Abstract
CD22 is currently recognized as a B cell-specific Siglec and has been exploited therapeutically with humanized anti-CD22 monoclonal antibody having been used against B cell leukemia. Herein, tissue-specific eosinophil mRNA microarray analysis identified that CD22 transcript levels of murine gastrointestinal (GI) eosinophils are 10-fold higher than those of lung eosinophils. In order to confirm the mRNA data at the protein level, we developed a FACS-based protocol designed to phenotype live GI eosinophils isolated from the murine lamina propria. Indeed, we found that jejunum eosinophils expressed remarkably high levels of surface CD22, similar to levels found in B cells across multiple mouse strains. In contrast, CD22 was undetectable on eosinophils from the colon, blood, thymus, spleen, uterus, peritoneal cavity and allergen-challenged lung. Eosinophils isolated from newborn mice did not express CD22 but subsequently upregulated CD22 expression to adult levels within the first 10 days after birth. The GI lamina propria from CD22 gene-targeted mice harbored more eosinophils than wild-type control mice, while the GI eosinophil turnover rate was unaltered in the absence of CD22. Our findings identify a novel expression pattern and tissue eosinophilia-regulating function for the “B cell-specific” inhibitory molecule CD22 on GI eosinophils.
Keywords: Eosinophils, mucosa, Siglec, CD22
Introduction
Eosinophils are multifunctional effect or leukocytes involved in a variety of allergic disorders including asthma and eosinophilic gastrointestinal diseases (EGIDs)(1, 2). Bridging the gap between innate and adaptive immunity, eosinophils have a pivotal role in TH2 inflammatory states. After exiting the bone marrow, circulating eosinophils home to the GI tract and become GI resident cells, a process orchestrated by eotaxin, its receptor CCR3 (3, 4) and the α4β7 integrin systems (5). In both mice and man, the GI tract harbors a large number of eosinophils in the lamina propria layer underneath the epithelium, forming the largest reservoir of eosinophils during homeostasis(6–8). It has been reported that GI eosinophils have a slower turnover rate compared to lung and blood eosinophils due to specific survival signaling (8). In contrast to circulating eosinophils, GI eosinophils have a more activated phenotype, measured by their degree of degranulation and expression of diverse cytokines(9, 10). It has been suggested that GI eosinophils are involved in mucosal anti-bacterial defense by actively releasing their toxic mediators and mitochondrial DNA (11). GI eosinophils are also causatively related to the pathogenesis of a variety of GI tract disorders, such as EGIDs(12), ulcerative colitis (13), Crohn’s disease (14)and intestinal tumors (15).
CD22 is a member of the sialoadhesin subclass of the Ig super family and is currently recognized as a B cell-specific surface glycoprotein expressed on both normal and malignant B cells throughout each stage of B cell development, except for plasma cells [for review, see (16)]. CD22 is generally an inhibitory co-receptor, serving as a molecular damper that negatively regulates B cell receptor (BCR) signaling, and is therefore critical for B cell activation, proliferation and survival (17–21). Following BCR crosslinking, the phosphorylated ITIMs in the cytoplasmic moiety of CD22 recruit effect or molecules such as SHP and SHIP-1, which negatively regulate the strength of BCR and CD19 downstream signaling (22). In addition to its cytoplasmic motifs, CD22 possesses 7 Ig-like domains in its extra cellular domain and may regulate cell-cell adhesion (23, 24). As a sialic acid-binding Ig-like lectin (Siglec), CD22 binds to its natural ligand 2, 6-linked sialic acid, which is present on the glycoproteins widely expressed on the surface of hematopoietic and non-hematopoietic cells (25). Given its B cell-specific expression pattern, CD22 has become one of the attractive target molecules for diagnosis and therapy of B-lineage acute leukemia (26), as well as other B-tropic autoimmune disorders such as systemic lupus erythematosus (SLE)(27)and Sjögren's syndrome (28).
Eosinophils have long been thought to participate in GI parasitic infections and allergic lung diseases; nonetheless, the transcriptional signature difference between GI and lung eosinophils has not been explored at a genome-wide level. In order to elucidate the physiological and pathological functions of eosinophils in a tissue-specific fashion, we examined differential gene expression of eosinophils isolated from the GI tract and lungs during homeostasis. Tissue-specific mRNA microarray analysis with mRNA extracted from flow-sorted GI and lung eosinophils indicated that, among the 513 genes with significantly altered expression between GI and lung eosinophils, expression of the pan-B cell marker CD22 is upregulated 10-fold in GI compared to lung eosinophils. This transcriptional difference was verified by qRT-PCR and flow cytometry at the protein level. We then used CD22 gene-deficient mice to examine a possible function of this inhibitory co-receptor in GI eosinophils and found that CD22 negatively regulates GI lamina propria eosinophilia. To our knowledge, this is the first report demonstrating that CD22 is highly expressed in eosinophils and functionally involved in regulating GI eosinophil levels.
Materials and Methods
Mice
C57BL/6N (CD22 +/+) mice were initially obtained from Charles River (Wilmington, MA), and CD22 gene-deficient mice (backcrossed to C57BL/6J for greater than 20 generations before arrival) were obtained from Jackson Laboratory (Bar Harbor, ME). BALB/c mice, germ-free Swiss Webster (SW) mice and non-germ-free SW mice were obtained from Taconic Farms Inc (Hudson, NY). CD2-IL-5 transgenic (tg) mice were obtained from Dr. Colin Sanderson (Institute for Child Health Research, Perth, Australia) and then backcrossed onto the BALB/c background for at least 20 generations. In all experiments, we used 2-3-month-old, age- and gender-matched mice, which were housed in specific pathogen-free conditions at Cincinnati Children’s Hospital Medical Center (CCHMC) under IUACAC-approved protocols. All mice were housed in a room with an ambient temperature of 22°C and a 12-hour light cycle.
