Abstract
Understanding the molecular mechanisms that shape an effective cellular response is a fundamental question in biology. Biochemical measurements have revealed critical information about the order of protein-protein interactions along signaling cascades, but lack the resolution to determine kinetics and localization of interactions on the plasma membrane. Furthermore, the local membrane environment influences membrane receptor distributions and dynamics, which in turn influences signal transduction. To measure dynamic protein interactions and elucidate the consequences of membrane architecture interplay, direct measurements at high spatiotemporal resolution are needed. In this review, we discuss the biochemical principles regulating membrane nanodomain formation and protein function, ranging from the lipid nanoenvironment to the cortical cytoskeleton. We also discuss recent advances in fluorescence microscopy that are making it possible to quantify protein organization and biochemical events at the nanoscale in the living cell membrane.
Introduction
In 1972, Singer and Nicholson proposed the Fluid Mosaic model, in which most membrane constituents diffuse rapidly and randomly about the two-dimensional surface of the lipid bi-layer (1). However, live cell imaging techniques such as single particle tracking have provided considerable evidence that many receptors and even lipids are restricted in lateral mobility. These observations, along with biochemical techniques, established a compartmentalized view of the plasma membrane, which focuses around three hypotheses of microdomain organization: lipid rafts (2), protein islands (3) and actin corrals (4). What remains to be understood is the specific contribution of these microdomains in regulating the signaling process.
There is mounting evidence for critical roles of the lipid nanoenvironment in regulating protein interactions. Favored interactions between certain types of lipids lead to their co-segregation in domains at the cell membrane, which led to the lipid raft theory. However, recent evidence is demonstrating that membrane organization is more complex than simple division of raft and non-raft regions. Also, proteins associated with the plasma membrane often undergo a lipid-based post-translational modification with the addition of an acyl chain to specific amino acids that can subsequently mediate the interaction of this protein with the lipids of the plasma membrane. Therefore, to fully characterize protein-protein interactions and understand the critical roles of lipids and membrane organization in regulating those interactions, it is important to study signaling events in living cells at high temporal and spatial resolution.
Biochemical principles regulating membrane nanodomain formation
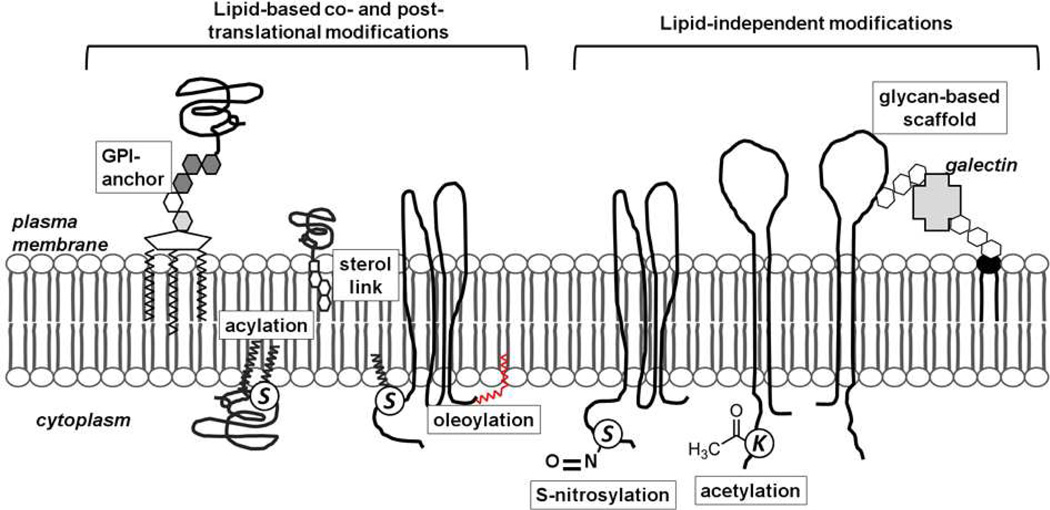
The formation of membrane nanodomains originates from lipid-lipid, lipid-protein and protein-protein based interactions, which implies the existence of a variety of biochemical principles that allow these interactions to occur at the molecular level. The major structural lipids in eukaryotic membranes are the glycerophospholipids that share a similar hydrophobic portion but have different polar headgroups that confer a specific molecular geometry to each phospholipid thus contributing to the regulation of membrane curvature. The other class of polar structural lipids is the sphingolipids. They contain two saturated hydrophobic chains that are longer and narrower than the phospholipids, pack tightly and confer rigidity to the lipid bilayer. The sphingolipids straight chains and headgroup spacing favor the intercalation of cholesterol, which further contributes to increasing the lipid packing density 5). These strong interactions between cholesterol and sphingolipids promote their co-segregation in domains at the plasma membrane, generally termed ‘rafts’. Lipid rafts can sequester specific signaling proteins and allow the formation of supra-molecular signaling complexes (6). Proteins that reside within cellular membranes have molecular features that allow them to be embedded in the highly hydrophobic milieu of the lipid membrane. For some membrane spanning proteins, the transmembrane domains typically consist of α-helices or β-sheets with their hydrophobic amino acid residues interfacing the hydrocarbon chains of the lipid bilayer. Alternatively, the association of proteins with the membrane can be mediated by specific co- or post-translational additions of lipid anchors such as the glycophosphatidylinositol (GPI) anchor, myristic acid tail, palmitic acid tail, etc (Fig. 1). Furthermore, membrane proteins often bear other non-lipid post-translational modifications (e.g. glycosylation, S-nitrosylation) that might mediate the interaction with specific signaling components or scaffold molecules thus contributing to the formation of functional membrane nanocompartments. The role of these non-lipid modifications in the organization of membrane nanodomains is still largely unexplored.
Figure 1. Biochemical principles regulating partitioning and nanoscale organization of membrane proteins.
The formation of membrane nanodomains originating from lipid-lipid, lipid-protein and protein-protein based interactions implies the existence of a variety of biochemical principles that allow these interactions to occur at the molecular level. Proteins associated with cellular membranes have molecular determinants that allow them to be embedded in the highly hydrophobic milieu of the lipid bilayer. Several forms of lipid-based modifications provide the proteins either permanently or transiently with the right membrane anchor. Non-lipid modifications further contribute to the fine-tuning of receptor function and subsequent signal transduction. The same protein can undergo different modifications, however the regulation and interplay of these modifications are still unknown.
Lipid nano-environment
Although the term ‘rafts’ remains controversial, the existence of lipid and protein nanodomains at the cell membrane is now widely accepted (7, 8). It should be noted that while liquid ordered domains readily assemble in artificial membranes, their existence in complex cellular membrane preparations has only recently been observed (9) and direct detection in intact cells has proven more challenging. However, non-overlapping nanodomains of the glycosphingolipids GM1 and GM3 as well as spatially distinct sphingomyelin (SM) rich clusters have been detected (10, 11), highlighting the compartmentalized nature of the plasma membrane.
