Abstract
Protein arginine deiminase activity (PAD) is increased in cancer, rheumatoid arthritis, and ulcerative colitis. Although the link between abnormal PAD activity and disease is clear, the relative contribution of the individual PADs to human disease is not known; there are 5 PAD isozymes in humans. Building on our previous development of F- and Cl-amidine as potent pan-PAD irreversible inhibitors, we describe herein a library approach that was used to identify PAD-selective inhibitors. Specifically, we describe the identification of Thr-Asp-F-amidine (TDFA) as a highly potent PAD4 inactivator that displays ≥15-fold selectivity for PAD4 versus PAD1 and ≥50-fold versus PADs 2 and 3. This compound is active in cells and can be used to inhibit PAD4 activity in cellulo. The structure of the PAD4•TDFA complex has also been solved and the structure and mutagenesis data indicate that the enhanced potency is due to interactions between the side chains of Q346, R374, and R639. Finally, we converted TDFA into a PAD4-selective ABPP and demonstrate that this compound, biotin-TDFA, can be used to selectively isolate purified PAD4 in vitro. In total, TDFA and biotin-TDFA represent PAD4-selective chemical probes that can be used to study the physiological roles of this enzyme.
Over the past decade, aberrant protein citrullination has emerged as an important post-translational modification (PTM) that is associated with human disease (1, 2). Although this PTM is best known to be elevated in rheumatoid arthritis (RA) (3), and to a lesser extent multiple sclerosis (MS) (4), recent data suggests that elevated protein citrullination is a hallmark of multiple additional human diseases including, ulcerative colitis (UC), ankylosing spondylitis (AS), osteoarthritis (OA), Crohn’s disease, glaucoma, and cancer (1–9). This PTM, which is generated by the Protein Arginine Deiminases (PADs), converts peptidyl-arginine into peptidyl-citrulline; as such thus this modification is alternatively termed citrullination or deimination. Among the five PAD isozymes, dysregulated PAD2 and PAD4 appear to be most associated with the aforementioned diseases. For example, PADs 2 and 4 are overexpressed in RA, MS, cancer, AS, OA, and glaucoma (3–9). Furthermore, mutations in PAD4 are associated with an elevated risk of developing both RA and Crohn’s Disease (7, 10). With respect to RA, a chronic, inflammatory, autoimmune disease that affects 1% of the world’s population (11, 12), one of the main connections between dysregulated PAD activity and this disease is the fact that RA patients produce anti-citrullinated peptide antibodies (ACPA) and these autoantibodies are highly specific for RA (13, 14). Additionally, the RA susceptibility locus (HLA-DRB*0401) binds with strong affinity to citrullinated peptides (15). With respect to MS, myelin basic protein, which is the main component of the myelin sheeth, is highly citrullinated and this increased citrullination is associated with the overexpression of both PAD2 and PAD4 (4, 16).
In addition to its roles in RA and MS, PAD4 is overexpressed in a number of tumors including breast adenocarcinomas, colorectal adenocarcinomas, and endometrial carcinomas, as well as a variety of cancer derived cell lines (1, 6). It has also been found that PAD4 is present in the blood of cancer patients and that its levels decrease after tumor resection (6). In total, these data suggest that PAD inhibition, and in particular the inhibition of PAD2 and PAD4, represents a novel therapeutic approach for a host of human diseases.
Previously, we reported the synthesis and characterization of two of the most potent PAD inhibitors, F- and Cl-amidine (17, 18). The structures of these compounds mimic the structure of Benzoyl Arginine Amide (BAA), a small molecule PAD substrate, except that the guanidinium group is replaced with a haloacetamidine warhead that covalently modifies an active site Cys that plays a critical role in nucleophilic catalysis. Inactivation likely proceeds via a multistep mechanism that involves nucleophilic attack by Cys645 (PAD4 numbering) on the iminium carbon of the warhead (19). Subsequent protonation of the resulting tetrahedral intermediate by His471 stabilizes this intermediate, which helps to facilitate the displacement of the halide, thus forming the inactivated thioether adduct. F- and Cl-amidine are active in cells and, most significantly, Cl-amidine reduces disease severity in animal models of RA, UC, and neuron degeneration (20–22), thereby helping to validate the PADs as therapeutic targets. However, both compounds inhibit all four of the active PAD isozymes (PAD6 activity has not been detected) with similar IC50 values (Table 1), thereby illustrating the need for PAD-specific inhibitors that can be used to address the relative contribution of individual PAD isozymes to human disease (18, 23–25). Additionally, such compounds will be useful chemical probes to dissect their physiological roles in normal cells. To develop such isoform-specific compounds, we describe herein the parallel solid phase synthesis and screening of a 264 member fluoroacetamidine containing peptide library, which led to the identification of TDFA as a highly specific PAD4 inactivator (Figure 1).
