Abstract
Neutrophils utilize immunoglobulins (Igs) to clear antigen, but their role in Ig production is unknown. Here we identified neutrophils around the marginal zone (MZ) of the spleen, a B cell area specialized in T-independent Ig responses to circulating antigen. Neutrophils colonized peri-MZ areas after post-natal mucosal colonization by microbes and enhanced their B-helper function upon receiving reprogramming signals from splenic sinusoidal endothelial cells, including interleukin 10 (IL-10). Splenic neutrophils induced Ig class switching, somatic hypermutation and antibody production by activating MZ B cells through a mechanism involving the cytokines BAFF, APRIL and IL-21. Neutropenic patients had fewer and hypomutated MZ B cells and less preimmune Igs to T-independent antigens, which indicates that neutrophils generate an innate layer of antimicrobial Ig defense by interacting with MZ B cells.
INTRODUCTION
Neutrophils are the first immune cells that migrate to sites of infection and inflammation to eliminate microbes and necrotic cells1. After sensing conserved molecular signatures associated with microbes and tissue damage, neutrophils activate defensive programs that promote phagocytosis, intracellular degradation, extracellular discharge of antimicrobial factors, and formation of neutrophil extracellular traps (NETs)2. These structures arise following cell death and consist of decondensed chromatin embedded with granular and cytoplasmic proteins that trap and kill microbes3. Neutrophils also release cytokines and chemokines that recruit monocytes to optimize antigen clearance4.
The long-held view that neutrophils exclusively function in the innate phase of the immune response has been challenged by studies showing that neutrophils also influence adaptive immunity by interacting with dendritic cells5. These innate immune cells present antigen to T cells after undergoing further maturation in response to neutrophil-derived cytokines such as tumor necrosis factor (TNF)6. Neutrophils also release interleukin-12 (IL-12), which promotes the polarization of naïve T cells into inflammatory T helper type-1 cells releasing interferon-γ (IFN-γ)2. In the presence of IFN-γ and other inflammatory cytokines, neutrophils also up-regulate the expression of antigen-loading major histocompatibility class-II molecules to acquire dendritic cell-like antigen-presenting function2.
Although there is growing evidence that neutrophils have an impact on the induction of T cell responses during infection, additional data show that neutrophils suppress T cell activation in the context of pregnancy and cancer2. Indeed, neutrophils are equipped with enzymatic systems such as inducible nitric oxide synthase (iNOS) and arginase that suppress T cells by generating nitrogen intermediates and depleting extracellular arginine, respectively7. Neutrophils would further regulate adaptive immunity by secreting IL-10 after sensing bacteria through Toll-like receptors (TLRs) and C-type lectin receptors8. Thus, neutrophils can either potentiate or down-modulate T cell responses in a context-dependent manner.
Neutrophils further crosstalk with the adaptive immune system by binding to B cell-derived immunoglobulin G (IgG) and IgA on opsonized microbes9,10. The ensuing activation of Fcγ and Fcα receptors regulates neutrophil effector functions1. Interestingly, neutrophils also produce B cell-activation factor of the TNF family (BAFF or BLyS) and a proliferation-inducing ligand (APRIL), two TLR-inducible B cell-stimulating factors related to the T cell molecule CD40 ligand (CD40L)11–14. In addition to promoting the survival and differentiation of B cells and Ig-secreting plasma cells11–14, BAFF and APRIL trigger IgM production and class switching from IgM to IgG or IgA independently of CD40L15–18. This T cell-independent (TI) pathway would enable antigen-sampling dendritic cells and other innate immune cells to enhance B cell responses at mucosal surfaces inhabited by commensal bacteria19.
TI Ig responses also occur in the marginal zone (MZ) of the spleen, a B cell area positioned at the interface between the circulation and the immune system20,21. B cells dwelling in the MZ are in a state of active readiness that enables them to mount prompt Ig responses to blood-borne antigens through a pathway that does not require a T cell-dependent (TD) germinal center reaction20–22. While some MZ B cell responses may occur upon translocation of commensal antigens across intact mucosal surfaces20, 21, 23–26, others follow systemic invasion by mucosal pathogens20, 21. In humans, MZ B cells have a circulating counterpart, contain mutated Ig genes and express surface IgM and IgD together with the memory molecule CD27 (refs. 21, 27, 28). These MZ B cells are different from hypermutated IgM-memory B cells, which emerge from a canonical germinal center reaction and express surface IgM and CD27 but not IgD22. The role of neutrophils in B cell activation and Ig production is unknown, but published studies show that these granulocytes home to the MZ in response to blood-borne bacteria29.
We show here that neutrophils colonized peri-MZ areas of the spleen in the absence of infection via a non-inflammatory pathway that became more prominent after post-natal mucosal colonization by bacteria. Compared to circulating neutrophils, splenic neutrophils expressed a distinct phenotype, formed MZ B cell-interacting NET-like structures, and elicited Ig class switching, somatic hypermutation and antibody production by activating MZ B cells through a mechanism involving BAFF and APRIL and the cytokine IL-21. Circulating neutrophils acquired B–helper function upon exposure to splenic sinusoidal endothelial cells (SECs) releasing signal transducer and activator of transcription 3 (STAT3)-dependent cytokines such as IL-10 in response to microbial signals. Patients with congenital neutropenia had fewer and hypomutated MZ B cells and produced less preimmune Igs to TI antigens. Thus, neutrophils may generate an innate layer of antimicrobial Ig defense by undergoing MZ B–helper reprogramming in the spleen.
RESULTS
Neutrophils colonize peri-MZ areas
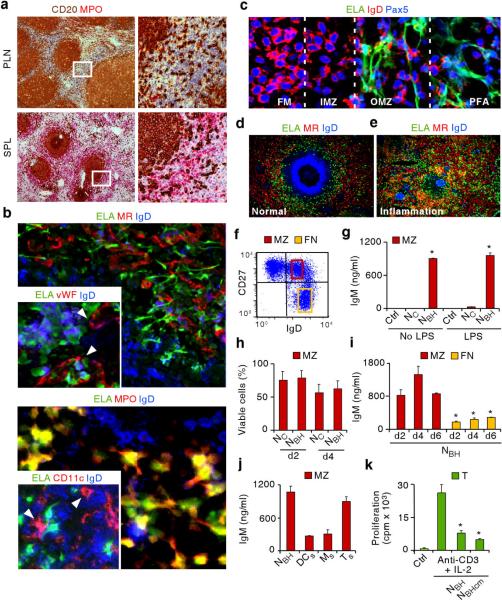
To determine whether neutrophils interact with B cells under homeostatic conditions, lymphoid organs from individuals with no inflammation or infection were stained for the B cell molecule CD20 and for the granulocytic enzyme myeloperoxidase by immunohistochemistry. Peripheral lymph nodes, tonsils and intestinal Peyer's patches contained no or few neutrophils, whereas some neutrophils were detected in the perifollicular area of mesenteric lymph nodes (Fig. 1a and Supplementary Fig. 1). Unexpectedly, even more neutrophils were detected in the perifollicular area of spleens that had no histological alterations (Fig. 1a and Supplementary Fig. 2a). As shown by light, fluorescence and confocal microscopy, splenic neutrophils expressed typical granulocyte molecules such as elastase, myeloperoxidase and the adhesion molecule CD15, and formed projections interacting with outer MZ and perifollicular B cells weakly expressing IgD (Fig. 1b,c and Supplementary Fig. 2b). Splenic neutrophils were also proximal to perifollicular SECs expressing the lectin mannose receptor (MR) and the coagulation protein von Willebrand factor, macrophages expressing the glycoprotein CD68 and the haptoglobulin receptor CD163, and dendritic cells expressing the integrin CD11c.
Figure 1. Neutrophils colonize the splenic MZ and function as NBH cells.