GI eosinophil isolation
Animals were euthanized by CO2 asphyxiation, and jejunums were excised 6 cm to 16 cm from the pylorus. Duodenum, ileum and colon were anatomically defined as 6cm distal to the pylorus, 10 cm proximal to the cecal sphincter and 5 cm distal to the cecum narrowing, respectively. The intestine segment was opened, washed with PBS and then subjected to EDTA buffer treatment for 15 minutes (1x HBSS with 10%FCS, 5mM EDTA, 40mM HEPES) at 37°C degree with agitation. Following a HBSS wash, tissues were minced into small pieces and treated with collagenase A (Roche) solution (2.4mg/mL in complete RPMI supplemented with 10% fetal calf serum) for 30 minutes at 37°C degrees with moderate agitation. The digest was passed through a 19-G needle three times and then two layers of gauze, and the filtered cell suspension was centrifuged at 300g for 5 minutes. After an additional wash with HBSS, the cells were suspended in appropriate volumes for flow cytometry staining or other experiments. The same procedure was applied to other solid tissues harboring eosinophils without modification, including the lung, stomach, and uterus.
Flow cytometry analysis of total GI lamina propria cells
Polychromatic flow cytometry compensation matrix setting was acquired in a pilot study with eosinophils from IL-5 tg mice. Single-cell suspensions of approximately 1 million total GI lamina propria cells were stained with markers for eosinophils and target proteins of interest using the 6 color FACS system. Antibodies/dyes (clone number) used in this study were purchased from Biolegend (CD22-AF647 OX-97 and isotype RTK-2071), Invitrogen (LIVE/DEAD Violet dye), BD (CD45-FITC 30-F11, Siglec F-PE E50-2440, CD11b-PECy7 M1/70, 7AAD) and R&D Systems (CCR3-PE 83103). Staining was performed on ice for 30 minutes in staining buffer (0.5%BSA, 0.01% NaN3 in 1x HBSS) with manufacturer-suggested titers, followed by a wash with the staining buffer. Stained cells were resuspended and subjected to analysis with the BD FACS Canto II flow cytometer. Raw flow cytometry data were analyzed by Flowjo software (Tree Star Inc., Ashland, OR). GI eosinophils were identified as 7AAD−CD45+Siglec-F+CD11b+ events. Quantification expression data were presented as ΔMFI (CD22 antibody MFI– isotype control MFI). For phosphoFACS, freshly isolated GI lamina propria cells were washed with PBS and rested in complete RPMI-1640 for 30 minutes and then kinetically subjected to the challenging media. Samples treated with vehicle media serve as control for baseline normalization. At the end of the incubation, cells were immediately fixed by adding equal volume of 4% formaldehyde and incubated at 37°C for 10 minutes, followed by ice-cold methanol permeabilization for 30 minute. Specific Thr202/Tyr204 p44/42 antibody (E10, Cell Signaling Technology, #4375) was used together with previously mentioned eosinophil markers to assess eosinophil MAPK activation. GI eosinophils were identified and gated as CD45-Siglec F-CD11b triple (+) events. Stimulated MFI values were normalized to vehicle treated samples for graphic presentation.
Flow-imaging analysis of GI eosinophils
Single-cell suspensions stained with different antibodies were prepared as described in the above-mentioned method and fixed with 2% formaldehyde in PBS. The antibody-stained samples were run through the ImageStreamX system (Amnis Corp. Seattle, WA) following the manufacturer’s instructions. Compensation matrix was established at the pixel level with single-stain controls. The image output was performed by analyzing the raw image data with the software of Amnis-IDEAS (Amnis Corp. Seattle, WA).
Genome-wide microarray analysis on sorted eosinophils and qPCR validation
Lung and GI eosinophils were isolated as mention in earlier sections, with the pulmonary circulation being perfused prior to isolation procedures. Live eosinophils were sorted as DAPI−CCR3+Siglec-F+CD45+CD4−CD8a−CD19−B220−side scatter high (SSChigh) cells from 10 animals using FACS Aria (BD). Total RNA from sorted eosinophils was extracted by a standard Trizol RNA isolation (Invitrogen) and subsequently column-purified with a RNeasy Mini Kit (QIAGEN). mRNA integrity was validated by the Agilent 2100 bio-analyzer (Agilent technologies, Santa Clara, CA). Eosinophil mRNA was amplified and labeled with the WT-Ovation Pico RNA Amplification System (NuGen, San Carlos, CA) and subjected to the GeneChip Mouse Gene ST 1.0 Array chip(Affymetrix), which covers the whole mouse genome with 28853 probe sets. Microarray expression analysis was performed at CCHMC’s Chip Core facility, and expression data were analyzed by the software of Gene spring GX 11 (Agilent Technologies). Briefly, the Affymetrix raw expression values were first filtered with the threshold of 400. Differential expression between GI and lung eosinophils was identified by a >2-fold change and a p value < 0.01 with a false discovery rate (FDR) correction. The same mRNA sample used for microarray analysis was reverse transcribed with the iScript cDNA Synthesis Kit (Bio-Rad, 170-8891), and real-time PCR was performed with the iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using a pair of primers spanning exons 4 and 5 of the CD22 transcript (forward 5’-GAAAATCCACCCGATACGTGC-3’; reverse 5’-TTGGAACGGTTTCTCCGAGAC-3’), resulting in a 115-bp amplicon.
OVA-induced GI inflammation model
As originally described by Brandt et al. (29), animals of sensitive strains (littermates of BALB/c-C57BL/6 F4 backcross) were initially sensitized with 100 μg of OVA/Alum twice, 14 days a part under aseptic conditions. Beginning on day 28, OVA (50mg in saline) or saline was administered by oral gavage to the animal every other day for a total of 7 times. Mice were sacrificed 24 hours after the last OVA challenge. Eosinophils were then isolated from the inflamed GI tract for CD22 expression analysis.