Table 1.
Summary of inactivators
| Inactivator | PAD1 | PAD2 | PAD3 | PAD4 |
|---|---|---|---|---|
| TDFA | ||||
| IC50 | 8.5 ± 0.8 | 71 ± 4.4 | 26 ± 7.4 | 2.3 ± 0.2 |
| KI (µM) | ND | ND | 180 ± 60 | 16 ± 10 |
| kinact (min−1) | ND | ND | 0.06 ± 0.009 | 0.4 ± 0.1 |
| Kinact/KI (M−1*min−1) | 1,700 | 500 | 400 | 26,000 |
| Fold Selectivity | 15 | 52 | 65 | 1 |
| TDCA | ||||
| IC50 | 2.8 ± 0.2 | 59 ± 6.6 | 32 ± 11 | 3.4 ± 0.5 |
| KI (µM) | 60 ± 20 | ND | 37 ± 16 | 34 ± 6 |
| kinact (min−1) | 1.2 ± 0.2 | ND | 0.03 ± 0.004 | 0.8 ± 0.1 |
| Kinact/KI (M−1*min−1) | 21,000 | 300 | 920 | 24,000 |
| Fold Selectivity | 1.1 | 80 | 26 | 1 |
| F-amidine | ||||
| IC50 | 30 ± 1.3 | 51 ± 9.0 | ≥ 350 | 22 ± 2.1 |
| KI (µM) | 110 ± 40 | ND | 290 ± 190 | 330 ± 90 |
| kinact (min−1) | 0.30 ± 0.03 | ND | 0.05 ± 0.01 | 1.0 ± 0.1 |
| Kinact/KI (M−1*min−1) | 2800 | 380 | 170 | 3000 |
| Fold Selectivity | 1.1 | 8 | 18 | 1 |
| Cl-amidine | ||||
| IC50 | 0.8 ± 0.3 | 17 ± 3.1 | 6.2 ± 1.0 | 5.9 ± 0.3 |
| KI (µM) | 62 ± 11 | ND | 28 ± 7.3 | 180 ± 33 |
| kinact (min-1) | 2.3 ± 0.1 | ND | 0.06 ± 0.005 | 2.4 ± 0.2 |
| Kinact/KI (M−1*min−1) | 37,000 | 1,200 | 2000 | 13,000 |
| Fold Selectivity | 0.4 | 11 | 7 | 1 |
Figure 1.
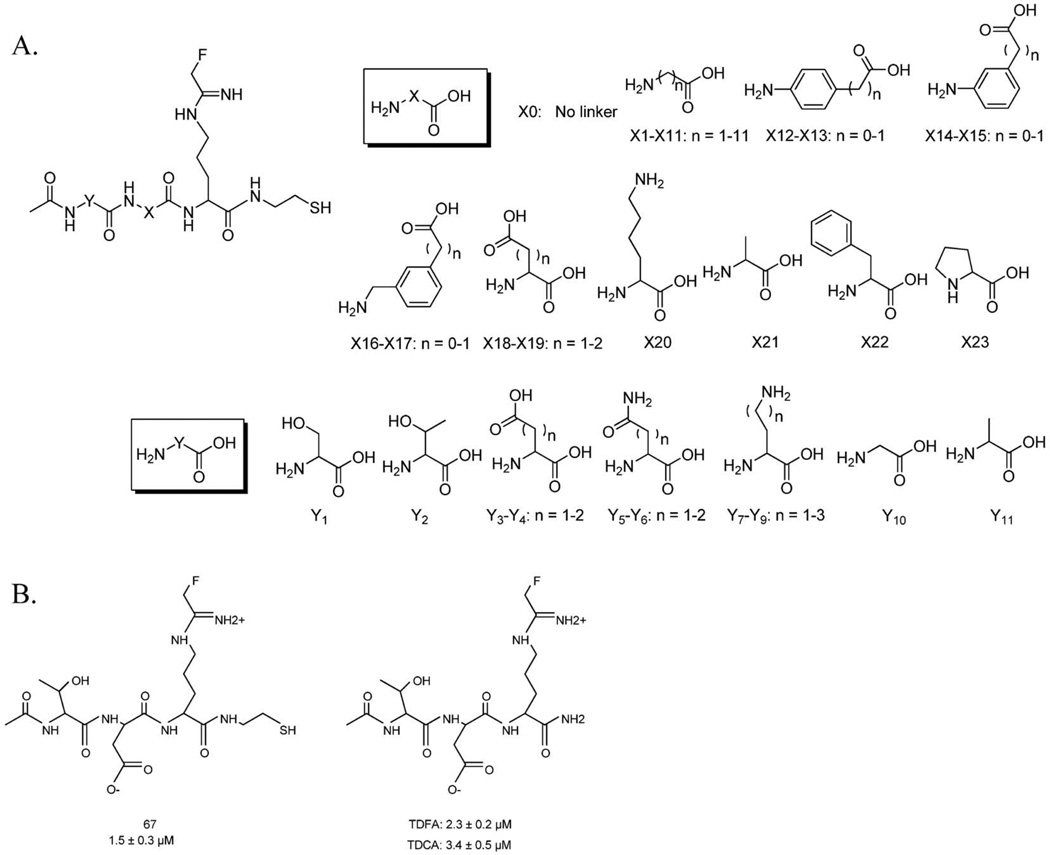
A. Structure of library compounds. B. Top hit from the library screen.
Inspired by the work of Lawrence and colleagues (26), the library was synthesized on cystamine-modified Tentagel resin, which allows for facile cleavage of the linker with DTT to facilitate in vitro screening of the inhibitors. The X and Y linkers present in the library (Figure 1A), which explore length, flexibility, aromaticity, polarity, and ionic interactions, were coupled to the resin using standard Fmoc-based solid phase peptide synthesis methods. In the first step of the synthesis (Scheme S1) Boc protected cystamine dihydrochloride was coupled to the Tentagel resin. Once the Boc group was removed, Fmoc-Orn(Dde) was coupled, the Fmoc group was removed and then the resin was divided into 24 tubes where the X linkers were subsequently coupled. Once again the Fmoc group was removed and each of the 24 tubes was divided into 11 wells of a 96-well filter plate. The N-terminus was acetylated upon removal of the Fmoc group and the Dde protecting group of Orn was removed with 2% hydrazine in DMF. The fluoroacetamidine warhead was then coupled to Orn, followed by deprotection of the side chains and cleavage from the resin