Abstract
A multitude of functions have evolved around cytosine within DNA, endowing the base with physiological significance beyond simple information storage. This versatility arises from enzymes that chemically modify cytosine to expand the potential of the genome. Some modifications alter coding sequences, such as deamination of cytosine by AID/APOBEC enzymes to generate immunologic or virologic diversity. Other modifications are critical to epigenetic control, altering gene expression or cellular identity. Of these, cytosine methylation is well understood, in contrast to recently discovered modifications, such as oxidation by TET enzymes to 5-hydroxymethylcytosine. Further complexity results from cytosine demethylation, an enigmatic process that impacts cellular pluripotency. Recent insights help us to propose an integrated DNA demethylation model, accounting for contributions from cytosine oxidation, deamination and base excision repair. Taken together, this rich medley of alterations renders cytosine a genomic “wild card”, whose context-dependent functions make the base far more than a static letter in the code of life.
In poker, the rules of the game can occasionally change. Adding a “wild card” to the mix introduces a new degree of variety and presents opportunities for a skilled player to steal the pot. Given that evolution is governed by the same principles of risk and reward that are common to a poker game, it is perhaps not surprising that a genomic “wild card” has an integral role in biology.
In the conventional view, the genome is a long polymer of A, C, G and T, which together define and differentiate organisms. However, it is increasingly clear that diversity within an organism is often governed by dynamic changes that take place within this scaffold (1). Here, we make the case that cytosine is the key residue that has taken on the role of genomic “wild card” in DNA. In particular, enzymes that chemically modify cytosine introduce a physiologically important layer of complexity to the genome, beyond that seen in the primary sequence.
Remarkably, modifications of every single position in the nucleobase of purines or pyrimidines in RNA have been described (2). Cytosine, for example, can be deaminated or methylated in many different non-coding RNAs to regulate various aspects of protein translation (3, 4). The mechanisms and physiologic significance of RNA cytosine modification have been discussed elsewhere and their scope continues to expand (5, 6, 7).
It is striking that relative to RNA, modifications of nucleobases within genomic DNA have been comparatively underappreciated. In this review, we examine the curious chemistry of cytosine and the DNA modifying enzymes that change its identity (Figure 1). We begin by examining the non-canonical ways in which genomic DNA fosters adaptability and variety. To understand how cytosine is the key to generating this genomic flexibility, we describe nature’s toolbox of enzymes for modifying the nucleobase and its analogs. Numerous modifications beyond cytosine methylation are now coming to the fore, including cytosine deamination, oxidation and demethylation. We examine the common thread that runs through these modifications: by influencing the identity of cytosine, a new degree of variety can be produced.
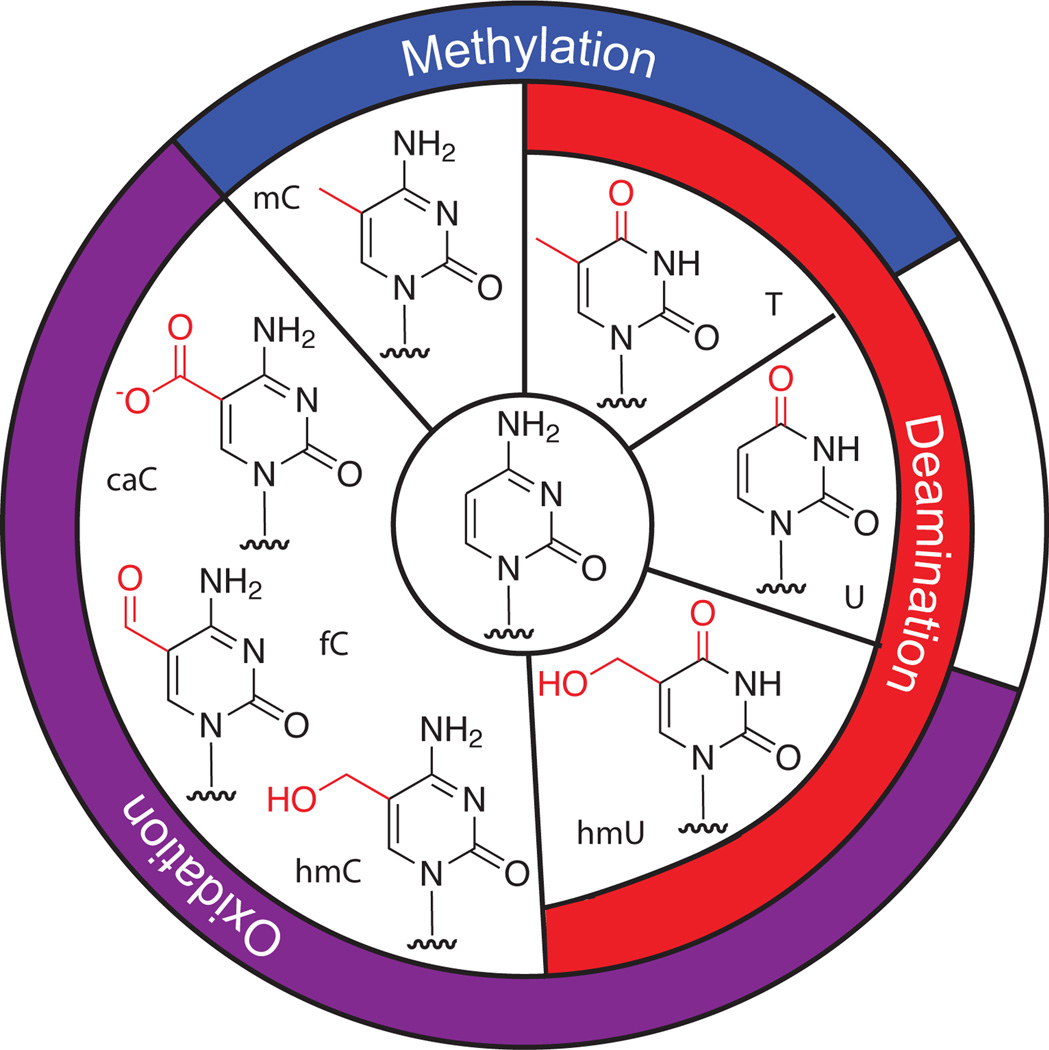
Figure 1. Cytosine as the Genomic “Wild Card”.
Within the context of the genome, cytosine can be modified by deamination, methylation, oxidation or demethylation to generate a series of analogs. In turn, these cytosine modifications influence coding sequences, gene expression and cellular identity. Amongst these analogs, enzymatic modifications can generate 5-methylcytosine (mC), 5-hydroxymethylcytosine (hmC), 5-formylcytosine (fC), 5-carboxycytosine (caC), 5-hydroxymethyluracil (hmU) and uracil (U) and thymine (T).
Adaptive Functions for the Genome
We typically think of the genome a stable, unchanging blueprint for life. However, as life demands variety and adaptability, many other “accessory” functions must also be hard-wired into the genome. For example, modification of DNA can help organisms distinguish self DNA from foreign DNA(8). In bacterial species, DNA methyltransferases have co-evolved with a partner restriction enzyme that shares the same sequence preference. Since only host DNA is methylated, this system allows for degradation of foreign DNA by the corresponding restriction enzyme. A second adaptive role for DNA is to mediate the expression or silencing of genes (9). While DNA modifications share this role with histone modification enzymes, all are needed in order to properly modulate transcriptional networks. Importantly, DNA modifying enzymes also allow for the reverse process to occur, “resetting” the genome for proper gametogenesis or reactivation of gene expression (10). Finally, the adaptive immune system demonstrates the importance of genomic malleability. The immunoglobulin (Ig) locus is a dramatic example of how the genome is pre-programmed to foster variety, through recombination and mutation that ultimately confer an adaptive advantage (11, 12).
