Abstract
The supraoptic nucleus (SON) is a particularly good model for the study of cell-specific gene expression since it contains two distinct neuronal phenotypes, the oxytocin (OXT) and vasopressin (AVP) synthesizing magnocellular neurons (MCNs). The MCNs are found in approximately equal numbers and selectively express either the OXT or the AVP gene in about 97% of the MCN population in the SON. An unresolved issue has been to determine what mechanisms are responsible for the highly selective regulation of the cell-type specific expression of OXT and AVP genes in the MCNs. Previous attempts to address this question used various bioinformatic and molecular approaches, which included using heterologous cell lines to study the putative cis-elements in the OXT and AVP genes, and the use of OXT and/or AVP transgenes in transgenic rodents. The data from all of the above studies identified a region <0.6kbp upstream of OXT exon I and about 3kb upstream of AVP exon I as being sufficient to produce cell-specific expression of the OXT and AVP genes, respectively, but failed to identify the specific cis-domains responsible for the MCN-specific gene expression. An alternative experimental approach to perform promoter deletion analysis in vivo, that is to use stereotaxic viral vector gene transfer into the SON in order to further dissect the cis-elements in the OXT and AVP genes, will be described here. This in-vivo method uses Adeno-Associated Viral (AAV) vectors expressing OXT-promoter deletion constructs and utilizes the enhanced green fluorescent protein (EGFP) as the reporter. The AAV constructs are stereotaxically injected into the rat brain above the SON and 2 weeks post injection the rats are sacrificed and assayed for EGFP expression. Using this method it has been possible to identify specific regions upstream of the transcription start site (TSS) in the OXT and AVP gene promoters which are responsible for conferring the cell-type specificity of the OXT and AVP gene expression in the SON.
Keywords: cell-type specific gene expression, transcription factors, transcription factor binding sites, magnocellular neurons, phenotype
The Magnocellular Neuron Phenotype
It is commonly understood that cell identity or phenotype is largely determined by the constellation of specific genes that are expressed by the specific cell-type. Several physiological differences have been described between the magnocellular oxytocin (OXT)- and vasopressin (AVP)-synthesizing neurons in the hypothalamus (1, 2), but the most prominent distinguishing feature between these phenotypes is their selective expression of the two neuropeptide genes (3). The supraoptic nucleus (SON) is an outstanding model for the study of cell-type specific gene expression since it contains principally two neuronal phenotypes, the OXT and AVP synthesizing magnocellular neurons (MCNs) which are found in approximately equal numbers in the SON. Although the earliest immunohistochemical (4) and in situ hybridization experiments suggested that expression of these genes in the MCNs was mutually exclusive (5), recent experiments using more sensitive morphological assays have shown that the OXT or the AVP genes are abundantly and selectively expressed in about 97% of the MCN population in the SON, but that about 3% of the MCNs in the normal SON have been shown to coexpress both peptides (6, 7). This suggests that the latter MCN population represents a third phenotype (see Fig.1A). Indeed, when the assay is made even more sensitive, such as in single cell PCR (Fig. 1B), it can be shown that virtually all the MCNs in the SON express both peptides, but at dramatically different levels (8). Quantitative analyses of this differential expression gives average mRNA ratios of 500:1 for the principal peptide versus the minor peptide in the OXT and AVP phenotypes and about 2:1 in the coexisting phenotype (9). Thus, cell-type specific expression of these peptide genes in the MCNs is a quantitative property and not an absolute one, comparable to the two orders of magnitude of selectivity usually seen for antibody and receptor binding to antigens and ligands, respectively.
Figure 1.

Magnocellular Neuronal Phenotypes in the rat SON.
A. Shows double-label immunofluorescent images for oxytocin (green) and vasopressin (red) containing magnocellular neurons (MCNs) in the rat SON. These two distinct MCN populations are the only neuronal phenotypes found in the SON. Most of the MCNs express only oxytocin or vasopressin, however, a small percentage of the MCNs (about 2–3%) are known to express both peptides (see arrows). Scale bar = 100µm. Abbreviations: OC, optic chiasm.
B. Oxytocin and vasopressin phenotypes of individual dissociated hypothalamic supraoptic magnocellular neurons were determined by RT-PCR. A photomicrograph is shown of an individual dissociated magnocellular neuron being picked up by a harvesting pipette touching the large cell body containing two large dendrites. Scale bar, 20µm. cDNAs from the individual cells were amplified by PCR with primers that are specific for oxytocin (OT), vasopressin (VP), or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The PCR products were then run on 1.5% agarose gels containing ethidium bromide.
A persistent and still unresolved fundamental issue has been to determine what mechanisms are responsible for the highly selective regulation of this cell-type specific expression of the OXT and AVP genes in the MCNs. Many previous attempts that have been made to address this question used various bioinformatic techniques and molecular approaches. These include using heterologous cell lines to identify the cis-elements in the OXT and AVP genes, and the use of OXT and/or AVP transgenes in transgenic rodents (3) to elucidate which DNA domains in the genes that are involved. The consensus of all of these studies is that a region <0.6kbp upstream of OXT exon I and about 3 kb upstream of AVP exon I is sufficient to produce cell-specific expression of the OXT and AVP genes, respectively. However, these studies failed to identify the specific cis-elements that were responsible for the MCN-specific gene expression [reviewed in Young and Gainer, (10) and Murphy and Wells, (11)].
In this paper, I briefly describe the history of studies of the molecular mechanisms that regulate cell-type specific OXT and AVP gene expression in the MCNs, and then present an alternative experimental approach to promoter deletion analysis in vivo that we have recently developed and used in our laboratory to study this issue in the SON. This in-vivo method uses adeno-associated viral (AAV) vectors, which contain OXT-and AVP-promoter deletion constructs fused to enhanced green fluorescent protein (EGFP) as the reporter, in order to elucidate the cell-type specific cis-elements in the OXT and AVP genes. The AAV constructs are stereotaxically injected into the rat brain above the SON and after two weeks post injection the rats are sacrificed and the MCNs are assayed for EGFP expression by immunohistochemistry. Using this method it is possible to identify specific regions upstream of the transcription start site (TSS) in the OXT and AVP gene promoters which are responsible for conferring the cell-type specificity of the OXT and AVP gene expression in the SON.
OXT and AVP: From Peptides to Genes
Following the discovery and complete characterization of the OXT and AVP by DuVigneaud in 1954 (12), attention turned to the mechanism by which these neurohypophysial peptides were synthesized in the hypothalamus. Pioneering studies by Howard Sachs and his colleagues showed that the synthesis of vasopressin in the brain required de novo protein synthesis (13), and they hypothesized that the AVP peptide was formed by the post-translational processing of a precursor protein (14). It is notable this prescient proposal preceded the discovery of proinsulin by three years (15). It was another 10 years before the first physical evidence for the existence of the OXT and AVP precursor proteins was reported (16, 17). Following the introduction of recombinant DNA methodology to the field by D. Richter and colleagues, there was rapid progress made in the elucidation of the sequences of the OXT and AVP cDNAs, genes, and precursor proteins (18–20). Subsequently, it was shown that the OXT and AVP genes in the mouse, rat, and human genomes are located on the same chromosome separated by a short (3.5 to 12 kbp) intergenic region, and are in opposite transcriptional orientations (21–25).
