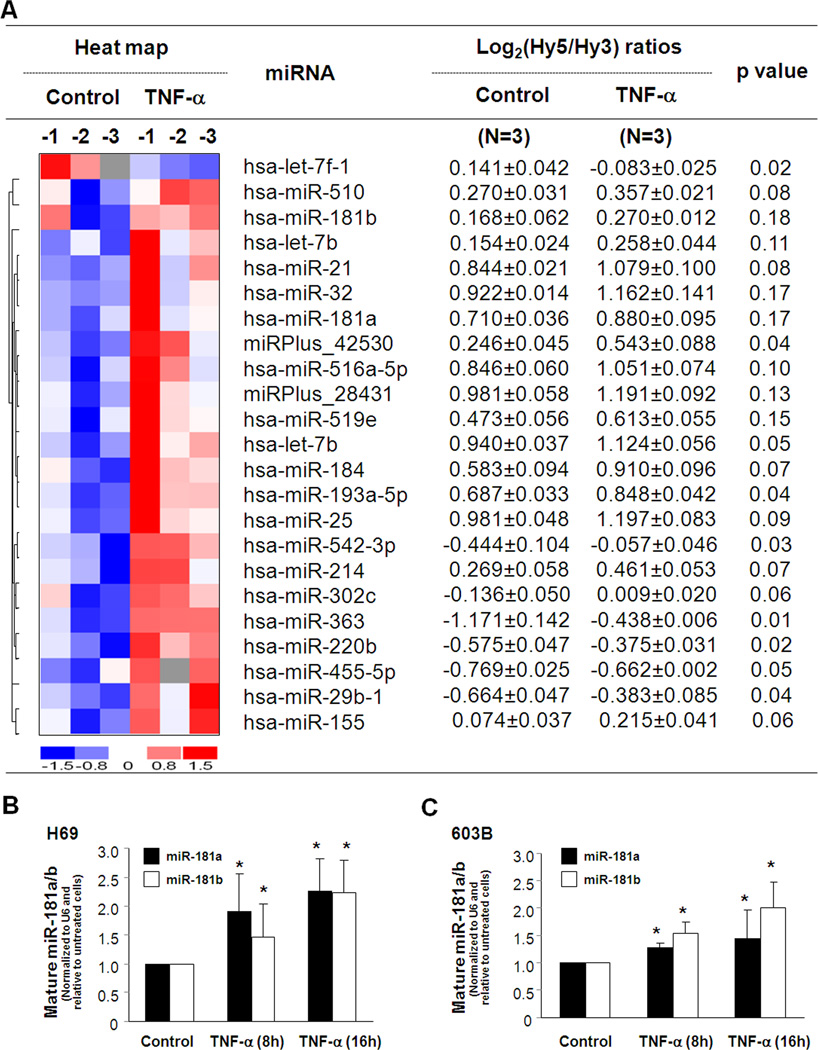
A total of 383 mature miRNAs were detected in the non-TNF-α-treated H69 cells using the miRCURY™ LNA human microRNAs analysis (Exiqon; Vedbaek, Denmark) (14,15). Among these expressed miRNAs, eight miRNAs were significantly upregulated following TNF-α treatment for 8h (p <= 0.05; Fig. 2A). Fourteen additional miRNAs, including miR-181a/b, showed a tendency to increase (0.05 < p <= 0.20; Fig. 2A). One miRNA, let-7f-1, was significantly downregulated in TNF-α-treated cells (Fig. 2A). All microarray data were described in accordance with MIAME guidelines and deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress, accession number: E-MEXP-2049). Real-time PCR analysis for selected mature miRNAs was performed to confirm the array results in H69 cells following TNF-α treatment. Increased expression of miR-181a/b, as shown in Fig. 2B and C, was detected in H69 and 603B cells following TNF-α treatment for 8h to 16h.
FIGURE 2.
TNF-α alters miRNA expression in cholangiocytes. miRNA expression profile detected in H69 cells following TNF-α (10 ng/mL) treatment for 8h by miRCURY™ LNA array (A). The left panel shows a heat-map of miRNAs that showed changes in their expression following TNF-α treatment. The horizontal axis indicates samples of non-TNF-α treated cells (n=3: Control-1, -2, and -3) and cells after exposure to TNF-α (n=3: TNF-α-1, -2, and -3). The right panel shows expression of miRNAs in H69 cells following TNF-α treatment. Cellular levels of miRNAs were presented as the log2 (Hy5/Hy3) ratios. p values are from the t’ test. hsa = Homo sapiens. B and C, Upregulation of mature miR-181a/b was confirmed by real-time PCR in H69 cells (B) and 603B cells (C) after 10 ng/mL TNF-α treatment for 8h and 16h, respectively. The amount of mature miRNAs was obtained by normalizing to the level of snRNA RNU6B. Data are expressed as the amount of miRNAs in the TNF-α-treated cells relative to the untreated controls, representative of three independent experiments. *, p < 0.05 ANOVA vs. the non-treated control.