In many forms of cell activation, including malignant transformation or pathogen invasion, the enzyme acid sphingomyelinase (SMase) hydrolyzes SM into ceramide, which is released within the cell membrane and alters its biophysical characteristics (12, 13). Therefore, changes in plasma membrane sphingolipid levels are likely to affect the function of signaling molecular complexes by altering the lipid nano-environment. Extensive atom-scale simulations of ternary raft mixtures containing cholesterol, sphingomyelin and phosphatidylcholine have shown nanoscale lateral heterogeneity and lateral pressure profiles clearly distinct from non-raft mixtures. Changes in lipid content might modify the lateral pressure profile in turn altering the function of certain classes of membrane proteins (14). Recent studies demonstrated that a voltage-gated potassium channel and its surrounding membrane lipids together represent a functional unit since the annular lipids were able to control the channel conformational switch from activated to resting state (15). Similarly, Coskun and colleagues demonstrated that the membrane lipid composition does not alter the ligand binding properties of the epidermal growth factor receptor (EGFR) but rather modulates its allosteric structural transition from inactive to a signaling competent dimer (16).
Another important lipid component of the plasma membrane is cholesterol that is known to play an essential role in regulating the biophysical properties of membrane proteins and lipids. By sequestering the downstream effector partners but not the β2-adrenergic receptor in lipid-ordered domains, cholesterol regulates the nanoscale organization of the receptor signaling machinery (17). Interestingly, cholesterol has been shown to induce a tilt in glycolipid headgroups ultimately modulating glycolipid-dependent surface recognition processes such as presentation of erythrocyte blood groups or sperm activation (18). Changes in cholesterol content at the cell membrane could therefore modulate glycolipid conformation by either masking or unveiling specific glycolipid sites, that in turn might affect interactions with other membrane molecules both in trans and in cis. Cholesterol is a major contributor to membrane fluidity. It has been shown that cholesterol coalesces with mobile FcεRI upon synapse formation, but avoids immobilized receptors, suggesting that membrane constituents are attracted to cholesterol-rich regions due to fluidity (19). Finally, cholesterol appears to be the glue that mediates raft-based interconnectivity at the nanoscale that might constitute the basis for large-scale raft coalescence observed upon cell activation (20).
Lipid rafts of variable biophysical properties and molecular composition have been found in plasma membrane preparations, indicating that biological membranes have the capacity to form lipid nanoenvironments of continuously variable size, composition and stability that represent the basic organizing principle of membrane compartmentalization (21). The formation of liquid-ordered domains at the plasma membrane rich in cholesterol and glycosphingolipids represents the dominant theory to explain raft existence (5). However, it is unlikely that the liquid-ordered domains alone give rise to the great variation in lipid and protein content exhibited by rafts. Recently, a ‘lipid matrix model’ of raft structure has been proposed that provides a plausible mechanism to explain how membrane rafts can achieve such a molecular complexity (22). The lipid matrix model takes into account the putative role of asymmetric sphingolipids (i.e. sphingolipids bearing N-acyl fatty acid tails of different length) in raft formation and function and envisages the existence of quasi-crystalline domains. It has been proposed that the liquid-ordered domains, formed by symmetric sphingolipids and cholesterol, may function as a matrix for recruiting raft proteins, including transmembrane proteins that stably reside at the cell membrane and are connected to the cytoskeleton. In addition, quasi-crystalline domains formed by asymmetric sphingolipids and phospholipids represent a matrix into which proteins tethered to the raft via GPI anchors or acyl chains can assemble and cluster. Additional interactions between the carbohydrate moiety of the glycolipids and the neighboring proteins adds yet another level of specificity that contributes to raft diversity (22).
Whereas increasing information is available about the lipids in the plasma membrane outer leaflet, the composition, organization and function of the inner leaflet are less well addressed. Phosphoinositides (PIPs) are concentrated at the cytosolic surface of membranes and become reversibly phosphorylated by PI-Kinases. Differentially phosphorylated PIPs display unique subcellular distribution with preferential localization to subsets of membranes (23). PIPs contribute to the unique negative charge of the inner leaflet, the bilayer asymmetry and, importantly, the differential targeting and trafficking of signaling proteins to the plasma membrane (24, 25). A recent elegant biophysical study by Lasserre et al. demonstrated the existence of highly dynamic lipid nanodomains in both the outer and the inner leaflets of the plasma membrane of T lymphocytes and the negative effect of raft alteration on the PI3-Kinase pathway (26). It remains to be established how receptors interacting with classical raft domains at the outer leaflet can signal across the membrane leaflet to these PIPs-enriched signaling domains.
The notion of lipid nanodomains with different molecular composition and physicochemical properties represents a clear advancement of our understanding of lipid rafts from undefined, elusive lipid platforms to fundamental biochemical entities responsible for membrane compartmentalization specificity.
Biochemical modifications of membrane proteins
The lipid and protein composition of many membrane domains has been extensively investigated. In spite of this, the underlying biochemical principles that determine how such a great variety of proteins associate with the plasma membrane and partition into specific nanodomains are still not entirely understood. To allow their embedding in the hydrophobic environment of the plasma membrane, proteins need dedicated moieties that can originate directly from their own amino acid sequence or from co- and post-translational modifications (Figure 1).
The most common lipid-based modifications found in membrane associated proteins are i) the addition of GPI anchors; ii) cysteine acylation also known as palmitoylation, iii) prenylation and myristoylation; and iv) the addition of sterol moieties at the C-terminus (27). While GPI anchors (28) and palmitoylation (29) are known to mediate protein partitioning into the tightly packed lipid rafts, prenylation (30) and myristoylation (31) seem to target proteins to less restrictive non-raft membrane areas. Furthermore, the addition of a GPI anchor, prenylation and myristoylation are stable co-translational modifications, whereas palmitoylation is dynamically regulated by enzymes and is therefore reversible (32). A typical example is the different raft affinity of the Ras protein isoforms that is dictated by the type of lipid anchor: while the doubly palmitoylated H-Ras strongly reside in lipid rafts, the prenylated K-Ras does not partition into lipid rafts, despite their significant homology in amino acid sequence (12, 33). Similarly, differential fatty acylation of membrane proteins has been shown to play an important role in T cell signaling, where localization and functional state of key signaling molecules, such as Lck and Fyn, and of co-receptors and adaptors involved in T cell activation have been shown to be regulated by the presence of specific lipid chains (34).
In addition to the unique sterol modification of the Hedgehog family proteins reported by Porter et al in 1996 (35), a novel form of fatty acid acylation has been recently documented for the lens integral membrane protein Aquaporin-0 (36). Combining direct tissue profiling by mass spectrometry of lens sections with proteomic analysis, Schey and colleagues observed that N- and C-terminus of Aquaporin-0 were modified by palmitoylation and oleoylation, respectively. Oleoylation represents the addition of oleic acid to a lysine residue via an amide linkage and appears to mediate the localization of Aquaporin-0 into lipid raft fractions (36). Future investigation may reveal further novel forms of protein modifications by fatty acid chains that direct membrane targeting and nanodomain partitioning.
A significant number of recent studies have focused on lipid-independent reversible redox modifications of specific cysteine residues as a new cell signaling mechanism (37). Nitric oxide produced from L-arginine by nitric oxide synthase enzymes can directly modify cysteines by covalent attachment resulting in the so-called S-nitrosylation. Important signaling molecules such as Ras (38, 39), β-catenin (40) and MyD88 (41) as well as a number of G protein-couple receptors have been found to be regulated by S-nitrosylation. Both acting on the cysteine residues, S-nitrosylation and palmitoylation are likely to have a dynamic interplay that could further fine-tune membrane receptor organization and cell signaling events. In addition, the nitric oxide has been shown to displace palmitate from proteins (42). The development of more sensitive techniques to monitor nitrosylation will most likely reveal novel molecular dynamics controlling the recruitment of proteins to the plasma membrane and modulating signal transduction (43).