Bioinformatic Predictions and Experimental Validations
One useful starting point for identifying the regulatory elements that might be involved in the cell-specific gene expression of the OXT and AVP genes is to first identify the evolutionarily conserved DNA sequences in these genes. Genomic sequences for OXT and AVP genes in many animal species are now available in the public database, and it is feasible to determine those DNA sequences in these genes that have been evolutionarily conserved. In order to do this we compared the DNA sequences in these genes between five mammalian species, rat, mouse, chimp, rhesus monkey, and human (Samal, Johnson, Gainer, in prep).As expected, there is a very high sequence conservation in the exons, especially in exons 1 and 2, and very low sequence conservation in the introns. The highest sequence conservation found in the non-coding DNA is about 500 bps 5’ upstream of the TSS, in the OXT and AVP gene promoters, and about 250 bps downstream of the gene bodies. Given the sequence conservation present in these non-coding regions, this suggests that significant regulatory sequences might be located in these domains, especially 500 bps 5’ upstream of the TSS (−500 bps) in both genes.
A number of investigators had previously noted the interspecies homologies in the promoter domain 5’ upstream of the TSS (19, 21, 24), and also that alignments of the OXT and AVP gene promoter sequences to one another showed that there were few if any similarities between their promoter domains, even within the same species. This differs dramatically from what is found by comparing their exons, e.g., a 96% homology is found for exon 2. Thus, the high sequence conservation found between species for the 500 bps 5’ upstream of the TSS in the OXT and AVP genes, and the absence of any similarities in sequence between the genes in this region is consistent with the view that regulatory elements directing their cell-type specific expression could reside in this 5’ domain. As a consequence of this hypothesis, transcription factor (TF) prediction programs were applied to the sequences that were 500 bps 5’ upstream of the TSS in both genes in order to identify putative TF motifs for further experimental study (26).
The preferred way to evaluate the relevance of these predicted transcription factor binding sites (TFBS) on the OXT and AVP promoters to the cell-type specific expression of these genes would be to do systematic promoter deletion experiments in homologous cell lines (i.e, cell lines derived from bona fide OXT and AVP expressing primary cells). This approach was very successful for the study of the GnRH gene and identified a number of regulatory elements that controlled its cell-type specific expression (27–29). Unfortunately, for OXT and AVP there are no homologous cell lines available that can serve as appropriate experimental surrogates for the OXT- and AVP-expressing MCNs found in vivo. As a result various investigators have studied this issue by doing experiments using heterologous cell lines. While such experiments obviously cannot determine whether the putative regulatory elements are actually involved in cell-type specific expression, they are able to validate the potential functionality of the sequence in the promoter. The results of such experiments which were largely conducted between 1990 to 2000 are reviewed in (3), and a graphic view of some of the regulatory elements present in the proximal promoter regions of the OXT and AVP genes is illustrated in Fig. 2.
Figure 2.
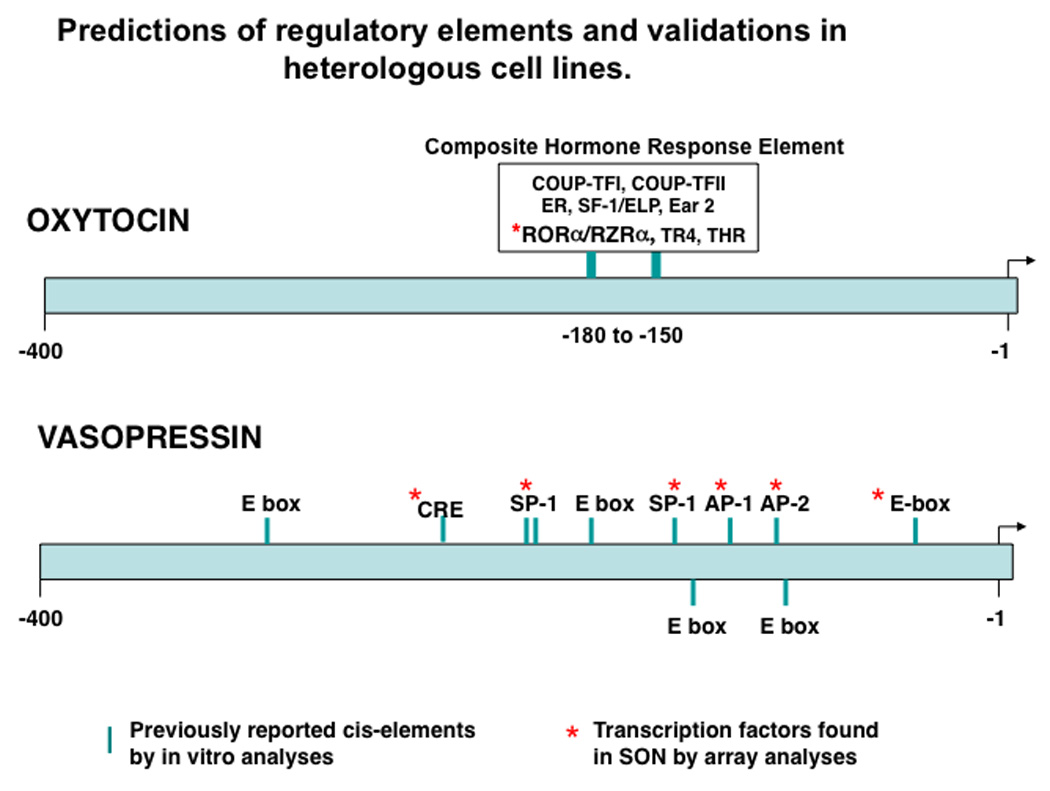
Regulatory elements in the OXT and AVP promoters 500bp upstream of their transcription start sites that are predicted by bioinformatic analyses and which were functionally validated by experiments using heterologous cell lines (based on data described in Burbach et al, (3). The asterisks show the transcription factors that bind to these elements which were detected in the SON by microarray analyses (based on data in Hindmarch(82); Mutsuga et. al, (83); and Yue et. al, (84)). Abbreviations: COUP-TF I and II:Chicken ovalbumin upstream promoter transcription factors I and II; ER: estrogen receptor; SF-1/ELP: steroidogenic factor; Ear2: V-erb-related protein 2 transcription factor; RORa/RZRa: retinoic acid receptor orphan receptor transcription factor family; THR: thyroid hormone receptor; CRE: camp response element; SP1: specificity protein 1 transcription factor; AP1: activator protein 1; AP2: activator protein 2, Ebox: DNA sequence, CANNTG/CACGTG,that binds helix loop-helix transcription factors (e.g., BMAL-CLOCK).
Experimental Models for the Study of Cell-type Specific OXT and AVP Gene Expression
Attempts to generate homologous AVP and OXT Cell lines by targeted oncogenesis
In view of the known effectiveness of using homologous cell lines as experimental models for promoter deletion studies relevant for cell-type specific expression analysis several efforts were made to generate such cell lines for AVP by targeted oncogenesis. The earliest attempt was made in 1987 by David Murphy and colleagues (30) who used 1.25 kbp of the bovine AVP promoter directly attached to the early region of the tumor virus, SV 40, encoding the large T-antigen. This transgene construct (termed AVP.SVER 1.25) was not expressed in the HNS, but was expressed in the anterior pituitary, where it produced tumors (30, 31). One interpretation for the failure of this construct to produce appropriate cell-specific expression is that it didn’t contain key regulatory elements normally found elsewhere in the AVP gene body. Indeed, the same 1.25 kbp bovine promoter when attached to a chloroamphenicol acetyl transferase (CAT) reporter produced ubiquitous expression of the construct in two lines of transgenic mouse lines, but also no notable expression in the hypothalamo-neurohypophysial system (HNS) (32). However, another more recent effort at targeted oncogenesis used a mouse AVP transgene which was known to produce excellent cell–type specific expression of a CAT reporter in the mouse brain (33). The transgene used to generate the oncogenesis was the same, but the CAT reporter was replaced by SV40 T antigen (34). The resulting transgenic mice contained many tumors in the brain, lymph nodes and spleen but again there was no expression in the magnocellular neurons in the hypothalamus.
