Abstract
Plasmodium falciparum Sir2A (PfSir2A), a member of the sirtuin family of nicotinamide adenine dinucleotide-dependent deacetylases, has been shown to regulate the expression of surface antigens to evade the detection by host immune surveillance. It is thought that PfSir2A achieves this by deacetylating histones. However, the deacetylase activity of PfSir2A is weak. Here we present enzymology and structural evidences supporting that PfSir2A catalyzes the hydrolysis of medium and long chain fatty acyl groups from lysine residues more efficiently. Furthermore, P. falciparum proteins are found to contain such fatty acyl lysine modifications that can be removed by purified PfSir2A in vitro. Together, the data suggest that the physiological function of PfSir2A in antigen variation may be achieved by removing medium and long chain fatty acyl groups from protein lysine residues. The robust activity of PfSir2A would also facilitate the development of PfSir2A inhibitors, which may have therapeutic value in malaria treatment.
Sirtuins are a family of enzymes known as nicotinamide adenine dinucleotide (NAD)-dependent deacetylases (1, 2). They regulate a variety of biological processes, including transcription and metabolism (3, 4). Plasmodium falciparum (P. falciparum) contains two sirtuins, PfSir2A and PfSir2B (5). It was shown that these two sirtuins regulate the expression of surface antigens to evade the detection by host immune surveillance (6, 7). Thus, inhibiting these sirtuins may help fight malaria. It was thought that PfSir2A and PfSir2B achieve this physiological function by deacetylating histones. In vitro studies on PfSir2A showed that it has deacetylase activity (8). However, the activity was weak compared to several other sirtuins (9), such as human Sirt1 and yeast Sir2. It was also reported that PfSir2A had ADP-ribosyltransferase activity (8). However, several reports questioned whether the ADP-ribosyltransferase activity of sirtuins was physiologically relevant since the measured activity of several sirtuins was weak (10, 11).
In addition to acetylation, lysine propionylation and butyrylation have been reported as posttranslational modifications that occur on proteins, including histones (12–14). Many fatty acyl-CoA molecules exist in cells as metabolic intermediates. If the shorter chain fatty acyl-CoA molecules (acetyl-CoA, propionyl-CoA, and butyryl-CoA) are used as acyl donors to modify proteins, it is possible that longer chain fatty acyl-CoA molecules can also be used to modify proteins. Given that PfSir2A’s deacetylase activity is weak, we set out to investigate whether longer chain fatty acyl lysine can be accepted as better substrates by PfSir2A.
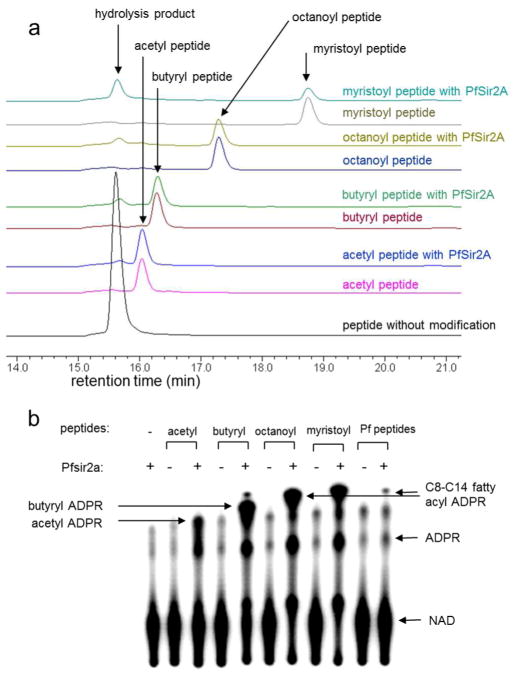
PfSir2A was expressed in E. coli and affinity purified to near homogeneity. For substrates, we synthesized histone H3 Lys9 (H3K9) peptides bearing acetyl, butyryl, octanoyl, and myristoyl groups on Lys9. A high-pressure liquid chromatography (HPLC) assay was used to monitor the activity of PfSir2A on these different acyl peptides. Interestingly, all four acyl peptides could be hydrolyzed (Figure 1a). The butyryl, octanoyl and myristoyl peptides could be hydrolyzed more efficiently than the acetyl peptide. The myristoyl peptides appeared to be hydrolyzed most efficiently.
