Abstract
The numerous regulatory roles of cellular RNAs suggest novel potential drug targets, but establishing intracellular drug-RNA interactions is challenging. Cisplatin (cis-diamminedichloridoplatinum(II)) is a leading anticancer drug that forms exchange-inert complexes with nucleic acids, allowing its distribution on cellular RNAs to be followed ex vivo. Although Pt adduct formation on DNA is well-known, a complete characterization of cellular RNA-Pt adducts has not been performed. In this study, the action of cisplatin on S. cerevisiae in minimal media was established with growth curves, clonogenic assays, and tests for apoptotic markers. Despite high toxicity, cisplatin-induced apoptosis in S. cerevisiae was not observed under these conditions. In-cell Pt concentrations and Pt accumulation on poly(A)-mRNA, rRNA, total RNA, and DNA quantified via ICP-MS indicate ~4–20 fold more Pt accumulation in total cellular RNA than in DNA. Interestingly, similar Pt accumulation is observed on rRNA and total RNA, corresponding to one Pt per (14,600 ± 1,500) and (5760 ± 580) nucleotides on total RNA following 100 and 200 µM cisplatin treatments, respectively. Specific Pt adducts mapped by primer extension analysis of a solvent-accessible 18S rRNA helix occur at terminal and internal loop regions and appear as soon as 1 hr post-treatment. Pt per nucleotide accumulation on poly(A)-mRNA is 4–6-fold lower than on rRNA, but could have consequences for low copy-number or highly regulated transcripts. Taken together, these data demonstrate significant accumulation of Pt adducts on cellular RNA species following in cellulo cisplatin treatment.
These and other small molecule-RNA interactions could disrupt processes regulated by RNA.
INTRODUCTION
As a modulator of gene expression at multiple levels, RNA is an important potential drug target (1–5). In addition to the well-defined functions of mRNA, tRNA, and rRNA, novel regulatory roles are continuously being defined in both transcription and translation (1, 6). Such findings include the discovery of siRNA, microRNA, piwi-interacting RNA, and long noncoding RNAs. Moreover, RNA damage and RNA-protein interactions have been linked to early events in disease and to programmed cell death (7–10). RNA targeting by small molecule interactions has the potential to influence these cellular pathways through both specific and nonspecific mechanisms.
While drug-RNA interactions have the potential to impact cell fate by disrupting RNA regulatory pathways, a challenging aspect for this field is assessing RNA-drug interactions and RNA accessibility in vivo. For this purpose, a covalent RNA-drug adduct is of value in quantifying target binding and following the fate of the targeted RNA. The inertness of metal-RNA adducts formed following treatment with Pt(II) anticancer drugs provides one method of monitoring small-molecule distribution on cellular RNA. Cisplatin (cis-diamminedichloridoplatinum(II)) is a potent antitumor agent that has had a particularly major clinical impact on the treatment of testicular and ovarian cancers. Currently, cisplatin and the structurally related carboplatin and oxaliplatin are used in the treatment regimes of 50–70% of cancer patients (11). In vivo, cisplatin-derived Pt(II) species form kinetically inert ‘covalent’ adducts with biomolecule targets (12, 13). Drug binding to adjacent purines on genomic DNA has been linked to cell cycle arrest at the G2 phase and the induction of programmed cell death, one foundation of antitumor activity (14). Non-genomic contributions to cisplatin’s therapeutic action are also under active investigation. In particular, cisplatin treatment can disrupt RNA-based processes such as splicing and translation (15–17). Targeting of non-DNA species, including RNA, by cisplatin may contribute to or sensitize a cell to the downstream effects of this drug, including the induction of apoptosis.
The distribution of Pt in different RNA species has not been previously determined in cellulo in eukaryotes. Here, we use Saccharomyces cerevisiae for in cellulo analysis of Pt adduct formation on mRNA, rRNA, and total RNA and DNA. S. cerevisiae was selected as a model and has been used previously for drug studies due to the high level of conservation between mammalian and yeast cellular processes, including components of cell-cycle regulation and mRNA turnover (18,19). We report the action of cisplatin on S. cerevisiae in minimal media based on growth curves, clonogenic assays, and tests for apoptotic markers. Despite high cytotoxicity, under the conditions studied cisplatin does not induce apoptosis. Estimated in-cell Pt concentrations and platinum accumulation on mRNA, rRNA, total RNA, and DNA were determined using inductively coupled plasma mass spectrometry (ICP-MS). Interestingly, while similar Pt accumulation was observed on rRNA and total RNA, significantly less accumulated on mRNA. Mapping by reverse transcription demonstrated specific Pt adduct formation on rRNA sequences conserved between yeast and humans. Taken together, these data highlight important differences in the relative accumulation of Pt on different RNA species and provide insight into the accessibility of cellular RNA to small, cationic molecules.
RESULTS AND DISCUSSION
Cisplatin treatment causes acute cell death
S. cerevisiae has been used previously to investigate cisplatin toxicity, including drug transport, DNA repair, and the genes involved in drug resistance and sensitivity (20–22). Within these reports, however, the sensitivity of S. cerevisiae to cisplatin treatment varies widely. In addition, although cisplatin causes apoptosis in mammalian systems (23), this topic had not been addressed for S. cerevisiae despite a growing body of work on yeast apoptotic-like cell death pathways (24,25). Therefore, we established cisplatin cytotoxicity in S. cerevisiae (strain BY4741) by growth and survival curves as well as with apoptotic markers.
