Abstract
To directly address whether regulating mRNA localization can influence animal behavior, we created transgenic mice that conditionally express Zipcode Binding Protein 1 (ZBP1) in a subset of neurons in the brain. ZBP1 is an RNA-binding protein that regulates the localization, as well as translation and stability of target mRNAs in the cytoplasm. We took advantage of the absence of ZBP1 expression in the mature brain to examine the effect of expressing ZBP1 on animal behavior. We constructed a transgene conditionally expressing a GFP-ZBP1 fusion protein in a subset of forebrain neurons and compared cocaine-cued place conditioning in these mice versus noninduced littermates. Transgenic ZBP1 expression resulted in impaired place conditioning relative to nonexpressing littermates, and acutely repressing expression of the transgene restored normal cocaine conditioning. To gain insight into the molecular changes that accounted for this change in behavior, we identified mRNAs that specifically immunoprecipitated with transgenic ZBP1 protein from the brains of these mice. These data suggest that RNA-binding proteins can be used as a tool to identify the post-transcriptional regulation of gene expression in the establishment and function of neural circuits involved in addiction behaviors.
Many mRNAs are transcribed rapidly in response to synaptic activity, the so-called Immediate Early Genes (IEGs), and these genes contribute to the molecular changes that lead to long-term synaptic plasticity (Kourrich et al. 2007). In response to a single dose of cocaine, mRNA levels for IEGs fos, jun, and zif268, which encode transcription factors, are increased and these increases are potentiated with repeated dosage (Hope et al. 1992; Moratalla et al. 1996). Chronic cocaine administration leads to the expression of the ΔFosB splice variant of FosB, a subunit of the AP-1 transcription factor (Nakabeppu and Nathans 1991; Hiroi et al. 1997). ΔFosB lacks the C terminus that inhibits transcriptional induction by other AP-1 subunits and is associated with an increased binding of AP-1 transcription sites in the striatum (Hope et al. 1994). Therefore, one mechanism through which cocaine potentiates IEG expression is by increasing the pool of activating AP-1 transcription factors.
Numerous studies have been performed on the trafficking and local translation of mRNA using neurons in culture. However, despite demonstration of the fundamental contribution of localized mRNA translation to synaptic plasticity, there are no studies about how mRNAs may be regulated at this level by exposure to drugs of abuse. Electrophysiological recording studies have long demonstrated a requirement for translation and new transcription in Long Term Potentiation, a measure of synaptic plasticity. Moreover, localization to the dendritic compartment depends on specific sequences found within these mRNAs. Based on these observations, it has been hypothesized that a subset of activity-regulated mRNAs is actively transported to synapses after synaptic activity, and localized translation of these has been proposed as a basic cellular mechanism required for synaptic plasticity (Rodriguez et al. 2008; Vuppalanchi et al. 2009; Wang et al. 2010; Sinnamon and Czaplinski 2011). Although the importance of mRNA localization and translational control mechanisms have been effectively demonstrated in vitro, few studies have examined how these post-transcriptional controls of gene expression in the cytoplasm contribute to behavior in vivo. Cultured neurons cannot replicate function within brain circuitry; therefore, in vivo studies in living animals provide the best opportunities to understand cellular functions within their context of the behaving brain. In vivo studies of neuron function often use neuron-specific transgenes in mice to investigate molecular pathways underlying brain function and behavior. For instance, transgenic mice expressing a mutant form of Calmodulin-dependent protein Kinase II subunit α (CaMKIIα) demonstrated an altered frequency response to stimulation in the theta range, along with impairment of spatial memory (Bach et al. 1995). Transgenic mice expressing a dominant-negative cAMP-dependent protein kinase (PKA) demonstrated its importance in the late phase of LTP, spatial memory, and contextual fear conditioning (Abel et al. 1997).
The striatum, including both the dorsal striatum and the nucleus accumbens, has been implicated in the rewarding and conditioned rewarding effects of addictive substances (Porrino et al. 2004; Fuchs et al. 2006; Volkow et al. 2006; Wong et al. 2006; Graybiel 2008; Robbins et al. 2008; Luscher and Malenka 2011). Natural rewards increase extracellular dopamine levels through the mesolimbic dopaminergic system that innervates the NAc (Koob 2000). Addictive drugs, such as cocaine, can increase the effective concentration of the dopamine released from the dopaminergic projections to the NAc by competitively inhibiting the dopamine transporter (Wu et al. 2001). This increase in dopamine has been linked to structural plasticity in neurons of the NAc, including long-lasting increases in dendritic length, spine density, and the number of branched spines (Robinson and Kolb 1997). Therefore, the cellular mechanisms of synaptic plasticity are likely to be critical for the effects of drugs of abuse. As a first step toward understanding how localized mRNA translation helps regulate synaptic plasticity after exposure to cocaine, we conditionally expressed a cytoplasmic RNA-binding protein, ZBP1, in NAc neurons to alter the cytoplasmic regulation of mRNAs. We report here that transgenic expression of ZBP1 reversibly impairs cocaine-cued place conditioning in these animals. To understand the molecular changes that account for the decrease in conditioning, we took advantage of the RNA-binding properties of the ZBP1 to isolate the mRNAs that copurified with it, and identify these using microarrays. This study provides the first demonstration that alterations in the post-transcriptional regulation of mRNA can effectively alter a behavioral response to an addictive drug.
Results
Generation and characterization of ZBP1-expressing transgenic mice
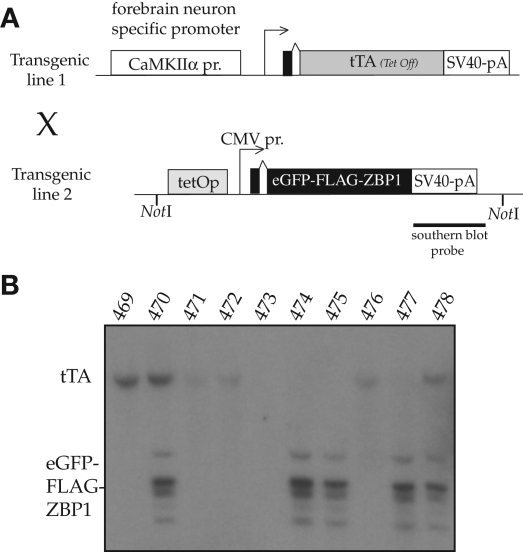
To probe the effect of altering post-transcriptional regulation of mRNA in behavioral response to drugs of addiction, we expressed transgenic ZBP1 protein specifically in forebrain neurons. Endogenous ZBP1 is not expressed in mature neurons; therefore, transgenic expression would not compete with endogenous protein for target binding (Leeds et al. 1997; Ioannidis et al. 2003, 2004; Perycz et al. 2011). A ZBP1 transgene was also likely to influence localized mRNA translation when expressed in neurons (Tiruchinapalli et al. 2003; Huttelmaier et al. 2005; Yao et al. 2006; Sasaki et al. 2010; Donnelly et al. 2011; Perycz et al. 2011; Welshans and Bassell 2011). To facilitate studies of transgene expression we used a GFP-ZBP1 fusion, with a FLAG epitope at the N terminus of ZBP1. These tags have no effect on ZBP1 protein function (Farina et al. 2003; Tiruchinapalli et al. 2003). We used a doxycycline-regulated transcription system to pharmacologically regulate the expression of ZBP1. We cloned the cDNA sequences encoding GFP-FLAG-ZBP1 into a plasmid containing the tetO tetracycline-regulated enhancer element (Fig. 1A). Nine independent transgenic lines were generated expressing GFP-ZBP1. Each of these lines was crossed with mice expressing the transcriptional trans-activator tTA (Tet-off), driven by the CaMKIIα promoter for expression of the transgene in the forebrain (Mayford et al. 1996).