in DTT containing HEPES buffer. Cleavage of the library members into a buffered solution allowed for these inhibitors to be assayed directly after cleavage. To initially screen the library, individual library members were pre-incubated with PAD4 for 15 min and then residual activity was measured at a saturating concentration of substrate. Under these conditions, cystamine modified F-amidine (FASH), the parent compound, yields 50% inhibition. Compounds that were more potent that FASH were considered hits. Although these conditions may discriminate against slow binding inhibitors, they provide a good balance between increases in potency that are due to increased affinity versus rate of inactivation, thereby enabling the selection of inhibitors with improved potency via an overall increase in the efficiency of inactivation. As can be seen in Figure S1, the results of the screen identified a number of compounds that appeared to be more potent than FASH.
The top 10 hits identified from the initial screen were synthesized on a larger scale, purified by HPLC, masses confirmed by MS, and IC50 values determined (Figure S2). Based on the results of the screen, inhibitor 67 was the most potent compound (IC50 = 1.5 ± 0.3 µM) (Figure 1B). Once identified, the amide version of 67, i.e., Thr-Asp F-amidine (TDFA), was synthesized and characterized (Figure 1B). For this compound, the synthesis was performed on Rink Amide resin in order to better mimic the structure of F-amidine (17, 18). Significantly, the IC50 values obtained for TDFA represents an ~10-fold enhancement in potency relative to F-amidine. Note that the IC50 values were almost identical for 67 and TDFA (1.5 ± 0.3 and 2.3 ± 0.2 µM, respectively), which indicates that the thiol moiety in 67 does not make a significant contribution to the observed enhancement in potency. Given that the replacement of the fluoro group in F-amidine with a chloro group led to the more potent Cl-amidine, we also synthesized the chloro containing analog of TDFA. Interestingly, the IC50 value for this compound, denoted Thr-Asp Cl-amidine (TDCA) (3.4 ± 0.5 µM), is not significantly different than that of TDFA (Table 1). We speculate that the lack of enhanced potency is due to an inability to properly position the warhead for nucleophilic attack on the iminium carbon, as we have previously shown that proper positioning of the warhead is critical for efficient enzyme inactivation (17, 19, 23).