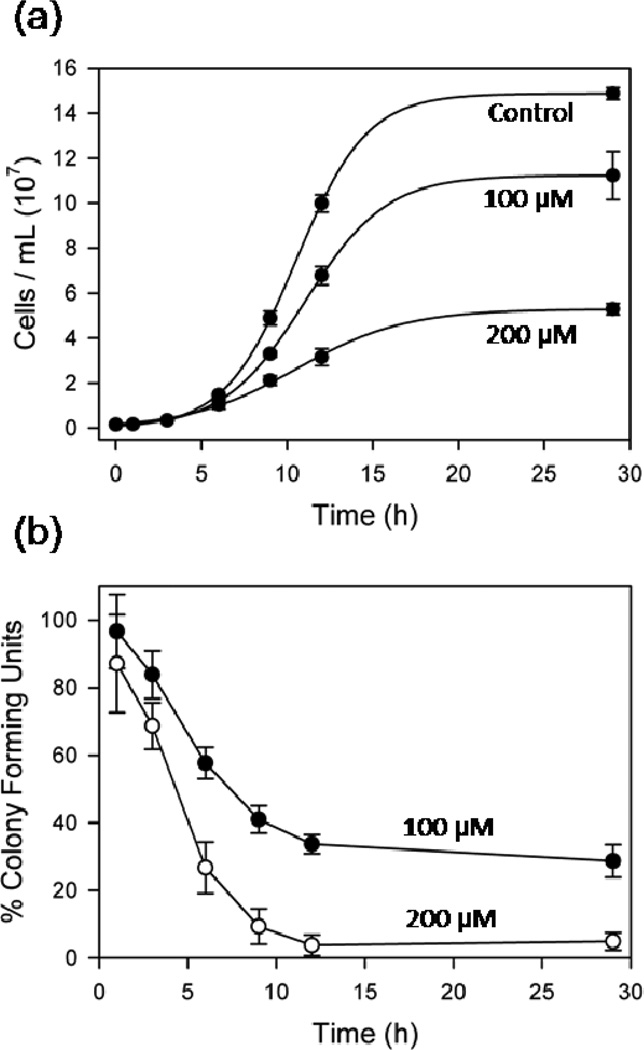
Cisplatin is activated by hydrolysis of the labile chlorido ligands, which in patients occurs upon exposure to relatively low intracellular [Cl−] (26). For cultured cells, however, significant cisplatin aquation may take place in the media. In rich media such as YEPD (yeast extract peptone dextrose), the highly reactive aquation products may interact with soft sulfur- and nitrogen-containing nucleophiles to effectively sequester the drug, which could be one reason for higher IC50 measurements than those observed in mammalian and cancer cell lines (e.g.,500 µM in S. cerevisiae in YEPD media (27) versus 2–40 µM for human cancer cell lines (28–30)). We therefore assayed drug toxicity in minimal SD (synthetic dextrose) liquid media (Figure 1a), and found a moderate (76 ± 8%) and severe (36 ± 1%) reduction in culture density at saturation for 100 and 200 µM drug, respectively. The effect of cisplatin on the viability of BY4741 cells, monitored by clonogenic assay (Figure 1b), shows a marked decrease in the number of dividing cells that begins after just 1–2 h incubation in the drug. The majority of irreversible cisplatin toxicity coincides with the onset of exponential growth that is observed in Figure 1a. We therefore chose 6 h (Table 1) as a relevant timepoint to investigate the distribution of cisplatin-derived Pt species on different RNAs within the cell.
Figure 1.
Cisplatin inhibits yeast growth and viability. (a) Exponential growth curves of BY4741 S. cerevisiae continuously treated with 0, 100, and 200 µM cisplatin in SD media. (b) Viability of cisplatin-treated yeast plated onto drug-free media,. Results presented as the means ± standard deviation from four (a) and three (b) independent experiments.
Table 1.
Cisplatin influence on S. cerevisiae
Measured at 6 h relative to control.
Cell death is not apoptotic
Cell death via apoptotic pathways has been reported for S. cerevisiae treated with several agents including anticancer drugs such as bleomycin and valproate, but has not been reported for metallodrugs such as cisplatin (24,25). We assayed cell cultures for hallmarks of apoptosis after continuous treatment with cisplatin for 6–12 h. Similar to mammalian systems, yeast apoptosis results in chromatin condensation and DNA fragmentation (31). DAPI DNA staining of BY4741 cells in 200 µM cisplatin at 6 h treatment showed significant differences from the control in chromatin morphology (Figure 2a). In almost all samples, nuclei were either fragmented and diffuse, or abnormally enlarged. These findings are consistent with an activation of apoptosis or an alternative programmed death pathway. Cell cycle arrest, previously reported in cisplatin-treated yeast and mammalian cultures (32, 33) was observed with an increase in both parent cell and bud size. Such examples of oncosis are generally associated with necrotic cell death (34), with some exceptions (35), and contrast with the reductions in cell sizes generally observed from slow cell division due to metabolic factors (36).
Figure 2.
Tests for apoptotic markers. (a) DAPI staining of yeast treated with 0 and 200 µM cisplatin for 6 h. (b) TUNEL assay of yeast treated with 0 and 200 µM cisplatin for 6 h, +/− 5 U DNase I. (c) Viability of BY4741, ΔAif1 and ΔYca1 treated with 200 µM cisplatin for 6 h, Inset: average cfu for control and cisplatin-treated cultures. Results presented as means ± standard deviation from three independent experiments.
Apoptosis was further assayed through terminal dUTP nick-end labeling (TUNEL) (31), to detect cleaved DNA. Despite disruptions in cell and chromatin morphology, cells were TUNEL-negative following treatment with 200 µM cisplatin for both 6 h (Figure 2b) and 12 h (data not shown). This suggests that under these conditions, cisplatin treatment disrupts normal chromatin segregation but is insufficient to initiate an apoptotic signal culminating in double-stranded DNA breaks.
The majority of yeast apoptosis pathways are mediated by YCA1, a type 1 metacaspase, or by AIF1, a homolog of mammalian apoptosis-inducing factor (24, 31). To determine if cisplatin-induced toxicity involves either of these pathways, we assessed cell viability in YCA1 and AIF1 deletion mutants treated with 200 µM cisplatin for 6 h. For both ΔYCA1 and ΔAIF1, no differences in cell viability were observed, indicating that neither protein is mediating cytotoxicity in yeast (Figure 2c). In summary, cisplatin-treated S. cerevisiae are undergoing a non-apoptotic form of cell death, but whether it is uncontrolled necrosis, a programmed necrosis, or alternate form of cell death cannot be determined by the present data (35).
