Abstract
A key consequence of regulatory T cell (Treg) suppression of CD4 T cells is the inhibition of IL-2 production, yet how Tregs attenuate IL-2 has not been defined. Current models predict a termination of TCR signaling, by disrupting T-APC contacts, or TCR-signal modification, through mechanisms such as cAMP. To directly define Treg effects on TCR signaling in CD4 T cell targets we visualized changes in nuclear accumulation of transcription factors at timepoints when IL-2 was actively suppressed. Nuclear accumulation of NFAT was highly dependent on sustained TCR signaling in the targets. However, in the presence of Tregs, NFAT and AP-1 signals were sustained in the target cells. In contrast, NFκB p65 was selectively attenuated. Thus Tregs do not generally terminate TCR signals. Rather, Tregs selectively modulate TCR signals within hours of contact with CD4 targets, independent of APC, resulting in the specific loss of NFκB p65 signals.
Introduction
Natural regulatory T cells (Tregs) counterbalance immunity by suppressing cell proliferation, survival, maturation, cytokine and/or chemokine production, and release of cytotoxic components from granules. This broad suppressive capacity is likely exerted by different mechanisms at different stages of immune activation (1, 2).
Fundamental to initial T cell activation is the receipt of signals that promote cytokine production, cell proliferation and cell survival. IL-2 is the first cytokine produced by naïve T cells and is critical for successful adaptive immunity (3). Upon TCR engagement the nuclear accumulation of NFAT, NFκB, and AP-1, in concert, drives early IL-2 transcription (4, 5). CD28 co-stimulatory signaling quantitatively changes TCR signaling: enhancing NFκB and AP-1 to promote transcription and stabilizing IL-2 mRNA (6, 7). Tregs suppress T cell activation by inhibiting cell proliferation and cytokine production, in particular early IL-2 production (8). We have mapped Treg suppression of IL-2 at the transcript and protein level to a tight kinetic window 6–10 hours after initial CD4 T cell activation (9). However the mechanism by which Tregs specifically abort IL-2 production remains unknown.
Tregs could negatively regulate T cell signals for IL-2 via CTLA-4/B7 (10), cAMP (11) or potentially by utilizing E3 ligases (12, 13). Alternatively, recent visualization of Treg suppressive events suggest Tregs could terminate T cell signals by disrupting the stability or duration of CD4 T cell-antigen presenting cell interactions (14, 15). To define the changes in T cell signaling in target CD4 T cells activated in the presence of Tregs we used multispectral imaging flow cytometry (Amnis Imagestream) to quantify the frequency of CD4 T cells with specific transcription factor (TF) nuclear accumulation. Tregs did not terminate T cell signaling at the time of IL-2 inhibition. Rather, signaling in targeted CD4 T cells was selectively modified by attenuation of nuclear NFκB but not NFAT and AP-1, through an APC-independent mechanism.
Materials and Methods
Mice and Antibodies
BALB/c mice (NCI) and Thy1.1 BALB/c mice were maintained in the pathogen-free animal facility at the University of Rochester Medical Center (Rochester, NY). Antibodies: mouse anti-NFAT1 IgG1 (Affinity BioReagents); mouse anti-NFAT2 IgG1, rabbit anti-p65 IgG, rabbit anti-c-Rel IgG, rabbit anti-c-Fos IgG, and rabbit anti-c-Jun IgG (Santa Cruz Biotechnology); FITC goat F(ab’)2 anti-rabbit IgG and FITC goat anti-mouse IgG1 (Southern Biotech); anti-Thy1.1 eFluor® 450 (eBioscience); anti-Thy1.2 PE (BD Pharmingen).
Cell purification
CD4 cells were isolated from spleen and lymph nodes. CD4+CD25-CD44low naïve T cells were sorted by FACSAria as a source of target CD4+ T cells or control T cells. Tregs were purified from CD4-enriched population using CD25+ MACS column (Miltenyi) (routinely >85% Foxp3+, with >85% suppression of CD4 proliferation at 1:1 target to Treg ratio). Antigen-presenting cells (APC) were isolated from spleen by complement lysis of Thy1.2-expressing T cells. Confirmatory experiments were performed using sorted CD4+CD25+Foxp3+/GFP+ cells from Foxp3/GFP reporter mice.
