Abstract
Periodontitis is a disease affecting the supporting structures of the teeth, which can eventually result in tooth loss. A three-dimensional (3D) tissue culture model was developed that may serve to grow a 3D construct that not only transplants into defective periodontal sites, but also allows to examine the effect of mechanical load in vitro. In the current in vitro study, green fluorescent protein labeled periodontal ligament (PDL) cells form rat incisors were embedded in a 3D matrix and exposed to mechanical loading alone, to a chemical stimulus (Emdogain; enamel matrix derivative [EMD]) alone, or a combination of both. Loading consisted of unilateral stretching (8%, 1 Hz) and was applied for 1, 3, or 5 days. Results showed that PDL cells were distributed and randomly oriented within the artificial PDL space in static culture. On mechanical loading, the cells showed higher cell numbers. Moreover, cells realigned perpendicular to the stretching force depending on time and position, with great analogy to natural PDL tissue. EMD application gave a significant effect on growth and upregulated bone sialoprotein (BSP) and collagen type-I (Col-I), whereas Runx-2 was downregulated. This implies that PDL cells under loading might tend to act similar to bone-like cells (BSP and Col-I) but at the same time, react tendon like (Runx-2). The combination of chemical and mechanical stimulation seems possible, but does not show synergistic effects. In this study, a new model was successfully introduced in the field of PDL-related regenerative research. Besides validating the 3D model to mimic an authentic PDL space, it also provided a useful and well-controlled approach to study cell response to mechanical loading and other stimuli.
Introduction
The periodontium (tissues surrounding teeth) comprises the periodontal ligament (PDL), the root cementum, and the alveolar bone. Periodontitis is an inflammatory disease caused by a microbial challenge in a susceptible host. Due to the inflammation, PDL, bone, and cementum are destroyed, which eventually might lead to tooth loss. Current treatments of periodontitis consist of removal of plaque and calculus, which indeed prevents further disease progression, but these treatments do not regenerate the lost tissues. Instead, histological studies have shown epithelial downgrowth, a mere reparative type of healing.1 Many recent studies focus on treatment strategies to achieve actual regeneration of the PDL rather than repair.
In vivo, the PDL is a well-organized flexible, fibrous suspension system connecting the dental root to the surrounding alveolar bone. PDL cells are continuously subjected to mechanical stress due to mastication, transmitting forces from the tooth to the alveolar bone. Research has shown that PDL cell behavior, including differentiation to great extent, is governed by such mechanical loading. Hence, the use of mechanical strain is one of the methods that are currently used to stimulate and differentiate PDL cells for tissue engineering purposes. Another option is the use of enamel matrix derivative (EMD, commercially available as Emdogain®, Straumann®), currently clinically used to regenerate periodontium. Enamel matrix proteins are secreted by Hertwig's epithelial root sheath during tooth development and are involved in cementum formation, preceding it in deposition order.2–4 Gestrelius et al.5 studied the effect of EMD on PDL cells in vitro, and concluded that EMD stimulated cellular proliferation, protein/collagen production, and formation of mineralized nodules in PDL cells. Several other authors corroborated the proliferative and stimulating effects of EMD on PDL cells.6,7
From a practical point of view, periodontal tissue engineering would include PDL cells that are expanded in vitro, placed into a resorbable scaffold, and after optimization can be subsequently delivered to a periodontal defect in vivo. Accordingly, it would be optimal to create an artificial, three-dimensional (3D) PDL space in which cells can be stimulated. In this way, the in vivo situation can be simulated in vitro, and the effect of strain or other stimuli on PDL cells can be studied.However, a controlled and easy applicable in vitro model that allows for the evaluation of multiple factors does not exist. Recently, a 3D model for studying ligament regeneration was described.8 However, it was mentioned that the experimental set-up was complicated, and the amount of tissue that could be created was very limited (only 20 μL).