miR-181a/b target PCAF 3'UTR, resulting in translational suppression in liver epithelial cells
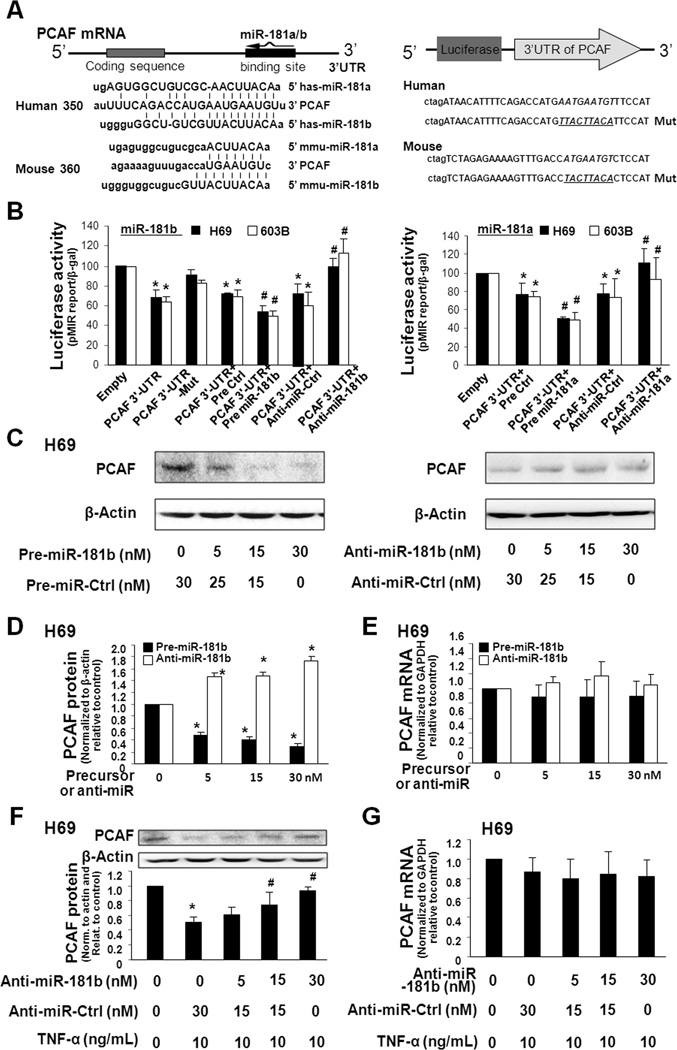
To test whether miRNA-mediated posttranscriptional regulation is involved in the downregulation of PCAF by TNF-α, we used the algorithms in the Targetscan 4.2 program (22) to screen these TNF-α-upregulated miRNAs. We found miR-181a/b with complementarity to PCAF 3’UTR, suggesting a potential for binding (Fig. 3A). We generated pMIR-REPORT luciferase constructs containing the human or mouse PCAF 3'UTR with the potential binding site (Fig. 3A). In addition, constructs with mutations at the putative binding site were also generated and used as the controls (Fig. 3A). H69 and 603B cells were transfected with these reporter constructs for 24h, followed by assessment of luciferase activity. As shown in Fig. 3B, luciferase activity was significantly decreased in cells transfected with the PCAF 3'UTR construct containing the potential binding site. No change in luciferase activity was observed in cells transfected with the mutant PCAF 3'UTR construct, suggesting endogenous translational repression of the construct with the PCAF 3'UTR. Anti-miR-181a/b markedly increased PCAF 3'UTR-associated luciferase reporter translation. In contrast, precursors to miR-181a/b further decreased luciferase reporter translation.
FIGURE 3.
PCAF is a target for miR-181a/b, and upregulation of miR-181a/b is involved in TNF-α-induced suppression of PCAF in cholangiocytes. A, The schematic of PCAF mRNA showed a potential binding site in its 3’UTR for miR-181a/b in humans and mice. The PCAF 3'UTR sequence covering the potential binding site for miR-181a/b was inserted into the pMIR-REPORT luciferase plasmid. A control plasmid with the mutant 3'UTR sequence was also generated for control. B, Binding of miR-181a/b to the potential binding site in the PCAF 3'UTR results in translational suppression. H69 and 603B cells were transfected with the pMIR-REPORT luciferase constructs and treated with the anti-miRs or precursors to miR-181a/b, or non-specific oligo control, for 24h followed by luciferase analysis. Mut = mutant; *, p < 0.05 ANOVA vs. the empty vector controls; #, p < 0.05 ANOVA vs. cells transfected with PCAF 3'UTR only. C to E, H69 cells were treated with various doses of miR-181b precursor or anti-miR-181b, followed by Western blot for PCAF protein (C and D) or PCR for PCAF mRNA (E). miR-181b precursor caused reciprocal alterations in PCAF protein, but not PCAF mRNA, in H69 cells. Representative Western blots are shown and densitometric ratio to β-actin is also presented. F and G, Anti-miR-181b inhibits TNF-α-induced suppression of PCAF protein without change in PCAF mRNA in H69 cells. Cells were transfected with the anti-miR-181b for 48h and then exposed to TNF-α treatment for 24h, followed by analysis of PCAF protein (F) and mRNA (G). Data in B-G are averages of three independent experiments. *, p < 0.05 ANOVA vs. the non-TNF-α-treated control; #, p < 0.05 ANOVA vs. the TNF-α-treated cells in B, C, D and F.
To test whether miR-181a/b are directly relevant to cellular PCAF protein level, we treated cells with anti-miRs or precursors to miR-181a/b for 48h and then measured PCAF protein content using Western blot. Transfection of H69 cells with the miR-181b precursor caused a dose-dependent decrease in PCAF protein level (Fig. 3C and D). No change in PCAF mRNA levels was found between the control cells and cells treated with miR-181b precursor (Fig. 3E), suggesting no effects on PCAF mRNA content. A dose-dependent increase in PCAF protein content, but not the message level, was identified in cells treated with anti-miR-181b (Fig. 3C–E). Anti-miR-181a and precursor to miR-181a also caused reciprocal alterations in PCAF protein level in H69 cells (Fig. S1). Precursors or anti-miRs to miR-181a/b displayed a similar effect on PCAF expression in 603B and AML-12 cells (Fig. S1).