Treg Suppression Assay
1×105 Thy1.1 naïve “target” CD4+ T cells were stimulated with anti-CD3 mAb (1 µg/ml) and APC (1×105) in co-culture with either 1×105 Thy1.2 control T cells (Ctrl T) or Thy1.2 Tregs. Cells were harvested for functional assays at various time points. In some experiments, Thy1.1 responder cells were pretreated with 1mM cAMP antagonist, Rp-8-Br-cAMPS (BioLog Life Science Institute) or the Src-kinase inhibitor PP1 (10 µM, Axxora) was added to cultures. In some experiments suppression was assayed following antibody-coated bead stimulation (16). M450 dynabeads (Invitrogen) were coated with anti-CD3 (2 µg per 25 µl beads) and anti-CD28 (2 µg per 25 µl beads). 4×104 antibody-coated beads were then used to stimulate 1×105 naïve target CD4+ T cells in co-culture with 1×105 Ctrl T or Tregs. At 12h cells were collected for p65 nuclear localization analysis (Imagestream) and IL-2 secretors by cytokine secretion assays (Miltenyi).
Flow cytometry
IL-2 secretors were detected by cytokine secretion assay kit (Miltenyi Biotec) according to manufacturers instructions. For phosphoflow, cells were fixed, permeabilized and stained with anti-pERK mAb according to manufacturers instructions (BD Bioscience).
Nuclear Translocation Analysis on Imagestream
Cells were fixed (1% paraformayldehyde), surface stained for Thy1.1/1.2, permeablized by 0.1% TritonX-100 (Sigma) and staining for NFκB p65, NFκB c-Rel, NFAT1, NFAT2, c-Fos or c-Jun. Nuclear dye Draq5 (5µM, Axxora) was added before analysis. Fluorescent images were visualized (>6,000 events per condition) on Imagestream (Amnis). A mask on the nucleus was created and within this area co-localization of transcription factors and nuclear dye was measured by similarity (IDEAS software, Amnis).
DNA-binding assay
Treg suppression assay was set-up as described above. At 6h, CD4+Thy1.1+ target cells were positive selected on Thy1.2, lysed and nuclear extracts prepared (Active Motif). p65 DNA-binding was quantified by TransAM™ NFκB p65 Transcription Factor Assay Kit (Active Motif). Briefly, 2 µg nuclear extracts were loaded into a 96-well plate coated with NFκB consensus sequence, followed by anti-p65 antibody and HRP detection at 450nm absorbance. Relative amount of p65 bound to DNA expressed as OD.
Results and Discussion
T cell signals and nuclear accumulation of transcription factors
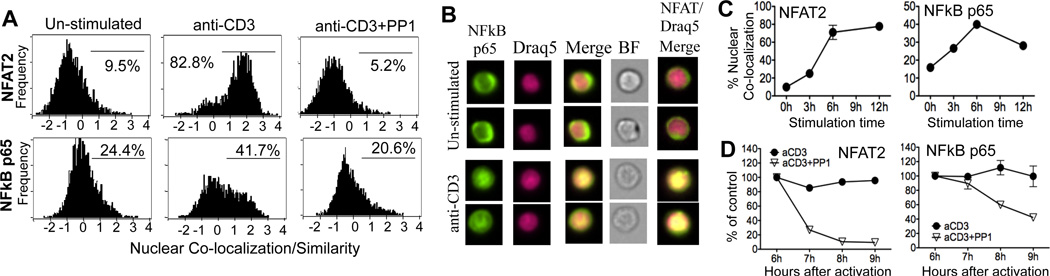
To accurately interpret Treg effects on target T cell signaling we first determined the relationship between continued T cell signaling and the nuclear accumulation of transcription factors (TF) key to IL-2 transcription. We performed single cell analysis of TF nuclear localization using multispectral imaging flow cytometry (17). Nuclear localization was defined by a positive “similarity” score, representing the correlation coefficient between two fluorescent signals: the relative co-localization of the nuclear dye Draq5 and the TF NFAT2 or NFκB p65 (Fig 1A, 1B). Unstimulated CD4+ T cells had a negative similarity score corresponding to the absence of nuclear NFAT2 and NFκB (Fig 1A, 1B). After 6h of activation, most CD4+ T cells were positive for nuclear NFAT2 (Fig 1A, 1B) and a proportion of CD4+ T cells exhibited nuclear localization of NFκB (Fig 1A, 1B). The Src kinase inhibitor, PP1, ablated the nuclear localization of NFAT2 and NFκB p65 showing the dependency on TCR/CD28-signaling (Fig 1A). Kinetically, nuclear NFAT2 and p65 peaked at 6h; NFAT2 was sustained through 12h while p65 declined (Fig 1C). To understand the dynamics of TF nuclear localization and TCR signaling we blocked Src kinase signaling at the peak of TF nuclear accumulation (6h) and followed the frequency of cells with a nuclear pool of NFAT2 and p65 (Fig 1D). Nuclear NFAT2 was very sensitive to termination of TCR signaling: an 80% loss in nuclear NFAT2 positive cells within 1h of PP1. In contrast, p65 was relatively stable in nucleus with only 10.36% loss of nuclear NFκB positive cells within 1h of PP1, possibly reflecting non-TCR signals sustaining nuclear NFκB p65. This single cell assay for detection of nuclear TFs provides a sensitive platform for the detection of possible perturbations in TCR signaling mediated by Tregs.