Aspergillus-induced allergic lung model
Protein extract of 100 μg Aspergillus Fumigatus (GREER, Lenoir, NC) was dissolved in sterile saline and administered into the airway of WT BALB/c mice by intranasal inhalation. A total of 9 challenges was given following a Monday-Wednesday-Friday regimen with 1 challenge/day for 3 weeks as previously reported (30).
Major Basic Protein (MBP) immunostaining and GI eosinophil quantitative morphometric analysis
The jejunum tissue was fixed in 4% paraformaldehyde in PBS, embedded in paraffin, cut into 5 μm transverse sections, and immunostained with anti-MBP antibody, a kind gift of Dr. James Lee (Mayo Clinic, Scottsdale, AZ) following common immunohistochemistry procedures. For morphometric analysis, all of the MBP labeled eosinophils on the whole transverse jejunum section were enumerated as the number of total eosinophil for this transverse section. To acquire the specific area of laminar propria on the same section, the digital micrograph of the whole transverse section was used to calculate areas of manually outlined lamina propria by Image-Pro PLUS software. (Media Cybernetics, L.P.) The lamina propria eosinophil density was calculated by dividing the above two values into the unit of eosinophil number / mm2.
GI eosinophil turnover assay
This assay was adopted from a previous publication (8). Briefly, animals were continuously fed with BrdU-containing water solution at a concentration of 80 mg/dL for 6 days, and GI eosinophil isolation was performed as described above. BrdU-positive eosinophils were detected using the BrdU APC Detection Kit from BD Pharmingen (BD Cat # 552598) in conjunction with the eosinophil markers CD45, Siglec-F and CD11b (as described above). The intranuclear staining was based on the manufacturer’s suggested protocols. After flow cytometry analysis, the gated eosinophil population was further gated by CD11c and BrdU with a no BrdU treatment control as an intensity reference. The CD11clowBrdUhigh sub-population represented newly migrated GI eosinophils(8).
Statistical analysis
Statistical significance was analyzed using a two-tailed student t-test in all instances except for the OVA sensitization study, in which a 2-way ANOVA was used. Data are graphed as mean ± standard error of the mean (SEM).
Results
GI eosinophils express a unique set of genes with 10-fold upregulation of CD22 transcript compared to lung eosinophils
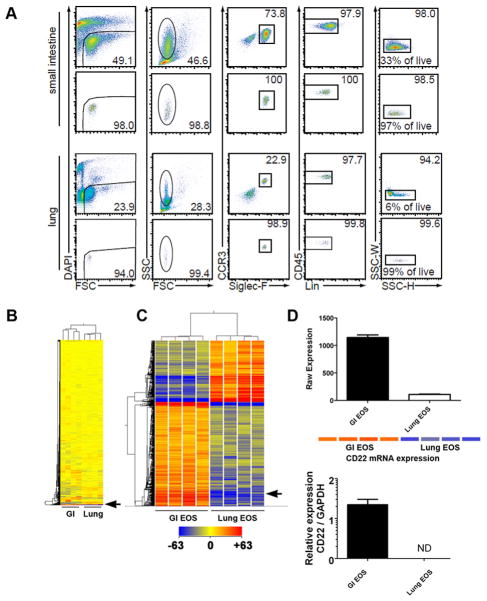
To address the physiological role of eosinophils in the GI tract and lung under homeostatic healthy conditions, we analyzed tissue-specific eosinophil gene expression patterns by genome-wide expression microarray analysis using mRNA isolated from FACS-sorted eosinophils from the small intestine or lung of naïve BALB/c mice. As shown in Figure 1A, we readily detected GI and lung eosinophil populations by multi-color FACS staining following purification. Live eosinophils were identified as DAPI-CCR3+Siglec-F+CD45+CD4−CD8a−CD19−B220−SSChigh cells. The eosinophil samples were sorted to a purity ranging from 93.4% to 98.7% (7 out of 8 samples had a purity of greater than 96%). Whole-genome expression profile analysis was performed on 4 normal lung and 4 normal intestine eosinophil mRNA samples. Among the 28,853 genes probed by the array, we found a cluster of 513 probe sets that was differentially regulated (fold change > 2, p< 0.01 post-Benjamini Hochberg FDR) between GI and lung eosinophils, with 319 being upregulated and 194 being downregulated (Figure 1B and 1C). Among the genes upregulated in GI eosinophils, CD22 was upregulated 10-fold as shown by the Affymetrix raw expression values (Figure 1D). Conventional qRT-PCR verified this microarray finding, demonstrating that CD22 transcripts were indeed strongly expressed by GI eosinophils as compared to the nearly undetectable levels in lung eosinophils (Figure 1D).
Figure 1. CD22 mRNA expression in murine GI eosinophils (EOS).
In A, live eosinophils were isolated under homeostatic conditions from the GI tract and lung by FACS sorting with the gating criteria set for DAPI−CCR3+Siglec−F+CD45+CD4−CD8a−CD19−B220−SSChigh cells. The percentage of each gate relative to the upper gate was printed on each plot. (FSC, forward scatter; SSC, side scatter, Lin, CD4-CD8a-CD19-B220) In B and C, mRNA isolated from GI and lung eosinophils was subjected to genome-wide expression microarray analysis. Among the 28,853 unique probe sets screened, 513 genes showed a >2-fold change with p<0.01 (post-FDR). Each lane represents eosinophil RNA isolated from a separate mouse (n = 4 per group). The position of CD22 on the heat diagram is shown with the arrow. In D, CD22 mRNA expression as determined by gene chip analysis Raw Expression value (top) and by real-time PCR (below) is given. In the upper panel of D, an expression heat map is also illustrated for visualization of upregulated bands in red color. In qPCR graph, CD22 expression in lung eosinophils was not detectable (ND).