In order to determine that both TDFA and TDCA irreversibly inhibit PAD4, an excess of each inhibitor was incubated with enzyme to form the PAD4•inactivator complex. After 20 h of dialysis, activity was measured. The results of these experiments showed no recovery of activity (Figure S3), thereby indicating that these compounds are irreversible inactivators. To ensure that the inability to recover activity was due to a modification of an active site residue, substrate protection experiments were performed. For these experiments, product formation was measured as a function of time with both TDFA and TDCA at two different concentrations of substrate. Consistent with the modification of the active site Cys, the rates of inactivation are considerably lower at the higher concentration of substrate (Figure S4).
To evaluate inhibitor selectivity, IC50 values were also determined for PADs 1–3 with both TDFA and TDCA (Table 1). Based on the IC50 values, TDFA is ≥ 4-fold selective for PAD4 relative to the other PADs. On the other hand, TDCA inhibits both PAD1 and PAD4 with ≥ 20-fold selectivity as compared to the other PADs. Given that IC50 values are dependent on both the substrate concentration as well as an isozyme’s affinity for that substrate, we also determined the kinact, KI, and kinact/KI values for all the isozymes to better gauge inhibitor selectivity. For PAD4, the KI, kinact, and kinact/KI values with TDFA are 16 ± 10 µM, 0.4 ± 0.1 min−1, and 26,000 M−1*min−1, respectively (Figure S5). For TDCA, the KI, kinact, and kinact/KI values are 34 ± 6 µM, = 0.8 ± 0.1 min−1, and 24,000 M−1*min−1, respectively (Figure S5). These values are consistent with the IC50 values in that they show that TDFA and TDCA are equipotent PAD4 inhibitors. The KI, kinact, and kinact/KI values for the remaining isozymes are summarized in Table 1. Based on these results, TDFA inhibits PAD4 with ≥ 15-fold selectivity as compared to the other PADs. Most impressively, this compound inhibits PAD4 over PAD2 with ≥ 52-fold selectivity. This shows an improved selectivity when compared to the first generation inhibitors, F- and Cl-amidine (Table 1). Interestingly, TDCA inhibits both PAD1 and PAD4 with ≥26-fold selectivity as compared to PAD3 and an impressive ≥ 80-fold selectivity over PAD2. The lack of selectivity between PADs 1 and 4 likely reflects the greater leaving group potential of the chloro group and the role that warhead positioning plays in enzyme inactivation.