Other antitumor agents including the DNA fragmenting bleomycin and the microtubule directed paclitaxel, as well as the ribosome targeting toxin ricin, appear to induce apoptotic markers in yeast (25, 37). From several reports, a stimulus can induce yeast apoptosis at low doses and necrosis at high doses (36,38). It is therefore plausible that cisplatin induces yeast apoptosis in a different treatment window than used here. It is also possible that, unlike the case for other toxins, yeast lacks a key component of a pathway through which cisplatin treatment triggers apoptosis. One candidate is the tumor suppressor p53, which has no yeast homologue (21). Significantly, mismatch repair pathways have been linked to p53-driven apoptosis in mammalian cell lines, while deletion of parallel MMR components in yeast does not influence cisplatin sensitivity (39). In mice, it has been demonstrated that cell cycle arrest occurs in a p53-independent manner, despite its requirement for the initiation of apoptosis (40). Thus, p53-independent cisplatin-induced cell toxicity pathways appear to be present that in yeast result in acute cell cycle arrest and disrupt chromatin morphologies, but not the hallmark DNA cleavage events associated with apoptosis. Similar phenotypes have been observed in S. cerevisiae following treatment with tunicamycin, an X-type agent that also causes cell-cycle arrest and the unfolded protein response, but death by non-apoptotic methods (41).
Intracellular Pt concentrations
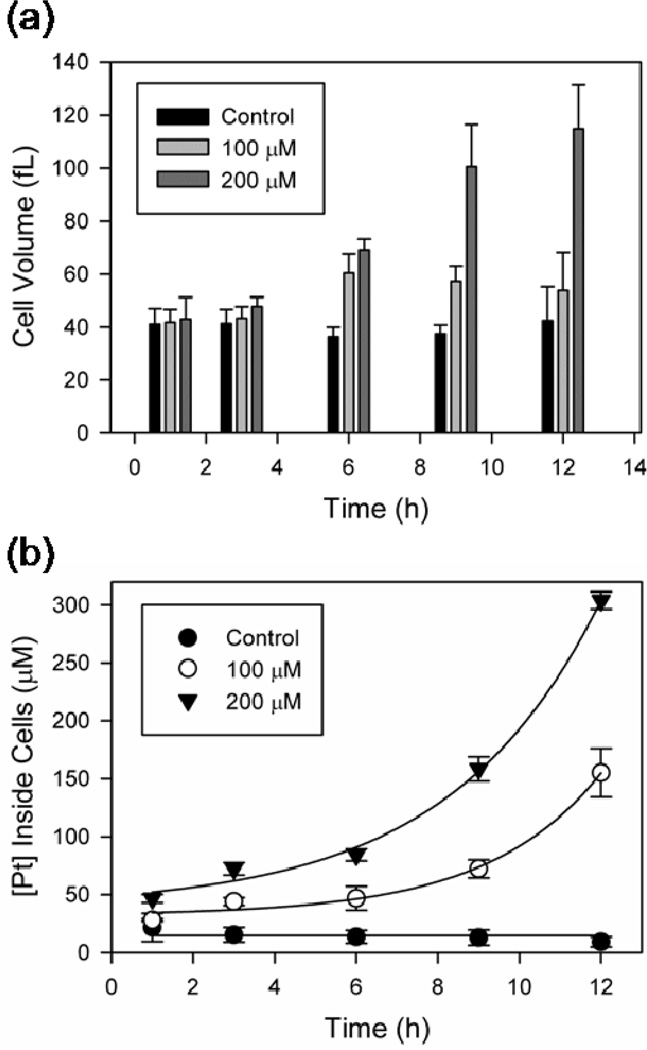
The concentration of Pt species inside a cell following cisplatin treatment is affected by a complex set of dynamics including passive diffusion, active transport, and active efflux from the cell (21,26). To assess intracellular Pt levels, the accumulation of Pt in whole yeast cells was measured by ICP-MS (Supplementary Figure S1). Values of 5.0 ± 0.6 × 106 and 2.1 ± 0.1 × 107 Pt/cell for 100 µM and 200 µM cisplatin treatment after 12 h reflect previous results of 7–30 × 106 Pt/cell for yeast incubated with 130 µM cisplatin for 18 h (42,43) and are in line with values for cisplatin-treated HeLa cells when differences in cell volume are taken into account (30,44).
The increased volumes of cisplatin-treated yeast cells were estimated for calculating intracellular Pt concentrations (Methods). An average volume of (~40 fL) is calculated for untreated yeast, consistent with previous measurements (34,45). The average size of 200 µM cisplatin-treated yeast continuously increases, while that of 100 µM cisplatin-treated yeast is consistent after 6 h, potentially reflecting differences in cell viability between these two treatment conditions at extended time points (Figure 3a). The resulting calculated in-cell Pt concentrations (Figure 3b) are 47 ± 10 and 84 ± 5 µM measured at 6 h for 100 and 200 µM cisplatin, respectively. At 12 h the in-cell Pt concentration exceeds the concentration of cisplatin in the media. This effect, observed previously for other anticancer metallodrugs (46), is consistent with both an active transport process (47) and the fact that these drugs produce kinetically inert adducts with cellular targets, placing drug binding under kinetic rather than thermodynamic control (48).
Figure 3.
Estimated cell volumes and in-cell Pt concentrations. (a) Average estimated cell volumes following treatment with 100 µM and 200 µM cisplatin (see Methods). (b) Calculated in-cell Pt concentrations based on Pt/cell ICP-MS measurements and the average estimated cell volumes. Results averaged from at least three independent experiments presented as means ± standard deviation.