Upregulation of miR-181a/b is involved in TNF-α-induced PCAF protein suppression
H69 and 603B cells were transfected with various doses of anti-miR-181b for 48h and then exposed to TNF-α for 24h, followed by Western blot for PCAF. The anti-miR-181b blocked the downregulation of PCAF protein in H69 and 603B cells induced by TNF-α treatment in a dose-dependent manner (Fig. 3F and Fig. S2). No significant change in PCAF mRNA levels was found in the cells following TNF-α stimulation with or without the treatment with anti-miR-181b (Fig. 3G). Anti-miR-181a had the same functions as anti-miR-181b (Fig. S2). Thus, anti-miR-181a/b could prevent the downregulation of PCAF protein induced by TNF-α. Coupled with the upregulation of miR-181a/b in cells following TNF-α treatment, the above data suggest that the relief of miR-181a/b-mediated translational repression is required for TNF-α-induced suppression of PCAF protein.
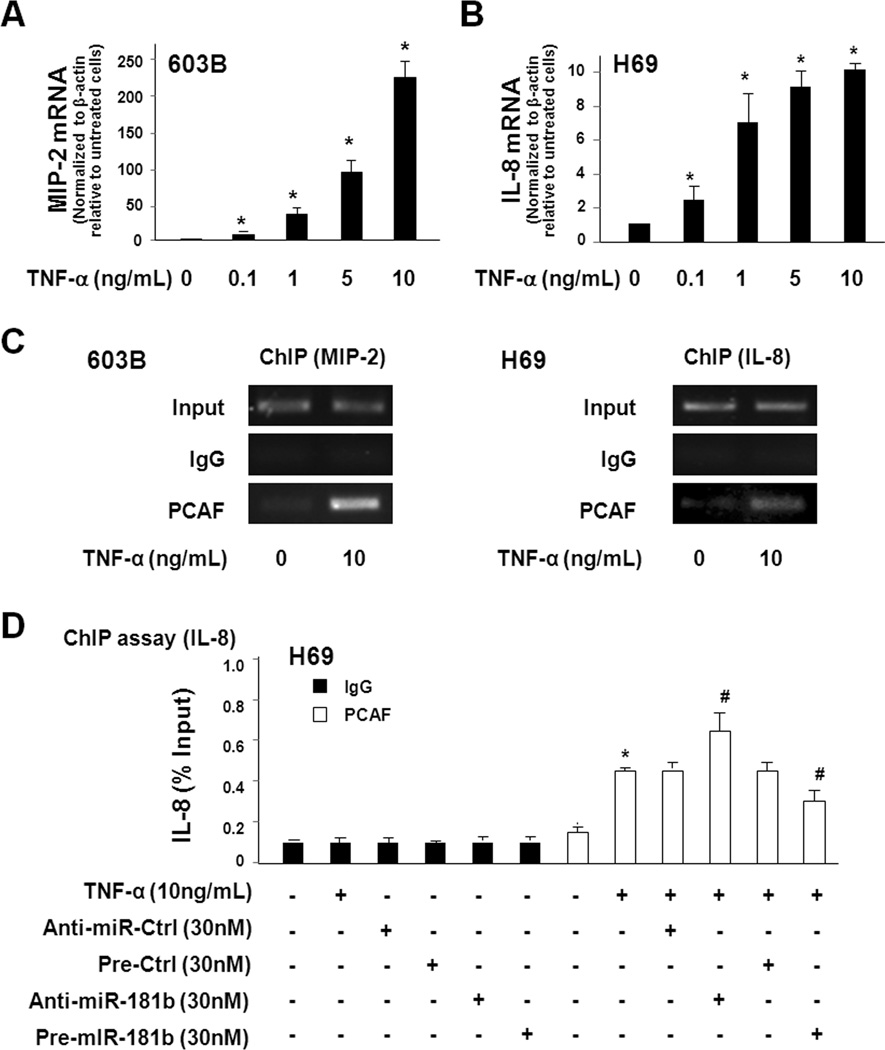
Promoter recruitment of PCAF is involved in TNF-α-induced transcription of IL-8 and MIP-2 genes
We exposed 603B and H69 cells to various doses of TNF-α for 1h. A dose-dependent increase of mRNA level for MIP-2 (for 603B) and IL-8 (for H69) was detected (Fig. 4A and Fig. 4B). ChIP analysis was then performed on cells after exposure to TNF-α for 1h. TNF-α stimulation increased recruitment of PCAF to the promoter region of the MIP-2 gene in 603B cells and the IL-8 gene in H69 cells (Fig. 4C). To test the effects of miR-181a/b on TNF-α-induced promoter recruitment of PCAF, H69 cells were transfected with the miR-181b precursor or anti-miR-181b for 48h and then exposed to TNF-α (10 ng/mL) for 1h following by ChIP analysis. Anti-miR-181b promoted TNF-α-induced PCAF promoter recruitment of the IL-8 gene (Fig. 4D). In contrast, miR-181b precursor partially suppressed TNF-α-induced PCAF promoter recruitment (Fig. 4D).
FIGURE 4.