Figure 1.
Construction of GFP-FLAG-ZBP1 transgenic mice. (A) Diagram of the transgenic expression constructs used for doxycycline-regulated ZBP1 expression. The tTA is expressed from a neuron-specific promoter, and the GFP-FLAG-ZBP1 protein is expressed only in the presence of the tTA protein. (B) Detection of transgenes in transgenic animals. Southern blot probed with the SV40 poly(A) signal produces bands that can identify both the ZBP1 transgene and the CaMKIIα-tTA transgene on a single blot (tTA = CaMKIIα-tTA, ZBP1 = EGFP-ZBP1). Bitransgenic animals were used to examine the expression of ZBP1 and the effect of this expression on behavior.
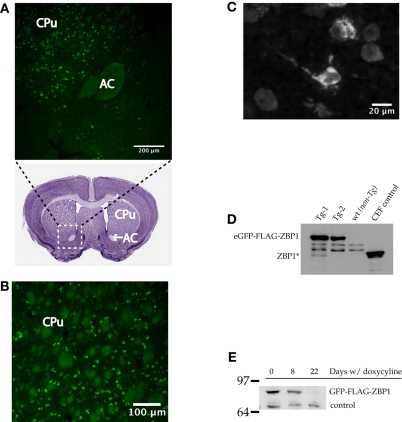
We analyzed fluorescence in whole brain slices to determine the expression pattern of the GFP-ZBP1 transgene in the nine transgenic lines. Line KL2169 only demonstrated visible GFP fluorescence in the striatum, a pattern of expression that has been reported for some transgenes in this driver line (Mayford et al. 1996). As expected, GFP-ZBP1 was cytoplasmic within neurons, strongly labeling the cytoplasm in the soma of neurons, but occasionally extending into proximal neuronal processes as well (Fig. 2C). Expression of GFP-ZBP1 was also analyzed by Western blotting of whole brain extracts (without cerebellum) using a polyclonal antibody raised against ZBP1 (Ross et al. 1997). A protein of the expected size for GFP-ZBP1 was observed only in the transgenic lines and was not present in nontransgenic littermate controls (Fig. 2D). As expected, no endogenous ZBP1 was detected (Leeds et al. 1997; Ioannidis et al. 2003, 2004). The other transgenic mouse lines showed similar ZBP1 expression patterns, and further analysis was performed using only KL2169 (data not shown). Treatment of line KL2169 with doxycycline (1 mg/mL in drinking water) suppressed ZBP1 expression after 22 d (Fig. 2E).
Figure 2.
Transgenic ZBP1 is expressed at high levels in the striatum. The GFP signal in brain coronal sections of bitransgenic animals was imaged to determine the expression of the transgene. (A,B) Expression in striatal subregions are shown at different magnifications. A Nissl-stained coronal section (brainmaps.org) approximate to the one imaged is shown below the GFP image in A, and the approximate region imaged in A is boxed with a white dashed line. The striatal structures, Anterior Commissure (AC) and Caudate Putamen (CPu), are labeled. The axon bundles of these structures are inherently autofluorescent, and therefore make a useful landmark for localization in the fluorescent images. (C) A high-magnification image shows subcellular localization of transgenic ZBP1 in transgenic brain. EGFP-FLAG-ZBP1 can be seen in proximal processes as well as the soma. (D) A Western blot using a polyclonal ZBP1 antibody demonstrates expression of the EGFP-ZBP1 fusion protein in bitransgenic brains (Tg-1 and Tg-2 are whole brain extracts from bitransgenic animals derived from two different founder EGFP-ZBP1 lines; [WT] Brain extract from wild-type littermate control mouse; [CEF] Chicken Embryonic Fibroblast cell extract as a control for western blotting). (E) Western blot using a polyclonal ZBP1 antibody demonstrates suppression of EGFP-FLAG-ZBP1 after 22 d of treatment with doxycycline, with visible suppression observed after 8 d of treatment. Brain extracts are from KL2169 mice prior to doxycycline (0 d), 8 d after doxycycline treatment, or 22 d after doxycycline treatment. The control band demonstrates the loading of protein in the 22-d treatment sample.
ZBP1 transgene expression reduces cocaine cue reactivity
Because GFP-ZBP1 expression was enriched in the striatum, a region implicated in many aspects of cocaine-regulated behaviors, we evaluated its effect on cocaine cue reactivity in a place-conditioning paradigm. Cocaine craving and seeking are powerfully controlled by environmental cues associated with cocaine administration, a process termed cue reactivity (Childress et al. 1993). Cue-conditioned place preference is a reliable way to evaluate cue reactivity in rodents (Bardo and Bevins 2000). Place conditioning is a useful model in evaluating effects on cue reactivity, because drugs that elicit or interfere with cued place conditioning have corresponding effects on drug self-administration and drug-seeking behaviors.