A steadily growing number of studies indicate acetylation of non-histone proteins as another important lipid-independent post-translational modification that can modulate multiple cellular processes from gene expression to receptor activity (44). Acetylation is the transfer of an acetyl group from acetyl-CoA to the e-NH2 group of the side chain of lysine residues by a lysine acetyltransferase. Predominantly known for its role in the regulation of gene expression, lysine acetylation is now recognized as an essential player in the regulation of cell activities such as cytoskeleton organization, cellular transport and protein stability (45). Interestingly, the involvement of acetylation in the regulation of membrane receptor function is also emerging. Acetylated tubulin has been shown to specifically interact with the cytoplasmic domain of the membrane-associated sodium pump NaATPase, which therefore acts as microtubule-membrane anchorage site. Furthermore, at the plasma membrane the prolactin receptor has been found to undergo cytoplasmic loop dimerization that depends on acetylation of multiple lysine sites along the loop and is essential to initiate the downstream signaling cascade (46).
All these competing reversible posttranslational modifications must be regulated by a complex interplay among different modifying enzymes and contribute to the dynamic regulation of nanoscale organization and function of a variety of membrane receptors. Together with the stable cotranslational alterations, these modifications represent additional tools used by the cell to fine-tune signal transduction.
Role of glycans in membrane compartmentalization
In eukaryotic cells, glycosylation is a widespread post-translational modification of secreted and membrane-anchored proteins as well as proteoglycans and glycolipids. Galectins, a family of galactose-specific animal lectins, bind and cross-link branches of specific N-glycans present on glycosylated molecules at the cell surface (47). In this way, galectins act as molecular organizers of the cell surface able to recruit proteins and lipids to compartments where homo- and heterotypic clustering can occur. This generates the so-called galectin scaffolds or lattices, the dynamics and composition of which are still unknown (48).
Recruitment of proteins to galectin lattices has been shown to prevent receptors from uncontrolled clustering and signaling (49). In fact, biophysical approaches such as FRET have allowed the visualization of Galectin-3 oligomerization, suggesting that this protein is indeed able to form small aggregates where certain receptors could be recruited (50). For example, the interaction of the T cell receptor with galectin-3 has been shown to negatively regulate T cell receptor response to antigens (51). Conversely, galectin lattice promotes EGFR signaling by sequestering the receptor away from negative regulatory Caveolin-1 (52). By binding to glycans present on raft associated glycolipids, galectins likely play an important role in regulating the communication between different types of membrane nanocompartments. Understanding the cross-talk between galectin lattices, lipid rafts and other types of membrane compartments in the regulation of receptor signaling represents one of the future challenges in membrane biology.
Imaging membrane compartmentalization
The biochemical events described above are responsible for dynamic molecular interactions that determine anchoring and detaching of proteins from scaffolds, the cytoskeleton, or other membrane proteins/lipids. This suggests that partitioning of membrane proteins is not a passive event, rather the restrictions placed on protein diffusion and localization regulate receptor accessibility to interaction partners, subsequently regulating signaling events. Therefore, understanding the contributions of microdomains to receptor function is needed to fully understand how signaling is regulated. Fluorescence imaging techniques (see Table 1) are providing new insights into membrane organization.
Table 1.
Fluorescence microscopy techniques for mapping of membrane organization or detecting protein-protein interactions
| Brief Description |
Quantitative Information |
Advantages | Limitations | |
|---|---|---|---|---|
| FRAP (70, 100) Fluorescence Recovery After Photobleaching | Fluorophores are bleached in a small region and recovery of signal is monitored over time. |
|
|
|
| FRET (81, 101, 102) Förster Resonance Energy Transfer | Energy transfer between donor and acceptor labels is dependent on distance between them, typically 1–10 nm. ET measured by changes in donor intensity or lifetime. |
|
|
|
| homoFRET (55, 56, 103) | Energy transfer between like fluorophores (i.e. GFP to GFP), detected by changes in polarization anisotropy. |
|
|
|
| SPT (63, 65, 67, 100) Single Particle Tracking | Direct tracking of individual protein trajectories. |
|
|
|
| FCS (70, 104) Fluorescence Correlation Spectroscopy | Analysis of intensity fluctuations generated as fluorescently-tagged proteins diffuse in and out of a stationary focal volume. |
|
|
|
| ICS (105, 106) Image Correlation Spectroscopy | Family of techniques similar to FCS, but intensity fluctuations are analyzed across an image or throughout a time series. |
|
|
|
| STED (53, 93, 107) Stimulated Emission Depletion | Gaussian excitation beam is overlaid with a doughnut-shaped beam that depletes emission from the outside ring of the excitation spot. |
|
|
|
| NSOM (8) Near-field Scanning Optical Microscopy | A scanned optical fiber excites and collects fluorescence near fiber tip. |
|
|
|
| Single Fluorophore Localization Microscopy (68, 107) | Localization of individual molecules with high precision to build a high resolution image. Variants include PALM, STORM and GSDIM. |
|
|
|
The elusive lipid rafts
As discussed above, the lipid nano-environment is considered to have a critical influence on cellular function. Despite the biochemical evidence for the existence of lipid rafts, the detection of these small and dynamic structures has been elusive. In 2009, Eggeling et al used STED-FCS to provide convincing evidence for cholesterol-driven compartmentalization (53). The sub-diffraction lateral resolution of the STED beam creates a smaller focal volume for FCS analysis than a traditional confocal beam. This enhanced resolution made it possible to determine that sphingolipids and GPI-APs are transiently trapped in cholesterol-dependent nanodomains (<20 nm). Another technique that can improve resolution of the FCS volume is NSOM. NSOM provides improved axial as well as lateral resolution, creating an even smaller FCS volume than STED. With this approach, Manzo et al have also detected heterogeneous behavior of sphingolipid diffusion that is consistent with compartmentalization (54).
Detection of raft-marker association on the cell surface has also demonstrated the ability of lipids to organize membrane proteins. Using homo-FRET, several groups have detected small clusters of the raft-marker GPI-AP (55, 56). Bramshueber et al (57) have examined GFP-GPI-AP and GM1 (lipid that marks raft domains) organization on living cells using an adaptation of FRAP to image the domains. They observed nanoscale clustering of these raft markers into stable platforms that were mobile and cholesterol dependent. NSOM is capable of directly mapping out the nanoscale organization of the membrane with ~ 100 nm resolution (8) (see Figure 2). Using NSOM, Van Zanten et al(20) found that cholera toxin-β (CTxB) binding to GM1 induces coalescence of CTxB-GM1 into nanodomains smaller than 120 nm. As expected, the classical non-raft marker CD71 did not colocalize with CTxB-GM1 and was randomly distributed. Interestingly, while raftophilic proteins (CD55, LFA-1 and GPI-AP) were found within close proximity to CTxB-GM1, the proteins did not mix, suggesting a recruitment of purported raft-associated proteins to GM1 that is stabilized by cholesterol-based interconnectivity. The clustering of proteins measured in these studies was found to be cholesterol-dependent, confirming cholesterol’s key role in domain formation at the nanoscale.