(a) Immunohistochemistry of peripheral lymph node (PLN) and spleen (SPL) stained for CD20 (brown) and myeloperoxidase (MPO, red). Boxes correspond to magnified right images. Original magnification, ×4 (left) and ×20 (right). (b–e) Immunofluorescence of normal (b, c, d) and inflamed hyper-IgD syndrome (HIDS) spleen (e) stained for elastase (ELA, green), MR, von Willebrand factor (vWF), MPO, CD11c or IgD (red), and IgD or Pax5 (blue). Original magnification, ×40 (b, c), ×63 (b smaller panels), and ×10 (d–e). Arrowheads point to sinusoids (b top smaller panel) and dendritic cells (b bottom smaller panel). FM, follicular mantle; IMZ, inner marginal zone; OMZ, outer marginal zone; PFA, perifollicular area. (f) Flow cytometry of IgD and CD27 on splenic CD19+ B cells. Yellow and red boxes indicate gates for follicular naïve (FN) IgDhighCD27− B cells and MZ IgDlowCD27+ B cells. (g) ELISA of IgM from splenic MZ B cells cultured for 7 d with medium (Ctrl), NC or NBH cells primed or not with LPS. (h) Flow cytometry of viable splenic MZ B cells cultured with NC or NBH cells for 2 or 4 d. (i) ELISA of IgM from splenic MZ and FN B cells cultured with NBH cells for 2, 4 and 6 d. (j) ELISA of IgM from splenic MZ B cells cultured with NBH cells, splenic dendritic cells (DCs), splenic macrophages (Ms) or splenic CD4+ T cells (Ts) for 6 d. (k) Proliferation of CD4+ T cells (T) cultured with medium alone (Ctrl) or anti-CD3 plus IL-2 in the presence or absence of NBH cells or NBH cell-derived conditioned medium (NBHcm). Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test). Data are from one of three experiments with similar results (a–f) or summarize three independent experiments (g–k).
Consistent with their possible role in homeostasis, perifollicular neutrophils were also detected in spleens from healthy rhesus macaques and mice (Supplementary Fig. 3a). Simian neutrophils expressed elastase and were positioned between MZ B cells and red pulp SECs, whereas mouse neutrophils expressed the granulocyte molecule Ly6G and surrounded a MZ area containing B cells weakly positive for IgD and macrophages strongly positive for the sialoadhesin receptor MOMA-1. Another indication of the role of neutrophils in homeostasis came from the analysis of spleens from patients with systemic inflammatory or infectious disorders, including systemic lupus erythematosus, hyper-IgD syndrome (an autoinflammatory disorder), HIV infection and sepsis. In these pathological spleens, neutrophils lost the selective perifollicular topography usually observed in normal spleens and instead extensively infiltrated follicular mantle and germinal center areas populated by B cells strongly positive and negative for IgD, respectively (Fig. 1d,e and Supplementary Fig. 3b). The aberrant topography of splenic neutrophils in inflamed spleens was associated with a partial or complete loss of the MZ. Thus, neutrophils colonize splenic peri-MZ but not follicular areas under homeostatic conditions.
Neutrophils have MZ B–helper function
Flow cytometry, light microscopy and electron microscopy did not show gross morphological and ultrastructural differences in circulating and splenic neutrophils (Supplementary Fig. 4a,b). Neutrophils amounted to 10%, 2%, 0.5% and 0.3% of total cells in spleens, mesenteric lymph nodes, peripheral lymph nodes and tonsils, respectively. In spleen and mesenteric lymph nodes, MZ B cells co-expressing IgD and the memory molecule CD27 were more abundant than in other lymphoid tissues and strongly expressed the complement receptor CD21 (Fig. 1f, Supplementary Fig. 4c, and data not shown). Enzyme-linked immunosorbent assay (ELISA) demonstrated induction of IgM secretion by MZ B cells exposed to splenic but not circulating neutrophils, which were therefore defined as B–helper (NBH) and conventional (NC) neutrophils, respectively (Fig. 1g). Priming with lipopolysaccharide (LPS) did not augment the IgM-inducing function of NBH cells, suggesting pre-existing activation. Viability assays indicated that the enhanced IgM-inducing function of NBH cells did not depend on their ability to promote better MZ B cell survival than NC cells (Fig. 1h). Accordingly, NBH cells apoptosis was equivalent to NC cells in long-term co-cultures in spite of showing better survival in short-term monocultures (Supplementary Fig. 5a).
NBH cells activated MZ B cells more effectively and more rapidly than follicular naïve B cells through both contact-dependent and contact-independent mechanisms (Fig. 1i and Supplementary Fig. 5b). The functional prominence of NBH cells was documented by the fact that they activated MZ B cells as effectively as splenic CD4+ T cells and more effectively than splenic macrophages and dendritic cells (Fig. 1j). The functional specificity of NBH cells was highlighted by their ability to induce contact-independent suppression of the proliferation of CD4+ T cells that were activated via the T cell receptor molecule CD3 and the cytokine IL-2 (Fig. 1k). Thus, NBH cells function as professional MZ B cell-helper cells and may suppress T cells to induce Ig responses in a TI manner.
Neutrophils induce Ig diversification and production
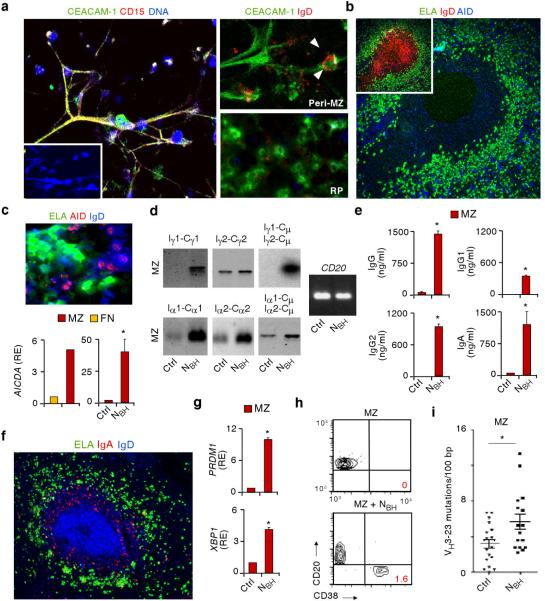
Confocal microscopy and wide-field microscopy showed that NBH cells expressing elastase, CD15 and a granulocytic microbial receptor known as carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM-1) formed DNA-containing NET-like projections that were in close contact with B cells expressing IgD (Fig. 2a and Supplementary Movie 1). Compared to red pulp NBH cells, outer MZ and perifollicular NBH cells had more abundant projections and often interacted with B cells expressing the DNA-editing enzyme activation-induced cytidine deaminase (AID) (Fig. 2b,c), a hallmark of ongoing Ig gene diversification in germinal center and some extrafollicular B cells16,17,30,31. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) showed that MZ B cells had more AICDA mRNA for AID than naive B cells but less AICDA than germinal center B cells, and demonstrated that MZ B cells up-regulated AICDA upon exposure to NBH but not NC cells (Fig. 2c and Supplementary Fig. 6a,b).
Figure 2. NBH cells interact with splenic MZ B cells to induce Ig CSR, SHM and production.
(a) Confocal microscopy of splenic NBH-B cell clusters stained for CEACAM-1, CD15 or IgD (red) and DNA (blue). Peri-MZ and red pulp (RP) NBH cells are magnified in right panels. Arrowheads point to a MZ B cell interacting with an extracellular projection from NBH cell. Original magnification, ×40 (left panel), or ×63 (right panels). (b) Immunofluorescence of a spleen stained for ELA (green), IgD (red) and AID (blue) with AID-positive B cells and NBH cells in peri-MZ and MZ areas (large panel) of an IgD+ primary follicle (small panel). Original magnification, ×10. (c) Top panel: immunofluorescence of NBH-B cell clusters stained for ELA (green), AID (red) and IgD (blue). Original magnification, ×63. Bottom panels: qRT-PCR of AICDA mRNA (encoding AID) from splenic follicular naïve (FN) and MZ B cells (left) and splenic MZ B cells cultured for 2 d in the presence of medium (Ctrl) or NBH cells (right). Results are normalized to PAX5 mRNA and are presented as relative expression (RE) compared with FN or MZ B cells incubated with medium alone. (d) Southern blot analysis of germline Iγ1-Cγ1, Iγ2-Cγ2, Iα1-Cα1 and Iα2-Cα2 transcripts and Iγ-Cμ and Iα-Cμ switch circle transcripts RT-PCR amplified from splenic MZ B cells cultured for 4 d with medium (Ctrl) or NBH cells. CD20 transcripts are a B cell-specific loading control. (e) ELISA of IgG, IgG1, IgG2 and IgA from MZ B cells cultured as in d for 7 d. (f) Immunofluorescence of spleens stained with ELA (green), IgA (red) and IgD (blue). Original magnification, ×10. (g) qRT-PCR analysis of PRDM1 (encoding Blimp-1) and XBP1 mRNAs from MZ B cells cultured as in e. Results are normalized to PAX5 mRNA and are presented as RE compared with MZ B cells incubated with medium. (h) Flow cytometry analysis of CD20 and CD38 on MZ B cells cultured as in e. (i) Number of VH3–23 gene mutations per 100 base pairs (bp) in MZ B cells cultured as in d for 12 d. Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test). Data are from one of three experiments with similar results (a,b, upper and bottom-left c panel, d,f,h) or summarize three independent experiments (bottom-right c panel, e,g,i).