Figure 1. TCR-dependent nuclear localization of transcription factors.
CD4 T cells were stimulated with anti-CD3/APC. A) At 6h cells were stained for NFκB p65 or NFAT2. Histograms gated on CD4+ T cells, and percentages of activation-induced nuclear accumulation shown. B) Representative cell images, Imagestream. C) Kinetic analysis. Graphs show mean and SEM of the percentage of cells with similarity scores above 0.5 from 3 independent experiments. D) CD4 cells were stimulated for 6h as in A) before addition of PP1. Cells were analyzed for nuclear TF and normalized to target cells in the absence of PP1; mean and SEM from 3 independent experiments.
Sustained NFAT and AP-1 signaling in the presence of Tregs
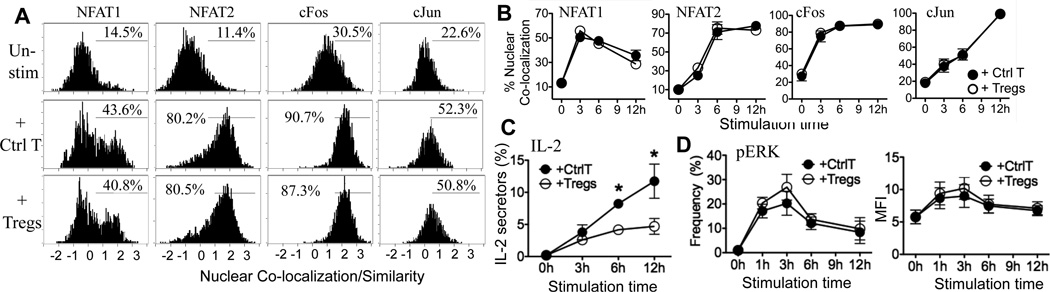
To determine if Tregs broadly extinguish target T cell signaling at the time of IL-2 inhibition, we first examined TCR-dependent nuclear NFAT in the target CD4+ T cells in co-culture with Tregs or Ctrl T, non-Treg CD4 T cells (Fig 2A, 2B). The presence of Tregs had no effect on the magnitude or timing of NFAT nuclear accumulation (Fig 2A, 2B), at timepoints when IL-2 production was suppressed (Fig 2C) (9). Thus IL-2 is downregulated despite ongoing TCR-dependent signals that support NFAT nuclear accumulation. Tregs also did not change nuclear accumulation of AP-1 components, cFos and c-Jun (Fig 2A, 2B). The AP-1 complex is largely regulated at the protein level, therefore we also analyzed c-Fos or c-Jun total protein in target T cells in co-culture with Tregs and found no change compared to targets cultured with Ctrl T (Fig S1, A).
Figure 2. CD4+ T cells sustained NFAT, AP-1 and Erk-signaling in the presence of Tregs.
