Abstract
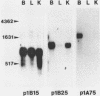
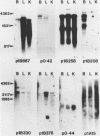
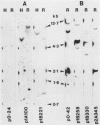
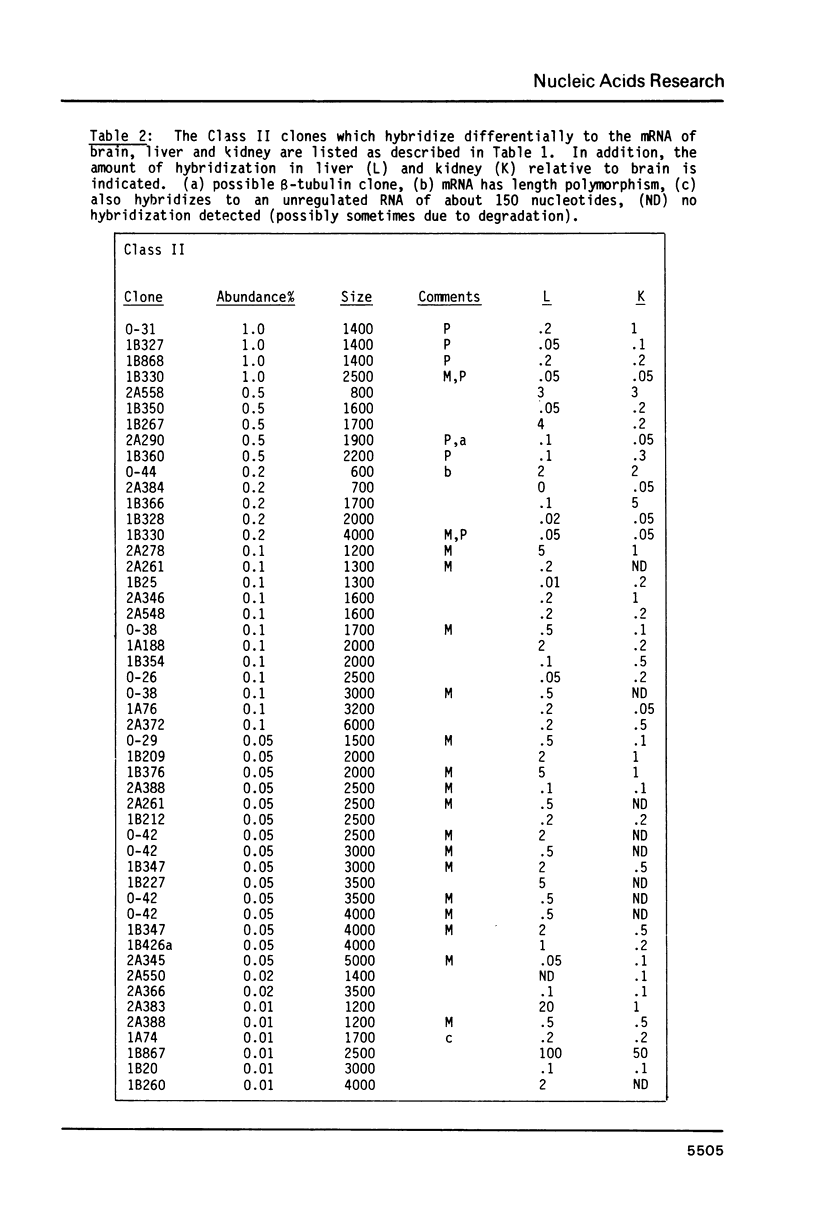
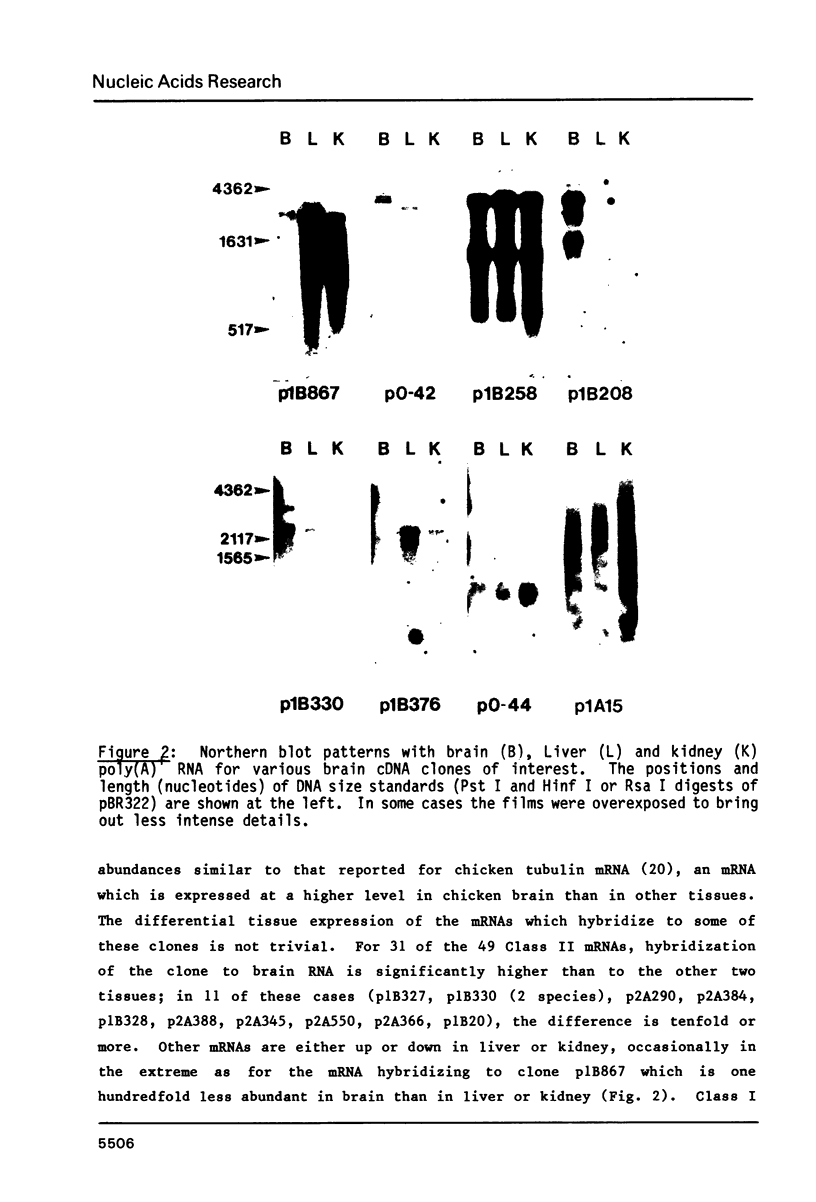
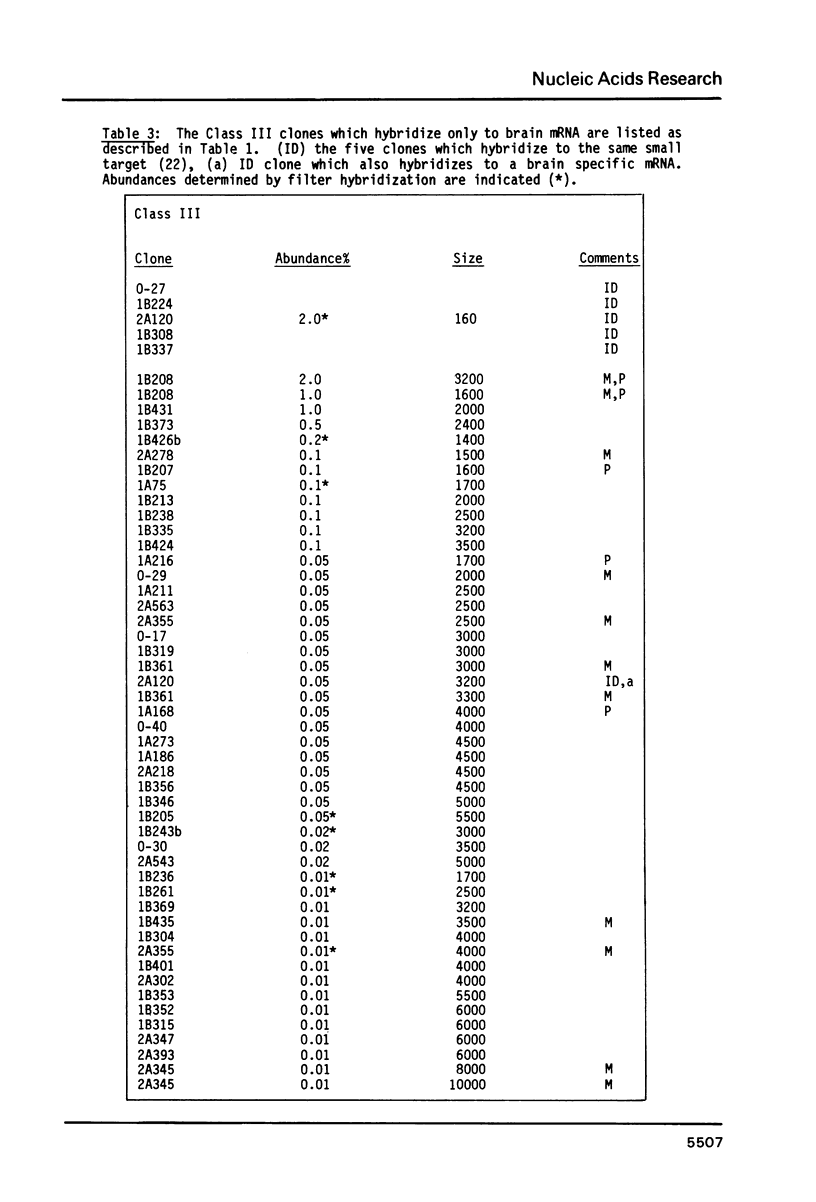
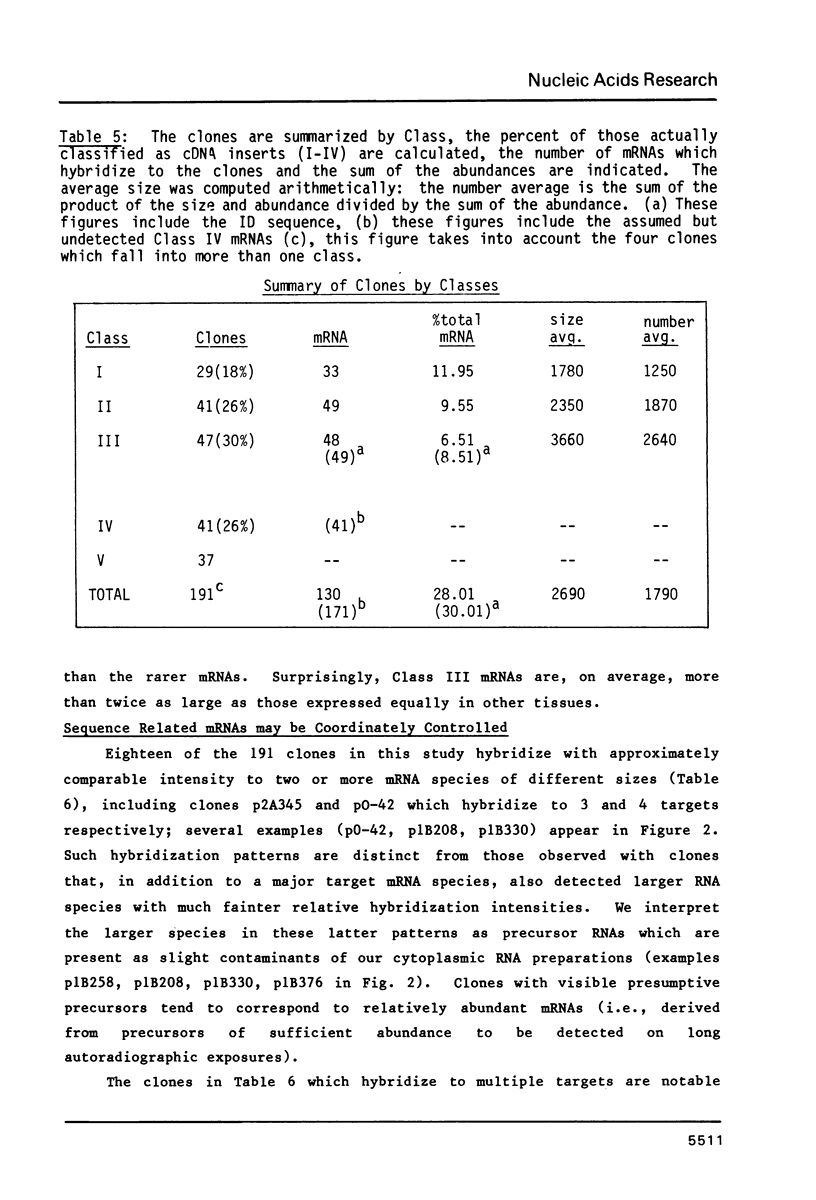
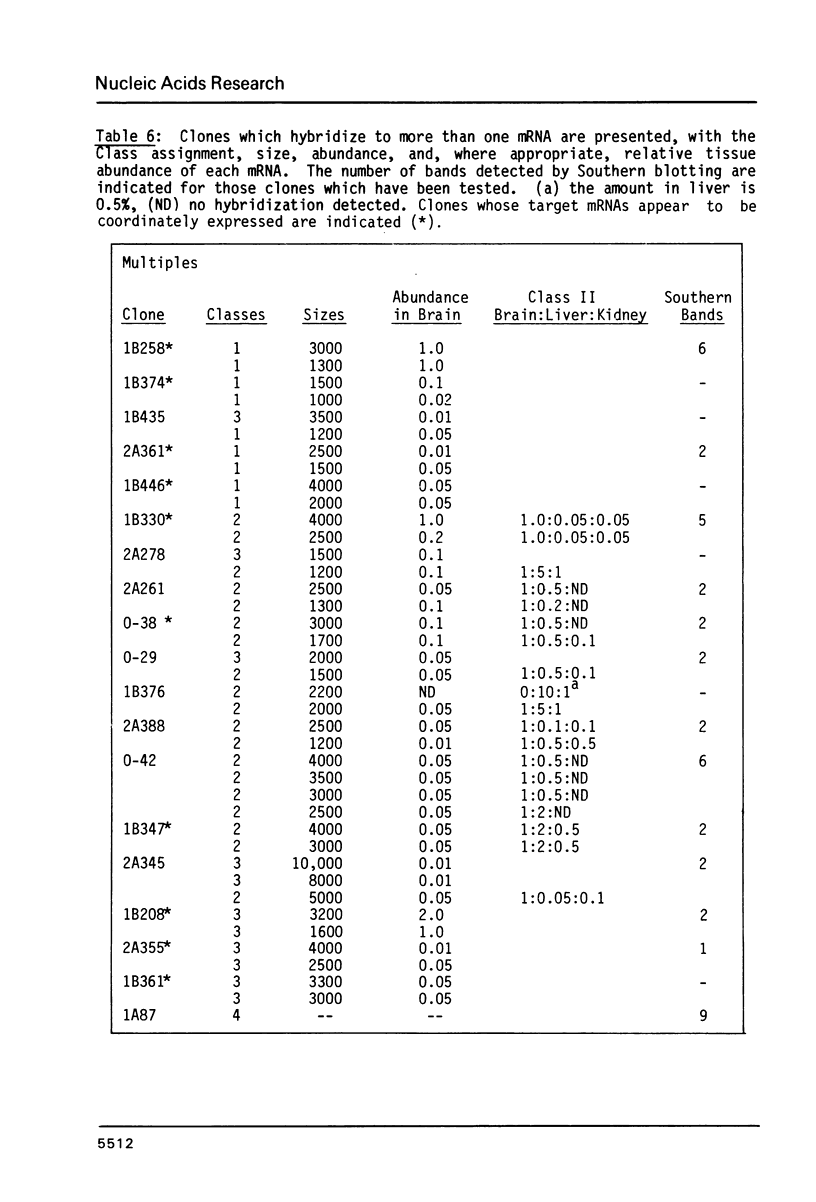
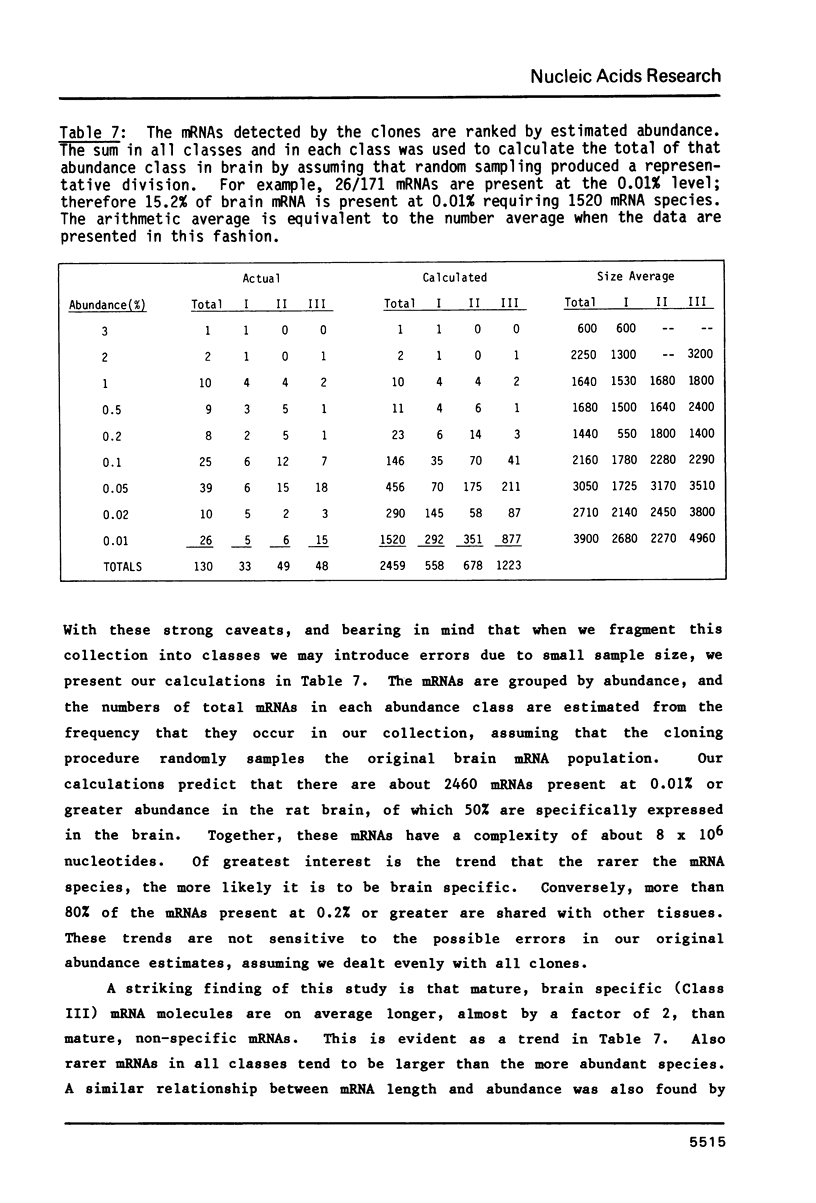
191 randomly selected cDNA clones prepared from rat brain cytoplasmic poly (A)+ RNA were screened by Northern blot hybridization to rat brain, liver and kidney RNA to determine the tissue distribution, abundance and size of the corresponding brain mRNA. 18% hybridized to mRNAs each present equally in the three tissues, 26% to mRNAs differentially expressed in the tissues, and 30% to mRNAs present only in the brain. An additional 26% of the clones failed to detect mRNA in the three tissues at an abundance level of about 0.01%, but did contain rat cDNA as demonstrated