Therefore, the major purpose is to develop a new, but relatively simple 3D model that can be used to examine fundamental aspects of PDL under loading and chemical stimulation, which in the long run can serve as construct to implant into periodontal defects. This type of research will enable us to gain better insight into the mechanisms that govern the formation of the PDL in vivo, but are also applicable to other tendon/ligament related fields. We hypothesize that PDL cells will be affected by mechanical loading, either or not in combination with EMD, in such a 3D model.
Materials and Methods
PDL fibroblasts
PDL fibroblasts were isolated from incisors of 40–43-day-old male Wistar rats. Extracted incisors were washed in phosphate-buffered saline (PBS) and subsequently washed thrice in minimum essential media (MEM)-α, containing 600 μg/mL gentamycin and 2.5 μg/mL fungizone (all from Gibco BRL, Life Technologies BV). A primary culture procedure of PDL cells was followed according to Mailhot and coworkers.9 Briefly, PDL was scraped from the middle third of the roots, avoiding contamination of epithelial or pulpal cells, by using a sterile scalpel. The freed portions of the PDL were minced and transferred to a T-25 flask, filled with 5 mL of basic medium (BM), that is, α-MEM containing 20% fetal calf serum (FCS), 60 μg/mL gentamycin, 0.25 μg/mL fungizone, and placed in a humidified atmosphere of 95% air, 5% CO2 at 37°C. Thereafter, medium was refreshed every 2 to 3 days. On sub confluency, cells were released with trypsin/EDTA (0.25% w/v crude trypsin, 1 mM EDTA, pH 7.2) and sub-cultured for 4 passages in T-75 flasks. The cells were counted using a Coulter® Counter (Coulter Electronics) and subsequently frozen until further use. For fluorescent cell observation, a similar cell type was derived from a green fluorescent protein (GFP) transgenic Sprague–Dawley rat.10,11 Cells of passage 4–6 were used in this study.
Alkaline phosphatase (ALP) activity was assessed to ensure that the PDL nature of the cells. Cells of three sources (PDL, gingival, and alveolar bone) from passage 4 were seeded in 24-well plates (2×104 cells/cm2) and cultured in osteogenic medium containing 10% FCS (Gibco), 50 μg/mL ascorbic acid (Sigma-Aldrich), 10 mM Na-β-glycerophosphate (Sigma), and 10−8 M dexametasone (Sigma). After 2 weeks of culture, medium was removed, and samples were washed twice with PBS. Subsequently, 1 mL MilliQ water was added into each well. After two freeze-thaw cycles, the supernatants were assessed for ALP activity. For this, 20 μL of 0.5 M 2-amino-2-methyl-1-propanol was added to 80 μL of sample. Next, 100 μL of p-nitrophenyl phosphate substrate solution was added. Mixtures were incubated at 37°C for 1 h, and ALP activity was measured at 405 nm using an ELISA microplate reader (Bio-Tek Instruments Inc). All PDL cells used in this study showed ALP activity intermediate to alveolar bone and gingival fibroblasts.
Medium containing EMD
For the experimental medium containing EMD (EM), in 90 mL of the previously described medium (BM) 0.3 mL EMD was dissolved. In detail, the plunger of the EMD syringe was carefully removed, subsequently 50 μL of EMD was taken out with a sterile pipette and added to the 90 mL medium (BM). Gentle rotations of the medium with the EMD were made until the EMD was completely dissolved, this method was repeated until all EMD (0.3 mL) was dissolved in the 90 mL medium. The final concentration of EMD was 100 μg/mL. This medium (EM) was used throughout the entire experiment, however, freshly prepared before each cell culture was started.
Substrates and coverslips
Elastomer silicone rubber dishes were prepared by mixing Elastosil A and Elastosil B (polydimethylsiloxane, Elastosil RT 601; Wacker-Chemie) in a 1:10 ratio. This mixture was poured into acrylic dish molds, left at room temperature for 2 h to remove air bubbles, and then placed in a furnace at 60°C for 24 h. Subsequently, the dishes were carefully removed from the template, cleaned with a 10% detergent (Liquinox®; Alconox Inc.) in MilliQ water, washed 10 times with pure MilliQ, rinsed with 100% ethanol, and autoclaved at 121°C for 15 min. Thereafter, the dishes were stored until further use. Immediately before cell seeding, a radio frequency glow discharge (RFGD) treatment (Harrick Scientific Corp.) for 5 min at 100 mTorr Argon was performed.