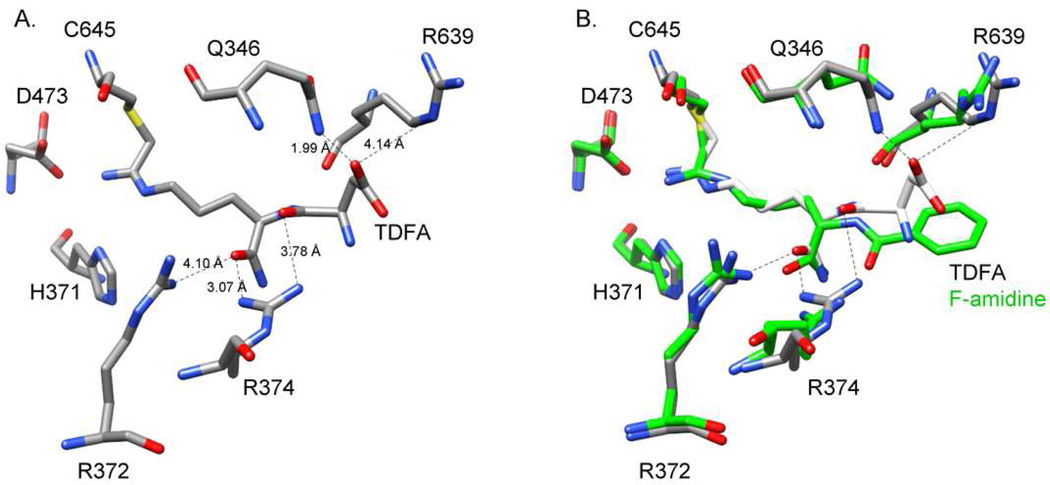
To explore the molecular basis for enhanced potency of TDFA and TDCA, we synthesized the corresponding substrate analog, i.e., a peptide with the sequence Ac-TDR-NH2, and evaluated its ability to act as a substrate for PADs 1, 2, 3, and 4. Interestingly, this peptide is an excellent substrate for both PADs 1 and 4, but a relatively poor PAD2 and 3 substrate (Table S1), which is consistent with the data obtained for the inhibitors. We also determined the structure of the PAD4•TDFA complex to 2.98 Å resolution. In comparison to the PAD4•F-amidine complex, the structures are virtually superimposable (rmsd = 0.367 Å for comparable 623 Cα). Although electron density was not detected for the Thr residue in TDFA, the position of the aspartyl group was well resolved and suggested that the enhanced potency observed with this compound is due to potential interactions between the Asp carboxylate and Q346, R374, and R639 (Figure 2). We confirmed that these residues play a role in TDFA binding by determining IC50 values for TDFA with the Q346A, R374Q, and R639Q mutants. The results indicate that the values are increased by 1.9-, 5.2-, and 2.9-fold, respectively (Table S2). Thus, the enhanced potency is most likely due to interactions between TDFA and both R374 and R639, as R374 is conserved in PADs 1 and 4 (it is a glycine in PADs 2 and 3 and an alanine in PAD6) and R639 is unique to PAD4 (R639 is a leucine in PADs 1 and 3, a phenylalanine in PAD2, and a glutamate in PAD6). This conclusion is supported by the fact that F-amidine is an equipotent inhibitor of PADs 1 and PAD4, whereas TDFA preferentially inhibits PAD4 over PAD1 by 15-fold. Although Q346 is also unique to PAD4, the lack of a more dramatic effect with the Q346A mutant is not altogether unsurprising as this structure represents a dead-end complex, and indicates that this residue contributes minimally to the binding of the initial encounter complex.
Figure 2.
Crystal Structure of PAD4 with TDFA bound. A. Active site residues of PAD4 with TDFA bound. B. Overlay of PAD4 with TDFA (elemental) and F-amidine (green) bound.
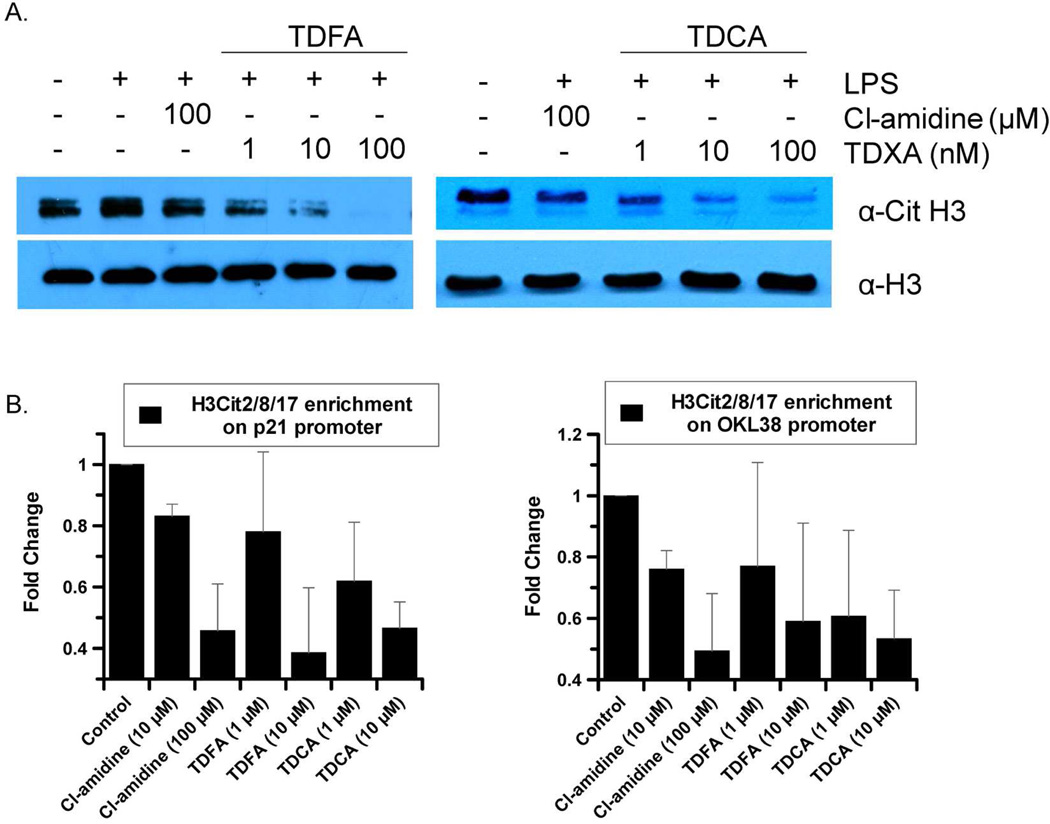