Figure 1.
PfSir2A could hydrolyze medium and long chain fatty acyl lysine more efficiently than acetyl lysine. a) Overlaid HPLC traces showing PfSir2A-catalyzed hydrolysis of different fatty acyl lysine peptides. Acyl peptides were used at 20 μM, PfSir2A at 1 μM, and NAD at 500 μM. The corresponding synthetic peptide without any acyl lysine modification (H3K9WW unmodified) was used as the control to indicate the position of the hydrolysis product formed. b) 32P-NAD assay could detect the presence of medium or long chain fatty acyl lysine modifications on P. falciparum proteins. PfSir2A were incubated with 32P-NAD and synthetic peptides bearing different acyl modifications. Negative controls were reactions without PfSir2A or peptides. The reactions were resolved by TLC and detected by autoradiography. With P. falciparum peptides (last two lanes), the acyl ADPR spot formed was similar to the C8–C14 acyl ADPR, suggesting that such fatty acyl groups were present and could be removed by PfSir2A.
To quantitatively compare the activity of PfSir2A on different acyl peptides, kinetic studies were carried out. The kinetics data (Table 1) suggested that acetyl H3K9 peptide was the least efficient substrate among the four acyl peptides tested, with a kcat/Km of 26 s−1M−1. The kcat/Km value for deacetylation was comparable to that reported by Sauve and coworkers (9). The butyryl, octanoyl, and myristoyl peptides were hydrolyzed with much higher catalytic efficiencies. In particular, the catalytic efficiencies for the hydrolysis of myristoyl peptide were more than 300-fold higher than that for the hydrolysis of acetyl peptide. The Km value for the myristoyl peptide was lower than 1 μM (PfSir2A was saturated with 2 μM of the myristoyl peptide. The Km value could not be accurately determined because of the detection limit at low substrate concentrations). The enzymology data demonstrated that PfSir2A preferentially hydrolyzes medium and long chain fatty acyl lysine.
Table 1.
Kinetics data for PfSir2A on different acyl peptides
| substrate | kcat (s−1) | Km for peptide (μM) | kcat/Km (s−1M−1) |
|---|---|---|---|
| H3K9 acetyl | 0.001 ± 0.0002 | 39 ± 9 | 2.6 × 101 |
|
| |||
| H3K9 butyryl | 0.001 ± 0.0002 | 8 ± 1 | 1.6 × 102 |
| H3K9 octanoyl | 0.001 ± 0.004 | 1.2 ± 0. 3 | 9.2 × 102 |
| H3K9 myristoyl | 0.01 ± 0.002 | <1.0a | >1.0 × 104 |
The Km value cannot be accurately determined due to the detection limit when substrate concentration was lower than 1 μM.