RT-PCR and Southern blot analysis were performed to study class switch recombination (CSR), a process that provides Igs with new effector functions by replacing the heavy chain (H) constant (C) region of IgM with that of IgG or IgA without changing antigen specificity30. Consistent with their increased AID expression, some MZ B cells expressed germline Iγ2-Cγ2, Iα1-Cα1 and Iα2-Cα2 transcripts as well as circle Iα1-Cμ and Iα2-Cμ transcripts (Fig. 2d), which are hallmarks of ongoing IgG2, IgA1 and IgA2 CSR, respectively16–18. In the presence of NBH cells, MZ B cells induced the expression of Iγ1-Cγ1, Iγ1-Cμ and Iγ2-Cμ mRNAs, and up-regulated the expression of Iγ2-Cγ2, Iα1-Cα1, Iα2-Cα2, Iα1-Cμ and Iα2-Cμ mRNAs, which indicated induction or enhancement of IgG1, IgG2, IgA1 and IgA2 CSR, respectively.
As shown by ELISAs, qRT-PCR, flow cytometry and fluorescence microscopy, NBH cells stimulated MZ B cells to produce IgA and IgG, including higher amounts of IgG2 than IgG1 (Fig. 2e,f). This effect was not elicited by NC cells, occurred more effectively in MZ than naive B cells, and correlated with the induction of PRDM1 and XBP1 mRNAs for the plasma cell-associated proteins Blimp-1 and XBP-1 and with the generation of plasma cells expressing the activation molecule CD38 but not CD20 (Fig. 2g,h and Supplementary Fig. 6c). Finally, DNA cloning and sequencing were performed to study somatic hypermutation (SHM), a process that mutates V(D)J exons encoding the antigen-binding variable (V) region of Igs30. Although typically occurring in germinal center B cells, SHM also targets MZ B cells, at least in humans21. In the presence of NBH cells, MZ B cells accumulated more mutations in the VH3–23 gene (Fig. 2i), which is often utilized by MZ B cells21. Thus, NBH cells may stimulate MZ B cells to undergo SHM in addition to CSR and plasma cell differentiation.
Neutrophils comprise two MZ B-helper subsets
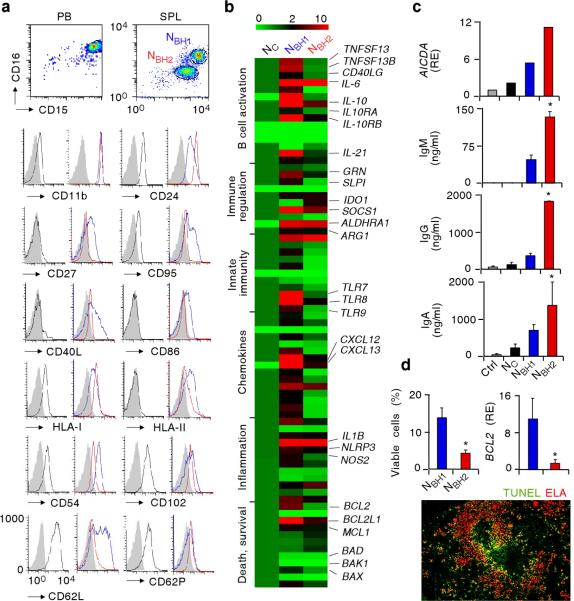
Flow cytometry, electron, light, wide-field or confocal microscopy, qRT-PCR arrays, ELISA, annexin-V assay, and terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assay were performed to further characterize NBH cells. We could identify two distinct NBH1 and NBH2 subsets of NBH cells based on their relative expression of multiple parameters, including CD15 and CD16 expression. Similar to NC cells, NBH cells showed typical granulocytic physical features and expressed canonical granulocytic molecules such as CD15 and the Fcγ receptor CD16 (Fig. 3a and Supplementary Fig. 7a). However, while NC cells had high CD15 and CD16 expression, NBH1 cells had intermediate CD15 and CD16 expression and NBH2 cells low CD15 and CD16 expression. Compared to NC cells, NBH1 and NBH2 cells expressed more CD11b and CD24, which are molecules that inhibit TLR signaling32,33, and more CD27, CD40L, CD86 (B7-2), CD95 (Fas), human leukocyte antigen-I (HLA-I) and HLA-II (Fig. 3a), which are molecules that indicate immune activation2,6. In addition, NBH1 and NBH2 cells expressed less CD54 (ICAM-1), CD62L (L-selectin), CD62P (P-selectin), and CD102 (ICAM-2) adhesion molecules (Fig. 3a), a phenotype consistent with endothelial adhesion and extravasation2,34. In spite of being morphologically and ultrastructurally similar to NBH2 cells, NBH1 were more activated than NBH2 cells, because they expressed more CD27, CD40L, CD86, CD95 and HLA-II but less CD24 (Fig. 3a and Supplementary Fig. 7b,c). This observation correlated with the persistence of NBH1 but not NBH2 cells in inflamed spleens with hypoplastic MZ (Fig. 3a and Supplementary Fig. 8).
Figure 3. NBH cells include NBH1 and NBH2 subsets distinct from NC cells.
(a) Flow cytometry of CD15, CD16, CD11b, CD24, CD27, CD95, CD40L, CD86, HLA-I, HLA-II, CD54, CD102, CD62L and CD62P on CD15highCD16high NC cells from peripheral blood (PB, black profiles), or splenic (SPL) CD15intCD16int NBH1 cells (blue profiles) and CD15lowCD16low NBH2 cells (red profiles). Filled gray profile, isotype control. (b) Gene expression profile of NC, NBH1 and NBH2 cells established by customized qRT-PCR arrays. Results are normalized to ACTB mRNA (encoding β-actin) and presented as relative expression compared with that of NC cells. Functional mRNA clusters and highly relevant mRNAs are indicated. TNFSF13, TNFSF13B, CD40LG, IL10RA, IL10RB, GRN, IDO1, ALDHRA1, ARG1, IL1B, NLRP3, NOS2 and BCL2L1 mRNAs encode APRIL, BAFF, CD40L, IL-10 receptor α, IL-10 receptor β, progranulin, IDO, RALDH, arginase I, IL-1β, NALP-3, iNOS and Bcl-xL, respectively. The color-ratio bar indicates high (red), low (green) and medium (black) gene expression intensity. (c) qRT-PCR of AICDA (encoding AID) and ELISA of IgM, IgG and IgA from MZ B cells cultured with medium (Ctrl), NC, NBH1 or NBH2 cells for 2 d (AICDA) and 4 d (Igs). Results are normalized to PAX5 mRNA (encoding Pax5) and are presented as relative expression (RE) compared with MZ B cells incubated with medium. (d) Flow cytometry of viable NBH1 and NBH2 cells cultured for 18 h with medium (top left panel), qRT-PCR of BCL2 mRNA (top right panel), and immunofluorescence of spleen stained for TUNEL-positive apoptotic DNA (green) and ELA (red). Results are normalized to ACTB mRNA (encoding β-actin) and are presented as RE compared with that of NC cells. Original magnification, ×10. Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test). Data are from one of three experiments with similar results (a, top c panel, bottom d panel) or summarize three independent experiments (b, mid-bottom c panels, top d panels).
Compared to NC cells, NBH1 and NBH2 cells had more mRNAs for B cell stimulating molecules such as BAFF, APRIL, CD40L and IL-21 (refs. 19, 35, 36), B cell chemoattractants such as CXCL12 and CXCL13 (ref. 19), immunoactivating receptors and cytokines such as TLR7, TLR8, IL-1β, IL-6, IL-8, IL-12 and TNF1,2,4,6, and immunoregulatory molecules such as IL-10, IL-10 receptor, arginase 1, retinaldehyde dehydrogenase 1 (RALDH1), iNOS, indoleamine-2,3-deoxygenase (IDO), suppressor of cytokine signaling 1 (SOCS1), progranulin and secretory leukocyte protease inhibitor (SLPI)1,32,33,37–39 (Fig. 3b and Supplementary Fig. 9). The presence of mRNA for CD40L was confirmed in NBH cells, but its expression was less than in splenic CD4+ T cells (Supplementary Fig. 10). Consistent with their increased activation state, NBH1 cells expressed higher amounts of many of the above mRNAs than NBH2 cells and had more mRNA for bcl-2, bcl-xL and mcl-1 anti-apoptotic proteins, but less mRNAs for bad and bak1 pro-apoptotic proteins40.