To evaluate cellular activity, we examined whether TDFA and TDCA could globally inhibit histone citrullination in HL-60 granulocytes (Figure 3). The results indicate that concentrations of TDFA and TDCA as low as 1 nM inhibit histone citrullination to a greater extent than 100 µM Cl-amidine. To evaluate histone citrullination on the p21 and OKL38 promoters, whose expression is known to be repressed by PAD4 (27, 28), chromatin immunoprecipitation (ChIP) experiments were performed with MCF-7 cells. Briefly Cl-amidine was diluted to a final concentration of 10 and 100 µM in media, whereas TDFA and TDCA were diluted to concentrations of 1 and 10 µM; PBS was used as a control. All compounds, as well as the PBS control were incubated for 48 h prior to harvesting for ChIP analysis. These results show that both TDFA and TDCA are at least 10-fold more potent than Cl-amidine (Figure 3). The enhanced cellular activity indicates that TDFA and TDCA are more potent than Cl-amidine both in vitro and in cell culture. It is also noteworthy that µM amounts of TDFA and TDCA were required to inhibit the citrullination of specific promoters, whereas nM amounts were required to inhibit global citrullination. A key difference between these studies is the incubation time (30 min versus 48 h for the global citrullination and ChIP assays, respectively), suggesting that TDFA and TDCA may be subject to time dependent degradation. Nevertheless, relative to the 10-fold enhancement in in vitro potency, the ≥100-fold enhancement to in vivo activity on the shorter time scale may be due to anyone of a number of factors including increased permeability.
Figure 3.
A. Western blot analysis of HL-60 granulocytes treated with either TDFA, TDCA or Cl-amidine. B. Chromatin Immunoprecipitation (ChIP) experiment to demonstrate the effects of TDFA and TDCA on H3Cit2/8/17 levels on the p21 and OKL38 promoters. Each ChIP experiment was conducted a minimum of three times with independent chromatin isolates to ensure reproducibility.
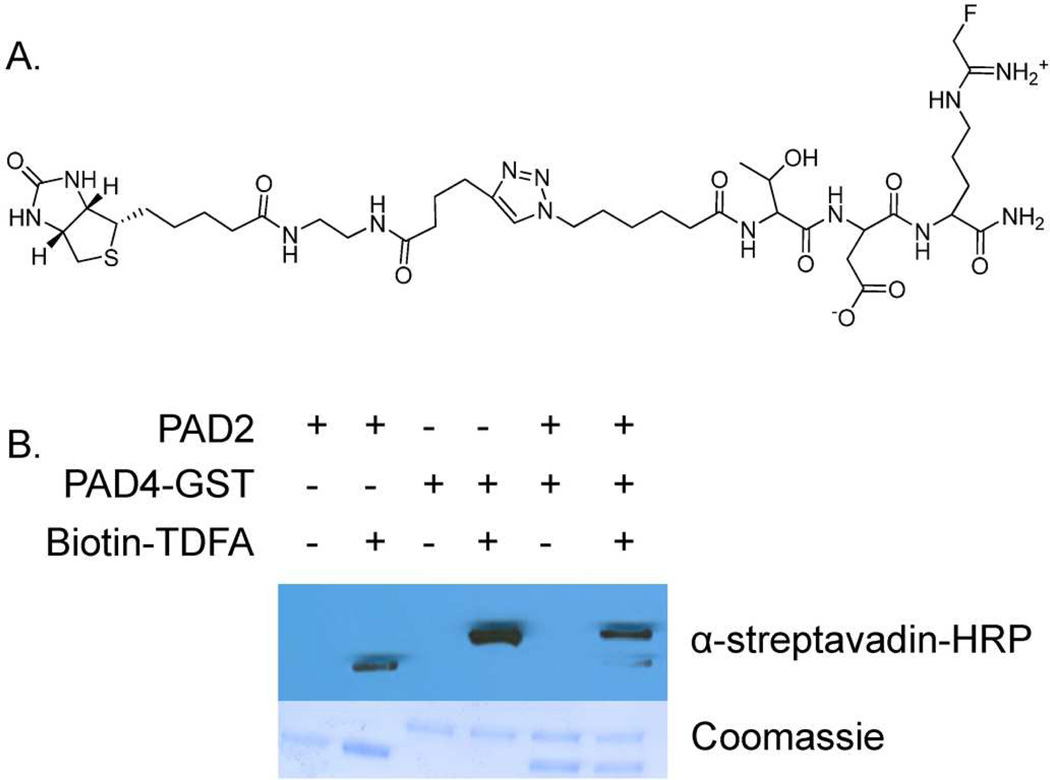
Since TDFA is selective for PAD4, a biotin-conjugated probe was synthesized (biotin-TDFA) to generate a PAD4-selective Activity-based Proteomic Probe or ABPP (Figure 4). Consistent with the fact that the N-terminal Thr does not interact with PAD4 in the crystal structure, the addition of the biotin moiety did not affect the IC50 values. To initially evaluate its ability to selectively isolate PAD4 over PAD2, biotin-TDFA was incubated with equimolar