GI eosinophils express CD22 protein at levels comparable to B cells
We next examined CD22 protein expression on GI eosinophils. Isolated total jejunum lamina propria cells from BALB/c mice were stained with eosinophil markers and anti-CD22 or isotype control antibody. Live eosinophils were identified as CD45+7AAD−SSChighCCR3+CD11b+ events. As shown in Figure 2A, murine jejunum eosinophils expressed similar levels of surface CD22 to jejunum B cells. Isotype control staining for each cell type did not reveal significant staining. We also performed B220 staining on the two above-mentioned populations, and the results indicated that the gated GI eosinophil population did not contain any B cell contamination. Moreover, cell sorting with the same gating strategy demonstrated that >95% of events in the eosinophil gate studied were eosinophils by morphological analysis (Figure 2B). As a control, we examined CD22 expression on eosinophils from multiple organs/tissues that harbor eosinophils under physiological conditions. As shown in Figure 2C, CD22 protein was specifically expressed on GI lamina propria eosinophils in the upper GI tract with the jejunum having the highest level.
Figure 2. CD22 protein expression on eosinophils.
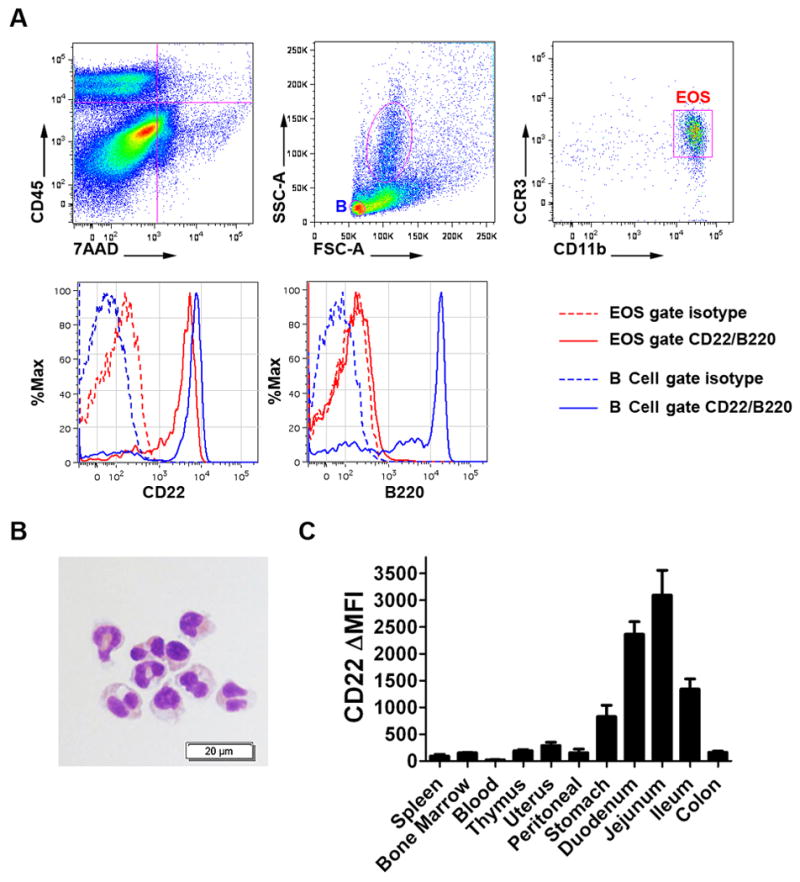
In A, live GI eosinophils were identified from the total isolated GI lamina propria cells with serial gating forCD45+7AAD-SSChighCCR3+CD11b+ cells (EOS in red). Compared with the B cell positive control (B in blue), GI eosinophils (red gate/histogram) express similar levels of CD22, which is shown by the difference between the isotype staining (dashed line) and CD22 staining (solid line). There was no contamination from B cells in the EOS gate as all events were B220 negative in the EOS. In B, FACS sorting with the same EOS gating strategy demonstrated that the events were eosinophils as assessed by morphologic features (circular nuclei, eosinophilic granules) following Diff-Quick staining of cytocentrifuged sorted cells. In C, CD22 expression on eosinophils from different tissues was assessed by ΔMFI (specific antibody MFI– isotype control MFI)with 3–4 mice per tissue group. All experiments were independently repeated at least twice. Data is presented as mean ± SEM.
Given the unexpected expression of CD22 by GI eosinophils, we aimed to further substantiate this finding. First, the specificity of the FACS-based analysis of tissue-derived eosinophils was ensured by analysis of CD22 gene-deficient mice, which did indeed reveal a lack of CD22 expression on GI eosinophils as assessed by Amnis IS-100 flow imaging. As shown in Figure 3B, wild type GI eosinophils (CD45+Siglec-F+CD11b+) demonstrated high levels of CD22 on the cell membrane, whereas CD22 signals in CD22−/−GI eosinophils (C57BL/6J)and isotype controls (BALB/c) were absent. Taken together, our results demonstrate that GI eosinophils express CD22 protein on their cell membranes.
Figure 3. GI eosinophil CD22 cell membrane expression wild type and CD22 deficient mice by FACS and flow imaging analyses.
In A, GI eosinophils were isolated from CD22+/+ and CD22−/− mice (C57BL/6), stained for CD22 and eosinophils markers, and gated as CD45+7AAD−SSChighCD11b+cells. Serial gating plots for eosinophil CD22 expression are shown. CD22 histograms from 4 mice of each genotype were overlapped to show consistency. This experiment was independently repeated three times. In B, flow imaging was used to characterize the expression pattern of CD22 on GI eosinophils. A single GI eosinophil was presented on 4 different channels (CD45, Siglec F, CD11b and CD22). The left panel shows GI eosinophil CD22 expression in CD22+/+ and CD22−/−C57BL/6J mice. The right panel shows GI eosinophil CD22 expression inCD22+/+BALB/c mice with anti-CD22 staining and isotype control staining as a reference.