Compared to NC cells, NBH1 and NBH2 cells induced more AICDA mRNA for AID and more IgM, IgG and IgA production in MZ B cells (Fig. 3c). In spite of showing worse survival, NBH2 cells had stronger MZ B-helper activity than NBH1 cells (Fig. 3c,d), which was likely due to the fact that NBH2 cells released more soluble BAFF, APRIL and IL-21 than NBH1 cells (Fig. 4a,b). NBH2 cells may also establish firmer and more extensive interactions with MZ B cells via post-apoptotic NETs. Consistent with this possibility and with their heightened activation state, NBH cells spontaneously formed NET-like projections, whereas NC cells did not (Supplementary Fig. 11 and Supplementary Movie 2). Moreover, apoptotic TUNEL-positive NBH cells likely corresponding to annexin V-positive NBH2 cells mostly occupied perifollicular areas adjacent to MZ B cells (Supplementary Fig. 12a,b). Thus, NBH cells include NBH1 and NBH2 subsets that are more activated and display more B-helper and regulatory properties than NC cells.
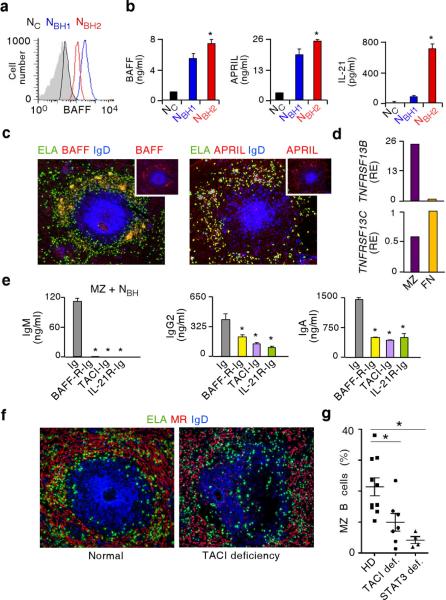
Figure 4. NBH cells activate MZ B cells via BAFF, APRIL and IL-21.
(a) Flow cytometry of BAFF on fresh NC, NBH1 and NBH2 cells (black, blue and red profiles, respectively; gray profile, isotype control). (b) ELISA of BAFF, APRIL and IL-21 from NC, NBH1 and NBH2 cells incubated with medium for 18 h. (c) Immunofluorescence of spleens stained for elastase (ELA, green), BAFF or APRIL (red) and IgD (blue). Top small panels highlight BAFF and APRIL staining patterns. Original magnification, ×10. (d) qRT-PCR of TNFRSF13B and TNFRSF13C encoding TACI and BAFF-R from splenic MZ and naive B cells. Results are normalized to ACTB mRNA (encoding β-actin) and are presented as relative expression (RE) compared with that of naive B cells. (e) ELISA of IgM, IgG2 and IgA from splenic MZ B cells cultured with NBH conditioned medium for 6 d in the presence of control Ig, BAFF-R-Ig, TACI-Ig or IL-21R-Ig antibodies. (f) Immunofluorescence of normal and TACI-deficient spleens stained for ELA (green), MR (red) and IgD (blue). Similar images were obtained from multiple follicles. Original magnification, ×10. (g) Flow cytometry of circulating MZ B cells from patients with deleterious TACI or STAT3 deficiency (def.) and age-matched healthy donors (HD). Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test; Mann-Whitney U test was used in g. Data are from one of three experiments with similar results (a,c,d,f) or summarize at least three independent experiments or measurements (b,e,g).
Neutrophils activate MZ B cells via BAFF, APRIL and IL-21
ELISA and flow cytometry showed that NBH1 and NBH2 cells expressed more surface BAFF and released more soluble BAFF, APRIL and IL-21 than NC cells (Fig. 4a,b). Fluorescence microscopy and qRT-PCR showed NBH cells expressing BAFF and APRIL as well as MZ B cells expressing mRNA for transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI) (Fig. 4c,d and Supplementary Fig. 13), an Ig-inducing receptor that binds both BAFF and APRIL18,19. Naive B cells expressed less mRNA for TACI than MZ B cells, but comparable mRNA for BAFF receptor (BAFF-R) (Fig. 4d), a survival-inducing receptor that binds BAFF but not APRIL13. Blocking BAFF and APRIL with soluble TACI-Ig or IL-21 with IL-21R-Ig abolished IgM production and impaired induction of IgG2 and IgA by conditioned medium from NBH cells (Fig. 4e). Blocking BAFF with soluble BAFF-R-Ig had a similar inhibitory effect. These experiments were performed with NBH cell conditioned medium, because Ig-containing decoy receptors may activate NBH cells via Fcγ receptors. Finally, patients with deleterious substitutions of TACI or STAT3, a protein that drives signals from multiple cytokines including IL-21 (ref. 35), showed poor MZ development and decreased circulating MZ B cells (Fig. 4f,g). These data indicate that NBH cells activate MZ B cells through a mechanism involving BAFF, APRIL and IL-21.
Splenic signals reprogram neutrophils
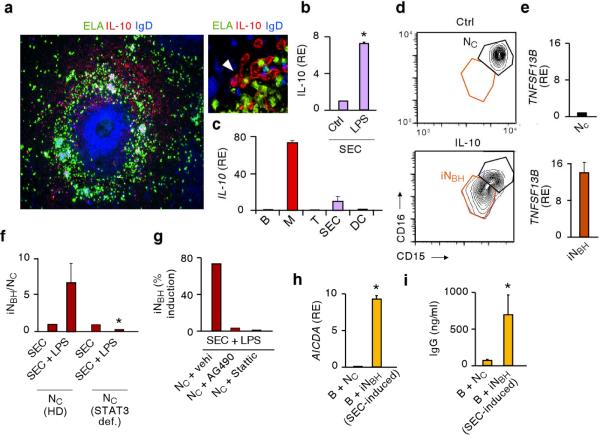
Given the close interaction of NBH cells with SECs, we wondered whether NC cells acquire NBH function in response to SEC-derived signals. Such signals may include IL-10, a cytokine that confers regulatory properties to neutrophils8. Fluorescence microscopy, confocal microscopy and flow cytometry showed IL-10 expression by perifollicular SECs expressing the adhesion protein CD31, the coagulation protein von Willebrand factor, the lymphoid protein CD8, and MR (Fig. 5a and Supplementary Fig. 14a–c). As shown by qRT-PCR and ELISA, SECs up-regulated the expression of IL-10-encoding mRNA and the secretion of IL-10 in response to microbial TLR ligands such as LPS (Fig. 5b and Supplementary Fig. 14d). SECs were not the only perifollicular source of IL-10, as macrophages produced even larger amounts of IL-10 (Fig. 5c and Supplementary Fig. 14e). NC cells exposed to IL-10 became inducible NBH-like cells (iNBH) cells that down-regulated the expression of CD15 and CD16, up-regulated the expression of mRNA for BAFF and APRIL, and activated the expression of AID-encoding mRNA in B cells (Fig. 5d,e and Supplementary Fig. 15). Also another splenic STAT3-activating stromal factor such as granulocyte monocyte-colony stimulating factor (GM-CSF) and microbial products such as LPS and zymosan induced some iNBH cells (Supplementary Fig. 15). This induction did not strictly correlate with apoptosis, but rather with expression of B cell-stimulating factors.
Figure 5. >NC cells acquire NBH-like properties upon exposure to SECs activated by microbial signals.
(a) Immunofluorescence of spleens stained for elastase (ELA, green), IL-10 (red) and IgD (blue). Arrow points to IL-10-expressing SECs. Original magnification, ×10 (left panel), or ×63 (right panel). (b and c) qRT-PCR of IL-10 mRNA from SECs incubated with medium (Ctrl) or LPS for 12 h (b) or from splenic CD19+ B cells (B), macrophages (M), CD4+ T cells (T), or dendritic cells (DC) (c). Results are normalized to ACTB mRNA (encoding β-actin) and presented as relative expression (RE) compared with SECs cultured with medium (b) or with B cells (c). (d) Flow cytometry of CD15 and CD16 on NC cells (black gate) and iNBH cells (red gate) from cultures carried out for 18 h with medium (Ctrl) or IL-10. (e) qRT-PCR of TNFSF13B mRNA encoding BAFF from NC and iNBH cells shown in d. Results are normalized to ACTB mRNA and presented as RE compared with NC cells. (f) Flow cytometry of the iNBH/NC cell ratio after migration of NC cells from individuals with STAT3 deficiency (def) or age-matched healthy donors across SECs exposed to LPS or medium for 4 h. (g) Flow cytometry of SEC-induced iNBH cells as in f with control vehicle (vehi, DMSO), AG490 (Jak2 inhibitor) or Stattic (STAT3 inhibitor). (h) qRT-PCR of AICDA mRNA from circulating unswitched (IgD+) B cells cultured for 2 d in the presence of NC cells or SEC-induced iNBH cells. qRT-PCR results are normalized to ACTB mRNA and presented as RE compared with B cells cultured with medium. (i) IgG ELISA from circulating IgD+ B cells cultured as in h for 7 d. Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test). Data are from one of three experiments with similar results (a,d,g) or summarize three independent experiments (b,c,e,f,h,i).