amounts of PADs 2 and 4 for 1 h. The reactions were quenched with 6× SDS-loading dye and separated via SDS-PAGE. Protein was then transferred to a nitrocellulose membrane and visualized with Streptavadin-HRP (Figure 3). Quantifying the bands using Image J, indicate that biotin-TDFA modifies PAD4 with 18-fold selectivity as compared to PAD2. These results suggest that biotin-TDFA will be a useful proteomic probe of PAD4 function. Future experiments to test its ability to selectively isolate PAD4 in cell culture are ongoing.
Figure 4.
A. Structure of biotin conjugated TDFA. B. Western blot showing selectivity of biotin-TDFA.
In summary, the synthesis and screening of a 264 compound library was performed using Fmoc-based solid phase synthesis. From this screen, a highly potent and selective PAD4 inhibitor was identified (TDFA) and readily converted into a PAD4 selective ABPP. This compound will undoubtedly prove to be a useful tool for studying the in vivo roles of PAD4 in both physiological and pathophysiological processes. Furthermore, a unique facet of our approach is our ability to rapidly convert a PAD selective inhibitor into an ABPP that can be used to selectively label/isolate a particular PAD.
Methods
Fmoc protected amino acids and resins were purchased from EMD (Gibbstown, NJ). PADs 1, 2, and 3 were purified by previously established methods (23). PAD4 was purified as previously described (29). The peptide Ac-TDR-NH2 was synthesized using the Fmoc approach and purified by reverse phase HPLC. Complete synthetic methods for the library and all other compounds described herein are included in the supporting information. Complete methods for the kinetic, inhibition and cellular activity assays, as well as the structural studies, are also included in the supporting information.
Supplementary Material
Abbreviations
- PAD
protein arginine deiminase
- Cit
citrulline
- BAEE
benzoyl L-arginine ethyl ester
- BAA
benzoyl L-arginine amide
- DTT
dithiothreitol
- TCEP
Tris(2-carboxyethyl)phosphine hydrochloride
- HEPES
N-(2-hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid)
- Orn
Ornithine
- TDFA
Threonine-Aspartate F-Amidine
- TDCA
Threonine-Aspartate Cl-amidine
- TDR
Ac-Threonine-Aspartate-Arginine-NH2
- ABPP
Activity based proteomic probe
Footnotes
This work was supported in part by The Scripps Research Institute and by NIH grant GM079357 to PRT.