Plastic coverslips were cut out of polystyrene petri-dishes (Greiner Bio-One GmbH) into square 27×24 mm (L×W mm) plates. The top of the plate was marked with a small indentation, and nine perforations (diameter 1 mm) were drilled, evenly distributed around the plate (Fig. 1A). Subsequently, the coverslips were cleaned and subjected to RFGD treatment as just described.
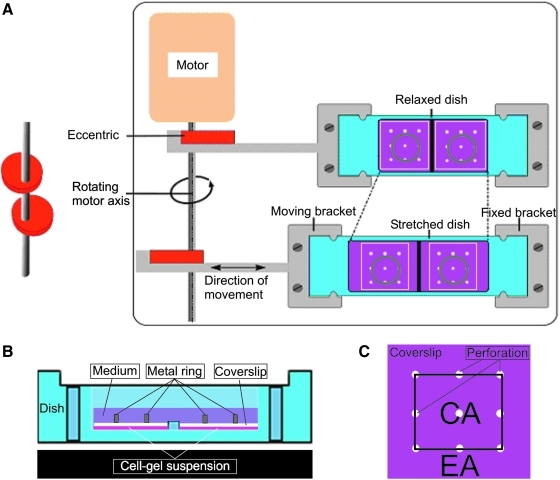
FIG. 1.
Experimental setup; (A) Top view of experimental setup; light blue represents the silicone dish; the white squares represent the plastic cover plates with nine holes/perforations (width×length; 27×24 mm, diameter of the perforation 1 mm); the gray circles represent the metal weights (diameter 15 mm, height 5 mm) to keep the gel in position; (B) Lateral view of a relaxed dish; in purple, the medium; in gray, the metal weight; in white, the plastic cover plate; and in pink, the cell-gel solution; (C) Top view of the coverslip; the area in between the holes was considered the central area (CA), and the area surrounding the holes was the edge area (EA).
Collagen gel containing PDL cells
On ice, and while continuously vigorously shaking, 20 μL of 10 times concentrated PBS was mixed with 3 μL 1N NaOH, 43 μL MilliQ, 124 μL of collagen solution (rat tail collagen type I; Becton Dickinson), and 10 μL cell solution containing 5×104 cells. A total of 180 μL of this solution was injected onto the bottom of the silicone dish, resulting in an average thickness of PDL space of 200 μm. Subsequently, the plastic cover slip with pores and a stainless steel holding ring (5 mm height×15 mm diameter, weight 1.0 g) were placed to secure the position of the gel (Fig. 1B). Thereafter, the dish was placed in a cell culture incubator to set the collagen gel at 37°C. After gelling for 30 min, 2.5 mL of BM or EM medium was added to each sample.
Loading regime/mechanical stimulation
To examine the effect of mechanical stimulation, unilateral cyclic stretch with an 8% magnitude and frequency of 1 Hz was applied for 1, 3, or 5 days, in a parallel direction to the longitudinal side of the silicone dish. The mechanical loading was applied intermittently, that is, 15 min of stretch and 15 min of rest for 8 h, after an initial static period of 16 h.12
Fluorescence microscopy for cell distribution, cell orientation, and cell number
To observe cell distribution and cell orientation, collagen gels containing GFP-PDL cells were observed with an automated fluorescent microscope (Axio Imager Microscope Z1; Carl Zeiss Micro Imaging GmbH) set at magnification of 20× at the time points 1 h after cell seeding, and on 1, 3, and 5 days of culture. For each sample, over 100 microscopic fields were randomly selected, and cells were examined and counted. For overall orientation with respect to the stretch direction, Image J software (Image J, National Institutes of Health) was used. Only cells that were extended not in contact with other cells and not in contact with the image perimeter were included in the measurement. Median angles and standard deviations were calculated, and groups were compared using an unpaired t-test (Instat version 3.05 GraphPad Inc). Angles were compared to the direction of the applied stretch, that is, a 90-degree angle means perpendicular to the stretch direction. When observed from above, cells were separately analyzed in the center and near the edges of the collagen gel (Fig. 1C). Thus, the area in between the holes was considered the central area (CA), and the area surrounding the holes was considered the edge area (EA) (Fig. 1C).