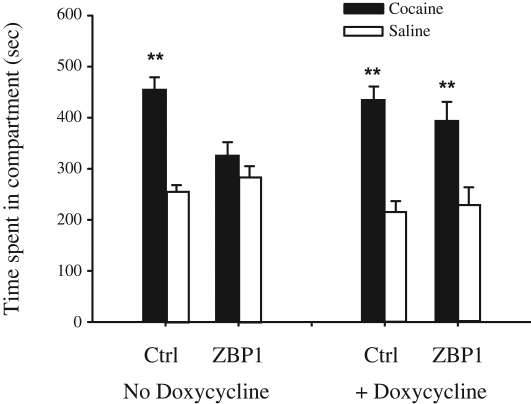
Cocaine conditioning increased the time that non-ZBP1 transgenic mice spent in the cocaine-trained compartment, demonstrating the effectiveness of cue conditioning as expected (Fig. 3, compare time spent in cocaine versus saline compartments for no doxycycline treatment control animals). However, the ZBP1 transgenic mice did not develop a strong preference for the cocaine-trained compartment (Fig. 3, compare time spent in cocaine versus saline compartments for no doxycycline treatment ZBP1 animals). To determine whether the reduced level of cue reactivity in transgenic mice was due to the presence of ZBP1 protein or a developmental effect of ZBP1 expression, we tested transgenic mice whose ZBP1 expression was acutely suppressed by doxycycline for 3 wk prior to the experiments, as well as throughout the conditioning and testing. Both control and ZBP1 transgenic groups where ZBP1 was suppressed demonstrated cocaine cue reactivity (Fig. 3, compare time spent in saline versus cocaine compartments for both control and ZBP1 animals with doxycycline). This result supports the conclusion that the reduced cocaine cue reactivity of transgenic animals in the previous experiment is dependent on the presence of the ZBP1.
Figure 3.
ZBP1 transgene expression impairs cocaine-cued conditioned preference. Cue reactivity is reflected by the increase in time mice spend in compartments in which they receive cocaine. Mice were tested without (No Doxycycline) or with doxycycline to suppress ZBP1 expression (Doxycycline). (**) P < 0.01 cocaine-paired versus saline-paired compartments, as determined by Newman-Keuls comparisons. n = 8–16/group.
Identification of mRNAs associated with GFP-ZBP1
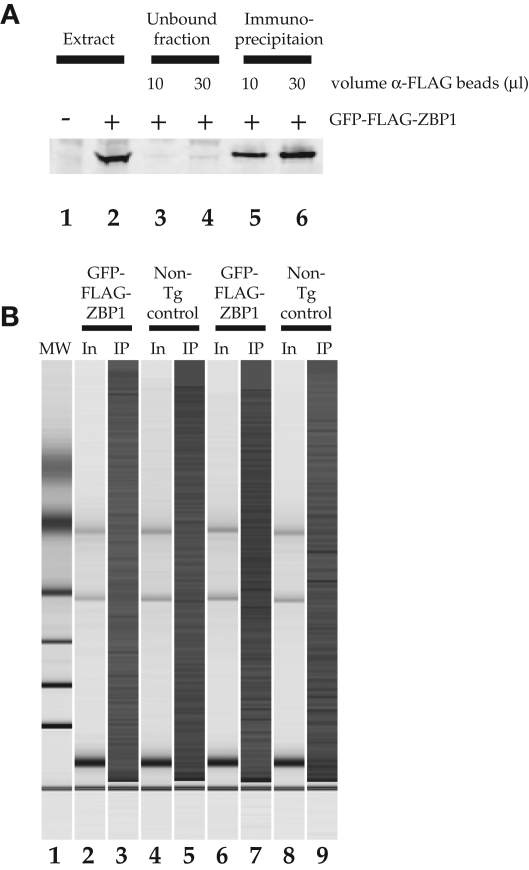
We hypothesize that decreased cue reactivity could be due to altered gene expression as a result of ZBP1 binding to mRNAs in mature neurons. We took advantage of the RNA-binding properties of ZBP1 to identify putative targets of GFP-ZBP1, whose expression could be affected. To determine which mRNAs are associated with transgenic GFP-ZBP1 in the brain in vivo we performed ribonucleoprotein-immunoprecipitation (RIP) followed by microarray of coprecipitated RNA (RIP-Chip). ZBP1 and its associated mRNAs form ribonucleoprotein (RNP) complexes that were isolated from forebrain extracts by immunoprecipitation with anti-FLAG affinity matrix. The FLAG-labeled GFP-ZBP1 was detectable in these extracts as a single protein of the expected size from transgenic mice that was absent in control mice (Fig. 4A). The protein was largely depleted from these extracts under the assay conditions, and at least 80% was recovered on the anti-FLAG resin, demonstrating that the protein was not degraded during immunoprecipitation. Equivalent control experiments were performed on nontransgenic mice of a similar age to ensure that RNA recovery was specific for the EGFP-ZBP1 protein. An aliquot of extract prior to immunoprecipitation was used to isolate total RNA for the input fraction. The remainder of each lysate was used for the immunoprecipitation, and RNA was extracted from the immunoprecipitates. We analyzed the quality of the RNA both in extracts and immunoprecipitates and found similar 28S/18S ribosomal RNA ratios in all of our extracts, suggesting that they were all of similar quality and suitable for microarray analysis (Fig. 4B).
Figure 4.
ZBP1-containing mRNP complexes are immunoprecipitated using FLAG antibodies. (A) Western blot of pre- and post-immunoprecipitation brain extracts using an anti-FLAG antibody demonstrates that intact protein is efficiently and quantitatively immunoprecipitated. (Lane 1) Brain extract littermate control; (lane 2) brain extract from FLAG-EGFP-ZBP1 expressing mouse; (lane 3) supernatant from immunoprecipitation using 10 µL of anti-FLAG beads; (lane 4) supernatant from immunoprecipitation using 30 µL of anti-FLAG beads; (lane 5) 20% of the bound fraction from immunoprecipitation using 10 µL of anti-FLAG beads; (lane 6) 20% of the bound fraction from immunoprecipitation using 30 µL of anti-FLAG beads. (B) Bioanalyzer results suggest that RNA is not significantly degraded and that RNA from each sample is similar. In addition, though the 28S and 18S rRNA bands are prominent in the extracts, these bands are not seen in the samples that were immunoprecipitated. Animals A and C are bitransgenic, while B and D are controls. (Lane 1) Ladder; (lane 2) extract A; (lane 3) elution A; (lane 4) extract B; (lane 5) elute B; (lane 6) extract C; (lane 7) elute C; (lane 8) extract D; (lane 9) elute D.
We used Affymetrix Mouse Genome 430 Plus 2.0 arrays to identify mRNAs that coimmunoprecipitated as part of an mRNP complex with GFP-ZBP1. Three independent experiments were performed to achieve statistical significance and to further decrease the likelihood of false positives. Of the nearly 40,000 transcripts represented on the Mouse Genome 430 Plus 2.0 microarray, 195 were at least fourfold enriched versus the transgenic input fraction and control immunoprecipitation (Supplemental Table 1). Nearly three times as many transcripts were enriched at least twofold in both conditions.