CD22 is upregulated on GI eosinophils in a postpartum fashion and is independent of intestinal flora
We next investigated what factors regulate GI eosinophil CD22 expression. In the case of B cells, LPS and IL-4 treatment has been shown to upregulate its surface CD22 expression (31). Therefore, we treated total splenocytes with LPS (25 μg/mL) and IL-4 (100ng/mL) in the presence of IL-5 (10ng/mL) for 24 hours. Eosinophils and B cells were identified by Siglec-F/CD11b double staining and CD19 staining, respectively. Although LPS and IL-4 upregulated the CD22 expression of spleen B cells by ~30%, they failed to enhance the CD22 expression on spleen eosinophils (Figure 4A).
Figure 4. Regulation of GI eosinophil CD22 expression as a function of perinatal age, GI bacterial colonization and cytokine exposure.

In A, whole splenocytes were cultured 24 hours in IL-5 (10ng/mL) with either IL-4 (100ng/mL) or LPS (25 μg/mL). The total cell population was stained for CD19 and eosinophil markers Siglec-F and CD11b, together with anti-CD22 or isotype control. CD22 expression on eosinophils and B cells was quantified by ΔMFI.In B, using the same staining strategy, CD22 expression was quantified by ΔMFI on GI eosinophils isolated from the intestine of mice at different time points after birth. In C and D, GI eosinophils were isolated from 2-month-oldgerm-free (germ -) and non-germ-free (germ +) Swiss Webster mice and stained for CD22 and eosinophil markers as mentioned above. In E, CD22 expression was assessed in GI eosinophils isolated from BALB/c WT and CD2-IL-5 transgenic (Tg) mice. In F, GI eosinophil CD22 expression was assessed following OVA-induced GI inflammation in CD22-deficient (CD22−/−) and wild type (CD22+/+) mice. Animals were intraperitoneally sensitized to OVA and intragastrically challenged with OVA. In G, BALB/c mice were challenged with Aspergillus fumigatus(Asp) or saline (Sal) intranasally to induce Th2-induced airway eosinophilia, and lung eosinophil CD22 expression was assessed in parallel to spleen and GI eosinophils.(* p < 0.05, ** p < 0.01, *** p < 0.001, two tailed t-test or ANOVA,NTC, non-treated controls)
A postpartum kinetic study of CD22 expression by GI eosinophils revealed that CD22 expression was not an early event during embryogenesis, but rather was upregulated to adult levels within the first 10 days after birth (Figure 4B). Since the kinetics of the CD22 postpartum upregulation resemble those of the development of GI commensal flora, we tested whether GI flora have a role in induction of CD22 expression on GI eosinophils. As shown in Figure 4C, GI eosinophils from 2-month-old germ-free mice were CD22 positive; in fact, the CD22 expression level was modestly increased on GI eosinophils in mice lacking intestinal bacteria (Figure 4D).
GI eosinophil CD22 expression is downregulated by eosinophil activation triggers
To examine if eosinophil activation signals affect CD22 expression, we compared CD22 expression levels on GI eosinophils from CD2-IL-5 tg and WT mice. Notably, over expression of IL-5 was associated with downregulation of CD22 expression (Figure 4E). To further confirm that CD22 is downregulated during eosinophil activation, CD22+/+ and CD22−/−mice were sensitized with OVA/Alum, orally challenged with saline or OVA to induce GI inflammation (29), and assessed for CD22 expression of GI eosinophils. OVA treatment downregulated GI eosinophil CD22 expression (Figure 4F). As a control, we measured CD22 expression on lung airway eosinophils following experimental asthma induction(30); CD22 expression was not increased on airway eosinophils in a setting of severe inflammation in the lung (Figure 4G).
CD22 negatively regulates GI lamina propria eosinophil levels
We hypothesized that CD22 would affect the eosinophil concentration in the GI tract as CD22 may negatively regulate eosinophil function and survival. Notably, flow cytometry analysis indicated that the percentage of eosinophils among total lamina propria cells was significantly increased in CD22−/− mice compared to WT controls (2.25 ± 0.14%; n = 6 vs. 2.83 ± 0.12%; n = 8, mean ± SEM, p < 0.01). The number of eosinophils among 100,000 total lamina propria cells isolated from jejunum tissue was significantly increased in the CD22-deficient mice (Figure 5A). In addition, with an independent methodology, we performed major basic protein (MBP) staining on CD22 +/+ and −/− jejunum sections and the subsequent morphometric quantification analysis revealed significantly increased eosinophilia in the GI lamina propria from CD22 −/− mice (Figure 5 B and C). We next examined if the augmented GI eosinophilia present in CD22−/− mice was a result of reduced eosinophil turnover. BrdU incorporation indicated that eosinophil turnover rates were not influenced by CD22 expression, as shown by the comparable percentage of newly synthesized/migrated BrdU+ eosinophils in the lamina propria of WT and CD22−/−mice (Figure 5D) (24.5 ± 1.2%; n =8 vs. 23.8 ± 1.2%; n = 8, mean ± SEM). In order to access the functional role of CD22 on GI eosinophil specific signaling strength, we utilized the phosphoFACS to quantify the MAPK phosphorylation by the eosinophil priming/survival cytokine IL-5 and eosinophil chemokine eotaxin-2. IL-5 exposure (10ng/mL) induced rapid and robust p44/42 MAPK phosphorylation in GI eosinophil within 10 minutes following stimulation, which diminished and returned to baseline by 30 minutes (Figure 5E). GI eosinophils isolated from CD22 +/+ and CD22 −/− mice exhibited comparable IL-5 induced MAPK activation. With the same method, we also tested the signaling strength of IL-5 induced p38 MAPK activation and eotaxin-2 induced p44/42 activation, and no phenotypic difference was observed between CD22 +/+ and CD22 −/− GI eosinophils (data not shown).
Figure 5. Effect of CD22 gene deficiency on GI lamina propria eosinophil levels.