Pt accumulation in different nucleic acids
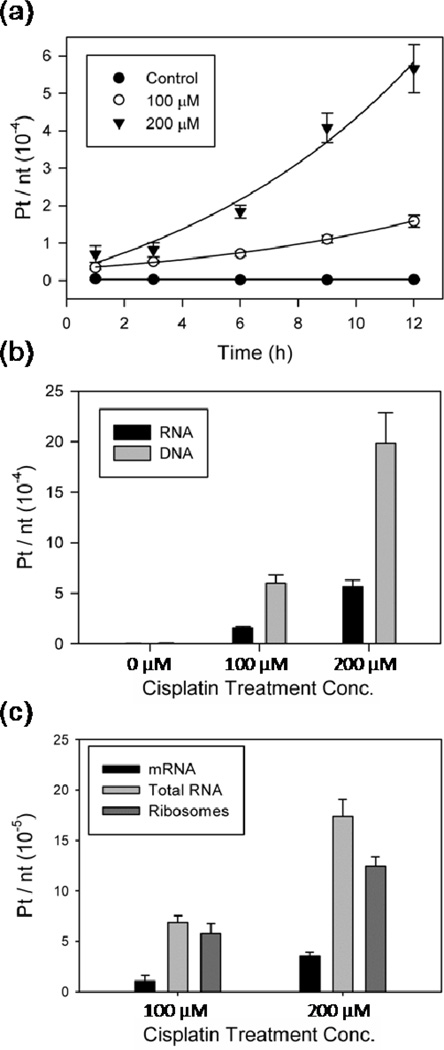
To lend insight into the exposure of cellular RNAs to intracellular Pt species, Pt adduct formation on total RNA extracted from cisplatin-treated yeast was quantified with ICP-MS. An exponential increase in Pt-RNA content was observed in parallel with the exponential increase in cellular Pt concentrations (Figure 4a), indicating that accumulation of Pt in RNA is proportional to whole-cell Pt accumulation. At 6 h, Pt accumulation corresponds to one Pt every 14,600 ± 1,500 and 5760 ± 580 nt for 100 and 200 µM cisplatin, respectively. For perspective, the yeast ribosome is roughly 5600 nt (49).
Figure 4.
Pt accumulation per nucleotide in total RNA, DNA, rRNA, and mRNA. (a) Total RNA from yeast treated with cisplatin. (b) Total RNA and genomic DNA at 12 h treatment. (c) mRNA, total RNA, and rRNA at 6 h. Data values provided in Supplementary Table S1, and presented as means ± standard deviation for at least three independent experiments.
A comparison of Pt accumulation on whole-cell RNA and DNA, performed following 12 h treatment with cisplatin (see Methods), yields ~ 3-fold more Pt bound to DNA than RNA on a per nucleotide basis (Figure 4b). Cellular distribution studies performed in human cell lines have observed an accumulation of cisplatin-derived Pt in the nucleus and nucleolus (50), consistent with the higher density of Pt on DNA. However, there is 10–50 fold more RNA in a yeast cell than DNA (51,52), resulting in ~4–20 fold more Pt accumulation in the total cellular RNA than in the total cellular DNA (Table 2).
Table 2.
Estimated Pt atoms accumulated in the total RNA or DNA of one yeast cella
| [Cisplatin] | ||
|---|---|---|
| 100 µM | 200 µM | |
| DNA (104) | 2 | 6 |
| RNA (104) | 7–34 | 24–120 |
Calculation based on the mass of DNA and RNA in one haploid S. cerevisiae cell (60)
Pt accumulation on poly(A)-mRNA and rRNA was compared in total cellular RNA harvested from yeast after 6 h of continuous cisplatin treatment. Poly(A)-messenger RNA was extracted with an mRNA Miniprep Kit while intact 25S and 18S rRNA were isolated by gel purification (Methods). Pt accumulation on rRNA is similar to that of total RNA on a per nucleotide basis, while significantly less accumulation per nucleotide is observed on poly(A)-mRNA (Figure 4c and Supplementary Table S1). Given that yeast RNA is 80% ribosomes, 15% tRNA, and 5% mRNA (51), it appears that Pt measured in total RNA is largely due to accumulation on ribosomal RNA. Assuming a statistical distribution of Pt adducts, these data indicate an average of 1 and 2 Pt adduct for every 3 ribosomes following 6 h treatments with 100 and 200 µM cisplatin, respectively.
In yeast, global protein synthesis is dramatically reduced as early as 2–4 hr post-cisplatin treatment, indicating a compromised translation machine, which may be a result of mRNA, tRNA, or rRNA modifications (27). In general, cationic aquated Pt species are expected to associate more readily with accessible sites in higher-order RNA structures, which present a larger electrostatic driving force (50). The enrichment of Pt on rRNA relative to poly(A)-mRNA (Figure 4c) may be due to the high negative charge density of the ribosome and may present a contributing factor to cisplatin’s cytotoxicity, although the probability of Pt adducts associating with higher-order regulatory structures amidst otherwise single-stranded regions of cellular mRNAs is yet unknown. Additional factors that remain to be determined include the influence of Pt-RNA adducts on mRNA surveillance pathways and message turnover (53).