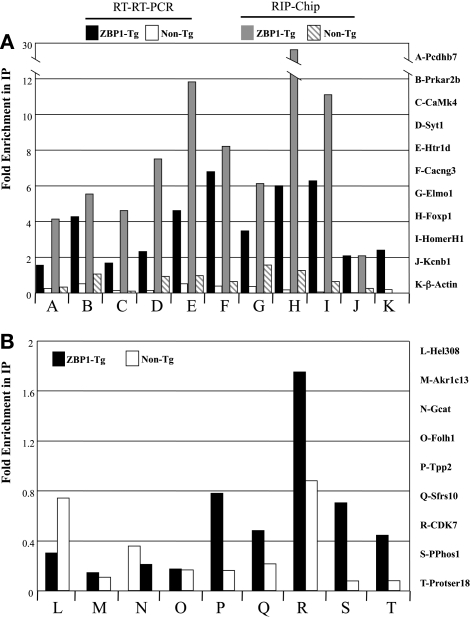
To validate our microarray data, we repeated the ZBP1 immunoprecipitation and used reverse transcription followed by Real Time PCR (RT–RT–PCR) analysis of a representative panel of 11 of the fourfold enriched mRNAs. Ten of these target mRNAs were enriched and eight were enriched at least twofold (Fig. 5A). These confirmed targets include protocadherin β7, Prkar2b (cAMP-dependent regulator protein kinase), Camk4 (calcium/calmodulin-dependent protein kinase 4), Syt1 (synaptotagmin 1), Htr1D (5-hydroxytryptamine (serotonin) receptor 1D), Cacnag3 (voltage-dependent calcium channel γ subunit 3), Elmo1 (engulfment and cell motility 1), FoxP1 (Forkhead box P1), HomerH1, and Kcnb1 (shab-related subfamily voltage-gated potassium channel). In addition, we assayed β-actin mRNA, a previously identified target for ZBP1, and found this mRNA to be more than twofold enriched in the ZBP1 immunoprecipitate. β-actin was not enriched in the microarray analysis, but this is likely due to its high expression level in the extract. High expression levels can limit the capacity for enhanced binding necessary for detection as enriched by the microarray method with our stringency protocol. We also used RT–RT–PCR to examine 10 mRNAs that were not enriched in the microarray analysis, and confirmed that nine of these demonstrated no enrichment (Fig. 5B). In summary, the microarray analysis reproducibly reflects mRNAs that are associated with the transgenic ZBP1 protein.
Figure 5.
RNAs found to be enriched by microarray analysis are also enriched when assayed by RT–RT–PCR. (A) mRNAs more than fourfold enriched by microarray were analyzed with RT–RT–PCR and the results plotted together. Fold enrichment is the ratio between RNA levels in the pre- and post-immunoprecipitation fractions for each mRNA. Fold enrichment from ZBP1 transgenic (ZBP1-Tg) and non-ZBP1 transgenic littermate control animals (Non-Tg) are indicated for both RT–RT–PCR and microarray (RIP-Chip). (B) Nine RNAs found not to be enriched by microarray analysis were also not enriched when assayed by RT–RT–PCR. The RT–RT–PCR ratios for these genes from ZBP1 transgenic (ZBP1-Tg) and nontransgenic control animals (Non-Tg) are indicated.
Discussion
Here, we report the establishment of ZBP1 transgenic mice capable of spatially and conditionally regulating transgene expression in subpopulations of forebrain neurons, including those involved in response to drugs of addiction. This transgenic RNA-binding protein was only visualized in the striatum. Previous use of this CaMKIIα driver has also observed in striatum-only expression for some transgenes (Mayford et al. 1996). The expression of ZBP1 in these neurons disrupted cocaine-conditioned cue preference, and this effect was due to the presence of the transgenic ZBP1 rather than a change in neuronal development as a result of ectopic expression of ZBP1. We propose that changes in gene expression of ZBP1-bound mRNAs in neurons of the striatum are the most plausible means to explain the effect of the transgene on behavior. One important result from these studies lies in the fact that a behavioral outcome such as cocaine-cued place conditioning can be directly controlled by the expression of a single RNA-binding protein in neurons.
The striatum, including the caudate-putamen and nucleus accumbens, is implicated in cocaine's cue reactivity (Everitt et al. 2008). Changes in gene expression within the striatum are functionally critical for cocaine cue reactivity (Hiroi et al. 1997; Carlezon et al. 1998; Agatsuma et al. 2010). Endogenous ZBP1 is not expressed in these areas in the mature brain; therefore, one explanation for the effect of transgenic ZBP1 on cue-conditioned preference could be dysregulation of mRNAs encoding factors involved in striatal synaptic plasticity such as Homer1. Cocaine also induces long-lasting alterations in dendritic length, spine density, and the number of branched spines in the striatum (Robinson and Kolb 1999). Localized translation of mRNA in this compartment is thought to be involved in these structural alterations (Eom et al. 2003); therefore, our data raise the possibility that regulating local translation of mRNAs in the Nucleus Accumbens may play a role in cocaine cue reactivity in vivo.
Using this in vivo system, we identified a pool of mRNAs in the forebrain that can physically interact with ZBP1, many of which have been implicated in synaptic plasticity. Previous data from microarrays suggest that ∼44% of the genes on an array are present in brain lysates (Brown et al. 2001). Because our assay was expected to contain ∼17,500 brain mRNAs, the 195 transcripts identified as associated with ZBP1 represent ∼1.1% of the RNAs expressed in the brain. This represents a slightly smaller subset of the population than has been found to associate with another mRNA-binding protein, Fragile X Mental Retardation protein (FMRP, which bound ∼4% of mRNA). However, these values are in the same order of magnitude and suggest that our result represents a reasonable estimate of the ZBP1-binding cohort (Ashley et al. 1993; Brown et al. 2001). The mRNAs we identified include cadherins, transcription factors, kinases, channels, receptors, and Ras family members (small GTPases that are involved in cellular signal transduction). Each of these gene families plays important roles in neuronal function, such as in regulating the strength of connections between neurons during plasticity, although other important cellular functions may also be controlled by these factors (Abel et al. 1997; Jungling et al. 2006; Pelkey et al. 2006; Kelly et al. 2007). Moreover, protein products of these families have non-uniform subcellular distributions, raising the possibility that the localization of their cognate mRNA supports their neuronal functions (Crino et al. 1998; Rosenblum et al. 2002; Morozov et al. 2003).