Thy1.1+ naïve CD4+ T cells were stimulated with anti-CD3/APC in co-culture with Thy1.2+ Ctrl T or Thy1.2+ Tregs. A) At 6h, cells were analyzed for nuclear NFAT1, NFAT2, cFos, and c-Jun by Imagestream. Histograms gated on Thy1.1+ target CD4+ T cells; percentages are the frequency of target T cells with nuclear TF. Representative plots from at least 3 experiments. B) Kinetics of nuclear NFAT and AP-1 in co-culture, frequency of Thy1.1+ targets. C) Frequency of IL-2 producers, * p<0.05 by paired student t test. D) pERK frequency, gated on Thy1.1+ targets. Mean and SEM from 3 independent experiments.
The sensitivity of nuclear NFAT to perturbations in TCR signaling (Fig 1D) and the absence of Treg-induced changes in nuclear NFAT (Fig 2) both suggest that CD4+ target T cell signaling remains largely intact in the presence of Tregs at the time of IL-2 regulation (Fig 2C). To confirm ongoing upstream signaling we used phospho-flow to measure kinase activation. Under our stimulation conditions we were unable to detect increases in p-PLCγ or p-Zap70 in the 6–12h timeframe. However, ERK signaling was readily detectable from 1h after anti-CD3/APC stimulation (Fig 2D). The presence of Tregs did not interfere with ERK signaling in the target T cells despite concomitant inhibition of IL-2 (Fig 2C, 2D).
Tregs attenuate nuclear accumulation of NFκB in CD4+ target T cells
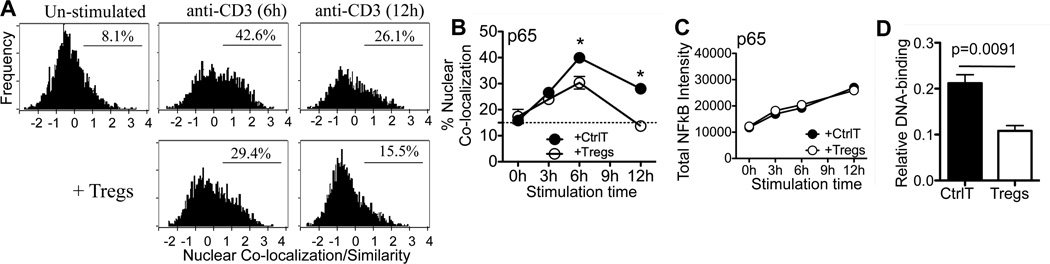
In contrast to NFAT and AP-1, Tregs significantly attenuated the frequency of target T cells with nuclear NFκB p65 at 6h after activation and was at down to un-stimulated levels by 12 h (Fig 3A, 3B). The reduction in nuclear p65 was not due to a decrease in total cellular p65 (Fig 3C), indicating differential generation or targeted degradation of NFκB p65 is unlikely. A NFκB DNA binding assay confirmed a Treg-induced loss in nuclear NFκB activity in CD4 targets (Fig 3D). Interestingly, Treg-attenuation of NFκB was selective for p65 and not seen for c-Rel (Fig S1, B) suggesting Treg may target the p65 complexed to IκBα, rather than the c-Rel-IκBβ complexes, predominant in naïve cells (18). IL-2 receptor signaling can regulate NFκB thus Tregs could modulate NFκB by limiting IL-2 availability (19, 20). Addition of exogenous IL-2 failed to rescue the Treg-mediated change in p65 (Fig S1, C). Cyclic AMP is also a known inhibitor of NFκB and a potential mediator of Treg suppression (11), however cAMP antagonists failed to block Treg suppression of p65 or IL-2 in our system (Fig S1, D–F). In contrast, increased CD28 signaling potently upregulates NFkB and was able to override Treg attenuation of NFκB p65 (Fig S2, A–C) consistent with CD28 signals bypassing Treg suppression (9).
Figure 3. Tregs attenuate nuclear NFκB in CD4+ T cells.
Cultures as in Fig 2. A) At indicated time points, cells were stained with anti-Thy1.1, NFκB p65 Ab, and Draq5. Representative histograms gated on the Thy1.1+ targets with percentage of targets with nuclear NFκB p65. B) Activation-induced nuclear NFκB in target CD4+ T cells in co-culture. C) Total fluorescence intensity of NFκB in the whole cell. B) and C) mean and SEM from 3 independent experiments, * p=<0.05 by paired student t test. D) DNA binding assay for p65. Thy1.1 target cells were isolated from Ctrl T or Treg co-culture at 6h. Nuclear extracts were assayed for amount of p65 bound to NFκB-binding consensus sequence. Representative data from one of two independent experiments.