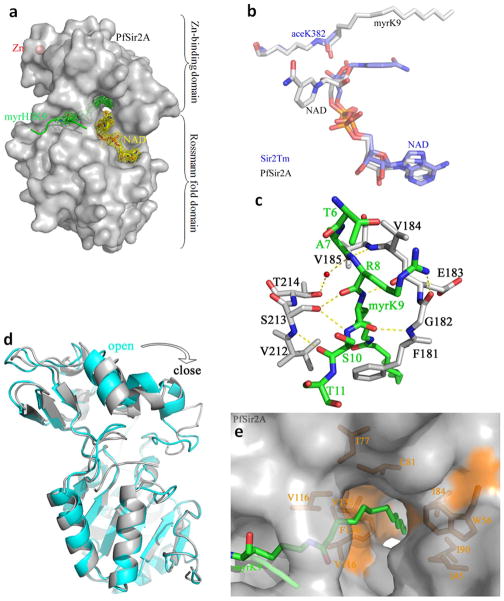
To understand the structural basis for PfSir2A’ preference for longer chain fatty acyl groups, we co-crystallized PfSir2A with an H3K9 myristoyl peptide to generate the PfSir2A-H3K9 myristoyl complex. The co-crystal was then soaked briefly in an NAD solution to obtain a ternary complex of PfSir2A with H3K9 myristoyl peptide and NAD. The structures were solved using molecular replacement with the deposited PfSir2A structure PDB 3JWP as the search model. The overall structure of PfSir2A was similar to other sirtuins, containing a small Zn-binding domain and a large Rossmann fold domain (Figure 2a) (15–18). The two substrates, H3K9 myristoyl peptide and NAD, bound to the cleft between the two domains. This binding mode of the two substrates was similar with the reported ternary complex structures of other sirtuins. For instance, the peptide substrates of the Thermotoga maritima Sir2 (Sir2Tm, PDB 2H4F, one of the first sirtuin ternary complex structures with both NAD and acetyl peptide bound) and PfSir2A superimposed well (Figure 2b) (19). The interactions between PfSir2A and H3K9 myristoyl peptide mainly came from main chain hydrogen bonds (Figure 2c), in agreement with other studies of sirtuins (19). Compared with the structure without any acyl peptide bound (PDB 3JWP), the binding of H3K9 myristoyl peptide to PfSir2A caused the Zn-binding domain to rotate clockwise to the Rossmann fold domain, so that PfSir2A moved from an open state to a close state which is similar to that observed in Sir2Tm (Figure 2d) (19). However, different from Sir2Tm, PfSir2A had a long open hydrophobic tunnel that accommodated the myristoyl group (Figure 2e). The hydrophobic tunnel was surrounded by several hydrophobic residues (Ile45, Trp56, Ile77, Ile80, Ile84, Ile90, Val116, Val135, Phe136, Ile178, and Leu181). This structure feature suggested that PfSir2A was optimized for recognizing fatty acyl groups.
Figure 2.
Structural basis for the recognition of myristoyl lysine by PfSir2A. a) Overall structure of PfSir2A. The Fo-Fc map at 1.6σ shows the H3K9 myristoyl peptide (green) and NAD (yellow) at the active site. b) The structural alignment between Sir2Tm (blue) and PfSir2A (grey). The positions of H3K9 myristoyl peptide and NAD in PfSir2A were similar to the positions of acetyl peptide and NAD in Sir2Tm. c) Hydrogen bonding interactions between the H3K9 myristoyl peptide (green) and PfSir2A (grey). d) Structural alignment between PfSir2A-AMP (cyan, PDB code: 3JWP) and PfSir2A-myrH3K9 showed that the binding of the substrate peptide myrH3K9 drove PfSir2A from an inactive open state to an active close state. e) PfSir2A had a long open hydrophobic tunnel which accommodated fatty acyl groups. PfSir2A surface representation: grey; myristoyl lysine: green; hydrophobic residues: orange.