Supporting Information Available
Complete methods. Supplementary Figure S1–S5 and Tables S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 3.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, Saxne T, Malmstrom V, Venables PJ. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58:2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 4.Wood DD, Ackerley CA, Brand B, Zhang L, Raijmakers R, Mastronardi FG, Moscarello MA. Myelin localization of peptidylarginine deiminases 2 and 4: comparison of PAD2 and PAD4 activities. Lab Invest. 2008;88:354–364. doi: 10.1038/labinvest.3700748. [DOI] [PubMed] [Google Scholar]
- 5.Cafaro TA, Santo S, Robles LA, Crim N, Urrets-Zavalia JA, Serra HM. Peptidylarginine deiminase type 2 is over expressed in the glaucomatous optic nerve. Mol Vis. 16:1654–1658. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CC, Isomoto H, Narumi Y, Sato K, Oishi Y, Kobayashi T, Yanagihara K, Mizuta Y, Kohno S, Tsukamoto K. Haplotypes of PADI4 susceptible to rheumatoid arthritis are also associated with ulcerative colitis in the Japanese population. Clin Immunol. 2008;126:165–171. doi: 10.1016/j.clim.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H, Toda T, Kimura N, Maruyama N. Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer's disease. J Neurosci Res. 2005;80:120–128. doi: 10.1002/jnr.20431. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya SK. Retinal deimination in aging and disease. IUBMB Life. 2009;61:504–509. doi: 10.1002/iub.184. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 11.Majithia V, Geraci SA. Rheumatoid arthritis: diagnosis and management. Am J Med. 2007;120:936–939. doi: 10.1016/j.amjmed.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Akil M, Amos RS. ABC of rheumatology. Rheumatoid arthritis--I: Clinical features and diagnosis. BMJ. 1995;310:587–590. doi: 10.1136/bmj.310.6979.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silveira IG, Burlingame RW, von Muhlen CA, Bender AL, Staub HL. Anti-CCP antibodies have more diagnostic impact than rheumatoid factor (RF) in a population tested for RF. Clin Rheumatol. 2007;26:1883–1889. doi: 10.1007/s10067-007-0601-6. [DOI] [PubMed] [Google Scholar]
- 15.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- 16.Wood DD, Moscarello MA. The isolation, characterization, and lipid-aggregating properties of a citrulline containing myelin basic protein. J Biol Chem. 1989;264:5121–5127. [PubMed] [Google Scholar]
- 17.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. A fluoroacetamidine-based inactivator of protein arginine deiminase 4: design, synthesis, and in vitro and in vivo evaluation. J Am Chem Soc. 2006;128:1092–1093. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Haloacetamidine-based inactivators of protein arginine deiminase 4 (PAD4): evidence that general acid catalysis promotes efficient inactivation. Chembiochem. 11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. N-α-Benzoyl-N5-(2-Chloro-1-Iminoethyl)-L-Ornithine Amide, a Protein Arginine Deiminase Inhibitor, Reduces the Severity of Murine Collagen-Induced Arthritis. J Immunol. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange S, Gogel S, Leung KY, Vernay B, Nicholas AP, Causey CP, Thompson PR, Greene ND, Ferretti P. Protein deiminases: New players in the developmentally regulated loss of neural regenerative ability. Dev Biol. 2011;355:205–214. doi: 10.1016/j.ydbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. Suppression of Colitis in Mice by Cl-Amidine: A Novel Peptidylarginine Deiminase (Pad) Inhibitor. Am J Physiol Gastrointest Liver Physiol. 2011;300:G929–G938. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol. 2011;300:G929–G938. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. N-alpha-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J Immunol. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TR, Lawrence DS. Acquisition of high-affinity, SH2-targeted ligands via a spatially focused library. J Med Chem. 1999;42:784–787. doi: 10.1021/jm980663f. [DOI] [PubMed] [Google Scholar]
- 27.Li P, Wang D, Yao H, Doret P, Hao G, Shen Q, Qiu H, Zhang X, Wang Y, Chen G. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene. 2010;29:3153–3162. doi: 10.1038/onc.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry. 2007;46:6578–6587. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leatherbarrow RJ. Grafit Ver 5.0. UK: Erathicus Software, Staines; 2004. [Google Scholar]
- 31.Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, Roberson MS, Kraus WL, Coonrod SA. Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate c-Fos Expression in Breast Cancer Cells. PLoS Genet. 2011;7:e1002112. doi: 10.1371/journal.pgen.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.