Rapid NFκB downregulation independent of APC
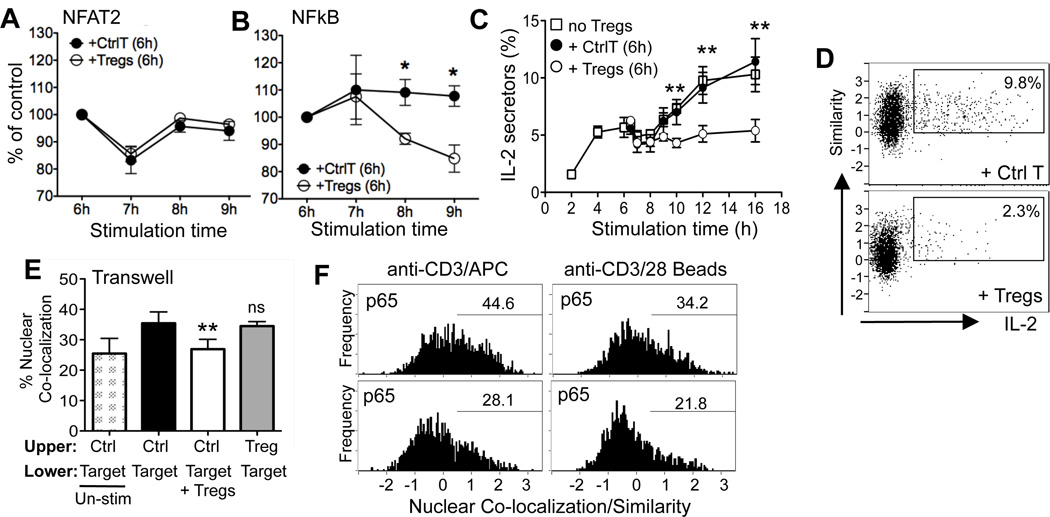
Full effector function requires continued signaling for at least 10h (21). To determine if Tregs can acutely regulate T cell activation we initiated CD4+ T cell cultures in the absence of Tregs and then added in freshly isolated Tregs or Ctrl T at 6h. Strikingly, Tregs attenuated nuclear NFκB and IL-2 production within 2 h of addition to the culture (Fig 4A–C) and was not due to acute IL-2 consumption (data not shown). It’s not known how the availability of individual TFs correlates with the magnitude of IL-2 production in CD4 T cells. Therefore to examine the relationship between the loss of nuclear NFκB and IL-2 attenuation we dual-labeled cells (Fig 4D). IL-2 production early after activation was found in a small fraction of CD4+ T cells within the population showing an activation-induced increase in nuclear p65 (positive similarity score, Fig 4D). However, there was no positive correlation between the degree of nuclear p65 (similarity score) and the amount of IL-2 produced (Fig 4D). The data suggests a threshold amount of nuclear NFκB is required for IL-2 gene competency but that the degree of nuclear NFκB does not control the magnitude of the IL-2 response. Therefore, by reducing the number of cells that reach this nuclear NFκB threshold (Fig 4D), Tregs appear to limit the number of IL-2 producing cells.
Figure 4. Acute regulation of NFκB independent of APC.
Naïve Thy1.1+ CD4 cells were stimulated with anti-CD3/APC. At 6h, Thy1.2+ Ctrl T or Thy1.2+ Tregs were added to the culture and the targets analyzed for nuclear NFAT2 (A) and NFκB (B). % of control cultures that did not have cells added at 6h; mean and SEM from 3 independent experiments. C) Frequency of IL-2 secretors after addition of Tregs or Ctrl T at 6h; mean and SEM from 3 independent experiments. D) Co-staining of p65 with IL-2 secretors at 12h, gated on Thy1.1+ targets cells and the frequency of IL-2 secretors with nuclear p65 shown. Representative plots from one of three independent experiments. E) Target cells loaded into lower chambers with anti-CD3/APC. Ctrl T cells or Tregs added to upper chamber or lower chambers. Frequency of targets cells with nuclear p65 at 12h. Mean and SEM from 3 independent experiments. F) CD4 cells were stimulated by either anti-CD3/APC or anti-CD3/CD28 coated beads with Ctrl T cells or Tregs. Representative data from one of two independent experiments. * p=<0.05, ** p=<0.005 by 2-tailed student t test.