NC cells migrating across LPS-activated SECs acquired iNBH cell properties via a process that was blocked by pre-exposing NC cells to inhibitors of Janus kinase 2 (Jak2) and STAT3 (Fig. 5f–g and Supplementary Fig. 16), two signal transducers activated by multiple cytokine receptors, including the IL-10 receptor. SEC-induced iNBH cells elicited not only AID-encoding mRNA, but also IgG production in MZ B cells (Fig. 5h,i). Finally, LPS-activated SECs produced not only IL-10, but also neutrophil-attracting chemokines such as IL-8 (CXCL8), CXCL1, CXCL2, CXCL3 and CXCL6 (Supplementary Fig. 17a,b), suggesting that SECs deliver both reprogramming and chemotactic signals to NC cells.
Neutrophils regulate Ig production to TI antigens
Having shown that NBH cells activated MZ B cells in vitro, we wondered whether NBH cells also modulated MZ B cells in vivo. Flow cytometry, ELISA and immunohistofluorescence were performed to study MZ B cells and steady-state serum Ig titers to TI antigens in patients with severe congenital neutropenia (SCN) caused by deleterious elastase substitutions40,41. Additional disease models included SCN with unknown molecular defects, SCN with substitutions in the Wiskott-Aldrich syndrome protein that regulates cell motility, neutropenia in Shwachman-Bodian-Diamond syndrome caused by substitutions of an unknown protein encoded by the SBDSP1 gene, neutropenia in warts-hypogammaglobulinemia-infections-myelokatexis syndrome caused by substitutions of the chemokine receptor CXCR4, cyclic neutropenia caused by deleterious elastase substitutions, chronic granulomatous disease caused by deleterious substitutions of the respiratory burst protein p91-PHOX, and leukocyte adhesion deficiency-1 caused by deleterious substitutions of the adhesion protein CD18 (refs. 40, 41).
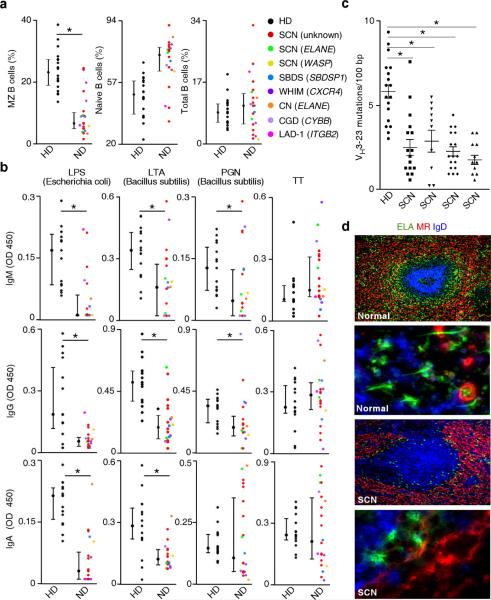
Compared to age-matched healthy individuals, patients with neutrophil disorders had conserved circulating total B cells expressing CD19 and naïve B cells expressing high IgD but not CD27 (Fig. 6a). In contrast, circulating MZ B cells expressing CD27 and weak IgD were decreased in most of these patients. Importantly, IgM, IgG and IgA to microbial TI antigens such as LPS, lipoteichoic acid (LTA), peptidoglycan (PGN), and galactose-α-1,3-galactose were decreased, even in patients with normal MZ B cells, whereas IgM, IgG and IgA to TD antigens such as tetanus or diphteria toxins and protein-conjugated capsular polysaccharides were normal (Fig. 6b and Supplementary Fig. 18a,b). IgM, IgG and IgA to capsular polysaccharides from Pneumococcus 9N, 14, 19F and 23F but not 1, 3, 4, 6B, 7F, 8, 9V, 12F, 18C and 19A were also decreased (Supplementary Fig. 18b and not shown). The reactivity of splenic MZ B cells to some of these TI antigens was confirmed in vitro (Supplementary Fig. 19).
Figure 6. NBH cells regulate MZ B cells and innate IgM, IgG and IgA responses to microbial TI antigens in vivo.
(a) Flow cytometry of circulating IgDlowCD27+ MZ B cells, IgDhighCD27− naive B cells and CD19+ total B cells from patients with neutrophil disorders (ND) and age-matched healthy donors (HD). SCN, severe congenital neutropenia; SBDS, Shwachman-Bodian-Diamond syndrome; WHIM, warts-hypogammaglobulinemia-infections-myelokatexis syndrome: CN, cyclic neutropenia; CGD, chronic granulomatous disease; LAD-1, leukocyte adhesion deficiency-1. ELANE, WASP, SBDSP1, CXCR4, CYBB and ITGB2 indicate genes encoding elastase, Wiskott-Aldrich syndrome protein, SBDS protein 1, CXCR4, p91-PHOX and CD18, respectively. (b) ELISA of IgM, IgG and IgA to LPS from Escherichia coli, LTA and PGN from Bacillus subtilis, or tetanus toxin (TT) from serum of patients and age-matched healthy donors as in a. OD, optical density. (c) Number of VH3–23 gene mutations per 100 base pairs (bp) in circulating MZ B cells from patients with SCN and an age-matched healthy donor. (d) Immunofluorescence of normal and SCN spleens stained for elastase (ELA, green), MR (red) and IgD (blue). Original magnification, ×10 (first and third panel from top) and ×63 (second and fourth panels from top). Error bars, median and percentile 25 and 75 (a, b) or s.e.m. (c); * P < 0.05 (Mann-Whitney U test). Data are from one of three experiments with similar results (d) or summarize multiple measurements (a–c).
A reduction of IgA and to some extent IgM and IgG to Lactobacillus plantarum, Haemophilus influenzae type-b, Salmonella typhimurium and Staphylococcus aureus but not Escherichia coli was also noted (Supplementary Fig. 20). Finally, patients with SCN had circulating MZ B cells with fewer VH3–23 gene mutations and their MZ was poorly developed and contained fewer NBH cells with shorter projections (Fig. 6c,d). iNBH cells obtained from some SCN cases showed conserved B-helper activity in vitro (Supplementary Fig. 21), suggesting that quantitative NBH cell defects may be more important than functional NBH cell defects in the impairment of MZ B cell responses observed in neutropenic patients. Thus, NBH cells may regulate MZ B cells and pre-immune Ig responses to TI antigens in vivo.
Splenic neutrophils involve mucosal microbes
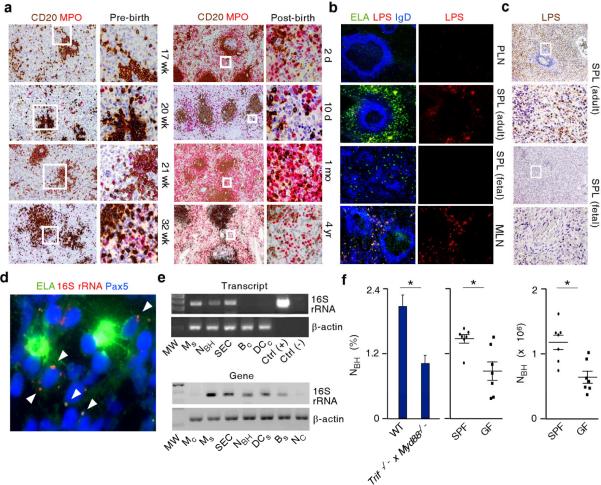
Systemic translocation of microbial products from mucosal surfaces influences the function of neutrophils25. Given the participation of microbial signals such as LPS in the reprogramming of NC cells by SECs and considering that NBH cells enhance innate Ig responses to microbial products that include LPS, we assessed the role of microbes in splenic colonization by NBH cells. Immunohistochemistry showed that splenic colonization by NBH cells started during fetal life but strongly increased as early as two days after birth (Fig. 7a), a time coinciding with mucosal colonization by bacteria. Fluorescence microscopy demonstrated that NBH cell-occupied perifollicular areas contained LPS in adult spleens and mesenteric lymph nodes, but not in fetal spleens and peripheral lymph nodes (Fig. 7b,c). Fluorescence in situ hybridization (FISH) combined with immunofluorescence showed bacterial 16S ribosomal RNA (16S rRNA) in splenic clusters of NBH cells and MZ B cells identified through the nuclear antigen Pax5 (Fig. 7d). 16S rRNA was also PCR amplified from splenic but not circulating NBH cells, macrophages, DCs, SECs and B cells (Fig. 7e). DNA sequencing confirmed the bacterial origin of PCR amplified products (not shown). Germ-free mice as well as mice lacking TLR signaling due to deletion of genes encoding MyD88 and TLR-associated protein TIR-domain-containing adapter-inducing interferon-β (TRIF) had fewer NBH cells expressing Ly6G and CD11b (Fig. 7f). Thus, we propose that TLR signals from mucosal commensals enhance splenic recruitment and reprogramming of NBH cells to enhance innate MZ B cell responses to highly conserved microbial TI antigens (Supplementary Fig. 22).