We hypothesize that ectopic expression of ZBP induces changes in gene expression within this pool of mRNAs, and it may be these changes that lead to the altered behavior. Consistent with this, ZBP1 transgene repression allowed the behavior to return to normal. One putative ZBP1 target, Homer1, negatively regulates cocaine-induced behavioral effects (Swanson et al. 2001; Ghasemzadeh et al. 2003a,b; Szumlinski et al. 2004). Homer1 protein localizes to dendrites and is sufficient to direct type 5 metabotropic glutamate receptors (mGluR5) to both dendrites and axons (Ango et al. 2000). In addition, Homer1 expression and its effects on localizing mGluR5 to neurites and synapses are regulated by neuronal activity (Ango et al. 2002). We hypothesize that a ZBP1-mediated increase in the distal localization of the mRNA for a scaffolding protein such as Homer1 might contribute to changes in synaptic plasticity through regulation of glutamate receptor signaling activity. However, it is clear that the localization, translation, and/or stability of another target mRNA may also be affected by ZBP1 expression. We also consider it possible that no single change in gene expression caused by ZBP1 expression contributes to the observed behavioral phenotype. Therefore, altered cue reactivity may result from the sum of several independent changes in gene expression yet to be characterized. For instance, several transcription factors were present on the list of ZBP1 targets, indicating that levels of these may be affected by ZBP1 expression, therefore suggesting that an altered transcriptional profile may be present in transgenic striatal neurons. Changes such as these are also a plausible contributor to the observed change in behavior.
Post-transcriptional control of gene expression is a target for intervention in treating addiction behaviors
Because our behavioral experiments have been performed in adult-behaving animals in an accepted paradigm that models aspects of addiction behaviors, they suggest that post-transcriptional control of gene expression in neurons contributes actively to the function of neuronal circuits involved in response to drugs of abuse. The phenotype in the conditioned place preference assay could be due to the effects of ZBP on either the acquisition, the expression, or both. A future challenge is to more precisely identify the way ZBP1 interferes with the various phases of conditioned place preference. Wild-type expression of ZBP1 in adult neurons or most other adult tissues cannot be detected, or, at a maximum, is below limits of sensitivity for detection. In the absence of wild-type ZBP1 it is not reasonable to conclude from our experiments that these target mRNAs are controlled by ZBP1 in a normal-behaving brain during cocaine-cued place conditioning. However, the expression of this transgene has a clear phenotypic consequence in this assay of cue reactivity. Our identification of mRNAs associated with the transgenically expressed protein is a powerful tool to identify sets of mRNAs whose expression may be able to influence this behavioral response to an addictive substance. Understanding the changes in gene expression that result from expressing ZBP1 therefore provide molecular targets that may be useful in developing therapies to improve the treatment of addiction, a widespread medical and social problem. Small molecule inhibitors designed to mimic the critical changes in gene expression that lead to the reduction in cue reactivity that we observe are clearly of therapeutic interest.
Materials and Methods
Plasmid construction
Klenow-treated EGFP-ZBP1 encoding the chicken ZBP1 mRNA was inserted into the EcoRV site of the MM400 vector using standard techniques to generate plasmid KL2070 (Mayford et al. 1996; Oleynikov and Singer 2003). Chicken ZBP1 protein is 98% identical to the mammalian ZBP1 protein. FLAG-tagged ZBP1 was inserted into BamHI/XbaI sites of pEGFP-C1 (Clontech) to generate plasmid KL2134. The EGFP-FLAG-ZBP1 fragment of KL2134 was Klenow treated and inserted into the EcoRV site of the MM400 vector as described above, producing plasmid KL2169. For genotyping, a 1.1-kb portion of the MM400 vector sequence including the SV40 3′ untranslated region and poly(A) was inserted into XhoI of pBluescript SK + (Stratagene) to generate plasmid KL1448.
Animal generation, genotyping, and maintenance
NotI fragments of KL2070 and KL2169 were isolated and injected into pronuclei of one-cell FVB embryos. Twenty-four hours later, embryos that divided were implanted in pseudopregnant foster females by oviduct transfer. These techniques, while quite common, have been reviewed in detail (Ittner and Gotz 2007). Genotyping of transgenic mice was performed by Southern blot. Genomic DNA was prepared by digesting tail fragments overnight, rocking at 55°C in 500 µL of tail buffer (100 mM Tris at pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl) with 100 µg/mL proteinase K. Digestion was followed by vortexing and centrifugation for 15 min at 16,100g and precipitation of the supernatant with an equal volume of isopropanol. DNA was then removed and resuspended by rocking overnight at 55°C in 100 µL of dH2O. After resuspension, DNA was precipitated before digestion with PvuII and gel loading (1% agarose with TBE). Gels were soaked while rocking in 0.25 M HCl for 10 min and rinsed three times in dH2O, followed by a 40-min incubation while rocking in 0.4 M NaOH. Transfer onto Hybond N + (Amersham) membranes was performed in 0.4 M NaOH overnight, and blots were prehybridized at 42°C in 2× SSC, 1% SDS, 10% Dextran sulfate, 50% formamide for at least 1 h prior to hybridization at 42°C overnight. Blots were washed with 2× SSC, 1% SDS at 42°C for 10 min, followed by one to two washes with 2× SSC, 1% SDS at 65°C for 20 min, and then one to two washes with 0.2× SSC, 1% SDS at 65°C for 30 min before exposing to Kodak Biomax XAR film (Kodak). The 1.1-kb XhoI fragment of KL1448 was used as a probe for genotyping founder animals and their progeny by Southern blotting. Progeny of the ZBP1 transgenic mice were backcrossed to the C57BL/6J background, and ZBP1 transgenics were also crossed with mice (line B) expressing the tTA transgene under control of the CaMKIIα promoter (Mayford et al. 1996). Animals were backcrossed for at least four generations before use in behavioral experiments. Because the 3′ polyadenylation sequence used for genotyping the ZBP1 transgenic mice also is present in CaMKIIα transgenic mice, identification of bitransgenic mice was performed with the same Southern blotting protocol used to identify animals containing the individual transgenes. In addition, PCR was used to detect the CaMKIIα transgene using JumpStart RED Taq ReadyMix (Sigma) and a thermocycler profile beginning with 3 min at 94°C, followed by 12 cycles of 94°C for 20 sec, 64°C for 30 sec, and 72°C for 35 sec, and then 25 cycles of 94°C for 20 sec, 58°C for 30 sec, and 72°C for 35 sec, and ending with 2 min at 72°C. PCR reactions included a tTA-specific primer pair with a 450-bp amplicon and a primer set amplifying the endogenous mouse gene Tcrd with an amplicon of 200 bp.