Mechanistically, Tregs could modulate NFκB directly in the target T cell or indirectly via the APC. A transwell experiment confirmed that NFκB was only modulated when Tregs were in close proximity to targets and APC (Fig 4E). To test the requirement for APC, we stimulated target T cells with anti-CD3/CD28 coated beads (16) with or without Tregs, using conditions where Tregs successfully inhibited IL-2 (Fig S2, D–E). In the absence of APC, Tregs retained the ability to inhibit nuclear NFκB in target T cells (Fig 4F). Tregs also retained the ability to attenuate NFκB in cultures with fixed APC (data not shown). Thus Tregs rapidly and selectively attenuate NFκB T cell activation signals in CD4 targets independent of the APC.
Our results suggest that models of Treg action whereby Tregs modulate the frequency or duration of T-APC conjugation (14, 15) cannot fully account for the early inhibition of IL-2. Rather, qualitative changes in T cell signaling, with a decrease in available nuclear NFκB, appear to underlie early suppressive events. Interestingly, an in vivo study using an NFκB-luciferase reporter also showed decreased pathogen-induced NFκB activation with Tregs present (22). The targeting of such a fundamental signaling pathway by Tregs suggests that attenuated NFκB may account for the ability of Tregs to modulate the activities of many cell types from mast cells to B cells (23) (1, 24). Lymphocytes may be particularly sensitive to Treg downregulation of NFκB p65 where co-stimulation or inflammatory cytokines are limited (self-antigen) but would override suppression when co-stimulatory signals, particularly CD28/NFκB or TNFα, were upregulated.
The downstream consequence of reduced NFκB in the CD4 T cells has not yet been established. We do not know if there is a direct linkage between Treg-attenuated NFκB and loss of IL-2. Once a threshold level of nuclear NFkB is achieved, the amount of nuclear NFκB p65 does not impact the magnitude of the IL-2 response. Therefore, Tregs appear to limit the number of T cells reaching that nuclear NFkB threshold. Loss of NFκB could modify expression of a number of anti-apoptotic molecules such as Bcl-2 and cell-cycle promoting molecules like CDK (25) leading to the indirect loss of early IL-2 producers due to a failure to support their survival. Alternatively, reduced NFκB could lead to an alteration in the balance of TFs with a predominant NFAT signal. NFAT signals trigger an anergy-related gene profile with the upregulation of E3 ligase-associate genes (26). Interestingly, we have shown that Tregs induce a unique transcriptional program in CD4 targets that shows the most overlap with ionomycin-induced anergy (27).
We show that Tregs do not terminate T cell activation signals, but instead induce a unique signaling program in CD4+ target T cells with sustained NFAT/AP-1 but significantly reduced nuclear NFκB p65. This early NFκB modulation is independent of the APC. We propose that Treg suppression occurs in distinct mechanistic phases with early modulation that can occur in an APC-independent fashion qualitatively altering T cell signaling and a later phase where modulation of the APC may terminate T-APC interactions. Thus the context in which CD4+ T cells encounter Tregs may make them differentially sensitive to these two phases and hence account for the heterogeneity, and controversy, in timing and proposed mechanisms of Treg suppression.
Supplementary Material
Acknowledgements
We thank the Fowell Lab for helpful discussion and Jim Miller for careful review of the manuscript. We also appreciate technical assistance from Angie Hughson, T. Bushnell (UR Flow Core) and T. George and R.A. DeMarco (Amnis).
Footnotes
This work was supported by grants to DJF: R01 AI070826 and U19 AI56390.