Enzymatic Modification of Cytosine and Related Analogs
We will describe the manner in which cytosine modifications modulate genomic potential, allowing DNA to serve as a stable, but malleable, reservoir of information. In order to examine the relevant biological pathways, we must first introduce the enzymes in nature’s toolbox for altering cytosine within DNA (Figure 2).
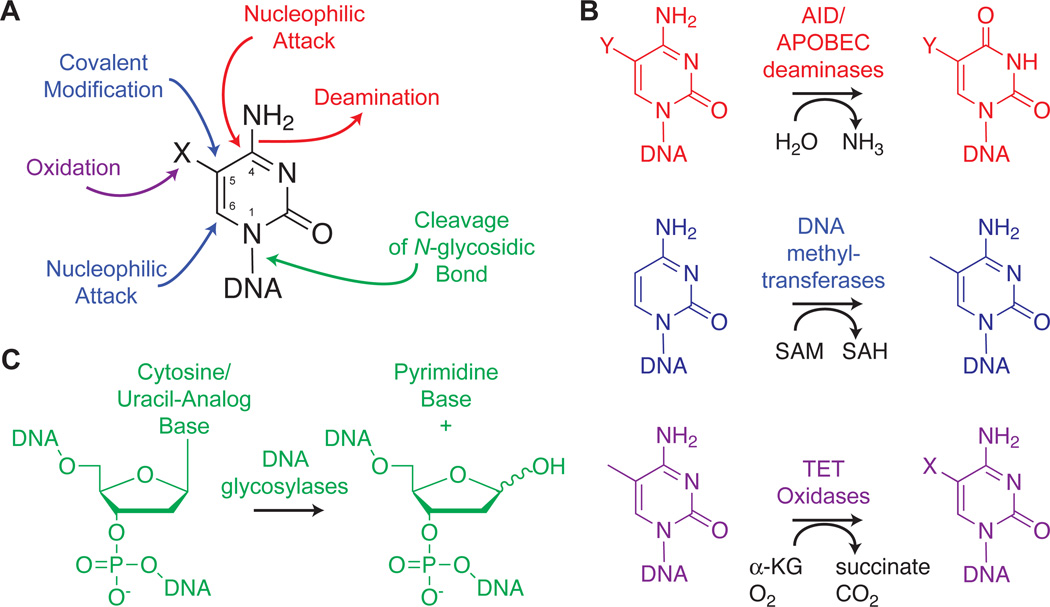
Figure 2. The Toolbox for Enzymatic Modification or Excision of Cytosine and Uracil Analogs.
(A) The cytosine nucleobase and its numbering are shown. DNA modifying enzymes target numerous positions for modification, exploiting the susceptibility of C4 or C6 to nucleophilic attack, the accessibility of C5 for alkylation or oxidation, and the cleavable sugar/base linkage for base excision repair. (B) The modifying enzymes include deaminases of the AID/APOBEC family, DNA methyltransferases and TET family oxidases. Y represents variable substitution at the 5-position of cytosine (unmodified, methyl or hydroxymethyl groups) in deamination, while X represents the variable oxidation state of the 5-methyl group in oxidation (hydroxymethyl, formyl or carboxyl groups). (C) DNA glycosylase enzymes can recognize uracil analogs and some modified cytosine bases, catalyzing hydrolysis of the N-glycosidic bond and excision of the base.
In duplex DNA, the C5 and C6 positions of cytosine lie in the major groove, unencumbered by Watson-Crick interactions. The electrophilic character of the C6 position makes it a key target of modifying enzymes. For example, DNA methyltransferases (DNMTs) transiently modify C6 by attack of an active site cysteine. Methylation results from the concerted addition of a methyl group derived from S-adenosylmethionine (SAM) to the C5 position (13, 14). The covalent intermediate breaks down, liberating the enzyme and generating genomic 5-methylcytosine (mC) (Figure 2B). Interestingly, in the absence of SAM, DNMTs can catalyze non-classical reactions, such as deamination at C4 (15, 16) or the addition of aldehydes to C5 (17), raising intriguing questions about the relevance of these non-classical functions in vivo.
The epigenetic impact of C5 methylation will be discussed later in this review, but it is important to note here that previously underappreciated oxidative modifications of mC are also possible. Physiologically, oxidation of mC is carried out by the TET family enzymes (Figure 2B), which belong to the Fe(II)/α-ketoglutarate-dependent oxygenase family that includes histone demethylases and the DNA damage repair enzyme AlkB (18, 19). Rao and colleagues initially discovered the TET family based on homology to a trypanosome enzyme known to catalyze oxidation of the exocyclic methyl group of thymine. Initially, TETs were shown to oxidize mC to 5-hydroxymethylcytosine (hmC) (18). However, more recent studies have revealed that TETs can catalyze iterative oxidation of mC. The products of iterative oxidation, 5-formylcytosine (fC) and 5-carboxylcytosine (caC), are stably detectable intermediates in genomic DNA from embryonic stem (ES) cells (20, 21). In total, the TET enzymes have provided a stable of new chemical handles whose impacts on transcriptional regulation and demethylation we will examine later in this review.
The C4 position of cytosine is relatively protected while engaged in Watson-Crick pairing, but in the context of single-stranded DNA, it becomes an important site for deamination by AID/APOBEC family enzymes (Figure 2B) (22). The mechanism of deamination involves activation of a zinc-bound water for nucleophilic attack at C4 and generation of a tetrahedral intermediate. An active site glutamate promotes deamination of C4 and the conversion of cytosine analogs into uridine analogs (23). In addition to deamination of unmodified cytosine, some studies have suggested that mC deamination can generate thymine (22, 24). However, the evidence surrounding this possibility is conflicting (25), and the full spectrum of AID/APOBEC activity against various cytosine analogs has not yet been clarified. These questions and their impact on diversity will be explored.
The distinction between genomic malleability and instability is subtle. Deamination of cytosine and 5-methylcytosine may cause transition mutations; deamination is therefore a very relevant threat to genome stability. In response, sophisticated DNA repair machinery has evolved to ensure the integrity of DNA (26), namely base excision repair enzymes (BER) and mismatch repair (MMR) enzymes. Interestingly, many of these “repair” enzymes are exploited to support cytosine’s role in generating diversity.
Several BER enzymes are worthy of particular attention, with uracil DNA glycosylase (UDG) standing out with a robust ability to excise uracil from DNA. Given the need to exclude uracil, UDG conspires with deoxyuridine triphosphatase to ensure the presence of thymine over uracil in DNA (27, 28). The only naturally occurring lesion that is efficiently targeted by UDG is uracil, though unnatural lesions such as 5-fluorouracil are also processed (29). Stringent selectivity against thymine occurs by enzymatic discrimination against bulky C5 substituents, while specific hydrogen bonding to a key active site asparagine residue selects uracil over cytosine (30, 31, 32). As we will note, in addition to its principal role in promoting DNA fidelity, UDG is exploited to generate diversity when uracil is purposefully introduced into the genome.