Figure 7. NBH cells colonize the spleen through a mechanism involving microbial signals.
(a) Immunohistochemistry of pre-natal and post-natal spleens stained for CD20 (brown) and MPO (red). Original magnification, ×4 (left panels) and ×20 (right panels). Boxes correspond to magnified right panels. (b) Immunofluorescence of peripheral lymph node (PLN), adult or fetal spleen (SPL), and mesentheric lymph node (MNL) stained for ELA (green), LPS (red) and IgD (blue). Boxes correspond to magnified right panels. Original magnification, ×10 (left panels) and ×40 (right panels). (c) Immunohistochemistry of adult or fetal SPL stained for LPS (brown). Boxes correspond to magnified mid and bottom panels. Original magnification, ×4 (top panels) and ×20 (bottom panels). (d) Immunofluorescence of spleens stained for elastase (ELA, green) and Pax5 (blue). FISH is for bacterial 16S rRNA (red). Arrows point to 16S rRNA in a NBH cell-MZ B cell cluster. Original magnification, ×63. (e) Bacterial 16S rRNA RT-PCR amplified from complementary DNA (cDNA, top gel) or PCR amplified from genomic DNA (gDNA, bottom gel) of SECs and splenic or circulating monocytes (Ms and Mc), dendritic cells (DCs and DCc), B cells (Bs and Bc), NC cells, and NBH cells. Total RNA from Escherichia coli and water from PCR mix were used as positive and negative controls, respectively. β-actin is a loading control. MW, molecular weight marker. (f) Flow cytometry of NBH cells from wild type (WT) and Trif−/− x Myd88−/− mice (left panel) or specific pathogen free (SPF) and germ free mice (mid and right panels). Error bars, s.e.m.; * P < 0.05 (one-tailed unpaired Student's t-test). Data are from one of three experiments with similar results (a–e) or summarize measurements from at least 5 mice per group (f).
DISCUSSION
We have shown here that neutrophils colonized splenic peri-MZ areas through a non-inflammatory pathway that became more prominent after post-natal mucosal colonization by bacteria. Splenic neutrophils expressed a distinct phenotype, formed MZ B cell-interacting NET-like structures, and elicited Ig class switching, somatic hypermutation and production by activating MZ B cells through a mechanism involving BAFF, APRIL and IL-21. Patients with congenital neutropenia had fewer and hypomutated MZ B cells and their serum contained less preimmune Igs to TI antigens, which indicates that neutrophils interact with MZ B cells to generate an innate layer of antimicrobial Ig defense.
Growing evidence shows that multiple granulocyte subsets such eosinophils and basophils enhance Ig production by B cells42,43. We found that neutrophils occupied splenic peri-MZ areas through a non-inflammatory process that accelerated after birth and involved mucosal colonization by bacteria. Compared to circulating NC cells, splenic NBH cells included NBH1 and NBH2 subsets that expressed B cell-stimulating factors such as BAFF, APRIL and IL-21 as well as B cell-attracting chemokines such as CXCL12 and CXCL13. NBH cells activated MZ B cells as efficiently as splenic T cells via contact-dependent and contact-independent mechanisms. This MZ B-helper activity was mostly associated with NBH2 cells, which indeed secreted more BAFF, APRIL and IL-21, a cytokine that plays a key role in Ig production35.
Compared to NC cells, NBH1 and to a lesser extent NBH2 cells also showed increased expression of TLRs, HLA-II, CD86, IL-1β, IL-6, IL-8, IL-12 and TNF, which suggests in situ activation by splenic signals. This activation was counter-balanced by an increased expression of regulatory molecules such as CD11b, CD24, SOCS1, IL-10, progranulin, SLPI, arginase, IDO and iNOS1, 32, 33, 37–39 and correlated with the capacity of NBH cells to induce contact-independent suppression of T cells like myeloid-derived suppressor cells do1,7. By exerting a dual B–helper T–suppressor function, NBH cells may maximize extrafollicular B cell responses to TI antigens while minimizing follicular B cell responses to TD antigens and inflammation. In the presence of inflammation caused by sepsis, HIV infection, hyper-IgD syndrome or systemic lupus erythematosus, NBH cells became disorganized and infiltrated lymphoid follicles, possibly to present antigen to T cells2. Infections and autoimmune disorders may also explain follicular infiltration by neutrophils in spleens from immunodeficient patients with deleterious TACI substitutions. Of note, inflammation was associated with loss of MZ tissue and NBH2 cells, the NBH subset with predominant MZ B-helper activity.
By expressing inhibitors of proteases, reactive oxygen species and TNF such as progranulin and SLPI1,37,39, NBH cells may afford to clear antigen and deliver B-helper signals without causing inflammatory tissue damage. In this regard, NBH cells occupied lymphoid sites characterized by continuous antigenic filtration, which included presence of bacterial 16S RNA. Although largely blocked by mesenteric lymph nodes44, some mucosal antigens undergo systemic translocation to influence the function of immune cells, including neutrophils23–25. Accordingly, spleens and mesenteric lymph nodes contained perifollicular LPS in addition to NBH cells, but LPS was only detectable after post-natal mucosal colonization by bacteria. By showing less NBH cells in MyD88-TRIF-deficient mice and germ-free mice, our data suggest that splenic filtration of microbial products triggers TLR-dependent recruitment, reprogramming and activation of NC cells. Microbe-independent signals may also play a role, because some NBH cells remained detectable in spleens not exposed to microbial signals.
In addition to enhancing SEC production of neutrophil-targeting chemokines such as IL-8, CXCL1, CXCL2, CXCL3 and CXCL6, microbial TLR signals facilitate the reprogramming of NC cells into NBH cells by inducing SEC release of STAT3-inducing mediators such as IL-10, a cytokine involved in the generation of non-inflammatory neutrophils8,45. IL-10 would promote reprogramming of NC cells as they interact with SECs to reach perifollicular and outer MZ areas. Additional IL-10 originated from perifollicular macrophages positioned nearby SECs and NBH cells. As suggested by published studies46, TLR signals would target not only SECs, but also MZ B cells, perhaps to enhance their responses to activation and chemotactic signals from NBH cells. Consistent with this possibility, TLR ligation up-regulates the BAFF and APRIL receptor TACI and the CXCL13 receptor CXCR5 on MZ B cells18,47.
Although expressing less BAFF and APRIL transcripts, NBH2 cells released more BAFF and APRIL proteins than NBH1 cells, which may relate to the fact that BAFF and APRIL require cleavage from inactive precursors to activate B cells. Thus, an NBH1 “preparatory” stage with increased gene transcription may be followed by an NBH2 stage with increased protein processing and release. Consistent with this interpretation, NBH2 cells stimulated MZ B cells more efficiently than NBH1 cells in spite of having a worse survival profile. NBH2 cells may further amplify their MZ B-helper activity by forming post-apoptotic NET-like projections3,7. These DNA-containing structures were more frequent in apoptotic peri-MZ NBH2 cells and might enhance TI Ig responses not only by trapping antigen, but also by delivering immunostimulating DNA to MZ B cells18,48. Yet, BAFF and APRIL expression seemed more important than apoptosis in determining the B-helper activity of NBH cells. Accordingly, MZ B cells expressed more TACI than naive B cells and thus responded more rapidly and effectively to NBH cells.