Promoter recruitment of PCAF is involved in TNF-α-induced transcription of MIP-2 and IL-8. A and B, TNF-α stimulation increases MIP-2 and IL-8 expression in a dose-dependent manner. 603B (A) and H69 (B) cells were exposed to various doses of TNF-α for 1h and expression of MIP-2 and IL-8 was quantified by real-time PCR. C, TNF-α induces promoter recruitment of PCAF to MIP-2 and IL-8 genes. Cells were exposed to TNF-α (10 ng/mL) for 1h and promoter recruitment of PCAF to MIP-2 and IL-8 genes was assessed by ChIP analysis using primers covering the NF-κB-binding region within the promoters of MIP-2 and IL-8 genes. D, Effects of functional manipulation of miR-181b on TNF-α-induced PCAF promoter recruitment to the IL-8 gene. H69 cells were treated with miR-181b precursor or anti-miR-181b for 48h and then exposed to TNF-α for 1h followed by ChIP analysis. The results were analyzed by real-time PCR and shown as the percentage of input. Data in A–D are averages of three independent experiments. *, p < 0.05 ANOVA vs. the non-treated control; #, p < 0.05 ANOVA vs. the TNF-α-treated cells.
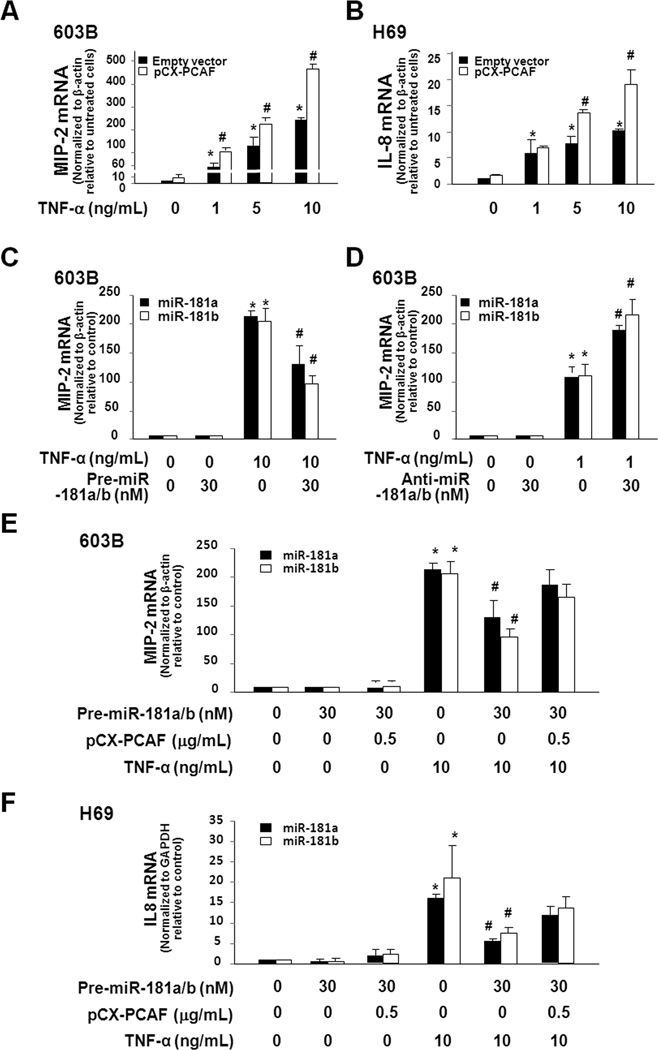
Functional manipulation of PCAF and miR-181a/b influences cell inflammatory responses to TNF-α
We transfected 603B and H69 cells with the full-length PCAF and then exposed them to TNF-α. A much higher mRNA level of MIP-2 (for 603B) and IL-8 (for H69) was detected in cells transfected with the full-PCAF (Fig. 5A and Fig. 5B). To test the effects of miR-181a/b on cellular inflammatory reactions to TNF-α, 603B cells were transfected with the precursors to miR-181a/b for 48h and then exposed to TNF-α (10 ng/mL) for 1h. Precursors to miR-181a/b could decrease the upregulation of MIP-2 mRNA induced by TNF-α (Fig. 5C). No significant difference was detected between the pre-miR-181a/b-treated and the non-treated control cells in the absence of TNF-α. When 603B cells were pretreated with anti-miR-181a/b for 48h and then exposed to TNF-α (1 ng/ml) for 1h, a marked increase in MIP-2 mRNA expression was detected (Fig. 5D). Overexpression of PCAF partially suppressed the inhibitory effects of miR-181 precursors on TNF-α-induced MIP-2 expression in 603B cells (Fig. 5E) or IL-8 expression in H69 cells (Fig. 5F). Precursors to miR-181a/b did not alter PCAF protein expression in cells transfected with the full-length PCAF (Fig. S3). These data suggest that functional manipulation of PCAF and miR-181a/b influences IL-8 and MIP-2 expression in response to TNF-α stimulation.
FIGURE 5.
Functional manipulation of miR-181a/b influences cell inflammatory responses to TNF-α through modulation of PCAF. A and B, Overexpression of PCAF enhances TNF-α-induced MIP-2 and IL-8 expression. 603B (A) and H69 (B) cells were transfected with the full-length PCAF or the empty control vector for 24h and then exposed to various doses of TNF-α for 1h. MIP-2 and IL-8 mRNA levels were quantified by real-time PCR. C and D, Effects of precursors and anti-miRs to miR-181a/b on TNF-α-induced MIP-2 expression in 603B cells. Cells were treated with precursors to miR-181a/b (C) or anti-miR-181a/b (D) for 48h and then exposed to TNF-α followed by real-time PCR. E and F, Overexpression of PCAF attenuated the inhibitory effects of precursors to miR-181a/b on TNF-α-induced MIP-2 and IL-8 expression. 603B (E) and H69 (F) cells were transfected with precursors to miR-181a/b and/or pCX-PCAF for 48h. Cells were treated with 10 ng/mL TNF-α for 1h, followed by real-time PCR analysis for IL-8 (for H69) or MIP-2 (for 603B). Transfection of cells with the full-length PCAF attenuated the inhibitory effects of precursors to miR-181a/b on TNF-α-induced upregulation of IL-8 and MIP-2. Data in A–F are averages of three independent experiments. *, p < 0.05 ANOVA vs. the untreated controls; #, p < 0.05 ANOVA between PCAF and empty vector transfected cells treated with the same concentration of TNF-α (in A and B), between pre-miR-181a/b treated and untreated cells following TNF-α stimulation (in C, E and F), and between anti-miR-181a/b treated and untreated cells following TNF-α stimulation (in D).