A second key DNA repair enzyme is thymine DNA glycosylase (TDG), which targets T:G mispairs that arise from deamination of mC in CpG motifs. Spontaneous deamination of mC produces thymine which, unlike uracil, is naturally occurring in DNA and therefore more challenging to recognize as a lesion (28). Furthermore, mC is an order of magnitude more prone to spontaneous deamination than cytosine (33, 34). These factors likely contribute to the increased mutation frequency at methylated CpG sequences in cancerous cells (35). A challenge lies in editing T:G mispairs: to repair this mutation without error, repair machinery much first recognize the mispair, and then specifically excise thymine and not guanine. TDG and the enzyme MDB4 are both capable of this activity. Mice deficient in MBD4 do exhibit increased C to T mutations and tumorigenesis (36, 37), although the embryonic lethality of the TDG knockout, and not MBD4, suggests additional important roles for TDG (38, 39).
Several features distinguish TDG from UDG. First, the enzyme actively recognizes the opposite strand G and a neighboring G, biasing activity towards T:G mismatches within CpG motifs (40). Second, the stability of the pyrimidine N-glycosidic bond, not simply the presence or absence of C5 substituents, impacts substrate preferences. In fact, TDG can not only cleave uracil-related nucleobases, but also modified cytosine residues whose N-glycosidic bond is destabilized, such as 5-fluorocytosine (41). Lastly, UDG knockout mice are viable and fertile, while the TDG knockout mice are embryonic lethal, standing as the only known DNA glycosylase with such a phenotype (38, 39, 42).
An additional BER enzyme that may contribute to diversity is single-stranded monofunctional DNA glycosylase (SMUG). This misnomer belies the fact that SMUG preferentially acts on double-stranded DNA and that it targets several uracil-related lesions (43). A water molecule adjacent to the C5 position provides a mechanism for selectively processing uracil. Intriguingly, a C5-hydroxymethyl substituent can replace this active site water (44), making 5-hydroxymethyluracil (hmU) a good substrate, with potential relevance to epigenetic reprogramming (45).
Deamination: Fostering Immunologic Diversity
The numerous DNA cytosine-modifying enzymes each play important physiologic roles in generating genomic variety. On its face, cytosine deamination is antagonistic to the primary function of DNA as a stable reservoir of information. However, when the process is highly targeted and controlled, purposeful deamination is used to yield beneficial mutations.
The foremost example of deamination as a means to diversity is demonstrated by the adaptive immune system (11, 23). The mature antibody pool is a collection of heterogeneous antigen-binding molecules produced through multiple diversity-generating mechanisms. Programmed recombination of gene segments (VDJ recombination) provides the initial repertoire of B-cells, each encoding a different surface-bound IgM molecule. However, this diversity is insufficient to yield the high-affinity interactions needed for robust immune responses. In a key transformation that occurs after exposure to antigen, B-cells in the germinal center are matured by two genome-altering processes: somatic hypermutation (SHM) and class switch recombination (CSR). In SHM, antibodies evolve from low-affinity to high-affinity by the introduction of mutations into their antigen-recognition loops at a rate 106 times that of spontaneous mutation. In CSR, the effector domain of the heavy chain is switched from IgM to yield the alternate isotypes IgA, IgE, or IgG.
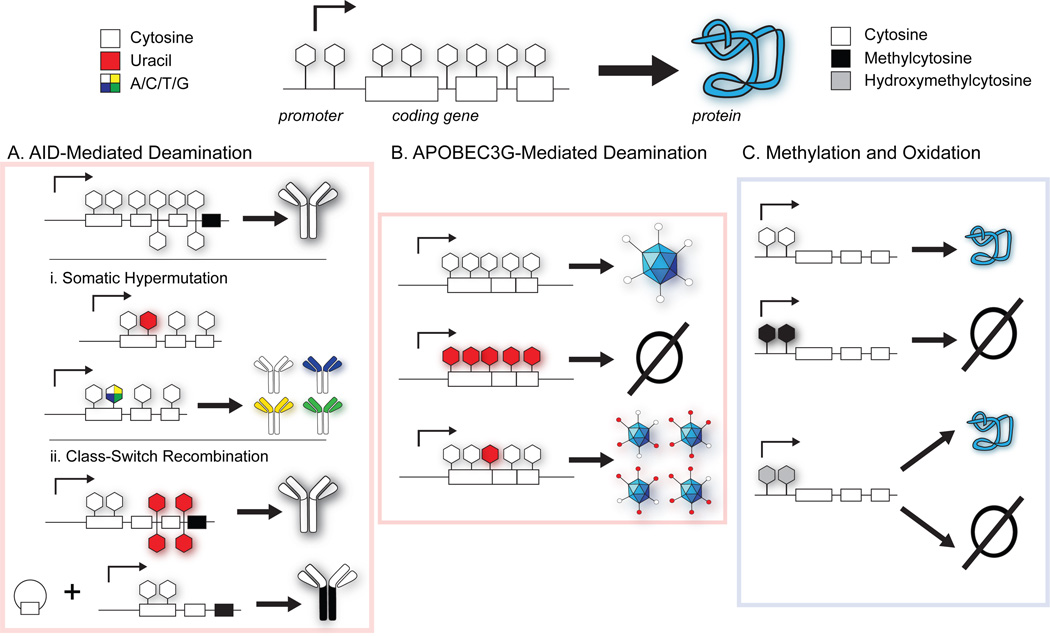
The DNA modifying enzyme activation-induced deaminase (AID) mutates key cytosines in the Ig locus to initiate the molecular events that lead to SHM or CSR (Figure 3A) (11, 23). AID expression is largely B-cell specific and restricted to germinal centers, the site of SHM and CSR (46). In SHM, AID introduces uracil into Ig locus DNA (47). The uracil lesions are then subjected to repair pathways involving UDG, mismatch repair enzymes, and low-fidelity, rather than high-fidelity, DNA polymerases, like DNA pol η. The DNA “repair” pathway is therefore co-opted to promote error-prone repair, resulting in hypermutation of antibody molecules. In CSR, AID targets cytosine residues that are on opposite strands in the switch regions immediately upstream of the various heavy chain loci encoding IgM, IgG, IgE or IgA. Clustered deamination on both DNA strands leads to double-stranded DNA breaks, which are resolved by recombination to result in isotype switching.
Figure 3. Cytosine modifications generate variety.
Cytosine typically serves as a stable reservoir of information, permitting gene expression and providing coding information. Deamination, methylation and oxidation all can alter the phenotype that results from the same starting genome. (A) Cytosine deamination in the immunoglobulin locus generates uracil. Error-prone repair of uracil results in localized mutations that increase antibody affinity in somatic hypermutation. Clustering of uracil bases leads to DNA breaks which are recombined, ultimately altering the antibody isotype. (B) Cytosine deamination of viral genomes by APOBEC3G. At high levels of deamination, retroviral restriction is achieved, while low-level mutagenesis can promote viral evolution and escape. (C) Cytosine methylation and hydroxymethylation regulate transcription. While methylation typically represses gene expression, the epigenetic role of hydroxymethylation is still being explored.
Given the fine line between genomic malleability and instability, an important factor in deamination by AID is appropriate targeting (48, 49). Hyperactive AID is associated with common oncogenic translocations as well as leukemic progression and drug resistance in chronic myeloid leukemia (50). AID is known to act throughout the genome, but preferentially acts at the Ig locus, with a balance between deamination and repair determining function (51). The mechanism by which the Ig locus is preferentially targeted remains enigmatic and is an important area of study, though some light has been shed on targeting at the local sequence level. Within the Ig locus, AID selectively targets hotspot sequences that are enriched in the antigen recognition loops and switch regions, thus promoting functional mutations over detrimental ones (52, 53).