Platinum adduct identification on rRNA
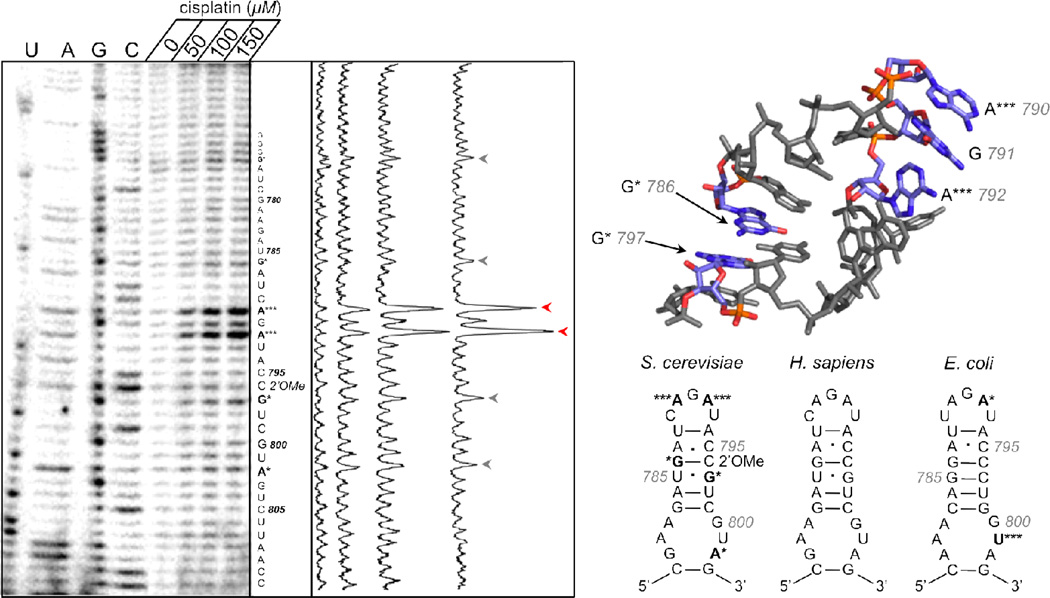
Specific locations of platinum adducts within S. cerevisiae ribosomal RNA were probed by RT primer extension, which stalls 3’ to adduct sites (54–56). Pt adducts may reflect the solvent accessibility and electrostatic potential of specific rRNA motifs. Using similar methods to probe for potential antibiotic sites, Rijal and Chow reported major platination sites within helix 24 of the E. coli small ribosomal subunit (56), revealing a significant RT stall at U801 that reflects Pt binding to adjacent guanines at positions 799 and 800 (Figure 5). Secondary binding was observed at A792, suggesting the formation of a 3’-GA-5’ adduct between G791 and A790. As an ideal antibiotic drug differentially targets bacterial and eukaryotic ribosomes, we compared platinum adducts in this region (helix 18 in S. cerevisiae), which is located in close proximity to the peptidyltransferase center (Supplementary Figure S2) and fully conserved between yeast and humans but not E. coli (Figure 5).
Figure 5.
Primer extension analysis of S. cerevisiae small ribosomal subunit helix 18. Primer extension analysis of RNA isolated from 6 h cisplatin-treated BY4741 (left) shows a dosage-dependant increase in termination intensity at the starred sites, indicating major (***) and minor (*) Pt binding sites. Dideoxy sequencing ladders denoted by U, A, G, and C. Experimental results are summarized on secondary structure of the S. cerevisiae helix 18 (right, lower panel). Results from Rijal and Chow (56) are summarized on the E. coli secondary structure (right, lower panel). E. coli numbering used for comparison. Major platinum binding sites and a potential crosslink between G797 and G786 are depicted on a helix 18 crystal structure (right, upper panel, PDB 3O30).
Primer extension was performed on total RNA extracted from yeast cultures treated with 0–150 µM cisplatin for 6 h. Figure 5 shows a dosage-dependant increase in termination intensity at several positions, predicting sites of platinum binding. Significant platinum adduct formation occurs at the purine-rich capping loop of helix 18, as evidenced by two major stops in the sequencing gel at positions A792 (***) and A790 (***) (Figure 5). Minor stalling is observed at A802 and G786. An additional minor stop site occurs directly on purine residue G797. This particular guanine lies 3’ to a 2’O-methylated cytosine residue, which is known to pause primer extension under certain conditions (62). Alternatively, stalling at this site may be the result of an interstrand crosslink between G797 and G786, whose N7 atoms are stacked favorably for such an interaction (Figure 5).
Certain RNA sites are expected to be more accessible and/or reactive. To detect early Pt accumulation on rRNA, helix 18 was probed after exposure to 100 µM cisplatin for 0, 1 and 3 h (Supplementary Figure S3). As expected, platinum targets the same purine residues as observed following 6 h treatment (Figure 5), showing transcription stops at positions A790 and A792 within the capping loop as early as 1 h post exposure to drug (Supplemental Figure 3). Stalling at A802 appears stronger at early timepoints, although its signal is minor in comparison to banding patterns observed within the terminal loop (A792 and A790) after 6 h cisplatin treatment. Importantly, at all timepoints, the reversal of the G783-C799 basepair in yeast and humans, which removes the preferred G799–G800 Pt binding site observed in E. coli, is sufficient to preclude Pt binding to G800 in yeast. Instead, binding within the stem is diverted to residue A802. These findings demonstrate that although aquated cisplatin products are highly reactive, they are remarkably sequence-specific in the context of complex RNA structures.
Conclusions
In this study, the ability of the anticancer drug cisplatin to form stable adducts with RNA was used to assess accumulation of Pt species on cellular RNA following drug treatment. Under the conditions of this study cisplatin toxicity was characterized by irreversible inhibition of cell division, but not apoptotic cell death. This suggests that in comparison with mammalian systems, a mediator of cisplatin-induced apoptotic pathways, such as p53, is lacking in S. cerevisiae. Comparison of Pt accumulation in RNA and DNA at 12 h shows ~4–20 fold more Pt accumulation in the total cellular RNA than in genomic DNA. Intact 25S and 18S ribosomal RNA accumulates the majority of cellular Pt, while significantly less accumulates on poly(A)-mRNA when compared on a per-nucleotide basis. Mapping by reverse transcription demonstrates that specific Pt adducts in eukaryotic ribosomes form in a dose- and time-dependent fashion, accumulating after just 1 h of treatment. Taken together, these data show significant accumulation of Pt adducts in eukaryotic RNA following treatment in cellulo with cisplatin, with significantly larger amounts of irreversible Pt-RNA adducts accumulating in ribosomal RNA as compared to mRNA, and a demonstrated specificity for particular binding sites in the eukaryotic ribosome. The results from these data emphasize potential for rapid and specific accumulation of small molecules on cellular RNA, with interactions that could impact complex RNA regulatory pathways.