In A, GI eosinophils were isolated by collagenase digestion and then subjected to 5 color flow cytometry analysis. The numbers of eosinophils were enumerated among 106 total lamina propria (GI LP) cells collected. Graph shown here is a representative experiment independently repeated three times. (** p < 0.01, two tailed t-test) In B, the jejunum section was stained with anti-major basic protein (MBP), and GI lamina-propria MBP (+) eosinophil density was assayed by morphometric analysis. (EOS/mm2GI LP, ** p < 0.01, two tailed t-test). In C, representative photomicrographs of the jejunum section MBP staining were shown, CD22 +/+ vs. CD22 −/− mice (Magnification 40X). In D, animals ingested BrdU-containing water (80mg/dL) for 6 days and were euthanized and analyzed for the percentage of BrdU+ GI eosinophils. The GI eosinophil populations shown on CD11c-BrdU double plots were sequentially gated out as CD45+, Siglec-F+ and CD11b+ events. (NTC, non-treated controls, n = 8 for each group, independently repeated twice) In E, by phosphoFACS, freshly isolated GI lamina propria cells were stimulated with IL-5 (10ng/mL) for indicated time points. The cells were then fixed, permeabilized and stained with p44/42 (Thr202/Tyr204) phospho-antibody together with eosinophil markers to assess eosinophil specific MAPK activation by IL-5. MAPK (p44/42) phosphorylation status of CD45-Siglec F-CD11b triple (+) eosinophils was represented in the histogram and quantified in the bar chart, CD22 +/+ vs. CD22 −/− GI eosinophils. (NTC, non-treated controls, n = 3 for each group, independently repeated twice).
Discussion
GI lamina propria eosinophils are a unique population of immunocytes that consists of the first line of mucosal defense in the GI mucosal barrier. Their expression profiles and specific functions remain to be elucidated, largely due to the difficulty in isolating these cells for subsequent expression and functional studies. Herein, we describe an isolation system that enabled us to examine these understudied cells from the lamina propria of the GI tract, uterus and stomach by permitting the isolation of viable and functionally intact eosinophils. With this system, we were able to show that eosinophils residing in different target organs express unique tissue-specific transcripts, presumably to suit disparate functions. It is important to note that although tissue specific eosinophil transcriptomes have been identified, verification of individual genes as we have done for CD22 is likely critical as contaminating cells in the expression analysis may have had a contributory role to the identified transcriptome. The current study is the first to demonstrate that CD22 is expressed on GI eosinophils, as shown by mRNA and protein analysis and confirmed by absent staining in CD22-deficient mice; this phenomenon can be observed across different mouse genetic backgrounds, namely BALB/c and C57BL/6J, with different propensities for TH1/TH2 polarization. GI eosinophil CD22 expression is an unexpected finding given that CD22 is known as an inhibitory receptor specifically expressed on B lymphocytes.
Although eosinophils are known to express the receptor for LPS and IL-4 (32, 33), which induce CD22 on B cells (31), these factors did not alter CD22 expression on GI eosinophils, suggesting that the CD22 co-receptor coupling and downstream signaling pathways in GI eosinophils are different from those in B cells. The postpartum CD22 expression kinetics data, in combination with the differential eosinophil CD22 expression in multiple tissues, suggest that the intestinal milieu developed postnatally (but not enteric commensal flora alone) contribute to the expression of CD22 on GI eosinophils. As shown by the tissue distribution and lung inflammation data, such a mechanism is not present in eosinophils from other tissues during homeostatic or inflammatory conditions. Notably, a unique feature of GI eosinophils is their activated phenotype. Multiple lines of evidence have shown that GI eosinophils are highly activated compared with blood eosinophils even in homeostatic mucosa (9, 10). Considering that CD22 is generally deemed to be an inhibitory co-receptor acting through its intracellular ITIM motif on B lymphocytes, it is likely that CD22 serves as a molecular damper in GI eosinophil responses, perhaps to prevent over-activation or to maintain their physiological state. By examining multiple factors associated with GI inflammation, specifically bacterial colonization, systemic IL-5 transgene overexpression and OVA-induced GI inflammation in mice, we demonstrated that expression of CD22 on GI eosinophils is downregulated following inflammation induced by innate (e.g. bacterial colonization) and adaptive (e.g. OVA) stimuli and by the molecular signaling induced by relevant cytokine cascades (e.g. IL-5 overexpression). The GI tract forms an environment wherein residential hematopoietic cells are subjected to multiple lines of biological, chemical and physical challenges. In this regard, downregulating an inhibitory receptor may initiate pro-inflammatory activity of GI eosinophils.
Collectively, we have identified that the canonical B cell marker CD22 is robustly expressed on murine GI eosinophils, which form a major eosinophil reservoir and may have key roles in GI mucosal defense. It is interesting to note that CD22 has recently been found to be expressed by other non-B cell populations including neurons (34), human basophils (35, 36), cultured human mast cells (37) and T cells(38). Notably, even within B cells, CD22 was recently shown to mediate receptor signaling events other than the BCR pathway (39–41), suggesting that CD22 could functionally regulate certain innate functions beyond the scope of BCR signaling. Although the GI tract has the largest reservoir of eosinophils in mice and man, there is a paucity of information regarding their homeostatic regulation, except for data concerning the key role of eotaxin-1 and its receptor CCR3, as well as the inhibitory receptor PIR-B (42). Herein, we have identified a novel checkpoint for controlling the baseline level of GI eosinophils involving a receptor not previously identified on eosinophils. Although the molecular control of GI eosinophil-specific expression has not been elucidated, it is interesting that CD22 expression in eosinophils is specific to the GI tract, implying a tissue-specific role. Indeed, early studies have already revealed a non-redundant role for CD22 in regulating the baseline level of GI eosinophil levels. It will certainly be interesting to examine the role of CD22 and its potential utility as a pharmacological target in eosinophilic inflammatory responses.