Though AID-catalyzed SHM and CSR are exemplars of purposeful cytosine deamination, they are not the only examples. AID is closely related to APOBEC enzymes, best known for their roles in restricting retroviruses such as HIV (54). One family member, APOBEC3G (A3G), acts as a kind of Trojan horse against HIV: it can be integrated into budding HIV virions and, upon infection of a new cell, works to damage the HIV genome. A3G deaminates the (−)-strand viral cDNA generated by reverse transcription, introducing a high frequency of uracil that impairs viral integration and disrupt essential viral proteins (Figure 3B). As a counterattack measure, lentiviral pathogens express Vif, a small accessory protein that targets A3G for ubiquitination and degradation (55). Intriguingly, even in the presence of Vif, A3G is occasionally packaged at low levels into HIV. This observation raises the possibility that low levels of A3G mutagenesis may in fact confer a survival advantage to HIV by yielding viral variants that can escape immune pressure or antiviral challenges (56). Indeed, sublethal mutagenesis and robust acquisition of resistance to antivirals has been demonstrated when HIV was cultured in the presence of cellular A3G (57, 58, 59). Thus, just as our immune system exploits cytosine deamination to generate variety via AID, viral pathogens, though primarily antagonized by A3G, also are able to control the deaminase to access beneficial genomic variety.
Methylation: Establishing Diverse Cell Lineages
While cytosine DNA deamination allows for “rewriting” the genome, cytosine methylation is known to modulate gene expression and cellular identity (Figure 3C). While this modification has been well studied, in the context of considering the role of cytosine in modulating genomic potential, certain aspects of this topic are worthy of reconsideration.
Cytosine methylation upstream of transcriptional start sites is a stable chemical modification associated with transcriptional repression in eukaryotic organisms (60). Cytosine methylation occurs predominantly in the context of CpG motifs. CpG motifs are disproportionately underrepresented in the human genome, occurring four times less frequently than would be predicted by a random distribution. Further, the motifs are highly enriched in specific regions designated as CpG islands (61). The non-random distribution of potential CpG methylation sites bolsters the notion that cytosine serves an important diversity-generating function.
CpG methylation alters transcriptional repression through multiple pathways, rooted in biophysical and biochemical changes that take place in the overall DNA structure (62). DNA methylation increases the melting temperature of duplex DNA, potentially decreasing promoter accessibility to RNA polymerase (63). Further, the C5 methyl group projects into the major groove of duplex DNA, providing a biochemical handle that can be interrogated by DNA binding proteins. The impact of methylation can be direct, abrogating binding of numerous transcription factors as one means to decreasing gene expression (60). Alternatively, transcriptional repression can be indirectly affected, via methyl-DNA binding proteins that subsequently recruit histone modifying enzymes (64). Functionally, cytosine methylation can restrain the inappropriate expression of genes; thus the identity and location of the modified cytosine shapes cellular function. In embryogenesis, methylation silences the transcription of lineage-specific genes (9). Pluripotency genes are similarly methylated upon differentiation to ensure the adoption of a lineage-specific cell fate (10). Methylation also impacts imprinting, the parental-specific regulation of gene expression of autosomal transgenes and endogenous genes (65). In contrast to embryogenesis, dysregulation of methylation may result in inappropriate silencing of tumor suppressor genes (66), a process that appears widespread in cancer (67). As a whole, the chemical modification of cytosine, as governed by DNMTs, plays an essential role in dictating the phenotypic outcome of the genome in a given cell.
Oxidation: Modulating the Genome?
An additional layer of complexity was revealed by the discovery that mC may be oxidized to hmC. This modification was first identified in bacteriophage genomes as a strategy to evade bacterial restriction endonucleases (68). The epigenetic landscape changed significantly when Rao and colleagues discovered the TET family of mC oxidase enzymes in mammals (18). Further studies have demonstrated that hmC is found throughout the body, albeit at a low frequency. In tissues where hmC is most enriched, the base comprises no more than 1% of all cytosines (69, 70). Much of the focus on hmC has surrounded its presence in embryonic tissues and stem cells. Indeed, several groups have described the presence of hmC in the paternal pronucleus of the fertilized egg (71, 72), and chromatin immunoprecipitation studies have shown an association between hmC and bivalent H3K4-H3K27 histone methylation, an epigenetic hallmark of key embryonic genes (73, 74). Though it is known that hmC levels in ES cells decrease during differentiation (75, 76, 77, 78), the modulation of hmC in adult tissues remains poorly understood. Within the genome, much like mC, hmC localizes upstream of transcription start sites, but it may also be found in intragenic bodies (74, 75).
Given that the discovery of eukaryotic hmC was so recent, work is ongoing to describe its functional significance. Initial reports implicated hmC as a “poised” intermediate on the path to cytosine demethylation, a topic we tackle in the next section (18, 79). However, the current data also strongly suggest that hmC, as a stable modification of cytosine, has its own epigenetic regulatory role with respect to modulating the genome (Figure 3C). From a biophysical perspective, hmC has been shown to partially alleviate the energetic barrier for melting mC-containing duplex DNA; Tm values are similar to those of free cytosine (63, 80). However, hmC appears enriched in the promoter region of a gene, a pattern that often correlates with transcriptional repression (74). Some DNA binding proteins like MeCP2 distinguish between mC and hmC, while others, like the maintenance methyltransferase factor Uhrf1, will bind both hmC and mC (81). This implies that the information encoded by hmC may dictate chromatin structure via mechanisms distinct from mC. This notion is strengthened by the observation that TET oxidases associate with Sin3A repressor complexes and histone deacetylases (82). At this time, early reports indicate that hmC may be a stable DNA modification that, like its precursor mC, causes transcriptional repression. Currently, it is unclear what impact intragenic hmC exerts; the base may disrupt methyl-binding domain interactions that remodel euchromatin to heterochromatin (83) or may activate transcription at alternative promoters (84). Clarifying these proposed epigenetic roles of hmC, in addition to its putative role in demethylation, is an important challenge ahead.
Demethylation: Combinatorial Modifications Reveal Genomic Potential
Cytosine methylation is critical for gene imprinting and cell lineage specification, as discussed above. The reverse of this process – the removal of the methyl group – allows cells to newly express previously repressed genes or to recover their totipotent potential. Until recently, this process of cytosine demethylation was thought to be a passive process in which replication without the action of maintenance DNMTs dilutes mC from DNA. However, mounting evidence suggests that replication-independent, “active” (enzymatic) demethylation occurs globally in totipotent cells (85, 86) and also in a locus-specific fashion within somatic cells (87, 88, 89, 90, 91). Active cytosine demethylation, therefore, has now been recognized as a crucial molecular process and is yet another example of the role of cytosine in modulating genomic potential.
Cytosine demethylation is relevant even at the earliest stages of mammalian development. Upon penetrating the zona pelucida, the paternal pronucleus is rapidly demethylated (85). Remarkably, the maternal pronucleus sits in the same cytoplasm and is exclusively demethylated via passive demethylation; the mechanism for such asymmetric demethylation remains unclear. Beyond the zygote and blastula stages, a subset of cells are induced to travel to the gonadal ridge and become primordial germ cells (PGCs). Although PGC genomes are widely methylated at the time they are designated, they are globally demethylated by the time they arrive at the gonadal ridge several days later (92). Given that maintenance DNMTs are expressed in PGCs, such global demethylation is assumed to require active demethylation.