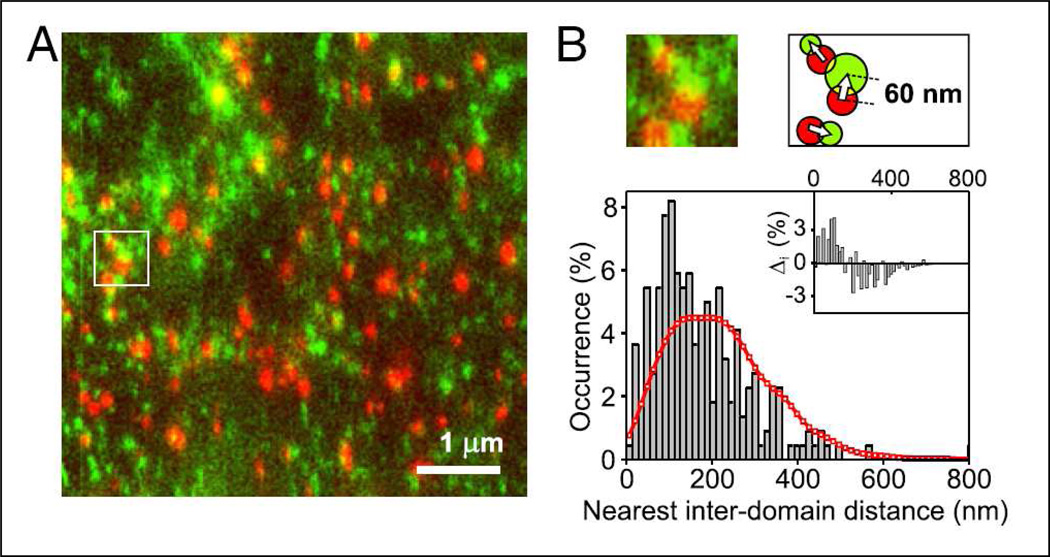
Figure 2. Mapping the membrane with NSOM.
(A) Representative dual color excitation/detection NSOM image of LFA-1 integrin nanoclusters (red) and GPI-APs (green) at the cell surface of fixed monocytes in the absence of ligand. (B) The cartoon shows how the distance between the center of mass of a fluorescent spot and its nearest neighboring spot is calculated. Nearest inter-domain distance distributions of LFA-1 nanoclusters to its closest GPI-AP (bars) together with simulations of random spatial distribution of LFA-1 nanoclusters with respect to GPI-APs (red). The inset corresponds to the difference (i in %) between experimental data and simulations. At shorter distances (cross-over point in i) both distributions are significantly different with P 0.01. These results demonstrated that LFA-1 nanoclusters, known to co-cap with large raft domains, are in fact spatially segregated but proximal to GPI-AP hotspots. Reproduced with permission from van Zanten et al (108).
Protein Islands
Douglas & Vale first demonstrated diffusional trapping of membrane proteins in discrete protein-defined microdomains (58). Using two-color imaging, they tracked individual LAT-GFP or Lck-GFP motion with respect to total CD2-mRFP. LAT and Lck were seen to diffuse rapidly in the non-CD2 regions, but undergo restricted diffusion upon entering a CD2-defined area, indicating that membrane proteins can be transiently trapped in membrane domains. In support of this evidence, electron micrographs of plasma membrane sheets labeled for signaling molecules revealed that proteins exist in distinct clusters surrounded by protein-free membrane (3, 59–61). Lillemeier and colleagues (3) termed these protein-rich regions “protein islands” and found that while all protein islands were enriched in cholesterol, some islands labeled with raft-markers and others with non-raft markers. Additionally, the protein islands require actin for stability. Using high speed PALM (Photoactivatable Localization Microscopy), TCR (T cell receptor) and LAT were shown to exist in stable yet distinct clusters on resting T cells, confirming in live cells that membrane proteins localize to distinct microdomains (62). These observations indicate that membrane partitioning is more complex than defining raft/non-raft compartments.
Actin-mediated receptor confinement
Single particle tracking studies have implicated membrane proximal actin structures in the formation of nanometer-sized ‘confinement zones’ that restrict lateral diffusion (4). Typically, these studies relied on chemical disruption of the actin cytoskeleton to correlate confinement with actin. In 2008, using two-color TIRF microscopy, the motion of individual quantum dot (QD)-tagged FcεRI was simultaneously imaged within the landscape of the membrane proximal GFP-actin bundles (63). This directly showed for the first time that actin indeed acts as a physical barrier to transmembrane protein diffusion. These observations defined a larger-scale actin organization than those described by Kusumi and colleagues (64), revealing a dynamic actin labyrinth with spatiotemporal scales on order of microns and seconds.
The ultimate question is whether this actin-dependent partitioning is a passive event or functions to alter protein behavior. Batista and colleagues have implicated actin as well as actin-binding proteins, ERM (ezrin-radixi-moesin), in regulation of B cell receptor (BCR) signaling. Using two-color TIRF microscopy, Treanor et al (65) observed that resting BCR diffusion is restricted by both actin and ezrin structures. Interestingly, they also observed that BCR can be constrained within actin rich regions, generating a population of receptors with reduced mobility. The disruption of the actin cytoskeleton induced cellular calcium response that correlated with increasing BCR diffusion. This work suggests a link between actin and ERM networks that partition receptors and prevent signaling. The partitioning may serve to sequester BCR from interactions partners such as active kinases or co-receptors. Alternatively, the compartmentalization may co-confine BCR with phosphotases that keep the receptor inactive. More recently, the same group has shown that ERM proteins are dephosphorylated upon BCR activation, which would alter BCR mobility and facilitate signaling (66). Together, these results indicate an active interplay between BCR and the actin network that controls BCR signaling.
Actin compartmentalization may also influence signaling events by increasing protein interactions. Two-color single QD tracking revealed that actin co-confines receptors, promoting receptor encounters. In the FcεRI system, actin co-confines resting receptors, maintaining them in close proximity for extended periods of time, thereby increasing the local receptor concentration (63). Actin was also shown to modulate FcεRI response to multivalent ligand binding, since disruption of actin increased the time for receptor immobilization upon antigen-induced crosslinking (63). A corresponding cytoskeleton confinement has been observed for the immunoreceptor CD36 on macrophages. Jaqaman et al showed that CD36 diffusion is constrained in linear channels that are actin and microtubule dependent (67). Receptor co-confinement in these domains leads to an increased local density of receptors by ~5-fold and promotes transient interactions between unliganded receptors.
Measuring protein dynamics and organization
Advancements in fluorescence microscopy techniques (Table 1) have made it possible to measure protein dynamics, aggregation state, and interactions on the living cell, facilitating measurements of biochemical parameters in situ. The family of image correlation techniques can determine average protein mobility, aggregation state and protein-protein interactions based on ensemble measurements. Imaging techniques that circumvent the diffraction limit, including NSOM (Figure 2) and localization microscopy, are capable of directly mapping out the nanoscale organization of the membrane (8, 68). Single molecule imaging, such as FRET imaging and multi-color single particle tracking (Figure 3), provides a view of protein behavior at the molecular-level (69).