by Southern blotting; this class probably represents rare mRNAs expressed in only some brain cells. Therefore, most mRNA expressed in brain is either specific to brain or otherwise displays regulation. Rarer mRNA species tend to be larger than the more abundant species, and tend to be brain specific; the rarest, specific mRNAs average 5000 nucleotides in length. Ten percent of the clones hybridize to multiple mRNAs, some of which are expressed from small multigenic families. From these data we estimate that there are probably at most 30,000 distinct mRNA species expressed in the rat brain, the majority of which are uniquely expressed in the brain.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantle J. A., Hahn W. E. Complexity and characterization of polyadenylated RNA in the mouse brain. Cell. 1976 May;8(1):139–150. doi: 10.1016/0092-8674(76)90195-1. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo M. S., Bishop J. O. Two classes of messenger RNA in cultured rat cells: repetitive sequence transcripts and unique sequence transcripts. J Mol Biol. 1974 Dec 25;90(4):649–663. doi: 10.1016/0022-2836(74)90530-0. [DOI] [PubMed] [Google Scholar]
- Chaudhari N., Hahn W. E. Genetic expression in the developing brain. Science. 1983 May 27;220(4600):924–928. doi: 10.1126/science.6189184. [DOI] [PubMed] [Google Scholar]
- Chikaraishi D. M. Complexity of cytoplasmic polyadenylated and nonpolyadenylated rat brain ribonucleic acids. Biochemistry. 1979 Jul 24;18(15):3249–3256. doi: 10.1021/bi00582a009. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Webb E. A., Cory S., Adams J. M. Molecular cloning of seven mouse immunoglobulin kappa chain messenger ribonucleic acids. Biochemistry. 1980 Jun 10;19(12):2702–2710. doi: 10.1021/bi00553a026. [DOI] [PubMed] [Google Scholar]