Several examples of locus-specific active demethylation suggest that this process is likewise important in the normal functioning of somatic cells. Fast methylation and demethylation cycling at the estrogen receptor promoter provide a notable example of locus-specific active demethylation (88, 89). Other studies in CD8+ T-cells illustrated that expression of IL-2 can be induced via replication-independent demethylation, suggesting a role for active demethylation in sustained immune responses (90). Finally, even neural plasticity is impacted by active demethylation as evidenced by changes at the promoter for brain-derived neurotrophic factor (91).
Although active demethylation is increasingly accepted as an important physiological process, its molecular basis remains controversial. Several DNA glycosylases have been described in Arabidopsis that can excise mC specifically; however, mammals appear to lack this activity (93). In the past several years, a wealth of new evidence has implicated several of the key cytosine modifying enzymes we have reviewed, particularly the AID/APOBEC deaminases, TET oxidases, and DNA glycosylases (94, 95, 96). Two major types of models have emerged: a deamination-initiated pathway (97, 98) and several variants of an oxidation-initiated pathway (17, 20, 21, 45, 95) (Figure 4).
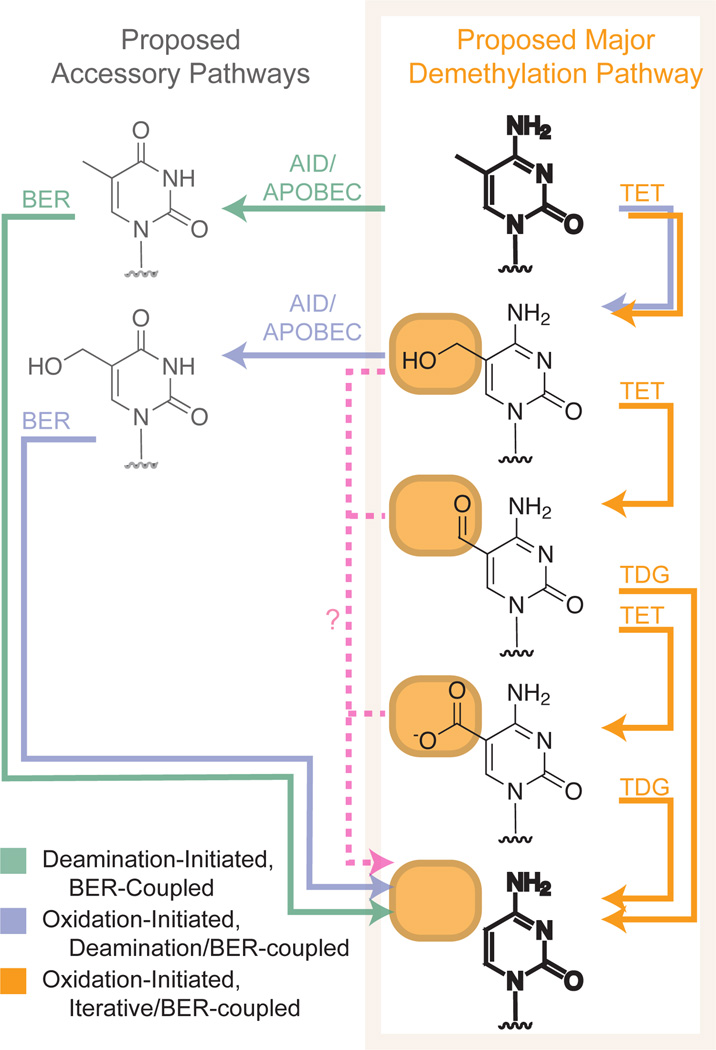
Figure 4. Integrated Model for Cytosine Demethylation.
Numerous mechanisms have been proposed for DNA demethylation, in which 5-methylcytosine (bold, top right) is converted to cytosine (bold, bottom right). Current evidence supports the existence of an iterative oxidation, BER-coupled pathway (orange) in embryonic stem cells. Though some evidence exists in favor of deamination-initiated, BER-coupled repair (green) and oxidation-initiated, deamination/BER-coupled (purple) pathways, important shortcomings of these routes make them more likely to serve accessory or tissue-specific roles. Enzymes which might directly removed the oxidized 5-substituent from intermediates in demethylation are possible but none have yet been clearly identified (pink).
In the deamination-initiated pathway, mC is first deaminated by an AID/APOBEC family member to yield thymine. The BER pathway subsequently recognizes the T:G mismatch and reverts the lesion to an unmodified cytosine. In support of the role AID/APOBEC enzymes may play in demethylation, AID-deficient PGCs were found to be more methylated than wild-type PGCs in a mouse model (99). In zebrafish embryos, coexpression of multiple AID/APOBEC members along with MBD4 caused global demethylation of the genome (100). AID was also shown to contribute to demethylation at key pluripotency loci such as the Nanog and Oct4 promoters in a heterokaryon system used to generate stem cells (101). Recent evidence that a TDG knockout is embryonic lethal supports the deamination-initiated pathway (38, 39), although not to the exclusion of the oxidation-initiated pathway, as we note below.
Several factors suggest that the deamination-initiated pathway is insufficient to fully explain demethylation, although this mechanism may indeed be an important accessory pathway towards that end. Deletion of AID is not embryonic lethal, as would be expected if this were the sole pathway for active demethylation (99). It is also hard to reconcile a prominent, genome-wide activity for AID with its known properties at the molecular level. While AID has indeed been shown to act outside of the Ig locus, this occurs several orders of magnitude less frequently than within the Ig locus (51). Furthermore, AID/APOBEC enzymes preferentially act on single-stranded DNA in particular sequence contexts (22, 52, 53), but most methylated, silenced loci are likely to be double-stranded in CpG contexts. In addition, although deaminases have been suggested to deaminate mC (24), such activity on mC is diminished relative to activity on cytosine (22). Therefore, the deamination-initiated pathway, although likely relevant in some instances, may not represent the major mechanism for demethylation.
The discovery of genomic hmC raised the possibility of oxidation-first pathways to demethylation (18). Despite the ongoing controversy, several observations bolster support for an oxidation-initiated mechanism. The striking prevalence of hmC in promoters suggests that TET oxidation of mC is likely to be an important step in demethylation (74, 75). TET knockdown in ES cells may decrease expression at loci involved in pluripotency, including Nanog (73, 74, 79, 82), and promoters undergoing active demethylation have also demonstrated a physiological association with TET (45). Finally, TET has also been shown to have a preference for binding at CpG nucleotides, where methylation is most relevant in humans (73, 82).
The route from hmC to cytosine is still under debate, but several potential pathways are worthy of consideration. These pathways can be characterized as deamination-coupled, BER-coupled or direct-reversion mechanisms. As yet, an enzyme capable of direct removal of the hydroxymethyl group from the 5-position of the base (dehydroxymethylation) has not been discovered; however, this is a mechanistically feasible reaction . Alternatively, hmC could be deaminated by AID/APOBEC enzymes to yield hmU, subsequently removed by an enzyme such as SMUG or TDG (41, 45). In this system, suggested to be active in neurons, overexpression of AID decreased endogenous hmC levels and both TET and AID contributed to demethylation at several neuron-specific promoters, although overall levels of demethylation were low (45). However, this proposed model relies on assumptions about the ability of AID/APOBEC enzymes to efficiently deaminate hmC. This activity has not yet been established, nor has sequencing revealed the presence of hmU as a detectable demethylation intermediate, although efficient removal of hmU from the genome may explain the latter point.