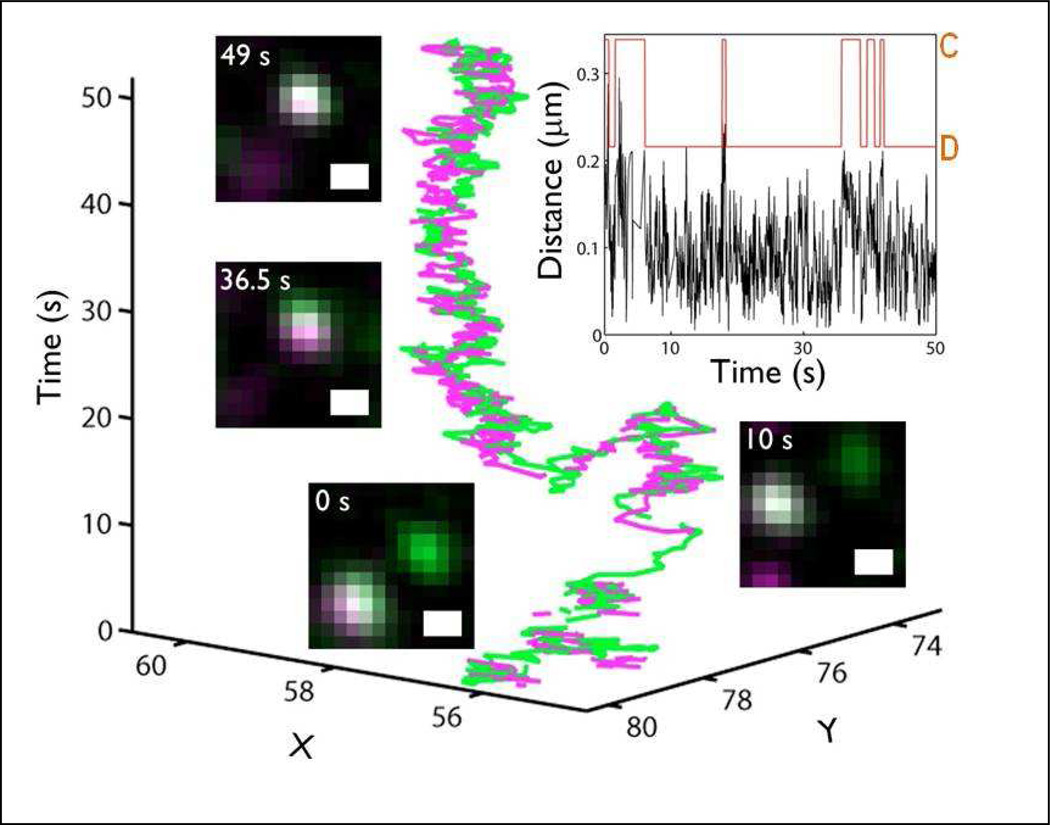
Figure 3. Capturing EGFR dimerization events.
Tracking of QD585-EGF-EGFR (green) and QD655-EGF-EGFR (magenta) complexes. Trajectory over time shows close proximity of the two ligand-bound receptors with correlated motion. Insets: Stills from the acquired time series show moments of high colocalization and times when the receptors separate. Top right: Plot of distance between the two receptors as a function of time demonstrates fluctuations in separation. This is captured by the 3-state HMM that identifies repeated transitions (orange line) between dimer (D) and domain confined (C) states. Image courtesy of Shalini Low-Nam and similar to Low-Nam et al (80).
Ensemble measurements of protein organization
Classical microscopy techniques continue to provide insight into biological systems. For example, a recent paper by Itano et al used two-color colocalization to show that the HIV-1 receptor DC-SIGN is organized in microdomains on the cell surface that are heterogeneous in composition, often mixing with other C-type lectins or clathrin (70). These results support the previous findings of DC-SIGN localization to microdomains using electron microscopy or NSOM (71, 72). Itano et al also combined FRAP, line scanning FCS and single QD tracking to show that DC-SIGN does not exchange readily between microdomains and that the mobility within the domains is low. Together, these studies demonstrate the power of integrating data from disparate techniques that cross multiple time and length scales to create a more complete picture of a protein behavior.
Recently, the family of ICS techniques has grown rapidly with new approaches and analysis methods. In 2009, the Gratton group demonstrated the ability to detect protein complex stoichiometry and dynamics of exchange using cross-correlation Number & Brightness and RICS analysis (73, 74). Nagy et al (75) used N&B to examine the distribution of EGFR on the plasma membrane. They found that EGFR exists as a monomer at low expression levels, but forms ligand-independent dimers at higher-levels of expression. Addition of EGF, lead to almost complete dimerization and the ultimate formation of higher order oligomers that associated with clathrin structures.
Spatial Intensity Distribution Analysis (SpIDA) has recently been developed by the Wiseman group. This technique analyzes fluorescence images based on fitting intensity histograms to determine protein concentration and aggregation (76). Since this analysis does not depend on spatial correlation, it can extract data from a single image and is not tied to the assumption that the sample is homogeneous. Swift et al (77) applied SpIDA to simultaneously monitor EGFR dimerization and internalization (concentration at the membrane) in response to transactivation via GPCR. They found a differential response depending on the specific GPCR involved; all GPCR transactivation induced EGFR dimerization, yet not all of the GPCRs induced rapid EGFR internalization.
Capturing Single Molecule Behavior
The ensemble measurements described above considered the average, steady state characteristics of proteins but do not address the stochastic nature of receptor encounters, which can only be appreciated though single molecule observations. Recently, several groups have reported methods to monitor receptor interactions at the single molecule level and determine dimerization kinetics. Using the coincidence of Cy3B- and Alexa488-labeled ligands, Hern et al (78) observed dimer formation and dissociation events as well as repeated interactions between the GPCR, M1 muscarinic acetylcholine receptor. Their results indicated that M1 exists in a dynamic equilibrium between monomer and dimer states. Analysis of the distribution of dimer durations determined an off rate of 1.3 s−1. Kasai et al (79) have used single molecule imaging to examine dimerization of another GPCR, the Formyl Peptide Receptor (FPR). In this work, they developed analysis methods to determine association and dissociation constants from single color data. They found that FPR also exists in a dynamic equilibrium with unliganded dimer lifetimes of 91 ms and found no significant change in the presence of ligand.
The previous two studies used organic dyes for labeling the GPCR. Low-Nam et al (80) have used two-color QD tracking to characterize EGFR dimerization (Figure 3). The use of QDs allows for tracking of receptors over longer times without potential artifacts due to photobleaching. In this study, EGFR was either labeled through ligand (QD-EGF) or with a non-activating camelid anti-erbB1 antibody fragment (QD-VHH). Since proteins are often co-confined in diffraction limited domains, correlated motion analysis was used to distinguish dimerization from colocalization. It was seen that resting receptors (QD-VHH) did not display correlated motion despite colocalization, while ligand-bound (QD-EGF) receptors demonstrated strong correlated motion. To quantify the kinetics of dimerization, a 3-state Hidden Markov Model was developed to extract transition rates between free, co-confined, and dimer states. The introduction of the co-confined state was required to accurately represent the data, indicating that co-confinement by microdomains plays an important role in receptor behavior. It was found that 2 ligand-bound receptors form more stable dimers than resting receptors, linking ligand occupancy to dimer stability. Furthermore, actin-based confinement was found to promote receptor dimerization (80).