The enzymology and structural data led to the hypothesis that PfSir2A may function to remove medium and long chain fatty acyl groups in malaria parasite. Protein lysine myristoylation has been reported to occur on several mammalian proteins (20, 21). To test whether malaria parasite proteins have medium or long chain fatty acyl modifications on lysine residues, a sensitive assay was developed to detect the presence of fatty acyl lysine in malaria parasites. With the use of 32P-NAD, the formation of fatty acylated ADP-ribose (ADPR) in PfSir2A-catalyzed defatty acylation of synthetic acyl peptides can be detected after thin-layer chromatography (TLC) separation and autoradiography. Longer chain fatty acyl ADPR species were more hydrophobic and thus moved faster than shorter chain fatty acyl ADPR species (Figure 1b). Notably, most NAD molecules were consumed when octanoyl and myristoyl peptides were incubated with PfSir2A, while there were still NAD molecules left when acetyl and butyryl peptides were incubated with PfSir2A. This observation confirmed the kinetic studies that octanoyl and myristoyl peptides were more efficient substrate for PfSir2A. When total protein extracts of malaria parasites were incubated with 32P-NAD and PfSir2A, the formation of a longer chain fatty acyl ADPR was detected. The position of the fatty acyl ADPR was similar to that formed in the reactions with synthetic octanoyl and myristoyl peptides, suggesting that fatty acyl group on P. falciparum proteins should have a similar chain length. The intensity of the fatty acyl ADPR spot was weak, suggesting that the concentration of the fatty acyl peptide in the P. falciparum protein extract was low. However, we could repeatedly detect this spot using the 32P-NAD assay. In addition, compared with the negative control without PfSir2A, the intensity for the acetyl ADPR spot did not increase. Therefore, PfSir2A’s deacetylase activity could not be detected using P. falciparum protein extract, but the activity of removing longer fatty acyl groups could be detected.
In summary, our enzymology and structural data demonstrated that PfSir2A was more efficient at removing medium and long chain fatty acyl groups than acetyl groups from peptides. Although it is known that other sirtuins can also hydrolyze propionyl and butyryl lysine, but the activity is typically weaker than the hydrolysis of acetyl lysine (22, 23). Therefore, our work demonstrates for the first time that longer fatty acyl lysines can be the preferred substrate for a sirtuin. The biochemical data suggest that P. falciparum proteins contain such fatty acyl lysine modifications, which can be removed by PfSir2A in vitro. The data imply that the biological function of PfSir2A may be achieved by its activity of removing medium and long chain fatty acyl groups. The detailed structures of the fatty acyl groups and the abundance of such modifications in comparison to the well-known acetyl lysine modification await future studies. The finding that PfSir2A could remove longer fatty acyl groups suggests that other sirtuins, especially those that have weak or no deacetylase activity, may also have this activity. Sirtuins should therefore be called “NAD-dependent deacylases”, instead of “NAD-dependent deacetylases”. The discovery of a robust activity for PfSir2A will also facility the development of PfSir2 inhibitors, which may have therapeutic value in treating malaria.
Methods
Cloning, expression, and purification of PfSir2A
PfSir2A gene was custom synthesized by Genscript. The sequence was codon optimized for overexpression in E. coli and cloned into pET-28a(+) vector with BamHI and XhoI restriction sites. The PfSir2A expression vector was then introduced into an E. coli BL21 with pRARE2. Successful transformants were selected by plating the cells on kanamycin (50 mg mL−1) and chloramphenicol (20 mg mL−1) luria broth (LB) plates. Single colonies were selected and grown in LB with kanamycin (50 mg mL−1) and chloramphenicol (20 mg mL−1) overnight at 37°C. On the following day the cells were then subcultured (1:1000 dilution) into 2 L LB with kanamycin (50 mg mL−1) and chloramphenicol (20 mg mL−1). The cells were induced with 500 μM of isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD600 of 0.6 and grown overnight at 15°C, 200 rpm. The cells were harvested by centrifugation at 6000 rpm for 10 minutes at 4°C (Beckman Coulter Refrigerated Floor Centrifuge) and passed through an EmulsiFlex-C3 cell disruptor (AVESTIN, Inc.) 3 times. Cellular debris was removed by centrifuging at 20000 rpm for 30 minutes at 4°C (Beckman Coulter). The protein was purified using gravity flow Ni-affinity chromatography (Sigma) and dialyzed into 25 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT, 10% (v/v) glycerol. The proteins were then aliquoted and kept frozen at −80°C. For crystallization, the His6 tag of PfSir2A was removed by overnight incubation at 4°C with 30 unit mL−1 of thrombin (Haematologic Technologies Inc.), followed by Ni-affinity column purification to remove any undigested PfSir2A. The tag-free PfSir2A was further purified on a Superdex™ 75 column (Bio-rad), dialyzed into 20 mM HEPES, pH 7.1, 20 mM NaCl, 5 mM DTT, 3% (v/v) glycerol, concentrated into 10 mg mL−1, frozen in liquid nitrogen, and stored at −80 °C for crystallization.