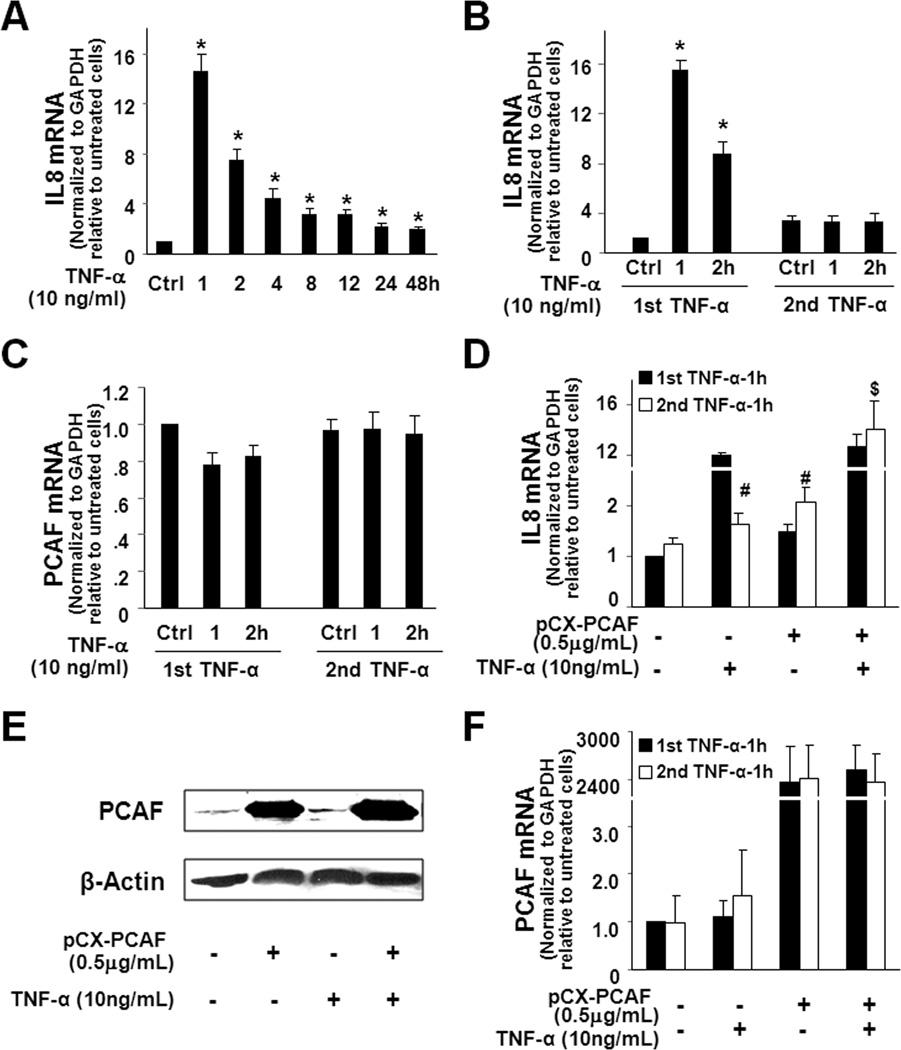
Suppression of PCAF provides negative feedback regulation to TNF-α-induced transcription of inflammatory genes in liver epithelial cells
Because promoter recruitment of PCAF facilitates transactivation of inflammatory genes induced by TNF-α, we speculated that TNF-α-induced suppression of PCAF would inhibit epithelial reactions in response to subsequent stimulation. To test this possibility, we measured the kinetics of IL-8 gene expression in H69 cells in response to TNF-α stimulation. TNF-α induced a time-dependent increase in IL-8 mRNA expression in H69 cells (Fig. 6A). Increase of IL-8 expression peaked at 1h (about a 14-fold increase) after addition of TNF-α and decreased gradually, but remained at a higher level than the non-treated cells for up to 48h after TNF-α stimulation (Fig. 6A). Interestingly, when the same amount of TNF-α was exposed to the cells that were pretreated with TNF-α for 24h, no further increase of IL-8 expression was detected (Fig. 6B), suggesting that TNF-α pretreatment can inhibit cell reactions to subsequent TNF-α stimulation. Consistent with our results in Fig. 1, no significant change in PCAF mRNA was measured in all the treated cells (Fig. 6C). Inhibition of TNF-α-induced expression of the COX-2 gene was also confirmed in TNF-α-pretreated H69 cells (Fig. S3). Similar results of TNF-α pretreatment on associated expression of MIP-2 by subsequent TNF-α stimulation were obtained in 603B cells (Fig. S4).
FIGURE 6.