METHODS
Cell cultures and treatments
S. cerevisiae strains BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0), yca1Δ (BY4741 yca1::kanMX4) and aif1Δ (BY4741 aif1::kanMX4) were gifts from the Stevens laboratory at the University of Oregon. Liquid cultures were grown on Synthetic Complete medium (SC) consisting of 0.67% yeast nitrogen base and 2% glucose supplemented with amino acids and nucleotide bases maintained in the dark at 30 °C with shaking at 200 rpm. Plated cells were grown on YEPD agar plates (1% yeast extract, 2% peptone, 2% glucose, and 2% agar). A 5 mM cisplatin (Sigma Aldrich) stock in 100 mM NaCl (stored in the dark for no more than a week) was used for all cisplatin treatments. Yeast cultures were pregrown to an OD600 of 5 then inoculated into 30 °C cisplatin-containing media to an OD600 of 0.075.
Culture growth, cell survival and cell size
Culture growth was measured by absorbance at 600 nm (1 AU600 = 2.0 × 107 cells/mL). Cell viability was measured by plating serial dilutions of treated and untreated yeast, at given growth times, onto drug-free YEPD agar plates (~250 cells/plate) and counting colonies after 3 d at 30 °C. The number of colony-forming units (cfu) was determined by dividing the cfu counts of treated cultures by those of untreated cultures (assumed to be 100%). Yeast cell radii were measured from differential interference contrast images obtained on a Carl Zeiss Axioplan 2 fluorescence microscope using a 100× objective and AxioVision software (Carl Zeiss, Thornwood, NY) and volumes were calculated by treating the yeast as spheres.
Nucleic acid extraction and purification
For measurements of Pt in total RNA ~1.2 × 108 cisplatin treated yeast cells were pelleted and RNA was extracted using the MasterPure RNA Purification Kit (Epicentre) according to manufacturer’s specifications. For both mRNA and rRNA samples total RNA was extracted from cisplatin treated cells according to the method of Motorin et al. (57). Poly(A)-mRNA was isolated using GenElute mRNA Miniprep Kit (Sigma), doing the binding and wash steps twice to ensure maximum mRNA purity. Ribosomal RNA was isolated from total RNA using 8% dPAGE. The 25S and 18S bands were visualized by brief staining with methylene blue, cut out and eluted with an Elutrap Electroelution System (Whatman), then desalted (3k Microsep Centrifugal Devices, Pall). DNA samples were purified as in Rose et al. including the optional RNase A treatment (58). Because Pt adducts on RNA are known to inhibit RNase activity (58) DNA was extracted at 12 h, a time at which there is 4–5 fold lower RNA content in yeast cell (52), in order to ensure sample purity. All pellets were collected at 4 °C and washed 3 times with deionized H2O before further processing.
Measurement of Pt content
Isolated RNA and DNA samples were desalted on in-house prepared G-25 sephadex spin columns (BioRad) and quantified by absorbance at 260 nm. Whole-cell and most nucleic acid samples were digested in 70% nitric acid (trace select, Fluka) for 2 hours at 65°C then diluted to 2% (v/v) nitric acid with milli-Q H2O. Pt content was determined by Inductively Coupled Plasma Mass Spectrometry (ICP-MS) using a Thermo VG PQExcell quadrupole ICP-MS equipped with a Gilson 222 autosampler at the W. M. Keck Collaboratory for Plasma Spectrometry (Oregon State University, Corvallis, Oregon). The instrument was calibrated for 194Pt, 195Pt and 196Pt by developing standard curves from a Pt standard (High Purity Standards). All measurements were done in triplicate using 115In as an internal standard.
Tests for apoptotic markers
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) was performed according to the method of Madeo et. al. (59) with the following modifications: cell walls were digested with 2.5 U zymolyase 100T (US biological) for 30 min at 30 °C in 1 mL sorbitol buffer (1.2 M sorbitiol, 0.5 mM MgCl2, 35 mM phosphate buffer pH 6.8), the permiabilization was carried out for 1 min, and the in situ cell death detection kit, fluorescein (Roche), was used with a 30 min incubation (full protocol Supplementary pS4). Positive control samples were incubated with 0.2, 1, and 5 U DNase I (Fermentas) for 1 h at 37 °C in the manufacturer’s rxn buffer. Images were acquired on a Carl Zeiss Axioplan 2 fluorescence microscope as described above. DAPI staining was performed by pelleting 1.0 × 107 yeast cells, washing 3x with PBS, fixing in 70% (v/v) EtOH for 30 min, washing with PBS, and then incubating with 0.5 µg/mL DAPI for 20 min (in the dark). The samples were then washed 3x with PBS and visualized immediately.
5’ End-labeling
The DNA primer designed for reverse transcription of helix 18 of the small yeast ribosomal subunit was purchased from Integrated DNA Technologies. γ-32P 5’ end-labeling was performed as previously described (53).
Reverse Transcription
Total RNA extracted from cisplatin-treated yeast cells was used as a template for RT. Yeast cells were inoculated at 5 × 106 cells/mL into cisplatin-containing SD-URA and grown for 6 h. Total RNA was isolated using a MasterPure Yeast RNA Extraction kit (Epicentre) and resuspended in 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA to a final concentration of 10 µg/µL. For primer extension, 1 µg of RNA template was annealed to 100 pmol of the specified 5’ end-labeled primer in the manufacturer’s reaction buffer and incubated in the presence of AMV Reverse Transcriptase (Fermantas) for 1.75 h at 42°C. The resulting cDNA products were diluted in loading buffer containing 0.005% (w/v) xylene cyanol and bromophenol blue and analyzed by 8% dPAGE. Bands were visualized using a GE phosphor screen in conjunction with a Storm phosphor screen imaging system and then quantified with ImageQuant 5.1 and normalized in Excel.