Up to five mice were housed per cage, and animals were maintained on a 14-h light:10-h dark cycle with food and water available ad libitum. For experiments that required the regulation of ZBP1 expression, doxycycline (Clontech) was added to the water at 1 mg/mL for at least 3 wk prior to the start of and throughout the experiments. Animal handling and use was in accordance with a protocol approved by the Animal Care and Use Committee of Albert Einstein College of Medicine and NIH guidelines.
Characterization of transgenic ZBP1 expression
ZBP1 expression was evaluated in adult mice, both by Western blotting and by imaging the GFP signal in transgenic mice. For Western blotting, protein extracts were prepared from whole brains without the cerebellum. Tissues were repeatedly rinsed with ice-cold PBS, 1 mM EGTA, 0.5 mM PMSF, and large tissue chunks were fragmented, followed by several successive washes and centrifugation of the tissue at 400g and 500g in the same wash buffer. After the final wash, an equal volume of lysis buffer (50 mM Tris at pH8.0, 150 mM NaCl, 5 mM MgCl2, 0.1 mg/mL Escherichia coli tRNA, 0.5 mM PMSF, 1 U/μL RNAseOUT (Invitrogen) 1× Complete Protease Inhibitor tablets (Roche), 0.5% NP40, 15 mM EDTA) was added to the tissue pellet, resuspended well, and resuspended samples were incubated for 5 min on ice. Samples were then centrifuged for 10 min at 10,000g, and the supernatant was removed from the pellet and recentrifuged. Protein content in the supernatant was quantified with a BCA assay (Pierce), and 5 or 10 µg in SDS sample buffer was loaded per lane. The NuPAGE (Invitrogen) system was used, and proteins were transferred onto Hybond ECL nitrocellulose membranes (Amersham) by wet blotting. Primary rabbit polyclonal antibody raised against full-length His-tagged recombinant ZBP1 was used at 1:4500, and anti-rabbit IRDye 800 (Rockland) secondary antibody was used at 1:5000. Signal was visualized using the Odyssey Infrared Imaging System (Li-Cor) and analyzed using IPLab software (BD Biosciences).
For direct imaging of EGFP signals, brains were frozen, embedded in OCT Tissue-Tex (Miles), and 8-μm sections were cut on a cryotome. For prefixed sections, dissected brains were incubated for 24 h at 4°C in 4% PFA (Electron Microscopy Sciences) prior to sectioning. Fluorescence was detected by imaging using an upright Olympus xenon-lamp fluorescence microscope.
Immunoprecipitation
After isolation of the forebrain from either transgenic or control animals, extracts were prepared as described above, a small aliquot was removed for analysis, and the remaining extract was added to 20 µL of a 50% slurry of anti-FLAG M2 resin (Sigma). Each forebrain extract was processed independently and incubated for 2 h at 4°C with shaking at 1000 rpm. Beads were collected by centrifugation at 500g, and the supernatant was removed. Beads were washed once quickly and then again for 5 min with rotating end over end in wash buffer (1× TBS, 0.05% NP40, 15 mM EDTA, 0.5 mM PMSF) including 0.1 mg/mL tRNA, followed by three 5-min washes with wash buffer that did not include tRNA. After collection, elution was performed by adding 50 µL of wash buffer and 50 µL of 2% SDS to the beads, followed by a 1-h incubation at 50°C. For isolation of RNA, the supernatant was extracted with an equal volume of phenol-chloroform, and the aqueous phase was precipitated overnight at −80°C with the addition of 20 µg of glycogen, 40% aqueous volume 3 M NaOAc, and 3× aqueous volume EtOH, followed by centrifugation and rinses with 80% EtOH.
Microarray analysis of immunoprecipitated RNAs
The quality of precipitated RNAs was assessed using an Agilent 2100 BioAnalyzer (Agilent). Affymetrix oligonucleotide microarrays were prepared, hybridized, and analyzed according to the manufacturer's instructions (www.affymetrix.com). Briefly, 50 ng of each immunoprecipitated RNA sample was annealed with oligo dT and incubated at 65°C for several minutes before placing on ice to produce double-stranded cDNA using the Affymetrix two-cycle cDNA synthesis kit as per the manufacturer's protocol. This cDNA was then converted to amplified RNA (aRNA) by in vitro transcription using MEGAscript T7 polymerase (Ambion). The aRNA was subsequently processed to double-stranded cDNA in a similar manner as the first-strand cDNA, except using random primers. Biotinylated UTPs were incorporated in the second in vitro transcription step (Affymetrix IVT Labeling Kit) to produce labeled, amplified cRNA. cRNA (15 µg) was fragmented with a metal-induced hydrolysis step to obtain 25- to 200-bp fragments that were hybridized to the Mouse Genome 430 Plus 2.0 GeneChip arrays according to the manufacturer's protocol. The system contains over 1 million probes and allowed us to analyze over 39,000 transcripts. After hybridization, chips were washed and stained with streptavidin-phycoerythrin (SAPE) before being scanned. The sensitivity of the assay was increased with an antibody amplification staining protocol with biotinylated goat IgG, followed by a second SAPE staining. The chip was then scanned, and images were analyzed qualitatively using GCOS software. Additional quantitative analysis to identify changes in gene expression was performed using GeneSpring (Silicon Genetics). For this analysis, values below 0.01 were set to 0.01, then each measurement was divided by the 50th percentile of all measurements from the sample. Each gene was divided by the median of its measurements in all samples. If the median was below 10, a value of 10 was used if the numerator was above 10. Otherwise, the measurement was not used. Affymetrix's Gene Chip Multiarray Averaging (GCRMA) was used as a normalization algorithm to further reduce false-positive detection. The list of targets was then filtered to find genes with good signal-to-noise ratios and was further filtered to use only genes labeled as present in at least one of two conditions. Finally, the list was filtered to identify genes with at least fourfold or twofold levels of enrichment in the ZBP1 immunoprecipitation. Three independent samples were run on separate microarrays for transgenic and nontransgenic animals.