A more recent model for efficient demethylation integrates several observations into a more appealing mechanism involving iterative oxidation directly coupled to BER. In several recent reports, the higher oxidation products of hmC, 5-formylcytosine (fC) and 5-carboxylcytosine (caC), were detected in the genome of ES cells (20, 21, 102). Furthermore, it was shown that fC and caC directly result from iterative oxidation of mC by TETs (20, 21). Based on the precedent of a related enzyme in pyrimidine salvage, Zhang and colleagues have proposed that an undiscovered decarboxylase could catalyze the regeneration of cytosine from caC (20). While the search for such an activity could be justified, support for a much more appealing model comes from He et. al. who revisit the dependence of demethylation on BER (103). These authors looked for DNA glycosylase activity against the higher oxidation products of mC. They found that the BER enzyme TDG recognizes and excises the highly oxidized caC nucleobase (21). Notably, no such activity was detected with MDB4. In line with their proposal, knockdown of TDG leads to an accumulation of caC in the genome of ES cells, while conversely TDG overexpression decreases caC content. An independent report from Maiti and Drohat has also subsequently confirmed that TDG excises fC and caC, while leaving hmC untouched (104). This proposed mechanism is consistent with the observation that TDG deficiency is embryonic lethal and leads to perturbed methylation patterns in embryogenesis (38, 39). While it has been assumed previously that a role for TDG in demethylation implicates a deamination-mediated pathway, this need not be the case; TDG can directly excise cytosine bases with weakened N-glycosidic bonds, as would likely be the case for fC and caC.
Although the field itself is rapidly evolving, we propose that these apparently disparate studies invoking deamination, oxidation and BER can be integrated into a more coherent model (Figure 4) (105). A gathering body of evidence supports important roles for the various TET isoforms in physiological niches where DNA demethylation is thought to be relevant. Though much remains to be resolved, disrupting expression leads to perturbed demethylation of paternal paternal pronuclei and embryonic demise in the case of TET3 (106), dysregulation of hematopoiesis in the case of TET2 (107, 108) and diminished embryonic growth of viable offspring in the case TET1 (109). These genetic findings couple with the biochemical studies to make a case for the TET enzymes as major regulators of DNA demethylation. We therefore suggest that an iterative oxidation-initiated/BER-coupled pathway could be a major route to demethylation, but that deaminase enzymes could serve an important accessory role to accelerate demethylation in certain physiological settings. This could occur because deamination would generate a uracil-related base, rather than a cytosine-related base, and the relevant BER enzymes are more efficient in excision of the products of deamination. This paradigm could explain the apparent contribution of deamination in heterokaryon systems (101), neurons (45), or settings where AID/APOBEC enzymes are overexpressed (45, 100). Together, a model invoking both major and accessory pathways accounts for the observations that TET, AID/APOBEC enzymes and BER enzymes all appear to contribute to demethylation, but that a predominant pathway is required in the setting of embryogenesis, where demethylation is critical to proper development and differentiation.
While the current evidence suggests that an iterative oxidation/TDG-coupled pathway plays a major role in cytosine demethylation, the model is far from resolved and several major gaps remain in our understanding (105). For instance, hmC accumulates to higher levels than fC and caC; what controls the extent of oxidative modification by TET? Next, although Xu and colleagues (21) propose a model where caC is the intermediate just prior to BER, Maiti and Drohat observe that fC is a better substrate for TDG than caC (104). What is the final oxidation intermediate prior to BER? Further, if BER is involved in lesion recognition, the process of reversion to cytosine would generate a basic sites and DNA nicks. Given the high load of lesions that would result from DNA cytosine methylation in CpG islands, how is genomic instability averted? There are also fundamental questions that remain regarding the proposed deamination-mediated, accessory pathway. For example, the biochemical plausibility of cytosine analogs as substrates for deamination by AID/APOBEC enzymes remains largely unassessed. Addressing these open questions will be essential to the ongoing debate over the mechanism of demethylation.
Appraising the Wild Card: Future Directions
Adaptability is essential to life, but it is counterbalanced by the need for genomic stability. We have made the case that cytosine modification provides mechanisms for adaptation, thus increasing the potential of the genome. Deamination of cytosine contributes to genetic variability by promoting purposeful mutations, as evidenced in the maturation of immune responses. Cytosine methylation or oxidation refines the genome by tailoring a gene program to a given cell lineage or altering gene expression in the face of environmental changes. Finally, multiple DNA-modifying pathways appear to collaborate to carry out cytosine demethylation, helping to establish a totipotent state in cells otherwise marked by methylation.
Although the a unique role for cytosine is increasingly evident, there are pressing questions that need to be explored. It is not immediately clear why cytosine is the base endowed with a special role in diversity generation. It is tempting to speculate that the pyrimidine base’s reactivity, coupled with thymine’s previously designated role in segregating DNA from RNA, allowed cytosine to fill this other niche. What is abundantly clear from the recent discovery of hmC, fC and caC is that the scope of cytosine modification is greater than previously appreciated. High sensitivity mass spectrometry has been key to the identification of novel DNA modifications, justifying an aggressive search for other such modifications (69, 102, 110). Given the advances in metabolomics, the use of labeled metabolites may provide additional mechanisms for detecting and tracking new DNA modifications.
Secondly, there are now several precedents to suggest we need to reevaluate the scope of reactions catalyzed by known DNA cytosine-modifying enzymes. TET enzymes, thought to catalyze hmC generation alone, now have been shown to produce fC and caC (20, 95); TDG, thought to act only on uracil analogs, can also excise oxidized cytosine analogs (21, 41, 104); and DNMT enzymes, thought to only catalyze methylation, can also add aldehydes (17). Resolving the complete catalytic repertoire of known DNA modifying enzymes is an important next step.
Thirdly, we should reinvigorate the search for novel enzymes that modify DNA, such as the proposed decarboxylase for caC (20, 95). Several appealing leads have already been suggested by bioinformatic analysis focused on discovering proteins with DNA-binding domains linked to known nucleotide modifying domains (111). New insights may also come from classical biochemical approaches for discovering proteins that interact specifically with DNA containing modified nucleobases.
Finally, and perhaps most critically, we are in need of novel chemical biology tools to detect site-specific modifications. Despite the wealth of information gained from methods such as bisulfite sequencing, we now know that these data need to be reinterpreted in the context of newly discovered modifications (112, 113). Several new methods have been developed to detect hmC in the genome, such as differential modification by glucosyltransferases, specific recognition of hmC and its adducts, and analysis of distinct electrical properties of modified DNA using nanopores (70, 74, 80, 114). Similar approaches are needed to fully catalog the products of deamination, iterative oxidation, and other modifications in the genome. Further, to assess the biological impact of these bases, we need methods to site-specifically control the identity of cytosine within the genome. We have tools to alter proteins within the complex milieu of the cell, but lack similar methods to explore the nature of the dynamic genome at the DNA level (1). With novel approaches at hand, we anticipate that fundamental insights into evolution and adaptation will come from exploring the “wild card” function of cytosine in the genome.
Acknowledgements
We are grateful to L. C. Wang, D. J. Krosky and C. F. Meyers for thoughtful commentary on the manuscript. R. M. Kohli is supported by the Rita Allen Foundation, the W. W. Smith Charitable Trust, and by an NIH/NIAID career development award (K08-AI089242).
Keywords
- Genomic Potential
Genomic potential refers to the number of different phenotypic outcomes that can result from the same starting template genome. These include changes in the protein coding sequence as well as changes in transcription of particular genes or pathways. Cytosine modifications can mediate both types of genomic variation.
- Cytosine Deamination
When the exocyclic amino group of cytosine is removed by hydrolytic deamination, catalyzed by the AID/APOBEC family of enzymes, a cytosine analog is changed into a uracil analog. Deamination is important in immune-pathogen interactions and may play a role in active DNA demethylation.