Transgenic Rodent Experimental Models
The failure to produce homologous cell lines to serve as model systems to study OXT and AVP cell-type specific gene expression led investigators to turn to transgenic rodent experimental models as an alternative. Russo et.al,(35) studied 14 different fusion genes in transgenic mice. One of these contained 2 kbp of the human AVP promoter linked to the human growth hormone gene. They found ubiquitous (ectopic) expression of this transgene throughout the mouse CNS, particularly in the cerebral cortex, and also in the hypothalamus and HNS neurons. However, this construct was no more specific or robust in its expression in the HNS than a metallothionin promoter that had been connected to the growth hormone reporter as a transgene. Interestingly, the CNS expression of the latter construct could be eliminated by removing all the introns from the growth hormone reporter gene (35), and only expression in tissues normally characteristic of metallothionin promoter expression (e.g., liver, pancreas, intestine, and kidney) was found with the latter construct. While all these observations focus on inappropriate (ectopic) expression of these transgenes, they also alert one to the general problems in the interpretation of transgenic data. Expression in a given tissue can reflect so-called “position effects” of the transgenes’ integration (36), enhancing influences of heterologous sequences (37), or silencing effects (38) that may derive from exogenous sequences present in the constructs. In some cases, the same reporter gene can act as an enhancer or as a repressor, depending upon the specific configurations of the DNAs in the construct. Habener et. al, (39) produced a transgenic mouse with a complex construct containing 800 bp of the mouse metallothionin I (MT) 5' flanking region (promoter) and 35 bp of its exon 1 connected through a 14 bp artificial DNA linker to the entire rat prepro AVP NP II gene. The authors reported the expression of the fusion gene in appropriate tissues (corresponding to the metallothionin promoter’s endogenous expression), but also in the HNS which normally does not express this gene. Thus, the above studies show that heterologous DNA and even the reporter’s DNA sequences in the transgene can interact with the endogenous genomic sequences to produce ectopic expression, thereby confounding any analysis regarding cell-type specificity. In spite of these limitations, several laboratories continued to employ the transgenic paradigm to investigate cell-type specific expression of the OXT and AVP genes.
The first report of robust cell-specific gene expression in the HNS of transgenic mice was made using a combined rat VP and OT (“minilocus”) construct (40, 41). The results were surprising in that although this construct contained about five times longer VP 5'-upstream sequence than OT 5'-upstream sequence, the rat OT transgene was robustly expressed (at between 10–30% of endogenous OT gene expression levels) in mouse OT cells only, whereas the VP gene was not expressed at all. The earliest successful transgenic rat studies on the VP gene (42–44) showed that by extending the 3'-downstream region of the VP gene to 3 kbp, it was possible to get cell-type specific expression of these transgenes in VP neurons only. All of these studies of various oxytocin and vasopressin constructs in transgenic mice and rats indicated that constructs containing genomic DNA from 0.5 to 9 kbp 5' upstream of the OT and VP genes but with no endogenous 3'-downstream sequences did not produce significant expression in the hypothalamic magnocellular neurons. This suggested that the intergenic region might contain cis-elements that are essential for their cell-specific gene expression in the magnocellular neurons. These experiments using transgenic rodents as systems to evaluate various deletions in the OXT and AVP genes continued to about 2003 at which time use of the transgenic model for cell-type specific expression analysis ceased. At this time, the consensus was that for the OXT gene inclusion of sequences 568bp upstream and 3.6kbp downstream of the gene body in the transgene produced cell-type specific expression, and for the Avp gene inclusion of 3.5kbp upstream and 2.1kbp downstream of the gene body produced cell-type specific expression (see Murphy and Wells, (11); and Young and Gainer, (10) for reviews of this subject).
An additional benefit of these transgenic studies is that they led to the production of transgenic lines of mice or rats that expressed the reporter EGFP specifically in either OXT or AVP neurons. These lines have served as very valuable experimental models for various physiological experiments(11, 43, 45–49).
Organotypic Culture Models and biolistic transfection
The transgenic rodent strategy for promoter deletion studies was abandoned in large part because of the excessive time and cost of the animal husbandry associated with this approach. An alternative in vitro approach was developed which used biolistic transfection of the MCNs in organotypic cultures of hypothalamus for deletion studies, and was used primarily to evaluate the downstream sequences in the transgenes that appeared to be needed for cell-type specific expression (50). These studies showed that the sequences 554 bp upstream of the TSS in the OXT gene, and only 430 bp downstream of exon III was sufficient to produce expression in the OXT MCNs, and that only 288bp upstream and 178bp downstream of the AVP gene were found to be necessary (50). These conclusions are shown in Fig. 3. The 178 bp domain downstream of exon III in the AVP gene appeared to contain similar motifs to those found in the 432bp domain downstream of exon III in the OXT gene (50), and further experiments showed that switching the positions of these two downstream elements produced equivalent expression in the OXT and AVP MCNs (unpublished data). From these observations, it was concluded that the key cis-elements that were responsible for the cell-type specific expression of the OXT and AVP genes were not located downstream but rather in the 5' flanking region upstream of the TSS in both genes.
Figure 3.
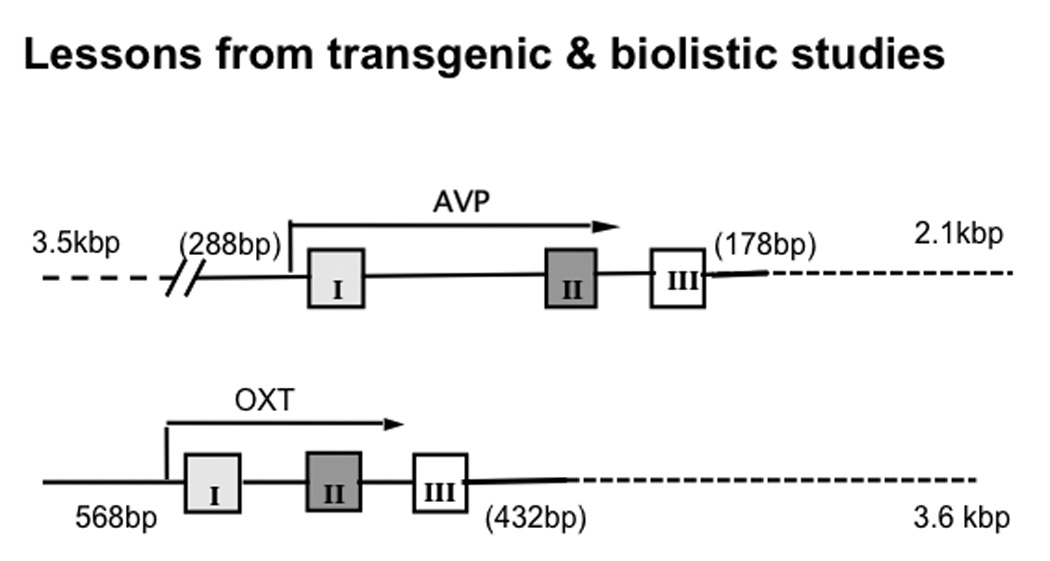
Summary of results from Transgenic and Organotypic Culture/Biolistics Studies (1987–2003). These constructs were found to produce cell-type specific expression in the Oxt and Avp MCNs in the SON. In transgenic rodents the Avp gene with 3.5kbp upstream and 2.1kbp downstream (dotted line) of the gene body produced cell-type specific expression, whereas using biolistics and organotypic culture only 288bp upstream and 178bp downstream were found to be neccessary. For the Oxt gene 568bp upstream and 3.6kbp downstream of the gene body in the transgene produced cell-type specific expression, whereas in biolistic experiments 568bp upstream and only 432bp downstream were found to be sufficient (see reviews by Young and Gainer, (10); and Murphy and Wells(11)).