Suppression of PCAF by miR-181a/b provides negative feedback regulation to TNF-α-induced expression of IL-8 in H69 cells. A, The kinetics of IL-8 mRNA expression in H69 cells in response to TNF-α stimulation. Cells were exposed to 10 ng/ml TNF-α for up to 48h, and IL-8 levels were measured by real-time PCR. B, TNF-α-pretreatment blocks IL-8 upregulation in H69 cells in response to subsequent TNF-α stimulation. An attenuated upregulation of IL-8 in response to TNF-α stimulation was detected in cells pretreated with TNF-α for 24h, compared with that of cells without TNF-α pretreatment. C, TNF-α-pretreatment does not change PCAF mRNA levels in H69 cells in response to subsequent TNF-α stimulation. D, Overexpression of PCAF attenuated the inhibitory effects of TNF-α-pretreatment on IL-8 upregulation in response to subsequent TNF-α stimulation. Cells were transfected with the full-length PCAF for 2h and exposed to the first TNF-α stimulation and followed by the second TNF-α stimulation 24h later. Cellular levels of IL-8 mRNA after TNF-α stimulation for 1h were quantified by real-time PCR. E, PCAF protein levels in cells transfected with the full-length PCAF in response to TNF-α. Cells were transfected with the full-length PCAF for 24h and then exposed to TNF-α for 1h. Representative Western blots were shown. F, Expression of PCAF mRNA in cells transfected with the full-length PCAF in response to TNF-α. Experiment design was the same as in D and PCAF mRNA levels were quantified by real-time PCR. Data in A-D and F are averages of three independent experiments. *, p < 0.05 ANOVA vs. the non-TNF-α treated controls (in A and B); #, p < 0.05 ANOVA vs. cells after the first TNF-α stimulation and $, p < 0.05 ANOVA vs. non-PCAF transfected cells after the second TNF-α stimulation (in D).
To further test that downregulation of PCAF accounts for the inhibition of cell response to secondary TNF-α stimulation, we transfected cells with the full-length PCAF and then measured expression of IL-8 or MIP-2 in TNF-α-pretreated cells in response to secondary TNF-α stimulation. Overexpression of PCAF restored cell response to transactivate the IL-8 gene in response to secondary TNF-α stimulation (Fig. 6D). Pretreatment of TNF-α did not alter expression of PCAF, at both the message and protein levels, in cells transfected with the full-length PCAF (Fig. 6E and 6F). Similar results were obtained for PCAF-dependent expression of MIP-2 in 603B cells (Fig. S4). These data indicate that suppression of PCAF provides a negative feedback loop to TNF-α-induced expression of inflammatory genes.
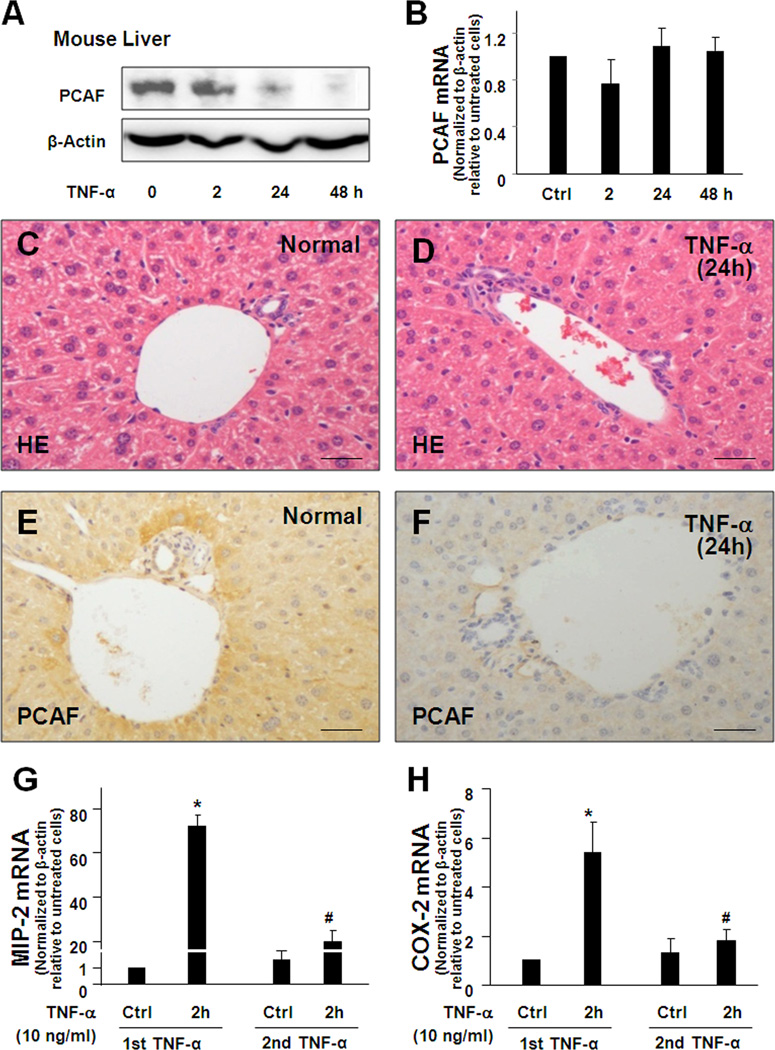
Expression of PCAF in liver epithelial cells in response to TNF-α stimulation and epithelial inflammatory reactions in response to secondary stimuli in mice
Consistent with in vitro results, we detected a significant decrease of PCAF protein content in the liver extract from mice following TNF-α i.p. injection (Fig. 7A). PCAF mRNA showed no significant change for up to 48h after TNF-α injection (Fig. 7B). Very mild inflammatory reaction of inflammatory infiltration was observed in the liver through H&E staining (Fig. 7C and 7D). Decrease of PCAF protein staining was detected by immunochemistry in hepatocytes and cholangiocytes in mice 24h following TNF-α i.p. injection (Fig. 7E and 7F).
FIGURE 7.