Supplementary Material
Acknowledgement
We thank A. Ungerer for assistance with the ICP-MS experiments at the W. M. Keck Collaboratory for Plasma Spectrometry at Oregon State University, L. Graham for assistance with protocols and imaging, and the Stevens laboratory for use of the Carl Zeiss Axioplan 2 fluorescence microscope. The BY4741, yca1Δ, and aif1Δ strains were a kind gift from the Stevens Laboratory. Funding from the NIH (GM058096 to VJD and 5T32GM007759 to MFO) and the University of Oregon is gratefully acknowledged.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 3.Chow CS, Bogdan FM. A structural basis for RNA-ligand interactions. Chem. Rev. 1997;97:1489–1514. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 4.Tor Y. Targeting RNA with small molecules. Chembiochem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 5.Tibodeau JD, Fox PM, Ropp PAR, Theil EC, Thorp HH. The up-regulation of ferritin expression using a small molecule ligand to the native mRNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:253–257. doi: 10.1073/pnas.0509744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattick JS. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 7.Kong QM, Lin CLG. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JT, Raines RT. Ribonucleases as novel chemotherapeutics: the ranpirnase model. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olmo N, Turnay J, González de Buitrago G, López de Silanes I, Gavilanes JG, Lizarbe MA. Cytotoxic mechanism of the ribotoxin alpha-sarcin - Induction of cell death via apoptosis. Eur. J. Biochem. 2001;268:2113–2223. doi: 10.1046/j.1432-1327.2001.02086.x. [DOI] [PubMed] [Google Scholar]
- 10.Jetzt AE, Cheng JS, Tumer NE, Cohick WS. Ricin A-chain requires c-Jun N-terminal kinase to induce apoptosis in nontransformed epithelial cells. Int. J. Biochem. Cell B. 2009;41:2503–2510. doi: 10.1016/j.biocel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson PJ, Sava G. Metal-based antitumour drugs in the post genomic era. Dalton Trans. 2006:1929–1933. doi: 10.1039/b601840h. [DOI] [PubMed] [Google Scholar]
- 12.Akaboshi M, Kawai K, Maki H, Akuta K, Ujeno Y, Miyahara T. The number of platinum atoms binding to DNA, RNA and protein molecules of HeLa-cells treated with cisplatin at its mean lethal concentration. Jpn. J. Cancer Res. 1992;83:522–526. doi: 10.1111/j.1349-7006.1992.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. Oncol. Rep. 2003;10:1663–1682. [PubMed] [Google Scholar]
- 14.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Ju J-F, Danenberg KD, Danenberg PV. Inhibition of pre-mRNA splicing by cisplatin and platinum analogs. Int. J. Oncol. 2003;23:785–789. [PubMed] [Google Scholar]
- 16.Rosenberg JM, Sato PH. Cisplatin inhibits in vitro translation by preventing the formation of complete initiation complex. Mol. Pharmacol. 1993;43:491–497. [PubMed] [Google Scholar]
- 17.Heminger KA, Hartson SD, Rogers J, Matts RL. Cisplatin inhibits protein synthesis in rabbit reticulocyte lysate by causing an arrest in elongation. Arch. Biochem. Biophys. 1997;344:200–207. doi: 10.1006/abbi.1997.0198. [DOI] [PubMed] [Google Scholar]
- 18.Mazzoni C, Falcone C. mRNA stability and control of cell proliferation. Biochem. Soc. Trans. 2011;39:1461–1465. doi: 10.1042/BST0391461. [DOI] [PubMed] [Google Scholar]
- 19.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida S, Herskowitz I. Genetic analysis of cisplatin resistance in yeast and mammals. In: Nitiss JL, Heitman J, editors. Yeast as a Tool in Cancer Research. The Netherlands: Springer; 2007. pp. 393–408. [Google Scholar]
- 21.Sinani D, Adle DJ, Kim H, Lee J. Distinct mechanisms for Ctr1-mediated copper and cisplatin transport. J. Biol. Chem. 2007;282:26775–26785. doi: 10.1074/jbc.M703973200. [DOI] [PubMed] [Google Scholar]
- 22.Liao C, Hu B, Arno M, Panaretou B. Genomic screening in vivo reveals the role played by vacuolar H+ ATPase and cytosolic acidification in sensitivity to DNA-damaging agents such as cisplatin. Mol. Pharmacol. 2007;71:416–425. doi: 10.1124/mol.106.030494. [DOI] [PubMed] [Google Scholar]
- 23.Fuertes MA, Castilla J, Alonso C, Perez JM. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr. Med.Chem. 2003;10:257–266. doi: 10.2174/0929867033368484. [DOI] [PubMed] [Google Scholar]
- 24.Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 25.Almeida B, Silva A, Mesquita A, Sarripalo-Marques B, Rodrigues F, Ludovico P. Drug-induced apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1436–1448. doi: 10.1016/j.bbamcr.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann RL. The effects of cisplatin and platinum (IV) on cell growth, RNA, protein, ribosome and DNA synthesis. Toxicol. Environ. Chem. 1988;17:139–151. [Google Scholar]
- 28.Beretta GL, Gatti L, Tinelli S, Corna E, Colangelo D, Zunino F, Perego P. Cellular pharmacology of cisplatin in relation to the expression of human copper transporter CTR1 in different pairs of cisplatin-sensitive and -resistant cells. Biochem. Pharmacol. 2004;68:283–291. doi: 10.1016/j.bcp.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Song IS, Savaraj N, Siddik ZH, Liu PM, Wei YJ, Wu CJ, Kuo MT. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol. Cancer Ther. 2004;3:1543–1549. [PubMed] [Google Scholar]
- 30.Zisowsky J, Koegel S, Leyers S, Devarakonda K, Kassack MU, Osmak M, Jaehde U. Relevance of drug uptake and efflux for cisplatin sensitivity of tumor cells. Biochem. Pharmacol. 2007;73:293–307. doi: 10.1016/j.bcp.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Frohlich K-U, Fussi H, Ruckenstuhl C. Yeast apoptosis—from genes to pathways. Semin. Cancer Biol. 2007;17:112–121. doi: 10.1016/j.semcancer.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Sorenson CM, Eastman A. Mechanism of cis-diamminedichloroplatinum(II)-induced cyto-toxicity - role of G2 arrest and DNA double-strand breaks. Cancer Res. 1988;48:4484–4488. [PubMed] [Google Scholar]