Microarray data accession
Eosinophil specific RNA microarray data has been deposited to the Gene Expression Omnibus (GEO) public repository under the accession code GSE33807. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33807)
Acknowledgments
The authors thank Shawna Hottinger and Joelle Rothenberg for editorial assistance and Dr. Ariel Munitz for scientific discussion.
Footnotes
This work was supported by NIH grants R37 A1045898 and R01 AI83450,CURED (Campaign Urging Research for Eosinophilic Disease), the Food Allergy Initiative, and the Buckeye Foundation.
References
- 1.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 2.Hogan SP, Rosenberg HF, Moqbel R, Phipps S, Foster PS, Lacy P, Kay AB, Rothenberg ME. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy. 2008;38:709–750. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 3.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest. 1999;103:1719–1727. doi: 10.1172/JCI6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurish MF, Humbles A, Tao H, Finkelstein S, Boyce JA, Gerard C, Friend DS, Austen KF. CCR3 is required for tissue eosinophilia and larval cytotoxicity after infection with Trichinella spiralis. J Immunol. 2002;168:5730–5736. doi: 10.4049/jimmunol.168.11.5730. [DOI] [PubMed] [Google Scholar]
- 5.Brandt EB, Zimmermann N, Muntel EE, Yamada Y, Pope SM, Mishra A, Hogan SP, Rothenberg ME. The alpha4bbeta7-integrin is dynamically expressed on murine eosinophils and involved in eosinophil trafficking to the intestine. Clin Exp Allergy. 2006;36:543–553. doi: 10.1111/j.1365-2222.2006.02456.x. [DOI] [PubMed] [Google Scholar]
- 6.Powell N, Walker MM, Talley NJ. Gastrointestinal eosinophils in health, disease and functional disorders. Nat Rev Gastroenterol Hepatol. 2010;7:146–156. doi: 10.1038/nrgastro.2010.5. [DOI] [PubMed] [Google Scholar]
- 7.Rothenberg ME, Mishra A, Brandt EB, Hogan SP. Gastrointestinal eosinophils. Immunol Rev. 2001;179:139–155. doi: 10.1034/j.1600-065x.2001.790114.x. [DOI] [PubMed] [Google Scholar]
- 8.Carlens J, Wahl B, Ballmaier M, Bulfone-Paus S, Forster R, Pabst O. Common gamma-chain-dependent signals confer selective survival of eosinophils in the murine small intestine. J Immunol. 2009;183:5600–5607. doi: 10.4049/jimmunol.0801581. [DOI] [PubMed] [Google Scholar]
- 9.Kato M, Kephart GM, Talley NJ, Wagner JM, Sarr MG, Bonno M, McGovern TW, Gleich GJ. Eosinophil infiltration and degranulation in normal human tissue. Anat Rec. 1998;252:418–425. doi: 10.1002/(SICI)1097-0185(199811)252:3<418::AID-AR10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Straumann A, Kristl J, Conus S, Vassina E, Spichtin HP, Beglinger C, Simon HU. Cytokine expression in healthy and inflamed mucosa: probing the role of eosinophils in the digestive tract. Inflamm Bowel Dis. 2005;11:720–726. doi: 10.1097/01.mib.0000172557.39767.53. [DOI] [PubMed] [Google Scholar]
- 11.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 12.Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID) J Allergy Clin Immunol. 2004;113:11–28. doi: 10.1016/j.jaci.2003.10.047. quiz 29. [DOI] [PubMed] [Google Scholar]
- 13.Wallon C, Persborn M, Jonsson M, Wang A, Phan V, Lampinen M, Vicario M, Santos J, Sherman PM, Carlson M, Ericson AC, McKay DM, Soderholm JD. Eosinophils express muscarinic receptors and corticotropin-releasing factor to disrupt the mucosal barrier in ulcerative colitis. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 14.Emanuel PO, Phelps RG. Metastatic Crohn's disease: a histopathologic study of 12 cases. J Cutan Pathol. 2008;35:457–461. doi: 10.1111/j.1600-0560.2007.00849.x. [DOI] [PubMed] [Google Scholar]
- 15.Legrand F, Driss V, Delbeke M, Loiseau S, Hermann E, Dombrowicz D, Capron M. Human eosinophils exert TNF-alpha and granzyme A-mediated tumoricidal activity toward colon carcinoma cells. J Immunol. 2010;185:7443–7451. doi: 10.4049/jimmunol.1000446. [DOI] [PubMed] [Google Scholar]
- 16.Tedder TF, Poe JC, Haas KM. CD22: a multifunctional receptor that regulates B lymphocyte survival and signal transduction. Adv Immunol. 2005;88:1–50. doi: 10.1016/S0065-2776(05)88001-0. [DOI] [PubMed] [Google Scholar]
- 17.Sato S, Miller AS, Inaoki M, Bock CB, Jansen PJ, Tang ML, Tedder TF. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 1996;5:551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 18.O'Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 19.Otipoby KL, Andersson KB, Draves KE, Klaus SJ, Farr AG, Kerner JD, Perlmutter RM, Law CL, Clark EA. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 1996;384:634–637. doi: 10.1038/384634a0. [DOI] [PubMed] [Google Scholar]
- 20.Nitschke L, Carsetti R, Ocker B, Kohler G, Lamers MC. CD22 is a negative regulator of B-cell receptor signalling. Curr Biol. 1997;7:133–143. doi: 10.1016/s0960-9822(06)00057-1. [DOI] [PubMed] [Google Scholar]