Expression of PCAF and inflammatory reactions in the liver in response to TNF-α stimulation in mice. A and B, TNF-α decreases PCAF expression in mouse liver. PCAF mRNA level showed no significant change in the liver tissues from mice following TNF-α administration. A significant decrease of PCAF protein content was detected in the liver extract from mice 24h following TNF-α i.p. injection. C and D, Very mild inflammatory reaction of inflammatory infiltration was observed in the liver through H&E staining. E and F, Decrease of PCAF protein staining was detected in hepatocytes and cholangiocytes in mice 24h following TNF-α i.p. injection by immunochemistry. G and H, Pre-injection of TNF-α for 24h attenuated the expression of MIP-2 (G) and COX-2 (H) in the liver extracts at 2h after secondary TNF-α injection, as measured by real-time PCR. *, p < 0.05 ANOVA vs. the controls; #, p < 0.05 ANOVA vs. liver extracts after the first TNF-α stimulation for 2h.
We detected an 80-fold increase in MIP-2 expression in the liver extracts at 2h after TNF-α injection. Expression of MIP-2 decreased to basal level at 24h following TNF-α injection (Fig. 7G). Consistent with in vitro results, only about a 20-fold increase of MIP-2 was detected at 2h after the second TNF-α administration in the liver obtained from mice with an initial TNF-α injection 24h previously (Fig. 7G). Similarly, inhibition of TNF-α-induced COX-2 upregulation was observed in the livers of mice pretreated with TNF-α for 24h (Fig. 7H).
DISCUSSION
Similar to that of protein-coding genes, expression of miRNA genes is regulated through both transcriptional and posttranscriptional mechanisms (12). Transcription of miRNA genes in epithelial cells is elaborately controlled and associated with nuclear transcription factor-mediated transactivation and transrepression. The primary transcripts of miRNAs (pri-miRNAs) then undergo further enzymatic cleavage to form mature miRNAs, and this maturation process involves several enzymes and associated proteins, including heterogenous nuclear RNA complex (hnRNP) proteins (23). Upon contact with TNF-α, TNF receptors activate downstream signaling pathways, such as the NF-κB and MAPK pathways, and mediate the transcription of a vast array of genes involved in cell survival and proliferation, inflammatory response, and anti-apoptotic factors (24,25). In this study, we found that TNF-α stimulation caused a distinct alteration in the expression profile of mature miRNAs in H69 cells. Interestingly, the majority of the miRNAs that were significantly altered in TNF-α-treated H69 cells are not NF-κB-responsive miRNAs, which we previously identified in H69 cells (14,15). Of these NF-κB-responsive miRNAs, only miR-21 and miR-155 showed a significant increase in TNF-α-treated H69 cells. Activation of the MAPK signaling pathway has also been reported to induce transcription of miRNA genes in different cell types in response to various stimuli (26). Moreover, activation of the MAPK/p38 pathway can phosphorylate hnRNP A1 and promote the cytoplasmic translocation of hnRNP A1 and miRNA maturation (27). We speculate that TNF-α-induced alterations in miRNA expression reflect the integrated result of activation of multiple interrelated signal pathways, including the NF-κB and MAPK pathways.
miRNA-mediated posttranscriptional gene suppression may be a critical component of the complex regulatory networks in epithelial inflammatory responses. The usual consequence of miRNA and mRNA interaction is the downregulation of protein expression by translational repression and/or mRNA cleavage (12,13). Whereas studies have predominantly focused on these TNF-α-upregulated effector molecules, upregulation of miRNAs in TNF-α-treated cells suggests to us that TNF-α may suppress gene expression through miRNA-mediated posttranscriptional gene repression, contributing to regulation of cell reactions in response to TNF-α stimulation. Indeed, our data demonstrate that TNF-α suppresses PCAF expression in liver epithelial cells both in vitro and in vivo. Moreover, TNF-α-induced suppression of PCAF appears to be associated with miR-181a/b-mediated posttranscriptional repression. Targeting of PCAF 3’UTR by miR-181a/b was confirmed using the luciferase reporter analysis. Functional manipulation of miR-181a/b caused reciprocal alterations in PCAF protein level in cultured hepatocytes or cholangiocytes. Importantly, inhibition of miR-181a/b function blocked TNF-α-induced PCAF suppression in these cells. Moreover, it appears that miR-181a/b suppress PCAF expression through inhibition of translation, not through induction of RNA degradation, as no change in PCAF mRNA levels was detected in cells following TNF-α stimulation or treatment with precursor or anti-miRs to miR-181a/b. Therefore, TNF-α may regulate expression of genes that are not directly regulated by TNF-α signaling at the transcriptional level through modulation of miRNA-mediated posttranscriptional gene suppression.
As one of the most common co-activators, PCAF can bridge the transcriptional factors to the transcriptional complex to access the appropriate level of gene activities in cells in response to extracellular stimuli (11). Different from other co-activators, PCAF has the capacity of HATs and, thus, can augment gene transcription through modulating chromatin remodeling. Our results of ChIP analysis confirmed the promoter recruitment of PCAF to the inflammatory MIP-2 and IL-8 genes in cells following TNF-α stimulation. Forced expression of PCAF enhances TNF-α-induced MIP-2 and IL-8 expression in liver epithelial cells. Because promoter recruitment of PCAF is usually gene-specific (28), it is of merit to investigate whether PCAF is involved in the transcriptional regulation of other TNF-α-regulated inflammatory genes.