- Hastie N. D., Bishop J. O. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976 Dec;9(4 Pt 2):761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- Kakidani H., Furutani Y., Takahashi H., Noda M., Morimoto Y., Hirose T., Asai M., Inayama S., Nakanishi S., Numa S. Cloning and sequence analysis of cDNA for porcine beta-neo-endorphin/dynorphin precursor. Nature. 1982 Jul 15;298(5871):245–249. doi: 10.1038/298245a0. [DOI] [PubMed] [Google Scholar]
- Land H., Schütz G., Schmale H., Richter D. Nucleotide sequence of cloned cDNA encoding bovine arginine vasopressin-neurophysin II precursor. Nature. 1982 Jan 28;295(5847):299–303. doi: 10.1038/295299a0. [DOI] [PubMed] [Google Scholar]
- Melli M. Clustering of the DNA sequences complementary to repetitive nuclear RNA of HeLa cells. J Mol Biol. 1975 Mar 25;93(1):23–38. doi: 10.1016/0022-2836(75)90357-5. [DOI] [PubMed] [Google Scholar]
- Meyuhas O., Perry R. P. Relationship between size, stability and abundance of the messenger RNA of mouse L cells. Cell. 1979 Jan;16(1):139–148. doi: 10.1016/0092-8674(79)90195-8. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Inoue A., Kita T., Nakamura M., Chang A. C., Cohen S. N., Numa S. Nucleotide sequence of cloned cDNA for bovine corticotropin-beta-lipotropin precursor. Nature. 1979 Mar 29;278(5703):423–427. doi: 10.1038/278423a0. [DOI] [PubMed] [Google Scholar]
- Rave N., Crkvenjakov R., Boedtker H. Identification of procollagen mRNAs transferred to diazobenzyloxymethyl paper from formaldehyde agarose gels. Nucleic Acids Res. 1979 Aug 10;6(11):3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Jay E., Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1976 Apr;3(4):863–877. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Tosi M., Pittet A. C., Fabiani L., Wellauer P. K. Tissue-specific expression of mouse alpha-amylase genes. J Mol Biol. 1980 Sep 5;142(1):93–116. doi: 10.1016/0022-2836(80)90208-9. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Bloom F. E., Lerner R. A. Common 82-nucleotide sequence unique to brain RNA. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4942–4946. doi: 10.1073/pnas.79.16.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Shinnick T. M., Bloom F. E. Identifying the protein products of brain-specific genes with antibodies to chemically synthesized peptides. Cell. 1983 Jul;33(3):671–682. doi: 10.1016/0092-8674(83)90010-7. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J., Hahn W. E. Sequence complexity of cDNA transcribed from a diverse mRNA population. Nucleic Acids Res. 1980 Sep 25;8(18):4259–4270. doi: 10.1093/nar/8.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. P., Buell G. N., Schimke R. T. Synthesis of double-stranded DNA complementary to lysozyme, ovomucoid, and ovalbumin mRNAs. Optimization for full length second strand synthesis by Escherichia coli DNA polymerase I. J Biol Chem. 1978 Apr 10;253(7):2483–2495. [PubMed] [Google Scholar]