Being strategically located at the interface with the circulation, MZ B cells are “geared” to rapidly respond to blood-borne antigens20,21. In addition to triggering rapid IgM secretion, NBH cells elicited AID expression as well as IgG and IgA CSR in MZ B cells, whereas NC cells did not. These findings correlated with the presence of ongoing CSR in some MZ B cells proximal to NBH cells. Of note, NBH cells induced IgG2 CSR, which typically occurs in B cell responses to TI antigens20,21. Unlike NC cells, NBH cells also triggered SHM, which could provide a mechanistic explanation to published studies showing that MZ B cells undergo SHM through an extrafollicular pathway that may not require T cells22,27,28. Finally, NBH cells promoted plasma cell differentiation and Ig production through a mechanism involving BAFF, APRIL and IL-21. Consistent with these data, patients with TACI or STAT3 deficiency had less MZ B cells. Of note, NBH cells exerted some of their MZ B-helper activity via a contact-dependent mechanism that may involve CD40L, a T cell molecule usually required for TD Ig responses in the germinal center of lymphoid folllicles19. Accordingly, patients with CD40L defects have decreased MZ B cells in addition to defective TD Ig responses21, which supports a role of CD40L in NBH cells.
The in vivo contribution of NBH cells to innate MZ B cell responses was indicated by the observation that patients with neutropenia or functional neutrophil defects had not only decreased and hypomutated MZ B cells, but also reduced serum IgM and IgG to TI antigens under steady-state conditions. This humoral deficiency is unlikely to originate from an increased microbial burden, which rather causes polyclonal B cell activation and hypergammaglobulinemia. Accordingly, serum IgM and IgG to TD antigens were not affected by neutrophil insufficiency, which further points to a specific involvement of NBH cells in preimmune TI Ig responses. In agreement with the IgA-inducing activity of NBH cells, neutrophil disorders were associated with decreased serum IgA to TI antigens and some mucosal bacteria. Although better known for its key role in mucosal immunity19, IgA also enhances systemic immunity by interacting with neutrophils9. Thus, NBH cells may cross-talk with MZ B cells to generate an innate line of IgA as well as IgM and IgG defense against systemic invasion by microbes breaching the mucosal barrier24,26. This implies that an insufficiency of NBH cells could contribute to the pathogenesis of systemic infections occurring in patients with neutrophil disorders. Conversely, harnessing NBH cells with specific adjuvants may enhance vaccine-induced Ig responses to poorly immunogenic TI antigens in healthy individuals.
ONLINE METHODS
Human subjects
Peripheral blood mononuclear cells were from buffy coats or fresh blood of healthy age-matched volunteers, patients with congenital neutrophil disorders (Supplementary Table 1), or with immunodeficiency caused by deleterious substitutions of TACI or STAT3 as described18,35. Spleens from individuals without clinical signs of infection or inflammation and normal histology were utilized for functional assays (Supplementary Table 2). Tonsils and lymph nodes were from individuals with follicular hyperplasia or cancer. Frozen and paraffin-embedded lymphoid tissues from healthy subjects, fetal demises, and patients with autoinflammatory disorders (HIDS), infection (HIV, sepsis), autoimmunity (lupus) or immunodeficiency (SCN, common variable immunodeficiency with deleterious TACI substitutions) were obtained from local repositories (Supplementary Table 3). The Institutional Review Board of Institut Municipal d'Investigació Mèdica, Sant Joan de Déu Hospital, Mount Sinai School of Medicine and Weill Medical College of Cornell University approved the use of blood and tissues.
Animals
Spleens were collected from rhesus macaques, Trif−/− x Myd88−/− mice and wild-type C57BL6/6 housed under specific pathogen-free or germ-free conditions. Animal experimentation was approved by The Institutional Review Board of Mount Sinai School of Medicine and Regierungspräsidium Freiburg.
Cells
Splenocytes were obtained by perfusion using phosphate buffer solution. NC and NBH cells were separated from whole blood or splenocytes using Histopaque-1077 (Sigma) gradient, followed by human neutrophil enrichment kit (Easysep; > 98% purity) purification. CD15highCD16high NC cells, CD15intCD16int NBH1 cells and CD15lowCD16low NBH2 cells were sorted by flow cytometry from whole blood or splenocytes with BD FACSAria II (BD Biosciences). IgDhighCD19+CD27− naïve B cells, CD19+IgDlowCD27+ MZ B cells, IgD−CD10+CD19+CD27+CD38+ germinal center B cells, unswitched IgD+ B cells, CD14+ monocytes, CD14+CD68+ macrophages, CD11c+HLA-IIhigh dendritic cells, and CD3+CD4+ T cells were sorted by flow cytometry or magnetically isolated (Miltenyi Biotec) from blood or splenic mononuclear cells. To isolate SECs, splenic samples were purified by enzimatic digestion in Hank's balanced salt solution (Lonza) containing 1 mg/ml collagenase-IV (Invitrogen) and 50 ng/ml DNAse-I (50 ng/ml) at 37°C for 45 min. CD8+MR+CD31+ SECs were sorted by flow cytometry after Ficoll Histopaque-1.077 gradient. Mouse splenocytes were obtained by processing splenic tissue through a 70-μm cell strainer.
Cultures and reagents
Cells were cultured in complete RPMI medium. Conditioned medium was obtained by culturing 24 h NBH cells in RPMI medium alone. IL-21R-Ig, BAFF-R-Ig, TACI-Ig or control-Ig (R&D) were used at 5 μg/ml, LPS and zymosan (InvivoGen) at 1 μg/ml, IL-10 and GM-CSF (Peprotech) at 50 ng/ml. Before adding IL-10 or GM-CSF, NC cells were incubated for 3 h in complete RPMI medium. B cells were co-cultured with neutrophils, macrophages, dendritic or T cells at a 1:1 ratio (1 × 106/ml). SECs were propagated in endothelial medium (Lonza). Trans-SEC migration assays were performed by seeding SECs for 48 h on 3-μm pore size membranes (BD Falcon) pre-coated with 0.1% gelatine (Sigma) in endothelial medium. Medium was replaced with RPMI before seeding NC cells and phenotype of transmigrated NC cells analyzed after 4 h. NC cells were pre-treated with 20 μM Stattic (SantaCruz), AG490 (EMD4Bioscences) or control DMSO for 2 h and washed. All reagents were endotoxin-free as established by a Limulus assay (GenScript).
Flow cytometry
Cells were incubated with Fc blocking reagent (Miltenyi Biotec) at 4 °C with Abs (Supplementary Table 4). Dead cells were excluded with 4'-6-diamidine-2'-phenylindole (DAPI) (Boehringer Mannheim) or 7-amino-actinomycin (7-AAD) (BD Pharmingen). Neutrophil survival was measured by Annexin-V Apoptosis Detection KitII (BD Pharmingen). Gates and quadrants were drawn to give ≤ 1% total positive cells in the isotype control. Cells were acquired using BD LSRII or FACSCantoII (BD) and analyzed by FlowJo (Tree Star).
Immunohistochemistry
Formalin-fixed and paraffin-embedded 3–4 μm tissue sections were stained using Bond Max Immunohistochemical Stainer (Leica Biosystems) and Kit Bond Polymer Refine Detection (Leica Biosystems) for CD20 and LPS (Supplementary Table 4) and Kit Bond Polymer Refine Red Detection (Leica Biosystems) for myeloperoxidase. Sections were counterstained with hematoxylin.
Immunofluorescence, confocal microscopy and FISH
Frozen tissues and cells were fixed as reported15,17 and stained with various combinations of Abs (Supplementary Table 4). TUNEL analysis was performed using the TACS2 TdT-DAB In Situ Apoptosis Detection Kit (Trevigen). EUB338 probe was used for FISH as reported49. Nuclear DNA was DAPI stained. Coverslips were applied with FluorSave reagent (Calbiochem) and images were acquired with a Zeiss Axioplan 2 microscope (Atto Instruments). Confocal images were generated with a TCS SPE inverted confocal microscope (Leica) by acquiring at least 12 different z-planes (Carl Zeiss Microimaging) with 0.3 μm z spacing. Three-dimensional views were reconstructed with Imaris and ImageJ software.
Giemsa and electron microscopy
Sorted neutrophils were stained for May-Grünwald Giemsa (Merck) or prepared for transmission electron microscopy as reported50. Blocks were examined under a CM100 transmission electron microscope (Philips).
ELISA
IgM, IgG and IgA to various antigen and bacteria were detected in M96-Nunc ELISA plates (Supplementary Table 5). Bacteria were grown to an optical density at 650 nm of 0.05–0.1, fixed with 0.5% formalin, and coated at 1:1000 dilution. Total IgM, IgG, IgA, BAFF, APRIL and IL-10 were detected as reported15–18. IL-8 and IL-21 were measured using a kit (PeproTech).
T cell proliferation assay
Peripheral blood CD4+ T cells were magnetically sorted (Miltenyi Biotec) and stimulated with 5 μg/ml anti-CD3 (clone SpvT3b M. López-Botet) and 50 U/ml of IL-2 (Chiron). 2 × 105 cells were seeded in 96-well plates with NBH cells or NBH conditioned medium for 3 d. Proliferative response was determined as described15–18.