For the cell number analysis, GFP-PDL cells were cultured for 1, 3, and 5 days. After each time period, samples were examined under the microscope as previously described, however now at a lower magnification (10×). The cell number in each field was calculated using Image J software. For each time point, 2 samples were used, and 40 microscopic fields were randomly taken from each sample. Nonspread cells were considered dead and subsequently excluded from the counting. The data were compared with a two-way analysis of variance (ANOVA) and post hoc Tukey testing (Instat version 3.05 GraphPad Inc).
3D image reconstruction
To observe cell morphology in 3D, collagen gels containing GFP-PDL cells were gently removed from the setup after 3 days of culture and transferred on to a custom-made polystyrene membrane (100 μm thick) created by solvent casting. These samples were observed using confocal laser scanning microscopy (Olympus FV1000) with a 63× oil immersion objective. Series of scanning images were made with an excitation wavelength of 488 nm and emission wavelengths of 550 nm. The single XY scan had an optical slice thickness of 1.5 μm. 3D projections were digitally reconstituted from stacks of confocal optical slices (Olympus, FV1000 version 1.6). Animations of top-down view were made by importing sequential images into Image J software. All observations were performed in duplicate for all groups.
Real-time polymerase chain reaction
Gene expression was studied after day 3 of incubation. The cell-gel constructs were collected, and the total RNA from the cultured cells was isolated using TRIzol® reagent (Invitrogen) according to the manufacturer's instruction. The RNA concentration was measured with NanoDrop (Nanodrop Technologies). After obtaining the mRNA, a first-strand reverse transcriptase polymerase chain reaction (PCR) was performed using the Superscript™ First-strand Synthesis System for RT-PCR (Invitrogen); according to the manufacturer's protocol.
The cDNA was then amplified, and specific gene expression was quantified in real-time PCR. For this reaction, 12.5 μL master mix, 2 μL DNA, 3 μL primer mix (1.5 μL forward primer and 1.5 μL reverse primer mixed), and 7.5 μL DEPC were added into the reaction system. Subsequently, the PCR was performed in a Real-Time PCR reaction apparatus with the desired temperatures. Primers were designed so as to avoid amplification of genomic DNA, and, therefore, each amplicon spans at least one intron (Table 1).
Table 1.
Rat Specific Primer Sequence for the Genes Used
| Forward (5′→3′) | Reverse (5′→3′) | NCBI reference sequence | Product BP size | |
|---|---|---|---|---|
| Collagen I | GAGCGATTACTACTGGATTGACCC | CAAGGAATGGCAGGCGAGAT | NM 053356 | 506 |
| C-fos | ATCCGAAGGGAAAGGAATAAGA | CAAGTCCAGGGAGGTCACAGA | NM 022197 | 246 |
| Cox-2 | TCCAACCTCTCCTACTACAC | GTTGCACGTAGTCTTCGATC | NM_017232.3 | 625 |
| BSP | GAAGCAGGTGCAGAAGGAAC | ACTCAACCGTGCTGCTCTTT | NM_012587.2 | 157 |
| Runx-2 | GAGCACAAACATGGCTGAGA | TGGAGATGTTGCTCTGTTCG | NM_053470.1 | 238 |
| GAPDH | CGATGCTGGCGCTGAGTAC | CGTTCAGCTCAGGGATGACC | XM_576394.3 | 413 |
The genes studied in the real-time quantitative PCR were coding for the three bone differentiation markers, that is, bone sialoprotein (BSP), Runx-2 transcription factor, and signaling molecules cyclooxygenase, the major extracellular matrix (ECM) protein collagen type-I, as well as the growth-factor-related transcription factor c-Fos. Relative gene expression was normalized to the household Gapdh gene expression. The expression of the tested genes was calculated via the 2−ΔΔCt method13 relative to the BM group. Statistical analysis was performed with a one-way ANOVA and post hoc Tukey testing using Instat.