Single molecule techniques have also been developed to examine receptor-ligand interactions. Huppa et al (81) used single molecule FRET (smFRET) to determine dissociation rates for the TCR-peptide-MHC complex in the context of the immunological synapse. When T cells with Cy3-scFv-labeled TCR (donor) were added to a planar lipid bilayer containing Cy5-peptide bound to MHC (pMHC), TCR-pMHC interactions resulted in measurable energy transfer between the FRET pair. The duration of the smFRET signal per interaction was quantified and from this TCR-pMHC dissociation rate (Kd) was determined. Importantly, the Kd measured in intact cells during synapse formation was found to be much higher than those determined from in vitro measurements of purified protein, highlighting the importance of cellular geometry in modulating protein behavior.
Outlook
More than a decade after the ‘discovery’ of lipid rafts (7, 82), membrane compartmentalization is now a well-recognized general mechanism of regulating receptor function and signal transduction, and its many facets are becoming increasingly known. Clearly, cells exploit a variety of biochemical actions to modulate the aggregation of proteins and lipids at the nano- and micro-scale. Newly emerging post-translational modifications of membrane proteins are likely to play a yet unknown role in membrane organization. The interplay among these modifications most likely represents an additional level of signal transduction regulation.
Understanding of the coupling mechanism(s) between outer and inner domains is still in its infancy. It remains to be established how the outer and inner lipid layers are organized with respect to each other, and whether they concertedly contribute to the regulation of membrane-associated receptor signaling. Trans-membrane proteins as well as cholesterol could be involved in linking the biochemical information at both sides of the plasma membrane. Recent developments in the area of artificial lipid membrane preparation have shown that stable asymmetric giant unilamellar vesicles can be obtained that will be instrumental for unraveling the molecular mechanisms behind inter-leaflet coupling (83).
At present, increasing evidence suggests that signaling nanodomains are also present in endosomes, putting forward the concept that endosomes are in fact intracellular signal transduction stations (84, 85). Interestingly, Albi and coworkers have recently demonstrated the existence of lipid domains in the cell nucleus and their role in regulating enzymes involved in vitamin D3 uptake, therefore influencing cell differentiation (86, 87). Although the nanodomains present in nuclear membranes and endosomes are likely to play important roles in numerous cellular processes (88), further investigation is needed to fully understand their properties and clarify their functions.
The current challenge in the membrane domain field is to understand how a cell integrates multiple biochemical strategies to induce, maintain or modify membrane compartments to regulate signal transduction in time and space. While the fluorescence microscopy techniques described here provide information on protein behavior that would not be accessible through conventional biochemical techniques, even higher spatiotemporal resolution will be needed to address this challenge. Enhancements in spatial resolution have rapidly progressed in the past decade with advancements in super-resolution techniques such as STED (68), PALM (68) and GSDIM (89). New advances in NSOM tip geometry are capable of enhancing the local electromagnetic fields to give ~30 nm axial resolution of protein localization on cell membranes (90). Super-resolution techniques are starting to close the temporal gap with high frame rate techniques using PALM (91, 92), STED (93) and Structured Illumination Microscopy (94). To understand the interplay between multiple proteins and lipids, higher multiplex imaging is needed. Hyperspectral microscopes that acquire the full emission spectrum of the sample rather than depending on filters (95), make it possible to increase the number of fluorophores that can be used to simultaneously image proteins and membrane markers. Many of these improvements depend, not only on improved instrumentation, but also on the generation of new fluorescent proteins, organic dyes and fluorescent nanoparticles that will increase multi-color capability and allow for longer/faster live cell imaging. Lipid labeling remains a specific challenge, but new strategies are being developed that are bringing new options for imaging of nanodomains and lipid-protein interactions (96–99). As imaging technologies continue to improve, cell biologists will be able to answer questions at spatiotemporal scales that were previously inaccessible. Integration of information from multiple disciplines, such as high resolution microscopy coupled to readouts of biochemical events, will ultimately provide a more complete description of how cell signaling is regulated.
Acknowledgements
We apologize to colleagues whose work could not be cited due to space limitations. We thank B. Wilson for critical reading of the manuscript and helpful suggestions and the reviewers for excellent suggestions. Work in our laboratories is supported by a Young Investigator Grant from the Human Frontier Science Program (HFSP) to DSL and AC; AC is the recipient of a Meervoud grant (836.09.002) from the Netherlands Organisation for Scientific Research (NWO) and DSL is supported by NSF CAREER Award MCB-0845062 and the UNM Spatiotemporal Modeling Center NIH P50GM085273.
Keywords
- Plasma membrane
The outer cell membrane made of a phospholipid bilayer that separates the cellular contents from the extracellular environment. Proteins embedded in the plasma membrane regulate responses to extracellular signals
- Signal transduction
Activation of a membrane protein, by ligand binding or other cues, initiates a cascade of protein-protein interactions that propagates the signal to the nucleus and produces a physiological response
- Protein biochemical modification
The chemical modification of a protein during or after its translation. This is achieved through the addition of biochemical functional groups such as lipids, carbohydrates, acetates, phosphates, etc., that change the chemical nature of certain amino acids and extend the range of functions of the protein
- Membrane partitioning
The formation of localized compartments on the plasma membrane due to lipid-lipid, lipid-protein, or protein-protein interactions
- Lipid nanodomains
Favored interactions between certain types of lipids lead to their co-segregation in domains at the cell membrane
- Actin Corrals
The restriction of membrane protein or lipid mobility by membrane proximal actin structure
- Protein Islands
Diffusional trapping of membrane proteins in discrete protein-rich microdomains
- Fluorescence microscopy
Optical imaging with sub-micron resolution using fluorescent-markers that allows for live cell imaging of dynamic cellular processes
References