While both the transgenic and in vitro biolistic studies provided valuable insights into regions of the AVP and OXT genes that could regulate their cell-type specific expression, neither of these experimental paradigms were sufficiently efficient or optimal for the continued study of multiple deletion constructs. As noted above, a significant problem for the transgenic approach is that the random insertion of the transgene often leads to artifacts such as insertional mutations, and ectopic expression due to enhancer trapping. The latter problem increases in probability when the construct is systematically shortened by the process of promoter deletion and possible insulator elements in the transgenes are removed. The biolistic transfection of organotypic cultures is limited by the very low probability that the gold particles coated with plasmids that are being randomly “shot” at the cultured slice explant will land in the cell nucleus of a MCN without damaging the cell. Moreover, in the biolistic transfection method the particles do not penetrate more than 100 um into the tissue, thereby making this method not useful for transfecting the ventrally located MCNs in the hypothalamus in vivo. Therefore, we turned to the more efficient viral vector method of gene transfer in vivo in order to do further promoter deletion studies in vivo. The viral vector method has the additional virtue of being able to be stereotaxically targeted by micoinjection to specific brain regions such as the SON, which has large numbers of OXT- and AVP-MCNs.
Viral Vector Gene Transfer Approaches
Viral vector methods have been routinely used as a means to deliver genes into the nervous system in vivo (51–55), and have been very effectively used for gene transfer into hypothalamic neurons (55–57)and in MCNs (58–60). In addition, viral vectors have been successfully used to study gene promoter domains that are involved in cell-specific gene expression in vivo (61–65). There are many choices of viral vectors that can be used for gene transfer to brain in vivo, and these include adenoviruses, adeno-associated viruses (AAV), herpes simplex viruses and retroviruses such as lentivirus (see Osten et al, (52)). We tested the transduction efficiencies of several of these in hypothalamus in our organotypic culture model and found that AAV was able to transduce OXT MCNs in vitro very effectively without any evidence of toxicity (66). A description of the AAV virus structure and its efficacy in hypothalamus in vitro is illustrated in Fig. 4. AAV vectors have also been specifically shown to be more efficient than lentiviral vectors in transducing neurons in the forebrain (67, 68). Consequently, we choose to use AAV vectors that contain promoter deletion constructs of the OXT and AVP gene promoters fused to EGFP reporters to transduce MCNs in the rat SON in vivo.
Figure 4.

Use of Adeno-Associated Virus (AAV) to study gene expression in MCNs in organotypic hypothalamic cultures.
Top: Genomic organization of AAV. The AAV genome produces four transcripts, Rep 40,52, 68 and 78 which are genes involved in replication, and three proteins, VP-1,-2 and-3 which are genes for forming the capsid. The ITRs (Inverted Terminal Repeat Sequences) are necessary for viral replication, rescue, packaging and integration.
Bottom: AAV transduction of hypothalamic slice explant cultures.
Slices were incubated in AAV-CMV-nLac-Z for 7 days and following this incubation, the tissues were fixed in 2 % paraformaldehyde and double immunostained for nuclear localized lac-z immunoreactivity (black) and oxytocin-neurophysin immunoreactivity (brown). A and B: Control slices (no AAV added). C and D: AAV transduced slices.(adapted from Kier et. al., (66)). The DAB (brown) stained OXT neurons shown in B and D are in the Accessory Nucleus in the hypothalamic slice.
OXT and AVP Promoter-Deletion Studies in vivo Using AAV
These in vivo experiments involve stereotaxic injection of the AAVs bearing various promoter deletions fused to EGFP reporters into rat SONs, and then allowing two weeks for expression of the EGFP. The rat brains are then perfusion fixed with paraformaldehyde, and double-label immunohistochemistry is performed on cryostat sections of the hypothalamus in order to assay the expression of the EGFP in either the OXT- or AVP-MCNs. Examples of the results of such experiments are shown in Fig. 5, where AAVs containing a 2.kbp promoter length for AVP and a 563kbp promoter length for OXT were injected into rat SONs and both produced cell-specific expression of the EGFP in the appropriate MCN phenotype. Fig 5A shows EGFP expression in the MCNs which co-localize with Avp-immunoreactivity (Avp-ir), but not in MCNs with with Oxt-immunoreactivity (Oxt-ir). Fig. 5B shows EGFP expression in the MCNs, which co-localize with Oxt-ir, but not those with Avp-ir (unpublished data). These experiments not only show that this a practical approach to promoter deletion analysis, but also for the first time show that a promoter length for AVP less than 3kbp can produce cell-type specific gene expression in vivo, and also confirms that the 563bp promoter length for OXT found effective in the transgenic studies also produce cell-type specific gene expression through the AAV route. Finally, we show in Fig. 5 that introns 1 and 2 and exons 2 and 3 in the OXT and AVP genes are not needed for cell-specific expression in the MCNs. In this regard, it should be noted that cell specific expression is maintained in AAV constructs that only have the OXT or AVP upstream promoter regions directly fused to the EGFP reporter and containing none of the exons, introns, including deletion of exon 1. (see V. Grinevich, this issue). These AAV experiments demonstrate, for the first time, that neither the exons nor the introns in the OXT or AVP genes are involved in the mechanisms producing their cell-type specific expression. It should also be noted that the constructs used in our deletion studies (see Fig 5) contained the intrinsic IGR elements that we previously determined in biolistic studies were necessary for expression in vitro (50). However, when AAV transduction in vivo is used these intrinsic IGR sequences can be replaced by an SV40 poly A signal sequence with no apparent change in efficacy (V. Grinevich, personal communication).
Figure 5.

Use of Adeno-Associated Virus (AAV) to study cell-type specific expression in (A) Avp and (B) Oxt MCNs in vivo. Stereotaxic injections of rAAV vectors containing construct with either Avp (in A) or Oxt (in B) promoters fused to EGFP reporters produced cell-specific expression of the EGFP in the injected SONs. A. Shows EGFP expression in the MCNs, which co-localize with Avp-immunoreactivity (Avp-ir), but not with Oxt-immunoreactivity (Oxt-ir). B. Shows EGFP expression in the MCNs, which co-localize with Oxt-ir, but not with Avp-ir. The IGR is the intergenic region sequence as defined in Fields et. al.(2003; ref 50). For the AVP construct the IGR was 181bp, for the OXT construct the IGR was 442bp. The ITRs (Inverted Terminal Repeat Sequences) are necessary for viral replication, rescue, packaging and integration (see Fig. 4 top).