Results
Cell distribution
One hour after seeding, cells appeared rounded in shape, and had not yet spread (Fig. 2A). One day later, most of the cells had attached to the collagen matrix and had started to spread (Fig. 2B). In time, cells became elongated and increased in number on visual inspection (Fig. 2C). Noticeably, at day 5, even cell colonies could be observed (Fig. 2D). At lower magnification, it was clear that the cells were evenly distributed within the artificial PDL space (Fig. 2E).
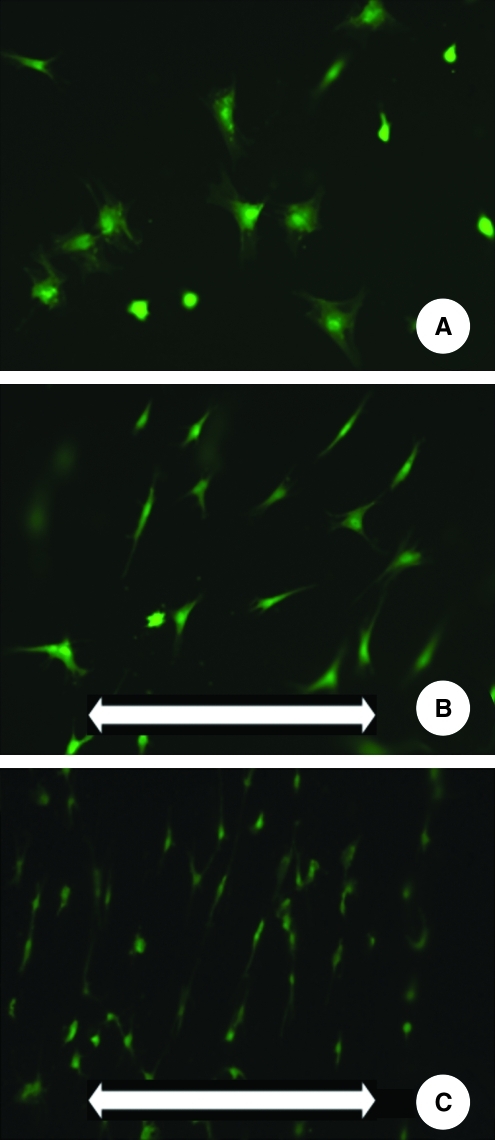
FIG. 2.
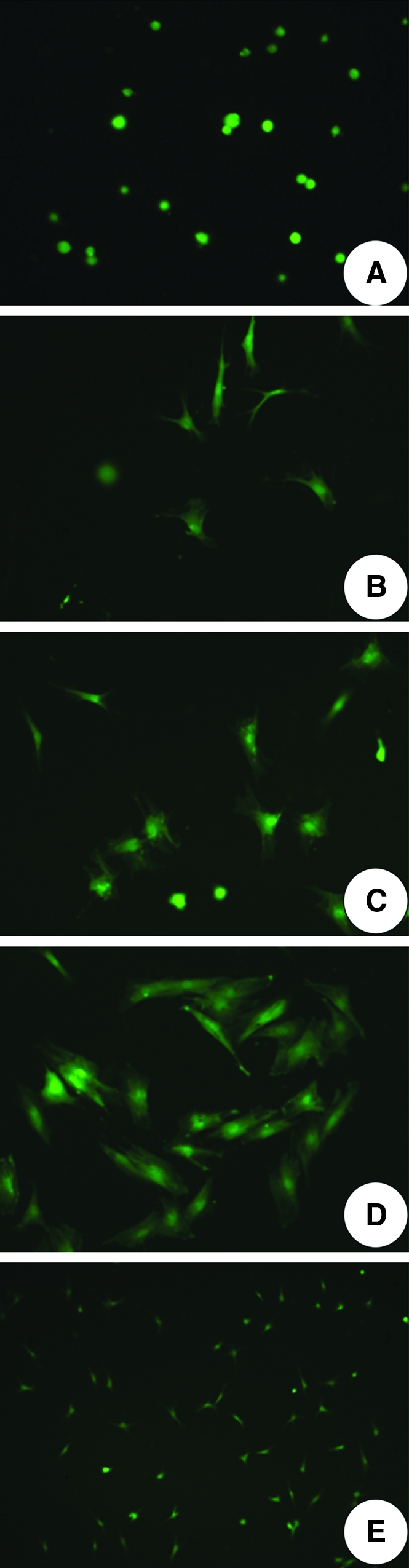
Cell distribution: (A) 1 h after cell seeding, original magnification 20×; (B) 1 day after cell seeding, 20×; (C) 3 days after cell seeding, 20×; (D) 5 days after cell seeding, 20×; (E) distribution of cells at day 3, 10×.