- 21.Walker JA, Smith KG. CD22: an inhibitory enigma. Immunology. 2008;123:314–325. doi: 10.1111/j.1365-2567.2007.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nitschke L, Tsubata T. Molecular interactions regulate BCR signal inhibition by CD22 and CD72. Trends Immunol. 2004;25:543–550. doi: 10.1016/j.it.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Nitschke L, Floyd H, Ferguson DJ, Crocker PR. Identification of CD22 ligands on bone marrow sinusoidal endothelium implicated in CD22-dependent homing of recirculating B cells. J Exp Med. 1999;189:1513–1518. doi: 10.1084/jem.189.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floyd H, Nitschke L, Crocker PR. A novel subset of murine B cells that expresses unmasked forms of CD22 is enriched in the bone marrow: implications for B-cell homing to the bone marrow. Immunology. 2000;101:342–347. doi: 10.1046/j.1365-2567.2000.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitschke L. CD22 and Siglec-G: B-cell inhibitory receptors with distinct functions. Immunol Rev. 2009;230:128–143. doi: 10.1111/j.1600-065X.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- 26.Robak T. Hairy-cell leukemia variant: recent view on diagnosis, biology and treatment. Cancer Treat Rev. 2011;37:3–10. doi: 10.1016/j.ctrv.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Traczewski P, Rudnicka L. Treatment of systemic lupus erythematosus with epratuzumab. Br J Clin Pharmacol. 2011;71:175–182. doi: 10.1111/j.1365-2125.2010.03767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saraux A. The point on the ongoing B-cell depleting trials currently in progress over the world in primary Sjogren's syndrome. Autoimmun Rev. 2010;9:609–614. doi: 10.1016/j.autrev.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Brandt EB, Strait RT, Hershko D, Wang Q, Muntel EE, Scribner TA, Zimmermann N, Finkelman FD, Rothenberg ME. Mast cells are required for experimental oral allergen-induced diarrhea. J Clin Invest. 2003;112:1666–1677. doi: 10.1172/JCI19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra A, Weaver TE, Beck DC, Rothenberg ME. Interleukin-5-mediated allergic airway inflammation inhibits the human surfactant protein C promoter in transgenic mice. J Biol Chem. 2001;276:8453–8459. doi: 10.1074/jbc.M009481200. [DOI] [PubMed] [Google Scholar]
- 31.Lajaunias F, Nitschke L, Moll T, Martinez-Soria E, Semac I, Chicheportiche Y, Parkhouse RM, Izui S. Differentially regulated expression and function of CD22 in activated B-1 and B-2 lymphocytes. J Immunol. 2002;168:6078–6083. doi: 10.4049/jimmunol.168.12.6078. [DOI] [PubMed] [Google Scholar]
- 32.Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, Matsushima K, Ohta K, Yamamoto K, Hirai K. Expression and function of Toll-like receptors in eosinophils: activation by Toll-like receptor 7 ligand. J Immunol. 2003;171:3977–3982. doi: 10.4049/jimmunol.171.8.3977. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Grabowski KA, Xin JP, Coleman J, Huang Z, Espiritu B, Alkan S, Xie HB, Zhu Y, White FA, Clancy J, Jr, Huang H. IL-4 induces differentiation and expansion of Th2 cytokine-producing eosinophils. J Immunol. 2004;172:2059–2066. doi: 10.4049/jimmunol.172.4.2059. [DOI] [PubMed] [Google Scholar]
- 34.Mott RT, Ait-Ghezala G, Town T, Mori T, Vendrame M, Zeng J, Ehrhart J, Mullan M, Tan J. Neuronal expression of CD22: novel mechanism for inhibiting microglial proinflammatory cytokine production. Glia. 2004;46:369–379. doi: 10.1002/glia.20009. [DOI] [PubMed] [Google Scholar]
- 35.Toba K, Hanawa H, Fuse I, Sakaue M, Watanabe K, Uesugi Y, Higuchi W, Takahashi M, Aizawa Y. Difference in CD22 molecules in human B cells and basophils. Exp Hematol. 2002;30:205–211. doi: 10.1016/s0301-472x(01)00791-3. [DOI] [PubMed] [Google Scholar]
- 36.Han X, Jorgensen JL, Brahmandam A, Schlette E, Huh YO, Shi Y, Awagu S, Chen W. Immunophenotypic study of basophils by multiparameter flow cytometry. Arch Pathol Lab Med. 2008;132:813–819. doi: 10.5858/2008-132-813-ISOBBM. [DOI] [PubMed] [Google Scholar]
- 37.Yokoi H, Myers A, Matsumoto K, Crocker PR, Saito H, Bochner BS. Alteration and acquisition of Siglecs during in vitro maturation of CD34+ progenitors into human mast cells. Allergy. 2006;61:769–776. doi: 10.1111/j.1398-9995.2006.01133.x. [DOI] [PubMed] [Google Scholar]
- 38.Sathish JG, Walters J, Luo JC, Johnson KG, Leroy FG, Brennan P, Kim KP, Gygi SP, Neel BG, Matthews RJ. CD22 is a functional ligand for SH2 domain-containing protein-tyrosine phosphatase-1 in primary T cells. J Biol Chem. 2004;279:47783–47791. doi: 10.1074/jbc.M402354200. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki N, Rademacher C, Paulson JC. CD22 Regulates Adaptive and Innate Immune Responses of B Cells. J Innate Immun. 2010 doi: 10.1159/000322375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuscano JM, Riva A, Toscano SN, Tedder TF, Kehrl JH. CD22 cross-linking generates B-cell antigen receptor-independent signals that activate the JNK/SAPK signaling cascade. Blood. 1999;94:1382–1392. [PubMed] [Google Scholar]
- 41.O'Reilly MK, Tian H, Paulson JC. CD22 is a recycling receptor that can shuttle cargo between the cell surface and endosomal compartments of B cells. J Immunol. 2011;186:1554–1563. doi: 10.4049/jimmunol.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munitz A, McBride ML, Bernstein JS, Rothenberg ME. A dual activation and inhibition role for the paired immunoglobulin-like receptor B in eosinophils. Blood. 2008;111:5694–5703. doi: 10.1182/blood-2007-12-126748. [DOI] [PMC free article] [PubMed] [Google Scholar]