Inflammation, while an essential physiological response to insult or injury, is potentially injurious to host tissues and is, therefore, a highly regulated process. A variety of extracellular and intracellular feedback pathways have evolved to prevent an inappropriate over-inflammatory response, including upregulation of several negative regulators, such as IRAK1, TRAF6, A20, and SIGIRR (29). Our data demonstrate that miR-181a/b-mediated suppression of PCAF may provide negative feedback regulation to liver epithelial cells in response to subsequent TNF-α stimulation. We found that pretreatment of cells with TNF-α can significantly attenuate epithelial expression of inflammatory genes in liver epithelial cells in response to secondary TNF-α stimulation both in vitro and in vivo. Pretreatment of cells with precursors to miR-181a/b showed an inhibitory effect on TNF-α-induced expression of inflammatory genes. Overexpression of PCAF can suppress the inhibitory effects of pretreatment with the precursors to miR-181a/b. Therefore, fine-tuning of the inflammatory reactions in hepatocytes and cholangiocytes in response to TNF-α stimulation may involve miR-181a/b-mediated suppression of PCAF. Such a negative feedback regulatory loop may function in concert with other regulatory mechanisms to ensure finely-controlled inflammatory responses in the liver. Recent studies have demonstrated the targeting of several negative regulators by inflammatory miRNAs, such as miR-146 targeting of IRAK1 and TRAF6 (26), miR-155 targeting of MyD88 and FADD (30), and miR-9 targeting of NFKB1 (31). Our work on miR-181a/b targeting of PCAF provides additional evidence that miRNAs are essential components in the feedback regulation of epithelial immune/inflammatory responses.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Guoku Hu for helpful and stimulating discussions and Ms. Barbara L. Bittner for her assistance in editing the manuscript. PCAF expression vector pCX-PCAF (Flag-tagged) was a gift from Dr. Tony Kouzarides (University of Cambridge, UK).
Footnotes
This work was partially supported by National Institutes of Health grants R01 AI071321 and U01 AI095532, and the Nebraska Tobacco Settlement Biomedical Research Program (LB692 and LB595) (to X-M.C).
The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Abbreviations used in this paper: PCAF, p300/CBP-associated factor; HATs, histone acetyltransferases; HDACs, histone deacetylases; miRNAs, microRNAs; TSA, trichostatin A; ChIP, Chromatin immunoprecipitation; 3’UTR, 3’-untranslated region.
REFERENCES
- 1.Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial iInfections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Alpini G, McGill JM, LaRusso NF. The pathobiology of biliary epithelia. Hepatology. 2002;35:1256–1268. doi: 10.1053/jhep.2002.33541. [DOI] [PubMed] [Google Scholar]
- 3.Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, Kaufmann SH, Gores GJ. Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 2000;106:1127–1137. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selmi C, Mackay IR, Gershwin ME. The immunological milieu of the liver. Sem. in Liver Dis. 2007;27:129–139. doi: 10.1055/s-2007-979466. [DOI] [PubMed] [Google Scholar]
- 5.Chen XM, O'Hara SP, LaRusso NF. The immunobiology of cholangiocytes. Immunol. Cell Biol. 2008;86:497–505. doi: 10.1038/icb.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Rahman I, Gilmour PS, Jimenez LA, MacNee W. Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol. Cell Biochem. 2002;234–235:239–248. [PubMed] [Google Scholar]
- 8.Kim SH, Kim DH, Lavender P, Seo JH, Kim YS, Park JS, Kwak SJ, Jee YK. Repression of TNF-alpha-induced IL-8 expression by the glucocorticoid receptor-beta involves inhibition of histone H4 acetylation. Exp. Mol. Med. 2009;41:297–306. doi: 10.3858/emm.2009.41.5.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G1124–G1136. doi: 10.1152/ajpgi.00091.2005. [DOI] [PubMed] [Google Scholar]
- 10.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly (ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 11.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 14.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000681. e1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu G, Gong AY, Liu J, Zhou R, Deng C, Chen XM. miR-221 suppresses ICAM-1 translation and regulates interferon-gamma-induced ICAM-1 expression in human cholangiocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;298:G542–G550. doi: 10.1152/ajpgi.00490.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, Chen XM. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 2009;183:1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am. J. Physiol. 1994;266:G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 19.Yahagi K, Ishii M, Kobayashi K, Ueno Y, Mano Y, Niitsuma H, Igarashi T, Toyota T. Primary culture of cholangiocytes from normal mouse liver. In Vitro Cell Dev. Biol. Anim. 1998;34:512–514. doi: 10.1007/s11626-998-0106-x. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bluthé RM, Pawlowski M, Suarez S, Parnet P, Pittman Q, Kelley KW, Dantzer R. Synergy between tumor necrosis factor alpha and interleukin-1 in the induction of sickness behavior in mice. Psychoneuroendocrinology. 1994;19:197–207. doi: 10.1016/0306-4530(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 22.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;14:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Gustin JA, Pincheira R, Mayo LD, Ozes ON, Kessler KM, Baerwald MR, Korgaonkar CK, Donner DB. Tumor necrosis factor activates CRE-binding protein through a p38 MAPK/MSK1 signaling pathway in endothelial cells. Am. J. Physiol. Cell Physiol. 2004;286:C547–C555. doi: 10.1152/ajpcell.00332.2002. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Monack DM, Kayagaki N, Wertz I, Yin J, Wolf B, Dixit VM. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 2005;21:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 28.Bennett MK, Osborne TF. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. U S A. 2000;97:6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibolet O, Podolsky DK. TLRs in the gut. IV. Negative regulation of Toll-like receptors and intestinal homeostasis: addition by subtraction. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1469–G1473. doi: 10.1152/ajpgi.00531.2006. [DOI] [PubMed] [Google Scholar]
- 30.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 31.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.