Cell orientation
In the static culture environment, the cells showed multipolar shapes with no evidence of preferential orientation (Fig. 3A). When mechanical loading was applied, cells changed their initial morphology and direction, that is, became elongated and oriented perpendicular to the strain direction (Fig. 3B). However, a distinct difference could be noticed between the CA (Fig. 3B) and the EA (Fig. 3C). More peripheral, the alignment was more prominent.
FIG. 3.
Cell morphological changes on mechanical loading: (A) Nonstretched sample, after day 1, original magnification 20×; (B) Stretched sample in CA, after 1 day, 20×; (C) Stretched sample in the EA, after day 1, 20×. The white arrow in the lower part of B and C indicates the direction of the applied loading.
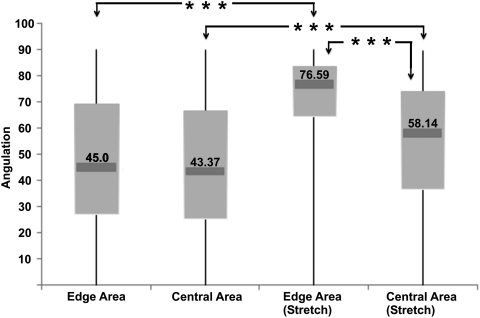
Besides visual inspection, cellular alignment was also quantified as shown in a Box-Whisker plot (Fig. 4). The chart shows that cells were randomly distributed in the nonstretched group. However, when subjected to mechanical loading, cells aligned perpendicularly to the stretching force. ANOVA testing showed that this cellular reorientation was significant compared with nonstretched groups, irrespective of the cell location (p<0.001). Noticeably observed from the angulation measurements, the cells close to the EA were more sensitive to the stretching force and more prone to alignment compared with cells in the CA (p<0.001). In addition, for both areas, there was a significant difference compared with the control group.
FIG. 4.
Box-Whisker plot showing cell orientation midpoint (median), and the first and third quartile (boxes), and the largest and smallest observations. A mean of 45° indicates no preferential orientation, whereas a higher angle indicates alignment perpendicular to the direction of force. Note that cells in the EA were more prone to alignment compared with the cells in the CA. ***p<0.001.
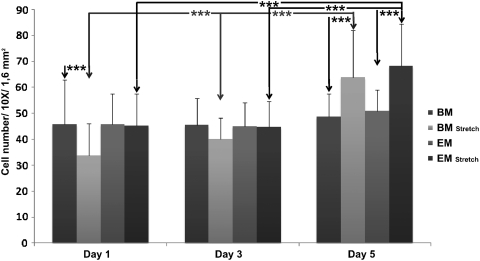
Cell number
Over time, cell numbers did not increase in the static groups (BM and EM) (Fig. 5). In contrast, when stretch was applied, both media showed an increased cell number over time (p<0.001). When comparing the groups at the individual time points, it became evident that at day 1 stretch gave significantly lower cell counts compared with the control in the BM group, but not in the EM group. This was compensated by proliferation, as on day 3 no statistical differences were observed anymore. Finally, on day 5, the mechanical loading resulted in a significantly higher cell number in both the BM and EM groups (p<0.001).