Quantitative PCR from immunoprecipitated RNAs
First-strand cDNAs were produced from RNAs isolated by immunoprecipitation using the SuperScript III First Strand Synthesis System (Invitrogen). The recommended protocol in the kit was followed, including incubation with random hexamer primers and dNTPs for 5 min at 65°C, followed by cDNA synthesis with 1× RT buffer, 5 mM MgCl2, 0.01 M DTT, and Superscript III RT for 50 min at 50°C and termination at 95°C for 5 min. Quantitative PCR reactions were in 10-μL volumes and included 2 ng/μL of cDNA, 5 µL of SYBR Green PCR Master Mix (Applied Biosystems), and 0.2 µM of each primer. PCR primers (22 bases) were used, with ∼50% GC content, and no more than two G's or C's in the four bases on the 3′ end. Amplicons were designed to be ∼150 bp and no more than 200 bp. Quantitative PCR was performed using 384-well plates in an ABI 7900HT thermocycler (Applied Biosystems). The cycle parameters included a 10-min hold at 95°C, followed by 40 cycles of denaturing for 15 sec at 95°C, and annealing/extension at 60°C for 1 min.
Behavioral analysis
Male transgenic EGFP-FLAG-ZBP1 mice and wild-type littermates from line KL2169 were used for cue conditioning experiments at the age of 3–6 mo. One group of mice of the two genotypes received no doxycycline; the other group received doxycycline from 3 wk prior to and during testing. Otherwise, the apparatus and the procedure were identical to those described in our previous work (Agatsuma et al. 2006, 2010; Zhu et al. 2007). The rectangular Plexiglas apparatus used for conditioning consisted of two large compartments (each 24.5 cm × 18 cm × 33 cm) with differences in wall patterns and ceiling light intensity as well as a central compartment (13 cm × 18 cm × 33 cm) separated from the two other compartments by guillotine doors (18 cm × 37 cm) as described. Cue conditioning experiments were performed over a period of 5 d. On day 1 (preconditioning), the mice were placed in the central compartment and were allowed to move freely between all of the compartments through the open guillotine doors for 15 min. On days 2–4 (conditioning), each animal was conditioned in two sessions per day, separated by 5 h. In each conditioning session, mice received subcutaneous injections of either 0.9% saline or 10 mg/kg of cocaine (cocaine hydrochloride salt, Sigma, dissolved in 0.9% saline) and were confined for 30 min in a single large compartment by the closed guillotine doors. Injection of saline has been demonstrated to produce no preference in either compartment in this assay (Agatsuma et al. 2006, 2010). Equal numbers of animals received pairings with cocaine in each compartment, and each animal received one cocaine injection and one saline injection per day. The order of injections was also balanced such that half of the animals received cocaine in the morning session and half in the afternoon session. On day 5 (testing), animals received no injection, and were placed in the central compartment. The mice were allowed to move freely for 15 min between all compartments via the open guillotine doors. An observer who was blinded to both genotype and treatment recorded the amount of time spent in each large compartment.
Acknowledgments
This work was supported by AR41480/GM084364 to R.H.S. and DA024330 to N.H. We thank Frank Doyle for assistance in reviewing microarray data.
Footnotes
[Supplemental material is available for this article.]
References
- Abel T, Nguyen PV, Barad M, Deuel TA, Kandel ER, Bourtchouladze R 1997. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell 88: 615–626 [DOI] [PubMed] [Google Scholar]
- Agatsuma S, Lee M, Zhu H, Chen K, Shih JC, Seif I, Hiroi N 2006. Monoamine oxidase A knockout mice exhibit impaired nicotine preference but normal responses to novel stimuli. Hum Mol Genet 15: 2721–2731 [DOI] [PubMed] [Google Scholar]
- Agatsuma S, Dang MT, Li Y, Hiroi N 2010. N-methyl-D-aspartic acid receptors on striatal neurons are essential for cocaine cue reactivity in mice. Biol Psychiatry 67: 778–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, Pin JP, Tu JC, Xiao B, Worley PF, Bockaert J, Fagni L 2000. Dendritic and axonal targeting of type 5 metabotropic glutamate receptor is regulated by homer1 proteins and neuronal excitation. J Neurosci 20: 8710–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, Robbe D, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L 2002. Homer-dependent cell surface expression of metabotropic glutamate receptor type 5 in neurons. Mol Cell Neurosci 20: 323–329 [DOI] [PubMed] [Google Scholar]
- Ashley CT Jr, Wilkinson KD, Reines D, Warren ST 1993. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science 262: 563–566 [DOI] [PubMed] [Google Scholar]
- Bach ME, Hawkins RD, Osman M, Kandel ER, Mayford M 1995. Impairment of spatial but not contextual memory in CaMKII mutant mice with a selective loss of hippocampal LTP in the range of the θ frequency. Cell 81: 905–915 [DOI] [PubMed] [Google Scholar]
- Bardo MT, Bevins RA 2000. Conditioned place preference: What does it add to our preclinical understanding of drug reward? Psychopharmacology 153: 31–43 [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487 [DOI] [PubMed] [Google Scholar]
- Carlezon WA Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, Nestler EJ 1998. Regulation of cocaine reward by CREB. Science 282: 2272–2275 [DOI] [PubMed] [Google Scholar]
- Childress AR, Hole AV, Ehrman RN, Robbins SJ, McLellan AT, O'Brien CP 1993. Cue reactivity and cue reactivity interventions in drug dependence. NIDA Res Monogr 137: 73–95 [PubMed] [Google Scholar]
- Crino P, Khodakhah K, Becker K, Ginsberg S, Hemby S, Eberwine J 1998. Presence and phosphorylation of transcription factors in developing dendrites. Proc Natl Acad Sci 95: 2313–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, et al. 2011. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J 30: 4665–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ 2003. Localization of a β-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci 23: 10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BJ, Belin D, Economidou D, Pelloux Y, Dalley JW, Robbins TW 2008. Review. Neural mechanisms underlying the vulnerability to develop compulsive drug-seeking habits and addiction. Philos Trans R Soc Lond B Biol Sci 363: 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RA, Branham RK, See RE 2006. Different neural substrates mediate cocaine seeking after abstinence versus extinction training: A critical role for the dorsolateral caudate-putamen. J Neurosci 26: 3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemzadeh MB, Permenter LK, Lake R, Worley PF, Kalivas PW 2003a. Homer1 proteins and AMPA receptors modulate cocaine-induced behavioural plasticity. Eur J Neurosci 18: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Ghasemzadeh MB, Permenter LK, Lake RW, Kalivas PW 2003b. Nucleus accumbens Homer proteins regulate behavioral sensitization to cocaine. Ann N Y Acad Sci 1003: 395–397 [DOI] [PubMed] [Google Scholar]
- Graybiel AM 2008. Habits, rituals, and the evaluative brain. Annu Rev Neurosci 31: 359–387 [DOI] [PubMed] [Google Scholar]
- Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ 1997. FosB mutant mice: Loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine's psychomotor and rewarding effects. Proc Natl Acad Sci 94: 10397–10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ 1992. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci 89: 5764–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ 1994. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron 13: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Ioannidis P, Mahaira L, Papadopoulou A, Teixeira MR, Heim S, Andersen JA, Evangelou E, Dafni U, Pandis N, Trangas T 2003. CRD-BP: A c-Myc mRNA stabilizing protein with an oncofetal pattern of expression. Anticancer Res 23: 2179–2183 [PubMed] [Google Scholar]
- Ioannidis P, Kottaridi C, Dimitriadis E, Courtis N, Mahaira L, Talieri M, Giannopoulos A, Iliadis K, Papaioannou D, Nasioulas G, et al. 2004. Expression of the RNA-binding protein CRD-BP in brain and non-small cell lung tumors. Cancer Lett 209: 245–250 [DOI] [PubMed] [Google Scholar]
- Ittner LM, Gotz J 2007. Pronuclear injection for the production of transgenic mice. Nat Protoc 2: 1206–1215 [DOI] [PubMed] [Google Scholar]
- Jungling K, Eulenburg V, Moore R, Kemler R, Lessmann V, Gottmann K 2006. N-cadherin transsynaptically regulates short-term plasticity at glutamatergic synapses in embryonic stem cell-derived neurons. J Neurosci 26: 6968–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MP, Isiegas C, Cheung YF, Tokarczyk J, Yang X, Esposito MF, Rapoport DA, Fabian SA, Siegel SJ, Wand G, et al. 2007. Constitutive activation of Gαs within forebrain neurons causes deficits in sensorimotor gating because of PKA-dependent decreases in cAMP. Neuropsychopharmacology 32: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G 2000. Drug addiction. Neurobiol Dis 7: 543–545 [DOI] [PubMed] [Google Scholar]
- Kourrich S, Rothwell PE, Klug JR, Thomas MJ 2007. Cocaine experience controls bidirectional synaptic plasticity in the nucleus accumbens. J Neurosci 27: 7921–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, Ross J 1997. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14: 1279–1286 [DOI] [PubMed] [Google Scholar]
- Luscher C, Malenka RC 2011. Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 69: 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science 274: 1678–1683 [DOI] [PubMed] [Google Scholar]
- Moratalla R, Elibol B, Vallejo M, Graybiel AM 1996. Network-level changes in expression of inducible Fos-Jun proteins in the striatum during chronic cocaine treatment and withdrawal. Neuron 17: 147–156 [DOI] [PubMed] [Google Scholar]
- Morozov A, Muzzio IA, Bourtchouladze R, Van-Strien N, Lapidus K, Yin D, Winder DG, Adams JP, Sweatt JD, Kandel ER 2003. Rap1 couples cAMP signaling to a distinct pool of p42/44MAPK regulating excitability, synaptic plasticity, learning, and memory. Neuron 39: 309–325 [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D 1991. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell 64: 751–759 [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey KA, Topolnik L, Lacaille JC, McBain CJ 2006. Compartmentalized Ca(2+) channel regulation at divergent mossy-fiber release sites underlies target cell-dependent plasticity. Neuron 52: 497–510 [DOI] [PubMed] [Google Scholar]
- Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J 2011. Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. J Neurosci 31: 5271–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrino LJ, Lyons D, Smith HR, Daunais JB, Nader MA 2004. Cocaine self-administration produces a progressive involvement of limbic, association, and sensorimotor striatal domains. J Neurosci 24: 3554–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW, Ersche KD, Everitt BJ 2008. Drug addiction and the memory systems of the brain. Ann N Y Acad Sci 1141: 1–21 [DOI] [PubMed] [Google Scholar]
- Robinson TE, Kolb B 1997. Persistent structural modifications in nucleus accumbens and prefrontal cortex neurons produced by previous experience with amphetamine. J Neurosci 17: 8491–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TE, Kolb B 1999. Alterations in the morphology of dendrites and dendritic spines in the nucleus accumbens and prefrontal cortex following repeated treatment with amphetamine or cocaine. Eur J Neurosci 11: 1598–1604 [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH 2008. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol 20: 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum K, Futter M, Voss K, Erent M, Skehel PA, French P, Obosi L, Jones MW, Bliss TV 2002. The role of extracellular regulated kinases I/II in late-phase long-term potentiation. J Neurosci 22: 5432–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ 2010. Phosphorylation of Zipcode Binding Protein 1 Is required for Brain-Derived Neurotrophic Factor signaling of local β-actin synthesis and growth cone turning. J Neurosci 30: 9349–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnamon JR, Czaplinski K 2011. mRNA trafficking and local translation: The Yin and Yang of regulating mRNA localization in neurons. Acta Biochim Biophys Sin 43: 663–670 [DOI] [PubMed] [Google Scholar]
- Swanson CJ, Baker DA, Carson D, Worley PF, Kalivas PW 2001. Repeated cocaine administration attenuates group I metabotropic glutamate receptor-mediated glutamate release and behavioral activation: A potential role for Homer. J Neurosci 21: 9043–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinski KK, Dehoff MH, Kang SH, Frys KA, Lominac KD, Klugmann M, Rohrer J, Griffin W 3rd, Toda S, Champtiaux NP, et al. 2004. Homer proteins regulate sensitivity to cocaine. Neuron 43: 401–413 [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ 2003. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Telang F, Fowler JS, Logan J, Childress AR, Jayne M, Ma Y, Wong C 2006. Cocaine cues and dopamine in dorsal striatum: Mechanism of craving in cocaine addiction. J Neurosci 26: 6583–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi D, Willis DE, Twiss JL 2009. Regulation of mRNA transport and translation in axons. Results Probl Cell Differ 48: 193–224 [DOI] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS 2010. Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ 2011. Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci 31: 9800–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DF, Kuwabara H, Schretlen DJ, Bonson KR, Zhou Y, Nandi A, Brasic JR, Kimes AS, Maris MA, Kumar A, et al. 2006. Increased occupancy of dopamine receptors in human striatum during cue-elicited cocaine craving. Neuropsychopharmacology 31: 2716–2727 [DOI] [PubMed] [Google Scholar]
- Wu Q, Reith ME, Kuhar MJ, Carroll FI, Garris PA 2001. Preferential increases in nucleus accumbens dopamine after systemic cocaine administration are caused by unique characteristics of dopamine neurotransmission. J Neurosci 21: 6338–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ 2006. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci 9: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Zhu H, Lee M, Agatsuma S, Hiroi N 2007. Pleiotropic impact of constitutive fosB inactivation on nicotine-induced behavioral alterations and stress-related traits in mice. Hum Mol Genet 16: 820–836 [DOI] [PubMed] [Google Scholar]