Further experiments showed that AAV vectors containing 448 bp, 325bp and 216bp (but not 100bp) upstream sequences of the OXT gene promoter can all support cell-specific OXT gene expression in OXT-MCNs but no expression in AVP MCNs (in preparation). AAVs containing the 100bp or 50bp upstream region can produce EGFP expression in the SON, but non-selectively in both the OXT-and AVP- MCNs, as might be expected of a “core promoter” region. Similar deletion experiments with the AVP gene have shown that AAV vectors containing 1.5kbp, 950bp, 543bp, and 288bp upstream sequences of the AVP gene promoter can all support cell-type specific AVP gene expression in AVP-MCNs but produce no expression in OXT MCNs (unpublished data). Fig. 6 summarizes the evolution of our views since 2003 about the regulatory domains in the OXT and AVP promoters that are responsible for the cell-type specific expression of these genes. Studies still in progress are evaluating whether AAVs containing 100bp or 50bp of the AVP upstream region lose their cell-type specific EGFP expression in the SON, in order to identify the “core promoter” region in the AVP gene.
Figure 6.

Comparisons of cis-domains in the Avp and Oxt gene promoters that are found to be important for their cell type specific expression before and after application of the AAV strategy.
Conclusions
One conclusion that can be clearly drawn from these studies is that the viral vector approach described here is a highly effective way to experimentally study cell-type specific gene expression in the central nervous system, and could easily be applied to any gene and brain region of interest. For example, it will be interesting to determine whether the regulatory elements identified from the study cell-type specific AVP gene expression in the SON, are the same or different in other AVP expressing regions such as the SCN, BNST or the amygdala.
As the regulatory elements that are responsible for the cell-type specific gene expression of the genes begin to be identified, then the next quest will be to determine the transcription factors (TFs) that bind to these elements (i.e, to the transcription factor binding sites, TFBSs). Table 1 shows some TFs that we have found in recent bioinformatic analyses to be predicted as present in the promoter domains that appear to be involved in cell-type specific expression (unpublished). Clearly further deletion experiments are needed on the OXT and AVP promoters in order to better define the specific 8–10 base sequences that usually constitute TFBSs. The only approximation to this level of analysis is our finding that the −216/−100 bp region in OXT gene promoter contains the key elements that determine its cell-specific expression in the OXT-MCNs. Given this information we hypothesize that: 1) there is a repressor element, (RE) in the −216 to −100 5’ upstream region of the OXT gene that inhibits its expression in AVP-MCNs, and 2) there may also be an activator in the −216 to −100 region of the OXT gene that could enhance OXT MCN expression.
Table 1.
Transcription Factor candidates for Cell-Type Specific Expression in the SON*
| AVP promoter | Oxt promoter |
|---|---|
| AP1 | ELF1 |
| AP2 | FOXO1 |
| Amt | HNF4a |
| Ets | PPARA |
| KROX | PPARG |
| MAF | RORa |
| Max | RXRa |
| MAZ | STAT3 |
| MZF1 | VDR |
| SP1 | |
| SP3 | |
| SP4 | |
| TGIF | |
| USF | |
| ZnF219 |
Based on Bioinformatic predictions in the critical DNA domains (Samal, Johnson & Gainer, unpublished) and supporting microarray data from Mutsuga et al (2004; 2005) Yue et al (2006) Hindmarch et al (2006; 2007) Qui et al (2007)
Interestingly, in the 1990s there were extensive reports from in vitro studies that used a variety of heterologous cell lines that identified this region in the OXT promoter as containing functional estrogen/retinoic acid receptor-like transcription factor binding sites (3, 69), and this nuclear hormone receptor(NHR)-rich region was termed the Composite Hormone Receptor Element (69) (see Fig. 3). These in vitro studies have shown that specific NHRs can act as either activators (70–73) (74, 75) or inhibitors (76–78) of OXT gene expression, and hence, these data are consistent with our hypothesis that elements in this 116 bp domain could regulate the activation or repression of OXT gene transcription in the OXT- or AVP-MCNs, respectively, depending on the specific NHRs being expressed in each of these cell-types.
In an effort to identify the specific NHRs that are expressed in the SON, Lopes de Silva and Burbach (79) found that five classical receptors and four orphan receptors were expressed in the SON, but only one of these, the thyroid hormone receptor-α (THRα) was found by in situ hybridization histochemistry to be expressed in the MCNs (80, 81). Unfortunately, there were no experiments reported that provide information as to whether the THRα is preferentially expressed in the OXT- or the AVP-MCNs. Experiments are currently in progress in our laboratory using laser capture microdissection of single OXT and AVP MNCs and real-time quantitative PCR to determine whether there is a selective OXT-versus AVP-MNC expression of THRα and other candidate NHRs.
Acknowledgements
This research was supported by the Intramural Research Program of the NINDS, NIH. I thank my many colleagues who contributed to this work, including Ray Fields, Todd Ponzio, Shirley House, Daniel Lubelski, Makoto Kawasaki, Madison Stevens, Omar Rashid, Yasmmyn Salinas, Yijun Shi, Babru Samal and Corey Johnson.
List of Abbreviations
- AAV
adeno-associated virus
- AVP
arginine vasopressin
- OXT
oxytocin
- EGFP
enhanced green fluorescent protein
- MCN
magnocellular neuron
- TSS
transcription start site
- TF
transcription factor
- TFBS
transcription factor binding site
- CAT
chloramphenicol acetyltransferase
- IGR
intergenic region
- THRα
thyroid hormone receptor-α
- RORA
orphan receptor RORalpha.
- Rep
replication genes inAAV
- Cap
capsid genes in AAV
- ITR
Inverted Terminal Repeat Sequences in AAV
References
- 1.Armstrong WE. Morphological and electrophysiological classification of hypothalamic supraoptic neurons. Prog Neurobiol. 1995;47(4–5):291–339. [PubMed] [Google Scholar]
- 2.Hatton GI. Oxytocin and vasopressin neurones: vive la difference! J Physiol. 1997;500(Pt 2):284. doi: 10.1113/jphysiol.1997.sp022019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burbach JP, Luckman SM, Murphy D, Gainer H. Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev. 2001;81(3):1197–1267. doi: 10.1152/physrev.2001.81.3.1197. [DOI] [PubMed] [Google Scholar]
- 4.Vandesande F, Dierickx K. Identification of the vasopressin producing and of the oxytocin producing neurons in the hypothalamic magnocellular neurosecretroy system of the rat. Cell Tissue Res. 1975;164(2):153–162. doi: 10.1007/BF00218970. [DOI] [PubMed] [Google Scholar]