RT-PCR, Southern blot, qRT-PCR and gene expression profiling
RNA was extracted and cDNA-retrotranscribed as reported15,17. Germline and switch circle transcripts were amplified by RT-PCR and hybridized with appropriate radiolabeled probes in Southern blott as reported15,17. qRT-PCRs were performed as described17,18,42 using specific primer pairs (Supplementary Table 6). Gene expression analysis was performed using a complete clustering algorithm by TIGR MeV (Dana Farber Cancer Institute).
Detection of bacterial 16S rRNA
Total RNA and genomic DNA were extracted as described15,17. cDNA from total RNA and genomic DNA were used as templates to amplify 16S rRNA using specific primer pairs (Supplementary Table 6). Total Escherichia coli RNA was used as positive control. PCR products were cloned, sequenced and identified as 16S rRNA using Ribosomal Database Project (Michigan State University) and BLAST (National Center for Biotechnology Information).
VH gene cloning, sequencing and mutation analysis
Total mRNA was isolated using PicoPure RNA kit (Applied Biosystems). After reverse transcription15,17, VH3-23-Cμ and VH3-23-Cγ transcripts were amplified using Cloned Pfu DNA polymerase (Stratagene) and primer pairs (Supplementary Table 6). PCR products were cloned into a pCR4 Blunt TOPO vector (Invitrogen) and screened by PCR using internal VH3-23 primers. Plasmids were isolated with Qiagen R.E.A.L. 96 well mini preps and sequenced (Genewiz, Inc.). Sequences were analyzed with IMGT (http://imgt.cines.fr/).
Statistical analysis
Values were expressed as mean ± standard error of the mean (s.e.m.). Statistical significance was assessed by one-tailed unpaired Student's t-test or the Mann-Whitney U test.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Farrés, J. Yélamos, A. Muntasell and M. López-Botet (Institut Municipal d'Investigació Mèdica-Hospital del Mar) for providing reagents and samples and for discussing data; N. Romo, S. Bascones, E. Ramirez and O. Fornas (Barcelona Biomedical Reasearch Park) for helping with cell sorting; S. Mojal for helping with the statistical analysis. Supported by Ministerio de Ciencia e Innovación grant SAF 2008-02725 to A. Cerutti; US National Institutes of Health grants R01 AI074378, P01 AI61093, U01 AI95613 and P01 096187 to A. Cerutti; EUROPADnet HEALTH-F2-2008-201549 to A. Cerutti; Juan de la Cierva post-doctoral fellowships to I.P. and G.M.; Sara Borrell post-doctoral fellowship to A. Chorny; pre-doctoral fellowships from the Instituto de Salud Carlos III and Ministerio de Ciencia e Innovación to C.B and M.G.; Yerkes base grant P51 RR00165 to G.S.; and Fondazione C. Golgi di Brescia, Associazione Immunodeficienze Primitive to A. Plebani.
Footnotes
AUTHOR CONTRIBUTIONS I.P. and M.C. designed and performed research, discussed data and wrote the paper; C.M.B. B.H. and K.C., designed and performed research; L. Cassis, M.G, L. Comerma, A. Chorny, M.S., W.X., G.M., A. Chiu, T.B. and S.C.G. performed research and discussed data; D.M.K., W.T., J.B.B., S.S., J.A.L., B.B., J.L., N.J., F.A., C. Diaz., N.T., A. Català, M.T., C.F., V.C., C.C., G.A.D., J.M.B., CM.F., G.S., C.C.R, M. Calvillo, C. Dufour., L.D.N., V.L., A.P., J.L.C., A.D., J.I.A., M.J., J.Y., N.M. and J.D. provided blood and tissue samples and discussed data; and A. Cerutti designed research, discussed data and wrote the paper.
COMPETING INTEREST STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 2.Soehnlein O. An elegant defense: how neutrophils shape the immune response. Trends Immunol. 2009;30:511–512. doi: 10.1016/j.it.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 5.Yang D, de la Rosa G, Tewary P, Oppenheim JJ. Alarmins link neutrophils and dendritic cells. Trends Immunol. 2009;30:531–537. doi: 10.1016/j.it.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gisbergen KP, Geijtenbeek TB, van Kooyk Y. Close encounters of neutrophils and DCs. Trends Immunol. 2005;26:626–631. doi: 10.1016/j.it.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Majlessi L, Deriaud E, Leclerc C, Lo-Man R. Coactivation of Syk kinase and MyD88 adaptor protein pathways by bacteria promotes regulatory properties of neutrophils. Immunity. 2009;31:761–771. doi: 10.1016/j.immuni.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Pasquier B, et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Tsuboi N, Asano K, Lauterbach M, Mayadas TN. Human neutrophil Fcgamma receptors initiate and play specialized nonredundant roles in antibody-mediated inflammatory diseases. Immunity. 2008;28:833–846. doi: 10.1016/j.immuni.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scapini P, et al. G-CSF-stimulated neutrophils are a prominent source of functional BLyS. J. Exp. Med. 2003;197:297–302. doi: 10.1084/jem.20021343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huard B, et al. APRIL secreted by neutrophils binds to heparan sulfate proteoglycans to create plasma cell niches in human mucosa. J. Clin. Invest. 2008;118:2887–2895. doi: 10.1172/JCI33760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay F, Schneider P. Cracking the BAFF code. Nat. Rev. Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 14.Schneider P, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J. Exp. Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Xu W, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat. Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 18.He B, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol. 2010;11:836–845. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerutti A, Chen K, Chorny A. Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 2011;29:273–293. doi: 10.1146/annurev-immunol-031210-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin F, Kearney JF. Marginal-zone B cells. Nat. Rev. Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 21.Weill JC, Weller S, Reynaud CA. Human marginal zone B cells. Annu.Rev. Immunol. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. [DOI] [PubMed] [Google Scholar]
- 22.Berkowska MA, et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118:2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krueger JM, et al. Peptidoglycans as promoters of slow-wave sleep. II. Somnogenic and pyrogenic activities of some naturally occurring muramyl peptides; correlations with mass spectrometric structure determination. J. Biol. Chem. 1984;259:12659–12662. [PubMed] [Google Scholar]
- 24.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 25.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas A, et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut. 2011;60:1506–1519. doi: 10.1136/gut.2010.224774. [DOI] [PubMed] [Google Scholar]
- 27.Weller S, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weller S, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc. Natl. Acad. Sci. U S A. 2001;98:1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balazs M, Martin F, Zhou T, Kearney J. Blood dendritic cells interact with splenic marginal zone B cells to initiate T-independent immune responses. Immunity. 2002;17:341–352. doi: 10.1016/s1074-7613(02)00389-8. [DOI] [PubMed] [Google Scholar]
- 30.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 31.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 32.Han C, et al. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Chen GY, Zheng P. CD24-Siglec G/10 discriminates danger- from pathogen-associated molecular patterns. Trends Immunol. 2009;30:557–561. doi: 10.1016/j.it.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiStasi MR, Ley K. Opening the flood-gates: how neutrophil-endothelial interactions regulate permeability. Trends Immunol. 2009;30:547–556. doi: 10.1016/j.it.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avery DT, et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 2010;207:155–171. doi: 10.1084/jem.20091706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerutti A, Puga I, Cols M. Innate control of B cell responses. Trends Immunol. 2011;32:202–211. doi: 10.1016/j.it.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–878. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 38.Mansell A, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat. Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 39.Tang W, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein C. Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu. Rev. Immunol. 2011;29:399–413. doi: 10.1146/annurev-immunol-030409-101259. [DOI] [PubMed] [Google Scholar]
- 41.Donadieu J, Fenneteau O, Beaupain B, Mahlaoui N, Chantelot CB. Congenital neutropenia: diagnosis, molecular bases and patient management. Orphanet J. Rare Dis. 2011;6:26. doi: 10.1186/1750-1172-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen K, et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat. Immunol. 2009;10:889–898. doi: 10.1038/ni.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu VT, et al. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat. Immunol. 2011;12:151–159. doi: 10.1038/ni.1981. [DOI] [PubMed] [Google Scholar]
- 44.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 45.De Santo C, et al. Invariant NKT cells modulate the suppressive activity of IL-10-secreting neutrophils differentiated with serum amyloid A. Nat. Immunol. 2010;11:1039–1046. doi: 10.1038/ni.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 47.Cinamon G, et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat. Immunol. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obata T, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci U S A. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerutti A, et al. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center-like phenotype differentiation in a human monoclonal IgM+IgD+ B cell line. J. Immunol. 1998;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.