- 1.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 5.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 8.van Zanten TS, Cambi A, Garcia-Parajo MF. A nanometer scale optical view on the compartmentalization of cell membranes. Biochim Biophys Acta. 2010;1798:777–787. doi: 10.1016/j.bbamem.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U S A. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiyokawa E, Baba T, Otsuka N, Makino A, Ohno S, Kobayashi T. Spatial and functional heterogeneity of sphingolipid-rich membrane domains. J Biol Chem. 2005;280:24072–24084. doi: 10.1074/jbc.M502244200. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Qin J, Chen ZW. Fluorescence-topographic NSOM directly visualizes peak-valley polarities of GM1/GM3 rafts in cell membrane fluctuations. J Lipid Res. 2008;49:2268–2275. doi: 10.1194/jlr.D800031-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ira, Johnston LJ. Sphingomyelinase generation of ceramide promotes clustering of nanoscale domains in supported bilayer membranes. Biochim Biophys Acta. 2008;1778:185–197. doi: 10.1016/j.bbamem.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Li X, Becker KA, Gulbins E. Ceramide-enriched membrane domains--structure and function. Biochim Biophys Acta. 2009;1788:178–183. doi: 10.1016/j.bbamem.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 14.Niemela PS, Ollila S, Hyvonen MT, Karttunen M, Vattulainen I. Assessing the nature of lipid raft membranes. PLoS Comput Biol. 2007;3:e34. doi: 10.1371/journal.pcbi.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Liu W, Anderson LY, Jiang QX. Lipid-dependent gating of a voltage-gated potassium channel. Nat Commun. 2:250. doi: 10.1038/ncomms1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coskun U, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci U S A. 108:9044–9048. doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pontier SM, Percherancier Y, Galandrin S, Breit A, Gales C, Bouvier M. Cholesterol-dependent separation of the beta2-adrenergic receptor from its partners determines signaling efficacy: insight into nanoscale organization of signal transduction. J Biol Chem. 2008;283:24659–24672. doi: 10.1074/jbc.M800778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingwood D, Binnington B, Rog T, Vattulainen I, Grzybek M, Coskun U, Lingwood CA, Simons K. Cholesterol modulates glycolipid conformation and receptor activity. Nat Chem Biol. 2011;7:260–262. doi: 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- 19.Carroll-Portillo A, Spendier K, Pfeiffer J, Griffiths G, Li H, Lidke KA, Oliver JM, Lidke DS, Thomas JL, Wilson BS, Timlin JA. Formation of a mast cell synapse: Fc epsilon RI membrane dynamics upon binding mobile or immobilized ligands on surfaces. J Immunol. 2010;184:1328–1338. doi: 10.4049/jimmunol.0903071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Zanten TS, Gomez J, Manzo C, Cambi A, Buceta J, Reigada R, Garcia-Parajo MF. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc Natl Acad Sci U S A. 2010;107:15437–15442. doi: 10.1073/pnas.1003876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn PJ. A lipid matrix model of membrane raft structure. Prog Lipid Res. 49:390–406. doi: 10.1016/j.plipres.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 24.de Keijzer S, Meddens MB, Kilic D, Joosten B, Reinieren-Beeren I, Lidke DS, Cambi A. Interleukin-4 Alters Early Phagosome Phenotype by Modulating Class I PI3K Dependent Lipid Remodeling and Protein Recruitment. PLoS One. 2011;6:e22328. doi: 10.1371/journal.pone.0022328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 26.Lasserre R, Guo XJ, Conchonaud F, Hamon Y, Hawchar O, Bernard AM, Soudja SM, Lenne PF, Rigneault H, Olive D, Bismuth G, Nunes JA, Payrastre B, Marguet D, He HT. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]
- 27.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 28.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 29.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 31.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 32.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 33.Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters (Review) Mol Membr Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bijlmakers MJ. Protein acylation and localization in T cell signaling (Review) Mol Membr Biol. 2009;26:93–103. doi: 10.1080/09687680802650481. [DOI] [PubMed] [Google Scholar]
- 35.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 36.Schey KL, Gutierrez DB, Wang Z, Wei J, Grey AC. Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry. 2010;49:9858–9865. doi: 10.1021/bi101415w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibiza S, Perez-Rodriguez A, Ortega A, Martinez-Ruiz A, Barreiro O, Garcia-Dominguez CA, Victor VM, Esplugues JV, Rojas JM, Sanchez-Madrid F, Serrador JM. Endothelial nitric oxide synthase regulates N-Ras activation on the Golgi complex of antigen-stimulated T cells. Proc Natl Acad Sci U S A. 2008;105:10507–10512. doi: 10.1073/pnas.0711062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raines KW, Cao GL, Lee EK, Rosen GM, Shapiro P. Neuronal nitric oxide synthase-induced S-nitrosylation of H-Ras inhibits calcium ionophore-mediated extracellular-signal-regulated kinase activity. Biochem J. 2006;397:329–336. doi: 10.1042/BJ20052002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thibeault S, Rautureau Y, Oubaha M, Faubert D, Wilkes BC, Delisle C, Gratton JP. S-nitrosylation of beta-catenin by eNOS-derived NO promotes VEGF-induced endothelial cell permeability. Mol Cell. 39:468–476. doi: 10.1016/j.molcel.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Into T, Inomata M, Nakashima M, Shibata K, Hacker H, Matsushita K. Regulation of MyD88-dependent signaling events by S nitrosylation retards toll-like receptor signal transduction and initiation of acute-phase immune responses. Mol Cell Biol. 2008;28:1338–1347. doi: 10.1128/MCB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker TL, Booden MA, Buss JE. S-Nitrosocysteine increases palmitate turnover on Ha-Ras in NIH 3T3 cells. J Biol Chem. 2000;275:22037–22047. doi: 10.1074/jbc.M001813200. [DOI] [PubMed] [Google Scholar]
- 43.Raju K, Doulias PT, Tenopoulou M, Greene JL, Ischiropoulos H. Strategies and tools to explore protein S-nitrosylation. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 45.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 46.Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, Zhang Z, Xu L, Hyun YM, Kim M, Zhuang S, Chin YE. Acetylation modulates prolactin receptor dimerization. Proc Natl Acad Sci U S A. 107:19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. Functions of cell surface galectin-glycoprotein lattices. Curr Opin Struct Biol. 2007;17:513–520. doi: 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demotte N, Stroobant V, Courtoy PJ, Van Der Smissen P, Colau D, Luescher IF, Hivroz C, Nicaise J, Squifflet JL, Mourad M, Godelaine D, Boon T, van der Bruggen P. Restoring the association of the T cell receptor with CD8 reverses anergy in human tumor-infiltrating lymphocytes. Immunity. 2008;28:414–424. doi: 10.1016/j.immuni.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Nieminen J, Kuno A, Hirabayashi J, Sato S. Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J Biol Chem. 2007;282:1374–1383. doi: 10.1074/jbc.M604506200. [DOI] [PubMed] [Google Scholar]
- 51.Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–739. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- 52.Lajoie P, Partridge EA, Guay G, Goetz JG, Pawling J, Lagana A, Joshi B, Dennis JW, Nabi IR. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J Cell Biol. 2007;179:341–356. doi: 10.1083/jcb.200611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 54.Manzo C, van Zanten TS, Garcia-Parajo MF. Nanoscale fluorescence correlation spectroscopy on intact living cell membranes with NSOM probes. Biophys J. 2011;100:L8–L10. doi: 10.1016/j.bpj.2010.12.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bader AN, Hofman EG, Henegouwen PMPVE, Gerritsen HC. Imaging of protein cluster sizes by means of confocal time-gated fluorescence anisotropy microscopy. Optics Express. 2007;15:6934–6945. doi: 10.1364/oe.15.006934. [DOI] [PubMed] [Google Scholar]
- 56.Sharma P, Varma R, Sarasij RC, Ira Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 57.Brameshuber M, Weghuber J, Ruprecht V, Gombos I, Horvath I, Vigh L, Eckerstorfer P, Kiss E, Stockinger H, Schutz GJ. Imaging of mobile long-lived nanoplatforms in the live cell plasma membrane. J Biol Chem. 2010;285:41765–41771. doi: 10.1074/jbc.M110.182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson BS, Pfeiffer JR, Oliver JM. FcepsilonRI signaling observed from the inside of the mast cell membrane. Mol Immunol. 2002;38:1259–1268. doi: 10.1016/s0161-5890(02)00073-1. [DOI] [PubMed] [Google Scholar]