- 5.Mohr E, Bahnsen U, Kiessling C, Richter D. Expression of the vasopressin and oxytocin genes in rats occurs in mutually exclusive sets of hypothalamic neurons. FEBS Lett. 1988;242(1):144–148. doi: 10.1016/0014-5793(88)81003-2. [DOI] [PubMed] [Google Scholar]
- 6.Kiyama H, Emson PC. Evidence for the co-expression of oxytocin and vasopressin messenger ribonucleic acids in magnocellular neurosecretory cells: simultaneous demonstration of two neurohypophysin messenger ribonucleic acids by hybridization histochemistry. Journal of neuroendocrinology. 1990;2(3):257–259. doi: 10.1111/j.1365-2826.1990.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 7.Mezey E, Kiss JZ. Coexpression of vasopressin and oxytocin in hypothalamic supraoptic neurons of lactating rats. Endocrinology. 1991;129(4):1814–1820. doi: 10.1210/endo-129-4-1814. [DOI] [PubMed] [Google Scholar]
- 8.Glasgow E, Kusano K, Chin H, Mezey E, Young WS, Gainer H. Single cell reverse transcription-polymerase chain reaction analysis of rat supraoptic magnocellular neurons: neuropeptide phenotypes and high voltage-gated calcium channel subtypes. Endocrinology. 1999;140(11):5391–5401. doi: 10.1210/endo.140.11.7136. [DOI] [PubMed] [Google Scholar]
- 9.Xi D, Kusano K, Gainer H. Quantitative analysis of oxytocin and vasopressin messenger ribonucleic acids in single magnocellular neurons isolated from supraoptic nucleus of rat hypothalamus. Endocrinology. 1999;140(10):4677–4682. doi: 10.1210/endo.140.10.7054. [DOI] [PubMed] [Google Scholar]
- 10.Young WS, 3rd, Gainer H. Transgenesis and the study of expression, cellular targeting and function of oxytocin, vasopressin and their receptors. Neuroendocrinology. 2003;78(4):185–203. doi: 10.1159/000073702. [DOI] [PubMed] [Google Scholar]
- 11.Murphy D, Wells S. In vivo gene transfer studies on the regulation and function of the vasopressin and oxytocin genes. J Neuroendocrinol. 2003;15(2):109–125. doi: 10.1046/j.1365-2826.2003.00964.x. [DOI] [PubMed] [Google Scholar]
- 12.Ragnarsson U. The Nobel trail of Vincent du Vigneaud. J Pept Sci. 2007;13(7):431–433. doi: 10.1002/psc.864. [DOI] [PubMed] [Google Scholar]
- 13.Takabatake Y, Sachs H. Vasopressin Biosynthesis. 3. In Vitro Studies. Endocrinology. 1964:75934–75942. doi: 10.1210/endo-75-6-934. [DOI] [PubMed] [Google Scholar]
- 14.Sachs H, Takabatake Y. Evidence for a Precursor in Vasopressin Biosynthesis. Endocrinology. 1964:75943–75948. doi: 10.1210/endo-75-6-943. [DOI] [PubMed] [Google Scholar]
- 15.Steiner DF, Cunningham D, Spigelman L, Aten B. Insulin biosynthesis: evidence for a precursor. Science. 1967;157(789):697–700. doi: 10.1126/science.157.3789.697. [DOI] [PubMed] [Google Scholar]
- 16.Gainer H, Sarne Y, Brownstein MJ. Biosynthesis and axonal transport of rat neurohypophysial proteins and peptides. J Cell Biol. 1977;73(2):366–381. doi: 10.1083/jcb.73.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gainer H, Sarne Y, Brownstein MJ. Neurophysin biosynthesis: conversion of a putative precursor during axonal transport. Science. 1977;195(4284):1354–1356. doi: 10.1126/science.65791. [DOI] [PubMed] [Google Scholar]
- 18.Ivell R, Richter D. Structure and comparison of the oxytocin and vasopressin genes from rat. Proc Natl Acad Sci U S A. 1984;81(7):2006–2010. doi: 10.1073/pnas.81.7.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruppert S, Scherer G, Schutz G. Recent gene conversion involving bovine vasopressin and oxytocin precursor genes suggested by nucleotide sequence. Nature. 1984;308(5959):554–557. doi: 10.1038/308554a0. [DOI] [PubMed] [Google Scholar]
- 20.Schmale H, Richter D. Single base deletion in the vasopressin gene is the cause of diabetes insipidus in Brattleboro rats. Nature. 1984;308(5961):705–709. doi: 10.1038/308705a0. [DOI] [PubMed] [Google Scholar]
- 21.Hara Y, Battey J, Gainer H. Structure of mouse vasopressin and oxytocin genes. Brain Res Mol Brain Res. 1990;8(4):319–324. doi: 10.1016/0169-328x(90)90045-f. [DOI] [PubMed] [Google Scholar]
- 22.Mohr E, Schmitz E, Richter D. A single rat genomic DNA fragment encodes both the oxytocin and vasopressin genes separated by 11 kilobases and oriented in opposite transcriptional directions. Biochimie. 1988;70(5):649–654. doi: 10.1016/0300-9084(88)90249-0. [DOI] [PubMed] [Google Scholar]
- 23.Richter D. Molecular events in expression of vasopressin and oxytocin and their cognate receptors. Am J Physiol. 1988;255(2 Pt 2):F207–F219. doi: 10.1152/ajprenal.1988.255.2.F207. [DOI] [PubMed] [Google Scholar]
- 24.Sausville E, Carney D, Battey J. The human vasopressin gene is linked to the oxytocin gene and is selectively expressed in a cultured lung cancer cell line. J Biol Chem. 1985;260(18):10236–10241. [PubMed] [Google Scholar]
- 25.Schmitz E, Mohr E, Richter D. Rat vasopressin and oxytocin genes are linked by a long interspersed repeated DNA element (LINE): sequence and transcriptional analysis of LINE. DNA Cell Biol. 1991;10(2):81–91. doi: 10.1089/dna.1991.10.81. [DOI] [PubMed] [Google Scholar]
- 26.Mohr E, Richter D. Sequence analysis of the promoter region of the rat vasopressin gene. FEBS letters. 1990;260(2):305–308. doi: 10.1016/0014-5793(90)80130-b. [DOI] [PubMed] [Google Scholar]
- 27.Kim HH, Wolfe A, Smith GR, Tobet SA, Radovick S. Promoter sequences targeting tissue-specific gene expression of hypothalamic and ovarian gonadotropin-releasing hormone in vivo. J Biol Chem. 2002;277(7):5194–5202. doi: 10.1074/jbc.M110535200. [DOI] [PubMed] [Google Scholar]
- 28.Iyer AK, Miller NLG, Yip K, Tran BH, Mellon PL. Enhancers of GnRH transcription embedded in an upstream gene use homeodomain proteins to specify hypothalamic expression. Mol Endocrinol. 2010;24(10):1949–1964. doi: 10.1210/me.2010-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novaira HJ, Yates M, Diaczok D, Kim H, Wolfe A, Radovick S. The gonadotropin-releasing hormone cell-specific element is required for normal puberty and estrous cyclicity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(9):3336–3343. doi: 10.1523/JNEUROSCI.5419-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy D, Bishop A, Rindi G, Murphy MN, Stamp GW, Hanson J, Polak JM, Hogan B. Mice transgenic for a vasopressin-SV40 hybrid oncogene develop tumors of the endocrine pancreas and the anterior pituitary. A possible model for human multiple endocrine neoplasia type 1. Am J Pathol. 1987;129(3):552–566. [PMC free article] [PubMed] [Google Scholar]
- 31.Stefaneanu L, Rindi G, Horvath E, Murphy D, Polak JM, Kovacs K. Morphology of adenohypophysial tumors in mice transgenic for vasopressin-SV40 hybrid oncogene. Endocrinology. 1992;130(4):1789–1795. doi: 10.1210/endo.130.4.1312426. [DOI] [PubMed] [Google Scholar]