References
- 1.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. 2008;9:239–244. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saparov A, Wagner FH, Zheng R, Oliver JR, Maeda H, Hockett RD, Weaver CT. Interleukin-2 expression by a subpopulation of primary T cells is linked to enhanced memory/effector function. Immunity. 1999;11:271–280. doi: 10.1016/s1074-7613(00)80102-8. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe SA, Zhou P, Dotsch V, Chen L, You A, Ho SN, Crabtree GR, Wagner G, Verdine GL. Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc. Nature. 1997;385:172–176. doi: 10.1038/385172a0. [DOI] [PubMed] [Google Scholar]
- 5.Macian F, Garcia-Rodriguez C, Rao A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 2000;19:4783–4795. doi: 10.1093/emboj/19.17.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acuto O, Michel F. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat Rev Immunol. 2003;3:939–951. doi: 10.1038/nri1248. [DOI] [PubMed] [Google Scholar]
- 7.Chen CY, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 8.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. Journal of Experimental Medicine. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sojka DK, Hughson A, Sukiennicki TL, Fowell DJ. Early kinetic window of target T cell susceptibility to CD25+ regulatory T cell activity. J Immunol. 2005;175:7274–7280. doi: 10.4049/jimmunol.175.11.7274. [DOI] [PubMed] [Google Scholar]
- 10.Bodor J, Fehervari Z, Diamond B, Sakaguchi S. ICER/CREM-mediated transcriptional attenuation of IL-2 and its role in suppression by regulatory T cells. Eur J Immunol. 2007;37:884–895. doi: 10.1002/eji.200636510. [DOI] [PubMed] [Google Scholar]
- 11.Bopp T, Becker C, Klein M, Klein-Hessling S, Palmetshofer A, Serfling E, Heib V, Becker M, Kubach J, Schmitt S, Stoll S, Schild H, Staege MS, Stassen M, Jonuleit H, Schmitt E. Cyclic adenosine monophosphate is a key component of regulatory T cell-mediated suppression. J Exp Med. 2007;204:1303–1310. doi: 10.1084/jem.20062129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wohlfert EA, Callahan MK, Clark RB. Resistance to CD4+CD25+ regulatory T cells and TGF-beta in Cbl-b−/− mice. J Immunol. 2004;173:1059–1065. doi: 10.4049/jimmunol.173.2.1059. [DOI] [PubMed] [Google Scholar]
- 13.Nurieva RI, Zheng S, Jin W, Chung Y, Zhang Y, Martinez GJ, Reynolds JM, Wang SL, Lin X, Sun SC, Lozano G, Dong C. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 2010;32:670–680. doi: 10.1016/j.immuni.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Q, Adams JY, Tooley AJ, Bi M, Fife BT, Serra P, Santamaria P, Locksley RM, Krummel MF, Bluestone JA. Visualizing regulatory T cell control of autoimmune responses in nonobese diabetic mice. Nat Immunol. 2006;7:83–92. doi: 10.1038/ni1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tadokoro CE, Shakhar G, Shen S, Ding Y, Lino AC, Maraver A, Lafaille JJ, Dustin ML. Regulatory T cells inhibit stable contacts between CD4+ T cells and dendritic cells in vivo. J Exp Med. 2006;203:505–511. doi: 10.1084/jem.20050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collison LW, Vignali DA. In vitro Treg suppression assays. Methods Mol Biol. 2011;707:21–37. doi: 10.1007/978-1-61737-979-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George TC, Fanning SL, Fitzgeral-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 18.McKarns SC, Schwartz RH. Biphasic regulation of Il2 transcription in CD4+ T cells: roles for TNF-alpha receptor signaling and chromatin structure. J Immunol. 2008;181:1272–1281. doi: 10.4049/jimmunol.181.2.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4(+) T cells. Nat Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 21.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 22.O'Mahony C, Scully P, O'Mahony D, Murphy S, O'Brien F, Lyons A, Sherlock G, MacSharry J, Kiely B, Shanahan F, O'Mahony L. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gri G, Piconese S, Frossi B, Manfroi V, Merluzzi S, Tripodo C, Viola A, Odom S, Rivera J, Colombo MP, Pucillo CE. CD4+CD25+ regulatory T cells suppress mast cell degranulation and allergic responses through OX40-OX40L interaction. Immunity. 2008;29:771–781. doi: 10.1016/j.immuni.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim HW, Hillsamer P, Banham AH, Kim CH. Cutting edge: direct suppression of B cells by CD4+ CD25+ regulatory T cells. J Immunol. 2005;175:4180–4183. doi: 10.4049/jimmunol.175.7.4180. [DOI] [PubMed] [Google Scholar]
- 25.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 26.Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 27.Sukiennicki TL, Fowell DJ. Distinct molecular program imposed on CD4+ T cell targets by CD4+CD25+ regulatory T cells. J Immunol. 2006;177:6952–6961. doi: 10.4049/jimmunol.177.10.6952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.