Synthesis of acyl peptides
Detailed procedure can be found in the Supporting Information. The identities of the peptides were confirmed using LCMS (LCQ Fleet, Thermo Scientific, Supplementary Figure 1);
HPLC assay and kinetics
Activity of PfSir2A was detected using HPLC. The reactions contained 20 mM of Tris pH 8.0, 1 mM DTT, 20 μM H3K9 modified peptide, 1 mM of NAD and 1 μM of PfSir2A and was incubated at 37°C for 1 hour. The reaction was quenched with 1 volume of 10% (v/v) TFA and spun down for 10 minutes at 18,000 g to separate the PfSir2A from the reaction. The supernatant was then analyzed by HPLC.
For kinetics, 1 mM of NAD, 20 mM Tris pH 8.0, 50 mM DTT, 0.5 μM (for butyryl, octanoyl and myristoyl H3K9 peptides) or 1 μM of PfSir2A (for acetyl H3K9 peptide) was used.. Peptide concentration used for H3K9 acetyl and butyryl were both 2, 4, 8, 16, 32, 64, 128, and 256 μM with an incubation time of 40 and 20 minutes, respectively. Peptide concentration used for H3K9 octanoyl was 1, 2, 4, 8, 16, 32, 64, and 128 μM with an incubation time of 15 minutes. Peptide concentration used for H3K9 myristoyl was 1, 2, 3, 4, 5, 6, 8, and 16 μM with an incubation time of 1 minute. The stock solutions of the different peptides were made in different solvents. H3K9 acetyl was stored in water while butyryl and octanoyl peptides were stored in 1:1 (v/v) DMSO:water. The myristoyl peptide was stored in DMSO. If only water was used to dissolve them, the peptides would stick to the plastic tubes used and led to errors in the concentrations. The amount of DMSO in the assays varied from 0 to 10% by volume. The quenched reactions were then analyzed by HPLC using a reverse phase analytical column (Sprite TARGA C18, 40 × 2.1 mm, 5 μm, Higgins Analytical, Inc.) with a 0% to 70% B gradient in 8 minutes at 1 mL/min. The acetyl peptide has a very close retention time to the unmodified peptide and a different column was used to separate the peaks (Kinetex XB-C18 100A, 75 × 4.60 mm, 2.6 um, Phenomenex). The product peak and the substrate peaks were quantified and converted to initial rates, which were then plotted against the modified peptide concentration and fitted using the Kaleidagraph program.
32 P-NAD assay
The reaction contained 50 mM Tris pH 8.0, 150 mM NaCl, 10 mM DTT, 60 μM H3K9 modified peptide, 0.1 μCi of 32P-NAD (American Radiolabeled Chemicals, ARP 0141–250 μCi) and 1 μM of Pfsir2A and was incubated at 37°C for 1 hour.
P. falciparum whole cell lysate (100 μL) was denatured with 6 M of Urea, 15 mM of DTT at 37°C for 15 minutes. Alkylation was carried out with 50 mM of iodoacetamide in the dark at RT for 1 hour. The solution was then diluted with 50 mM Tris pH 7.4 cotaining 1 mM CaCl2 so that the final concentration of urea was less than 0.75 M. Trypsin was added at 0.1 μg/μL and the solution was incubated overnight at 37°C. The digest was quenched with 10% TFA (v/v, final pH of 2–3) and desalted with a Waters C18 Sep-Pak column. The peptides were eluted 5 times with 1 mL of 90% ACN/0.1% TFA (v/v) and lyophilized. The peptides were then solubilized in 50 μL of water and 1 μL was used in the 32P-NAD assay under conditions described above. The reaction was incubated at 37°C for 15 minutes and 1 μL of the reaction mixture was spotted onto a polyester backed silica plate (100 μm thick, Waters). After the spots were dried, the plate was developed in 30:70 (v/v) 1M ammonium bicarbonate:95% ethanol. The plate was dried and exposed overnight in a PhosphorImaging screen (GE Healthcare). The signal was detected using a STORM860 phosphorimager (GE Healthcare).