- 32.Ang HL, Ungefroren H, De Bree F, Foo NC, Carter D, Burbach JP, Ivell R, Murphy D. Testicular oxytocin gene expression in seminiferous tubules of cattle and transgenic mice. Endocrinology. 1991;128(4):2110–2117. doi: 10.1210/endo-128-4-2110. [DOI] [PubMed] [Google Scholar]
- 33.Jeong SW, Castel M, Zhang BJ, Fields RL, Paras P, Arnheiter H, Chin H, Gainer H. Cell-specific expression and subcellular localization of neurophysin-CAT-fusion proteins expressed from oxytocin and vasopressin gene promoter-driven constructs in transgenic mice. Exp Neurol. 2001;171(2):255–271. doi: 10.1006/exnr.2001.7785. [DOI] [PubMed] [Google Scholar]
- 34.Lee JW, Park JH, Kim KS, Lee EJ, Kim MO, Kim SH, Jeong SW, Kim CW, Lee HJ, Kang KS, Chang KT, Hyun BH, Ryoo ZY. Vasopressin-SV40 T antigen expression in transgenic mice induces brain tumor and lymphoma. Biochem Biophys Res Commun. 2003;302(4):785–792. doi: 10.1016/s0006-291x(03)00225-0. [DOI] [PubMed] [Google Scholar]
- 35.Russo AF, Crenshaw EB, 3rd, Lira SA, Simmons DM, Swanson LW, Rosenfeld MG. Neuronal expression of chimeric genes in transgenic mice. Neuron. 1988;1(4):311–320. doi: 10.1016/0896-6273(88)90079-7. [DOI] [PubMed] [Google Scholar]
- 36.Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990:6679–6714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 37.Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5(5):566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 39.Habener JF, Cwikel BJ, Hermann H, Hammer RE, Palmiter RD, Brinster RL. Metallothionein-vasopressin fusion gene expression in transgenic mice. Nephrogenic diabetes insipidus and brain transcripts localized to magnocellular neurons. J Biol Chem. 1989;264(31):18844–18852. [PubMed] [Google Scholar]
- 40.Young WS, 3rd, Reynolds K, Shepard EA, Gainer H, Castel M. Cell-specific expression of the rat oxytocin gene in transgenic mice. J Neuroendocrinol. 1990;2(6):917–925. doi: 10.1111/j.1365-2826.1990.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 41.Belenky M, Castel M, Young WS, 3rd, Gainer H, Cohen S. Ultrastructural immunolocalization of rat oxytocin-neurophysin in transgenic mice expressing the rat oxytocin gene. Brain Res. 1992;583(1–2):279–286. doi: 10.1016/s0006-8993(10)80034-4. [DOI] [PubMed] [Google Scholar]
- 42.Grant FD, Reventos J, Gordon JW, Kawabata S, Miller M, Majzoub JA. Expression of the rat arginine vasopressin gene in transgenic mice. Mol Endocrinol. 1993;7(5):659–667. doi: 10.1210/mend.7.5.8100353. [DOI] [PubMed] [Google Scholar]
- 43.Waller S, Fairhall KM, Xu J, Robinson IC, Murphy D. Neurohypophyseal and fluid homeostasis in transgenic rats expressing a tagged rat vasopressin prepropeptide in hypothalamic neurons. Endocrinology. 1996;137(11):5068–5077. doi: 10.1210/endo.137.11.8895381. [DOI] [PubMed] [Google Scholar]
- 44.Zeng Q, Carter DA, Murphy D. Cell specific expression of a vasopressin transgene in rats. J Neuroendocrinol. 1994;6(5):469–477. doi: 10.1111/j.1365-2826.1994.tb00610.x. [DOI] [PubMed] [Google Scholar]
- 45.Young WS, 3rd, Iacangelo A, Luo XZ, King C, Duncan K, Ginns EI. Transgenic expression of green fluorescent protein in mouse oxytocin neurones. J Neuroendocrinol. 1999;11(12):935–939. doi: 10.1046/j.1365-2826.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhang BJ, Kusano K, Zerfas P, Iacangelo A, Young WS, 3rd, Gainer H. Targeting of green fluorescent protein to secretory granules in oxytocin magnocellular neurons and its secretion from neurohypophysial nerve terminals in transgenic mice. Endocrinology. 2002;143(3):1036–1046. doi: 10.1210/endo.143.3.8700. [DOI] [PubMed] [Google Scholar]
- 47.Ueta Y, Fujihara H, Serino R, Dayanithi G, Ozawa H, Matsuda K, Kawata M, Yamada J, Ueno S, Fukuda A, Murphy D. Transgenic expression of enhanced green fluorescent protein enables direct visualization for physiological studies of vasopressin neurons and isolated nerve terminals of the rat. Endocrinology. 2005;146(1):406–413. doi: 10.1210/en.2004-0830. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki H, Kawasaki M, Ohnishi H, Otsubo H, Ohbuchi T, Katoh A, Hashimoto H, Yokoyama T, Fujihara H, Dayanithi G, Murphy D, Nakamura T, Ueta Y. Exaggerated response of a vasopressin-enhanced green fluorescent protein transgene to nociceptive stimulation in the rat. J Neurosci. 2009;29(42):13182–13189. doi: 10.1523/JNEUROSCI.2624-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibata M, Fujihara H, Suzuki H, Ozawa H, Kawata M, Dayanithi G, Murphy D, Ueta Y. Physiological studies of stress responses in the hypothalamus of vasopressin-enhanced green fluorescent protein transgenic rat. J Neuroendocrinol. 2007;19(4):285–292. doi: 10.1111/j.1365-2826.2007.01532.x. [DOI] [PubMed] [Google Scholar]
- 50.Fields RL, House SB, Gainer H. Regulatory domains in the intergenic region of the oxytocin and vasopressin genes that control their hypothalamus-specific expression in vitro. J Neurosci. 2003;23(21):7801–7809. doi: 10.1523/JNEUROSCI.23-21-07801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10(2):302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Osten P, Grinevich V, Cetin A. Viral vectors: a wide range of choices and high levels of service. Handb Exp Pharmacol. 2007 178;:177–202. doi: 10.1007/978-3-540-35109-2_8. [DOI] [PubMed] [Google Scholar]
- 53.Papale A, Cerovic M, Brambilla R. J Neurosci Methods. 1. Vol. 185. 2009. Viral vector approaches to modify gene expression in the brain; pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 54.Lim ST, Airavaara M, Harvey BK. Viral vectors for neurotrophic factor delivery: a gene therapy approach for neurodegenerative diseases of the CNS. Pharmacol Res. 2010;61(1):14–26. doi: 10.1016/j.phrs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Pol AN, Ozduman K, Wollmann G, Ho WS, Simon I, Yao Y, Rose JK, Ghosh P. Viral strategies for studying the brain, including a replication-restricted self-amplifying delta-G vesicular stomatis virus that rapidly expresses transgenes in brain and can generate a multicolor golgi-like expression. J Comp Neurol. 2009;516(6):456–481. doi: 10.1002/cne.22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garza JC, Kim CS, Liu J, Zhang W, Lu X-Y. Adeno-associated virus-mediated knockdown of melanocortin-4 receptor in the paraventricular nucleus of the hypothalamus promotes high-fat diet-induced hyperphagia and obesity. The Journal of endocrinology. 2008;197(3):471–482. doi: 10.1677/JOE-08-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Backer MW, Fitzsimons CP, Brans MA, Luijendijk MC, Garner KM, Vreugdenhil E, Adan RA. An adeno-associated viral vector transduces the rat hypothalamus and amygdala more efficient than a lentiviral vector. BMC Neurosci. 2010:1181. doi: 10.1186/1471-2202-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bienemann AS, Martin-Rendon E, Cosgrave AS, Glover CP, Wong LF, Kingsman SM, Mitrophanous KA, Mazarakis ND, Uney JB. Long-term replacement of a mutated nonfunctional CNS gene: reversal of hypothalamic diabetes insipidus using an EIAV-based lentiviral vector expressing arginine vasopressin. Mol Ther. 2003;7(5 Pt 1):588–596. doi: 10.1016/s1525-0016(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 59.Geddes BJ, Harding TC, Hughes DS, Byrnes AP, Lightman SL, Conde G, Uney JB. Persistent transgene expression in the hypothalamus following stereotaxic delivery of a recombinant adenovirus: suppression of the immune response with cyclosporin. Endocrinology. 1996;137(11):5166–5169. doi: 10.1210/endo.137.11.8895393. [DOI] [PubMed] [Google Scholar]