- Cytosine Methylation
DNA methyltransferase enzymes introduce a methyl group at the C5 position of cytosine to generate 5-methylcytosine. This modification is well understood to lead to transcriptional repression.
- Cytosine Oxidation
The important epigenetic base 5-methylcytosine can be oxidized by TET family enzymes at the exocyclic methyl group to generate 5-hydroxymethylcytosine and higher oxidation products. These modifications are stably detectable in the genome and play a role in regulating gene expression and cellular identity.
- Base Excision Repair
This DNA repair process is initiated by a DNA glycosylase that breaks the N-glycosidic bond between the sugar and the nucleobase, excising unwanted nucleobases. The product is called an abasic site and can be processed by an enzymatic pathway that restores an unmodified base at the site of excision.
- Active DNA Demethylation
Demethylation of cytosine residues that is carried out by an enzymatic pathway that acts independent of DNA replication during cellular division. This term stands in contrast to passive demethylation, where methylated DNA is diluted through rounds of replication in the absence of maintenance DNA methyltransferases.
References
- 1.Kohli RM. Grand Challenge Commentary: The Chemistry of a Dynamic Genome. Nat. Chem. Biol. 2010;6:866–868. doi: 10.1038/nchembio.471. [DOI] [PubMed] [Google Scholar]
- 2.Grosjean H. Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour. In: Grosjean H, editor. DNA and RNA Modification Enzymes. Austin: Landes Bioscience; 2009. pp. 1–18. [Google Scholar]
- 3.Gerber AP, Keller W. RNA Editing by Base Deamination: More Enzymes, More Targets, New Mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 4.Motorin Y, Lyko F, Helm M. 5-Methylcytosine in RNA: Detection, Enzymatic Formation and Biological Functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gott JM, Emeson RB. Functions and Mechanisms of RNA Editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 6.Ishitani R, Yokoyama S, Nureki O. Structure, Dynamics, and Function of RNA Modification Enzymes. Curr. Opin. Struct. Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 7.He C. Grand Challenge Commentary: RNA Epigenetics? Nat. Chem. Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 8.Bickle TA, Kruger DH. Biology of DNA Restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christophersen NS, Helin K. Epigenetic Control of Embryonic Stem Cell Fate. J. Exp. Med. 2010;207:2287–2295. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng S, Jacobsen SE, Reik W. Epigenetic Reprogramming in Plant and Animal Development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of Activation-Induced Cytidine Deaminase, the Engraver of Antibody Memory. Adv. Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 12.Bassing CH, Swat W, Alt FW. The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell. 2002;109 Suppl:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 13.Santi DV, Garrett CE, Barr PJ. On the Mechanism of Inhibition of DNA-Cytosine Methyltransferases by Cytosine Analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 14.Goll MG, Bestor TH. Eukaryotic Cytosine Methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 15.Yebra MJ, Bhagwat AS. A Cytosine Methyltransferase Converts 5-Methylcytosine in DNA to Thymine. Biochemistry. 1995;34:14752–14757. doi: 10.1021/bi00045a016. [DOI] [PubMed] [Google Scholar]
- 16.Shen JC, Rideout WM, 3rd, Jones PA. High Frequency Mutagenesis by a DNA Methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 17.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-Methyltransferases Add Aldehydes to DNA. Nat. Chem. Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 18.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loenarz C, Schofield CJ. Expanding Chemical Biology of 2-Oxoglutarate Oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 20.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet Proteins can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-Mediated Formation of 5-Carboxylcytosine and its Excision by TDG in Mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-Induced Cytidine Deaminase Deaminates Deoxycytidine on Single-Stranded DNA but Requires the Action of RNase. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The Biochemistry of Somatic Hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 24.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-Induced Cytidine Deaminase Deaminates 5-Methylcytosine in DNA and is Expressed in Pluripotent Tissues: Implications for Epigenetic Reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 25.Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, Scharff MD, Martin A. Methylation Protects Cytidines from AID-Mediated Deamination. Mol. Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--Occurrence, Consequences and Repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 27.Olinski R, Jurgowiak M, Zaremba T. Uracil in DNA--its Biological Significance. Mutat. Res. 2010;705:239–245. doi: 10.1016/j.mrrev.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Poole A, Penny D, Sjoberg BM. Confounded Cytosine! Tinkering and the Evolution of DNA. Nat. Rev. Mol. Cell Biol. 2001;2:147–151. doi: 10.1038/35052091. [DOI] [PubMed] [Google Scholar]
- 29.Grogan BC, Parker JB, Guminski AF, Stivers JT. Effect of the Thymidylate Synthase Inhibitors on dUTP and TTP Pool Levels and the Activities of DNA Repair Glycosylases on Uracil and 5-Fluorouracil in DNA. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savva R, McAuley-Hecht K, Brown T, Pearl L. The Structural Basis of Specific Base-Excision Repair by Uracil-DNA Glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 31.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal Structure and Mutational Analysis of Human Uracil-DNA Glycosylase: Structural Basis for Specificity and Catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 32.Stivers JT, Drohat AC. Uracil DNA Glycosylase: Insights from a Master Catalyst. Arch. Biochem. Biophys. 2001;396:1–9. doi: 10.1006/abbi.2001.2605. [DOI] [PubMed] [Google Scholar]
- 33.Duncan BK, Miller JH. Mutagenic Deamination of Cytosine Residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Mathews CK. Effect of DNA Cytosine Methylation upon Deamination-Induced Mutagenesis in a Natural Target Sequence in Duplex DNA. J. Biol. Chem. 1994;269:7066–7069. [PubMed] [Google Scholar]
- 35.Cooper DN, Youssoufian H. The CpG Dinucleotide and Human Genetic Disease. Hum. Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 36.Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG Mutability and Tumorigenesis in MBD4-Deficient Mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 37.Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, Edelmann W. Mbd4 Inactivation Increases C->T Transition Mutations and Promotes Gastrointestinal Tumor Formation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic Lethal Phenotype Reveals a Function of TDG in Maintaining Epigenetic Stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 39.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA Glycosylase is Essential for Active DNA Demethylation by Linked Deamination-Base Excision Repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal Structure of Human Thymine DNA Glycosylase Bound to DNA Elucidates Sequence-Specific Mismatch Recognition. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of Human Thymine DNA Glycosylase Depends on N-Glycosidic Bond Stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA Glycosylase (UNG)-Deficient Mice Reveal a Primary Role of the Enzyme during DNA Replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 43.Liu P, Burdzy A, Sowers LC. Substrate Recognition by a Family of Uracil-DNA Glycosylases: UNG, MUG, and TDG. Chem. Res. Toxicol. 2002;15:1001–1009. doi: 10.1021/tx020030a. [DOI] [PubMed] [Google Scholar]
- 44.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase SMUG1. Mol. Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 45.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class Switch Recombination and Hypermutation Require Activation-Induced Cytidine Deaminase (AID), a Potential RNA Editing Enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 47.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil Residues Dependent on the Deaminase AID in Immunoglobulin Gene Variable and Switch Regions. Nat. Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavri R, Nussenzweig MC. AID Targeting in Antibody Diversity. Adv. Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 49.Kothapalli NR, Fugmann SD. Targeting of AID-Mediated Sequence Diversification to Immunoglobulin Genes. Curr. Opin. Immunol. 2011;23:184–189. doi: 10.1016/j.coi.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, Hofmann WK, Jumaa H, Groffen J, Heisterkamp N, Martinelli G, Lieber MR, Casellas R, Muschen M. The B Cell Mutator AID Promotes B Lymphoid Blast Crisis and Drug Resistance in Chronic Myeloid Leukemia. Cancer. Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two Levels of Protection for the B Cell Genome during Somatic Hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 52.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A Portable Hotspot Recognition Loop Transfers Sequence Preferences from APOBEC Family Members to Activation-Induced Cytidine Deaminase. J. Biol. Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohli RM, Maul RW, Guminski AF, McClure RL, Gajula KS, Saribasak H, McMahon MA, Siliciano RF, Gearhart PJ, Stivers JT. Local Sequence Targeting in the AID/APOBEC Family Differentially Impacts Retroviral Restriction and Antibody Diversification. J. Biol. Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and Related Cytidine Deaminases in the Host Response to Viral Infection. Adv. Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 55.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and Intrinsic Immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillai SK, Wong JK, Barbour JD. Turning Up the Volume on Mutational Pressure: Is More of a Good Thing always Better? (A Case Study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G Contributes to HIV-1 Variation through Sublethal Mutagenesis. J. Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. Human APOBEC3G-Mediated Editing can Promote HIV-1 Sequence Diversification and Accelerate Adaptation to Selective Pressure. J. Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mulder LC, Harari A, Simon V. Cytidine Deamination Induced HIV-1 Drug Resistance. Proc. Natl. Acad. Sci. U.S. A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klose RJ, Bird AP. Genomic DNA Methylation: The Mark and its Mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Deaton AM, Bird A. CpG Islands and the Regulation of Transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuks F. DNA Methylation and Histone Modifications: Teaming Up to Silence Genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 63.Thalhammer A, Hansen AS, El-Sagheer AH, Brown T, Schofield CJ. Hydroxylation of Methylated CpG Dinucleotides Reverses Stabilisation of DNA Duplexes by Cytosine 5-Methylation. Chem. Commun. (Camb) 2011;47:5325–5327. doi: 10.1039/c0cc05671e. [DOI] [PubMed] [Google Scholar]
- 64.Li E. Chromatin Modification and Epigenetic Reprogramming in Mammalian Development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Sasaki H. Genomic Imprinting in Mammals: Its Life Cycle, Molecular Mechanisms and Reprogramming. Cell Res. 2011;21:466–473. doi: 10.1038/cr.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herman JG, Baylin SB. Gene Silencing in Cancer in Association with Promoter Hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 67.Tsai HC, Baylin SB. Cancer Epigenetics: Linking Basic Biology to Clinical Medicine. Cell Res. 2011;21:502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyatt GR, Cohen SS. The Bases of the Nucleic Acids of some Bacterial and Animal Viruses: The Occurrence of 5-Hydroxymethylcytosine. Biochem. J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective Chemical Labeling Reveals the Genome-Wide Distribution of 5-Hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the Mammalian Zygote is Linked with Epigenetic Reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 72.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the Paternal Genome upon Fertilization Involves Genome-Wide Oxidation of 5-Methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual Functions of Tet1 in Transcriptional Regulation in Mouse Embryonic Stem Cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-Wide Mapping of 5-Hydroxymethylcytosine in Embryonic Stem Cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic Regulation of 5-Hydroxymethylcytosine in Mouse ES Cells and during Differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 76.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive Enzymatic Quantification of 5-Hydroxymethylcytosine in Genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruzov A, Tsenkina Y, Serio A, Dudnakova T, Fletcher J, Bai Y, Chebotareva T, Pells S, Hannoun Z, Sullivan G, Chandran S, Hay DC, Bradley M, Wilmut I, De Sousa P. Lineage-Specific Distribution of High Levels of Genomic 5-Hydroxymethylcytosine in Mammalian Development. Cell Res. 2011 doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet Proteins in 5mC to 5hmC Conversion, ES-Cell Self-Renewal and Inner Cell Mass Specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of Methylcytosine from Hydroxymethylcytosine in DNA Molecules. J. Am. Chem. Soc. 2010;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-Hydroxymethylcytosine by the Uhrf1 SRA Domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and Hydroxymethylcytosine in Transcription and DNA Methylation Fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-Wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved Role of Intragenic DNA Methylation in Regulating Alternative Promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the Zygotic Paternal Genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 86.Yamazaki Y, Mann MR, Lee SS, Marh J, McCarrey JR, Yanagimachi R, Bartolomei MS. Reprogramming of Primordial Germ Cells Begins before Migration into the Genital Ridge, Making these Cells Inadequate Donors for Reproductive Cloning. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. DNA Demethylation in Hormone-Induced Transcriptional Derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 88.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient Cyclical Methylation of Promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 89.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA Methylation of a Transcriptionally Active Promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 90.Bruniquel D, Schwartz RH. Selective, Stable Demethylation of the Interleukin-2 Gene Enhances Transcription by an Active Process. Nat. Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 91.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent BDNF Gene Regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 92.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic Reprogramming in Mouse Primordial Germ Cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 93.Gehring M, Reik W, Henikoff S. DNA Demethylation by DNA Repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 94.Ooi SK, Bestor TH. The Colorful History of Active DNA Demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 95.Wu SC, Zhang Y. Active DNA Demethylation: Many Roads Lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu JK. Active DNA Demethylation Mediated by DNA Glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fritz EL, Papavasiliou FN. Cytidine Deaminases: AIDing DNA Demethylation? Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chahwan R, Wontakal SN, Roa S. Crosstalk between Genetic and Epigenetic Information through Cytosine Deamination. Trends Genet. 2010;26:443–448. doi: 10.1016/j.tig.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 99.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-Wide Erasure of DNA Methylation in Mouse Primordial Germ Cells is Affected by AID Deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA Demethylation in Zebrafish Involves the Coupling of a Deaminase, a Glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming Towards Pluripotency Requires AID-Dependent DNA Demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angew. Chem. Int. Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 103.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-Wide Reprogramming in the Mouse Germ Line Entails the Base Excision Repair Pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maiti A, Drohat AC. Thymine DNA Glycosylase can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: Potential Implications for Active Demethylation of CpG Sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nabel CS, Kohli RM. Demystifying DNA Demethylation. Science. 2011;333:1229–1230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 106.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The Role of Tet3 DNA Dioxygenase in Epigenetic Reprogramming by Oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 107.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 Loss Leads to Increased Hematopoietic Stem Cell Self-Renewal and Myeloid Transformation. Cancer. Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and is a Recurrent Event during Human Lymphomagenesis. Cancer. Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 109.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is Dispensable for Maintaining Pluripotency and its Loss is Compatible with Embryonic and Postnatal Development. Cell. Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Munzel M, Globisch D, Carell T. 5-Hydroxymethylcytosine, the Sixth Base of the Genome. Angew. Chem. Int. Ed Engl. 2011;50:6460–6468. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- 111.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of Novel Families of Enzymes Involved in Oxidative and Other Complex Modifications of Bases in Nucleic Acids. Cell. Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The Behaviour of 5-Hydroxymethylcytosine in Bisulfite Sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin SG, Kadam S, Pfeifer GP. Examination of the Specificity of DNA Methylation Profiling Techniques Towards 5-Methylcytosine and 5-Hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nomura A, Sugizaki K, Yanagisawa H, Okamoto A. Discrimination between 5-Hydroxymethylcytosine and 5-Methylcytosine by a Chemically Designed Peptide. Chem. Commun. (Camb) 2011;47:8277–8279. doi: 10.1039/c1cc12131f. [DOI] [PubMed] [Google Scholar]