Crystallization, data collection, and structural refinement
PfSir2A was mixed with 10 equivalent of H3K9 myristoyl, diluted into 3 mg mL−1 with crystallization buffer, and incubated on ice for 30–60 minutes. Crystals were grown at room temperature with hanging drop vapor diffusion method at the condition of 16% (w/v) PEG 3350, 0.1 M NaF, 7% (v/v) formamide. PfSir2A-H3K9 myristoyl co-crystals were soaked in the cryoprotectant solution (18% (w/v) PEG 3350, 0.1 M NaF, 10% (v/v) formamide, 15% (v/v) glycerol) with 10 mM NAD for 2–10 minutes at room temperature immediately before data collection. All data were collected at Cornell High Energy Synchrotron Source F2 station. The data were processed using the programs HKL2000 (24). Using the program Molrep from the CCP4 suite of programs (25), the structures were solved by molecular replacement with PfSir2A-AMP structure (PDB code: 3JWP) as the search template. Refinement and model building were performed with REFMAC5 and COOT from CCP4. The X-ray diffraction data collection and structure refinement statistics were shown in Supplementary Table 1. Atomic coordinates and structure factors were deposited in the Protein Data Bank under accession codes 3U3D and 3U31 for PfSir2A-myrH3K9 and PfSir2A-myrH3K9-NAD, respectively.
Supplementary Material
Acknowledgments
This work is supported in part by NIH R01GM086703 (H.L.), NIH RR01646 (Q.H.) and Hong Kong GRF766510 (Q.H.). A.Y.Z. is a CBI training grant trainee (NIH T32 GM08500).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 3.Imai S-i, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends in Pharmacological Sciences. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 6.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 7.Tonkin CJ, Carret ClK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LMd, Scherf A, Ivens A, Speed TP, Beeson JG, Cowman AF. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e1000084. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an Unusual Sirtuin with Dual Histone Deacetylase and ADP-Ribosyltransferase Activity. Eukaryotic Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French JB, Cen Y, Sauve AA. Plasmodium falciparum Sir2 is an NAD+-dependent deacetylase and an acetyllysine-dependent and acetyllysine-independent NAD+ glycohydrolase. Biochemistry. 2008;47:10227–10239. doi: 10.1021/bi800767t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 11.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogs and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. N-Lysine Propionylation Controls the Activity of Propionyl-CoA Synthetase. J Biol Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Lin Y, Darwanto A, Song X, Xu G, Zhang K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J Biol Chem. 2009;284:32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Mol Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 16.Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Mol Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 17.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 18.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 19.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson FT, Bursten SL, Locksley RM, Lovett DH. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. J Exp Med. 1992;176:1053–1062. doi: 10.1084/jem.176.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevenson FT, Bursten SL, Fanton C, Locksley RM, Lovett DH. The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proc Natl Acad Sci U S A. 1993;90:7245–7249. doi: 10.1073/pnas.90.15.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith BC, Denu JM. Acetyl-lysine Analog Peptides as Mechanistic Probes of Protein Deacetylases. J Biol Chem. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 23.Bheda P, Wang JT, Escalante-Semerena JC, Wolberger C. Structure of Sir2Tm bound to a propionylated peptide. Protein Science. 2011;20:131–139. doi: 10.1002/pro.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.