- 60.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J Cell Sci. 2007;120:2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 62.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treanor B, Depoil D, Gonzalez-Granja A, Barral P, Weber M, Dushek O, Bruckbauer A, Batista FD. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 2010;32:187–199. doi: 10.1016/j.immuni.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treanor B, Depoil D, Bruckbauer A, Batista FD. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J Exp Med. 2011;208:1055–1068. doi: 10.1084/jem.20101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaqaman K, Kuwata H, Touret N, Collins R, Trimble WS, Danuser G, Grinstein S. Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell. 2011;146:593–606. doi: 10.1016/j.cell.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- 69.Lidke DS, Wilson BS. Caught in the act: quantifying protein behaviour in living cells. Trends Cell Biol. 2009;19:566–574. doi: 10.1016/j.tcb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itano MS, Neumann AK, Liu P, Zhang F, Gratton E, Parak WJ, Thompson NL, Jacobson K. DC-SIGN and influenza hemagglutinin dynamics in plasma membrane microdomains are markedly different. Biophys J. 2011;100:2662–2670. doi: 10.1016/j.bpj.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Bakker BI, de Lange F, Cambi A, Korterik JP, van Dijk EM, van Hulst NF, Figdor CG, Garcia-Parajo MF. Nanoscale organization of the pathogen receptor DC-SIGN mapped by single-molecule high-resolution fluorescence microscopy. Chemphyschem. 2007;8:1473–1480. doi: 10.1002/cphc.200700169. [DOI] [PubMed] [Google Scholar]
- 72.Cambi A, de Lange F, van Maarseveen NM, Nijhuis M, Joosten B, van Dijk EM, de Bakker BI, Fransen JA, Bovee-Geurts PH, van Leeuwen FN, Van Hulst NF, Figdor CG. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol. 2004;164:145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Digman MA, Wiseman PW, Choi C, Horwitz AR, Gratton E. Stoichiometry of molecular complexes at adhesions in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2170–2175. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Digman MA, Wiseman PW, Horwitz AR, Gratton E. Detecting protein complexes in living cells from laser scanning confocal image sequences by the cross correlation raster image spectroscopy method. Biophys J. 2009;96:707–716. doi: 10.1016/j.bpj.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci U S A. 2010;107:16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godin AG, Costantino S, Lorenzo LE, Swift JL, Sergeev M, Ribeiro-da-Silva A, De Koninck Y, Wiseman PW. Revealing protein oligomerization and densities in situ using spatial intensity distribution analysis. Proc Natl Acad Sci U S A. 2011;108:7010–7015. doi: 10.1073/pnas.1018658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swift JL, Godin AG, Dore K, Freland L, Bouchard N, Nimmo C, Sergeev M, De Koninck Y, Wiseman PW, Beaulieu JM. Quantification of receptor tyrosine kinase transactivation through direct dimerization and surface density measurements in single cells. Proc Natl Acad Sci U S A. 2011;108:7016–7021. doi: 10.1073/pnas.1018280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JE, Lazareno S, Molloy JE, Birdsall NJ. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, Fujiwara TK, Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, vanvBergen en Henegouwen PMP, Wilson BS, Lidke DS. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand-binding. Nat Struct Mol Biol. doi: 10.1038/nsmb.2135. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactionsin situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 83.Chiantia S, Schwille P, Klymchenko AS, London E. Asymmetric GUVs prepared by MbetaCD-mediated lipid exchange: an FCS study. Biophys J. 100:L1–L3. doi: 10.1016/j.bpj.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fehrenbacher N, Bar-Sagi D, Philips M. Ras/MAPK signaling from endomembranes. Mol Oncol. 2009;3:297–307. doi: 10.1016/j.molonc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 86.Bartoccini E, Marini F, Damaskopoulou E, Lazzarini R, Cataldi S, Cascianelli G, Gil Garcia M, Albi E. Nuclear Lipid Microdomains Regulate Nuclear Vitamin D3 Uptake and Influence Embryonic Hippocampal Cell Differentiation. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cascianelli G, Villani M, Tosti M, Marini F, Bartoccini E, Magni MV, Albi E. Lipid microdomains in cell nucleus. Mol Biol Cell. 2008;19:5289–5295. doi: 10.1091/mbc.E08-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 89.Lalkens B, Testa I, Willig KI, Hell SW. MRT letter: Nanoscopy of protein colocalization in living cells by STED and GSDIM. Microsc Res Tech. 2011 doi: 10.1002/jemt.21026. [DOI] [PubMed] [Google Scholar]
- 90.van Zanten TS, Lopez-Bosque MJ, Garcia-Parajo MF. Imaging individual proteins and nanodomains on intact cell membranes with a probe-based optical antenna. Small. 2010;6:270–275. doi: 10.1002/smll.200901204. [DOI] [PubMed] [Google Scholar]
- 91.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 93.Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MG. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sinclair MB, Haaland DM, Timlin JA, Jones HD. Hyperspectral confocal microscope. Appl Opt. 2006;45:6283–6291. doi: 10.1364/ao.45.006283. [DOI] [PubMed] [Google Scholar]
- 96.Haberkant P, Schmitt O, Contreras FX, Thiele C, Hanada K, Sprong H, Reinhard C, Wieland FT, Brugger B. Protein-sphingolipid interactions within cellular membranes. J Lipid Res. 2008;49:251–262. doi: 10.1194/jlr.D700023-JLR200. [DOI] [PubMed] [Google Scholar]
- 97.Haberkant P, van Meer G. Protein-lipid interactions: paparazzi hunting for snap-shots. Biol Chem. 2009;390:795–803. doi: 10.1515/BC.2009.074. [DOI] [PubMed] [Google Scholar]
- 98.Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem Biol. 2009;4:581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 99.Yap MC, Kostiuk MA, Martin DD, Perinpanayagam MA, Hak PG, Siddam A, Majjigapu JR, Rajaiah G, Keller BO, Prescher JA, Wu P, Bertozzi CR, Falck JR, Berthiaume LG. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res. 2010;51:1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marguet D, Lenne PF, Rigneault H, He HT. Dynamics in the plasma membrane: how to combine fluidity and order. Embo J. 2006;25:3446–3457. doi: 10.1038/sj.emboj.7601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 102.Robia SL, Campbell KS, Kelly EM, Hou Z, Winters DL, Thomas DD. Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ Res. 2007;101:1123–1129. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tramier M, Coppey-Moisan M. Fluorescence anisotropy imaging microscopy for homo-FRET in living cells. Methods Cell Biol. 2008;85:395–414. doi: 10.1016/S0091-679X(08)85017-0. [DOI] [PubMed] [Google Scholar]
- 104.He HT, Marguet D. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu Rev Phys Chem. 2011;62:417–436. doi: 10.1146/annurev-physchem-032210-103402. [DOI] [PubMed] [Google Scholar]
- 105.Digman MA, Gratton E. Lessons in fluctuation correlation spectroscopy. Annu Rev Phys Chem. 2011;62:645–668. doi: 10.1146/annurev-physchem-032210-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochemistry and Biophysics. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 107.Leung BO, Chou KC. Review of super-resolution fluorescence microscopy for biology. Appl Spectrosc. 2011;65:967–980. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 108.van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia-Parajo MF. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci U S A. 2009;106:18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]