- 60.Ideno J, Mizukami H, Honda K, Okada T, Hanazono Y, Kume A, Saito T, Ishibashi S, Ozawa K. Persistent phenotypic correction of central diabetes insipidus using adeno-associated virus vector expressing arginine-vasopressin in Brattleboro rats. Mol Ther. 2003;8(6):895–902. doi: 10.1016/j.ymthe.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 61.Chen H, McCarty DM, Bruce AT, Suzuki K. Gene transfer and expression in oligodendrocytes under the control of myelin basic protein transcriptional control region mediated by adeno-associated virus. Gene Ther. 1998;5(1):50–58. doi: 10.1038/sj.gt.3300547. [DOI] [PubMed] [Google Scholar]
- 62.Geller SF, Ge PS, Visel M, Greenberg KP, Flannery JG. Functional promoter testing using a modified lentiviral transfer vector. Mol Vis. 2007:13730–13739. [PMC free article] [PubMed] [Google Scholar]
- 63.Kuroda H, Kutner RH, Bazan NG, Reiser J. A comparative analysis of constitutive and cell-specific promoters in the adult mouse hippocampus using lentivirus vector-mediated gene transfer. The journal of gene medicine. 2008;10(11):1163–1175. doi: 10.1002/jgm.1249. [DOI] [PubMed] [Google Scholar]
- 64.Li C, Hirsch M, Carter P, Asokan A, Zhou X, Wu Z, Samulski RJ. A small regulatory element from chromosome 19 enhances liver-specific gene expression. Gene Ther. 2009;16(1):43–51. doi: 10.1038/gt.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B, Li J, Fu FH, Chen C, Zhu X, Zhou L, Jiang X, Xiao X. Construction and analysis of compact muscle-specific promoters for AAV vectors. Gene Ther. 2008;15(22):1489–1499. doi: 10.1038/gt.2008.104. [DOI] [PubMed] [Google Scholar]
- 66.Keir SD, House SB, Li J, Xiao X, Gainer H. Gene transfer into hypothalamic organotypic cultures using an adeno-associated virus vector. Exp Neurol. 1999;160(2):313–316. doi: 10.1006/exnr.1999.7236. [DOI] [PubMed] [Google Scholar]
- 67.Doherty FC, Schaack JB, Sladek CD. Comparison of the efficacy of four viral vectors for transducing hypothalamic magnocellular neurosecretory neurons in the rat supraoptic nucleus. J Neurosci Methods. 2011;197(2):238–248. doi: 10.1016/j.jneumeth.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Backer MW, Brans MA, Luijendijk MC, Garner KM, Adan RA. Optimization of adeno-associated viral vector-mediated gene delivery to the hypothalamus. Human gene therapy. 2010;21(6):673–682. doi: 10.1089/hum.2009.169. [DOI] [PubMed] [Google Scholar]
- 69.Burbach JP. Regulation of gene promoters of hypothalamic peptides. Front Neuroendocrinol. 2002;23(4):342–369. doi: 10.1016/s0091-3022(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 70.Adan RA, Walther N, Cox JJ, Ivell R, Burbach JP. Comparison of the estrogen responsiveness of the rat and bovine oxytocin gene promoters. Biochem Biophys Res Commun. 1991;175(1):117–122. doi: 10.1016/s0006-291x(05)81208-2. [DOI] [PubMed] [Google Scholar]
- 71.Adan RA, Cox JJ, van Kats JP, Burbach JP. Thyroid hormone regulates the oxytocin gene. J Biol Chem. 1992;267(6):3771–3777. [PubMed] [Google Scholar]
- 72.Adan RA, Cox JJ, Beischlag TV, Burbach JP. A composite hormone response element mediates the transactivation of the rat oxytocin gene by different classes of nuclear hormone receptors. Mol Endocrinol. 1993;7(1):47–57. doi: 10.1210/mend.7.1.8383287. [DOI] [PubMed] [Google Scholar]
- 73.Richard S, Zingg HH. The human oxytocin gene promoter is regulated by estrogens. J Biol Chem. 1990;265(11):6098–6103. [PubMed] [Google Scholar]
- 74.Richard S, Zingg HH. Identification of cis-acting regulatory elements in the human oxytocin gene promoter. Mol Cell Neurosci. 1991;2(6):501–510. doi: 10.1016/1044-7431(91)90017-i. [DOI] [PubMed] [Google Scholar]
- 75.Chu K, Zingg HH. Activation of the mouse oxytocin promoter by the orphan receptor RORalpha. J Mol Endocrinol. 1999;23(3):337–346. doi: 10.1677/jme.0.0230337. [DOI] [PubMed] [Google Scholar]
- 76.Burbach JP, Lopes da Silva S, Cox JJ, Adan RA, Cooney AJ, Tsai MJ, Tsai SY. Repression of estrogen-dependent stimulation of the oxytocin gene by chicken ovalbumin upstream promoter transcription factor I. J Biol Chem. 1994;269(21):15046–15053. [PubMed] [Google Scholar]
- 77.Chu K, Zingg HH. The nuclear orphan receptors COUP-TFII and Ear-2 act as silencers of the human oxytocin gene promoter. J Mol Endocrinol. 1997;19(2):163–172. doi: 10.1677/jme.0.0190163. [DOI] [PubMed] [Google Scholar]
- 78.Lipkin SM, Nelson CA, Glass CK, Rosenfeld MG. A negative retinoic acid response element in the rat oxytocin promoter restricts transcriptional stimulation by heterologous transactivation domains. Proc Natl Acad Sci U S A. 1992;89(4):1209–1213. doi: 10.1073/pnas.89.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopes da Silva S, Burbach JP. The nuclear hormone-receptor family in the brain: classics and orphans. Trends Neurosci. 1995;18(12):542–548. doi: 10.1016/0166-2236(95)98376-a. [DOI] [PubMed] [Google Scholar]
- 80.Bradley DJ, Towle HC, Young WS., 3rd Spatial and temporal expression of alpha- and beta-thyroid hormone receptor mRNAs, including the beta 2-subtype, in the developing mammalian nervous system. J Neurosci. 1992;12(6):2288–2302. doi: 10.1523/JNEUROSCI.12-06-02288.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopes da Silva S, Cox JJ, Jonk LJ, Kruijer W, Burbach JP. Localization of transcripts of the related nuclear orphan receptors COUP-TF I and ARP-1 in the adult mouse brain. Brain Res Mol Brain Res. 1995;30(1):131–136. doi: 10.1016/0169-328x(94)00289-q. [DOI] [PubMed] [Google Scholar]
- 82.Hindmarch C, Yao S, Beighton G, Paton J, Murphy D. A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc Natl Acad Sci U S A. 2006;103(5):1609–1614. doi: 10.1073/pnas.0507450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mutsuga N, Shahar T, Verbalis JG, Brownstein MJ, Xiang CC, Bonner RF, Gainer H. Selective gene expression in magnocellular neurons in rat supraoptic nucleus. J Neurosci. 2004;24(32):7174–7185. doi: 10.1523/JNEUROSCI.2022-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yue C, Mutsuga N, Verbalis J, Gainer H. Microarray analysis of gene expression in the supraoptic nucleus of normoosmotic and hypoosmotic rats. Cell Mol Neurobiol. 2006;26(4–6):959–978. doi: 10.1007/s10571-006-9017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