- 33.Grossmann KF, Brown JC, Moses RE. Cisplatin DNA cross-links do not inhibit S-phase and cause only a G2/M arrest in Saccharomyces cerevisiae. Mutat. Res. 1999;434:29–39. doi: 10.1016/s0921-8777(99)00011-7. [DOI] [PubMed] [Google Scholar]
- 34.Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J. Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenberg T, Carmona-Gutierrez D, Buttner S, Tavernarakis N, Madeo F. Necrosis in yeast. Apoptosis. 2010;15:257–268. doi: 10.1007/s10495-009-0453-4. [DOI] [PubMed] [Google Scholar]
- 36.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: A regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XP, Baricevic M, Saidasan H, Tumer NE. Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae. Infect. Immun. 2007;75:417–428. doi: 10.1128/IAI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q, Zhou B. Copper and manganese induce yeast apoptosis via different pathways. Mol. Biol. Cell. 2007;18:4741–4749. doi: 10.1091/mbc.E07-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lage H, Dietel M. Involvement of the DNA mismatch repair system in antineoplastic drug resistance. J. Cancer Res. Clin. Oncol. 1999;125:156–165. doi: 10.1007/s004320050258. [DOI] [PubMed] [Google Scholar]
- 40.Zamble D, Jacks T, Lippard SJ. p53-dependent and –independent responses to cisplatin in mouse testicular terato carcinoma cells. PNAS. 1998;95:6163–6168. doi: 10.1073/pnas.95.11.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudgeon DD, Zhang N, Ositelu OO, Kim H, Cunningham KW. Nonapoptotic Death of Saccharomyces cerevisiae Cells That Is Stimulated by Hsp90 and Inhibited by Calcineurin and Cmk2 in Response to Endoplasmic Reticulum Stresses. Eukaryot. Cell. 2008;12:2037–2051. doi: 10.1128/EC.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burger H, Capello A, Schenk PW, Stoter G, Brouwer J, Nooter K. A genome-wide screening in Saccharomyces cerevisiae for genes that confer resistance to the anticancer agent cisplatin. Biochem. Bioph. Res. Co. 2000;269:767–774. doi: 10.1006/bbrc.2000.2361. [DOI] [PubMed] [Google Scholar]
- 43.Schenk PW, Brok M, Boersma AWM, Brandsma JA, Den Dulk H, Burger H, Stoter G, Brouwer J, Nooter K. Anticancer drug resistance induced by disruption of the Saccharomyces cerevisiae NPR2 gene: a novel component involved in cisplatin- and doxorubicin-provoked cell kill. Mol. Pharmacol. 2003;64:259–268. doi: 10.1124/mol.64.2.259. [DOI] [PubMed] [Google Scholar]
- 44.Moran U, Phillips R, Milo R. Snapshot: key numbers in biology. Cell. 2010;141:1262–1262. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Tamaki H, Yun C-W, Mizutani T, Tsuzuki T, Takagi Y, Shinozaki M, Kodama Y, Shirahige K, Kumagai H. Glucose-dependent cell size is regulated by a G protein-coupled receptor system in yeast Saccharomyces cerevisiae. Genes Cells. 2005;10:193–206. doi: 10.1111/j.1365-2443.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- 46.Bergamo A, Messori L, Piccoli F, Cocchietto M, Sava G. Biological role of adduct formation of the ruthenium(III) complex NAMI-A with serum albumin and serum transferrin. Inv. New Drugs. 2003;21:401–411. doi: 10.1023/a:1026243000320. [DOI] [PubMed] [Google Scholar]
- 47.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bancroft DP, Lepre CA, Lippard SJ. Pt-195 NMR kinetic and mechanistic studies of cis-diamminedichloroplatinum and trans-diamminedichloroplatinum(II) binding to DNA. J. Am. Chem. Soc. 1990;112:6860–6871. [Google Scholar]
- 49.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, MÜller KM, Pande N, Shang Z, Yu N, Gutell RR. The Comparative RNA Web (CRW) Site: An Online Database of Comparative Sequence and Structure Information for Ribosomal, Intron, and Other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapman EG, Hostetter AH, Osborn MF, Miller AL, DeRose VJ. Binding of kinetically inert metal ions to RNA: The case of Pt(II) In: Sigel A, Sigel H, Sigel RKO, editors. Metal Ions in Life Sciences: Structural and Catalytic Roles of Metal Ions in RNA. Cambridge: Royal Society of Chemistry; 2011. pp. 347–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ju Q, Warner JR. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast. 1994;10:151–157. doi: 10.1002/yea.320100203. [DOI] [PubMed] [Google Scholar]
- 52.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Bio. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 54.Chapman EG, DeRose VJ. Enzymatic Processing of Platinated RNAs. J. Am. Chem. Soc. 2010;132:1946–1952. doi: 10.1021/ja908419j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papsai P, Snygg AS, Aldag J, Elmroth SK. Platination of full length tRNA(Ala) and truncated versions of the acceptor stem and anticodon loop. Dalton Trans. 2008;38:5225–5234. doi: 10.1039/b719542g. [DOI] [PubMed] [Google Scholar]
- 56.Rijal K, Chow CS. A new role for cisplatin: probing ribosomal RNA structure. Chem. Commun. 2009;1:107–109. doi: 10.1039/b816633a. [DOI] [PubMed] [Google Scholar]
- 57.Motorin Y, Muller S, Behm-Ansmant I, Brantant C. Identification of modified residues in RNAs by reverse transcription-based methods. Method. Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 58.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: a Laboratory Course Manual. NewYork: Cold Spring Harbor Laboratory Press; 1990. Yeast DNA miniprep; pp. 140–142. [Google Scholar]
- 59.Madeo F, Frohlich E, Frohlich K-U. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman F. Getting started with yeast. Method. Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.