FIG. 5.
Cell number data: the average cell number after 1, 3, and 5 days of culture. Values are represented as mean±standard deviation. 1.6 mm2 represents the total surface measured for a picture taken. ***p<0.001.
3D confocal scanning microscopy
The 3D morphology of cells in the collagen gel was also studied using confocal scanning microscopy. After 1 day of culture, cells were distributed in multilayers for both the nonstretched (Supplementary Video S1; Supplementary Data are available online at www.liebertonline.com/tec) and stretched conditions (Supplementary Video S2). In the XY axis plane, cells converted their direction perpendicular to the applied strain force in the stretched group (Supplementary Video S2). In a 3D observation, cells at the inner part of the gel showed a bipolar shape with spherical nucleoli, surrounded by a few short filopodia (Supplementary Video S3). On the contrary, cells on both solid surfaces (Supplementary Video S4) (silicon bottom and coverslip) showed mainly spread multipolar cytoplasm branches surrounding a flat nucleus. Within those surface attached cells, some short slender cytoplasmic projections toward the central part of the collagen gel (artificial PDL) could also be found.
Gene expression
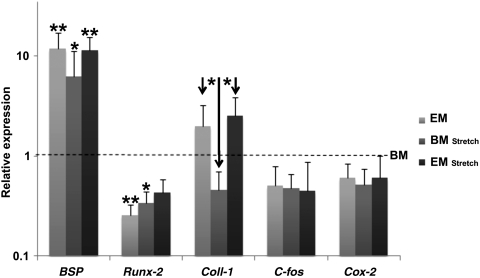
The expression on RNA level of the tested genes was measured relative to the BM group at day 3 (Fig. 6). All three other groups showed a higher expression of BSP compared with BM, indicating that both mechanical loading and the chemical stimulus EMD influenced gene expression. Both EM and BM medium in combination with stretch gave significantly lower Runx-2 (relative) expression levels than the control group. For collagen I expression, a lower expression of the BM stretch group was detected when compared with EM and EM stretch. For both C-fos and Cox-2, no significant differences were detected.
FIG. 6.
Influence of cyclic strain and EMD on gene expression. Values were normalized to GAPDH and relative to the BM group (Black dotted line). Note that there are significant effects on gene expression for BSP, Runx-2, and Coll-1 with stretching force and EMD. *p<0.05; **p<0.01. EMD, enamel matrix derivative; BM, basic medium.
Discussion
Periodontitis is characterized by the loss of both the soft and hard tissues surrounding teeth, which can eventually lead to tooth loss. Complete and functional periodontal regeneration involves the restoration of the complete specific morphology composed of the PDL in between the tooth and root socket surface. The cells within the PDL tissue play a central role in this periodontal healing process. Considering the complexity of the PDL and the effect of different stimuli, it is crucial to develop controlled and easy to apply in vitro models to investigate PDL (cell) activity. Many researchers have focused on cell behavior in a two-dimensional (2D) environment, whereas in vivo, the PDL composes a 3D environment in which cells are surrounded by the ECM. The aim of our study was to develop a simple 3D model that can be used to examine fundamental aspects of PDL under loading as well as chemical stimulation and in the long run perhaps can be used for the preparation of a construct for periodontal defect regeneration.
Our exact same system has been well described in 2D cell culture regarding cell compatibility, attachment, differentiation, etc.14 This system was now further developed toward a 3D model. Regarding the presented set-up, initially multiple adaptations were tested before the presented model could be used. The natural situation comprises of two hard tissue surfaces, of the bone and root. The coverslip represents one of these hard tissues. Still, cover plates without perforations were initially used and appeared to limit cell survival; thus, they had to be provided with openings for better cell nutrition and oxygen exchange. Still, cells sometimes appeared to aggregate preferentially near the holes on visual inspection of the fluorescence images. For future further experiments, it can be advantageous to use a rigid but permeable coverslip to allow for totally equal conditions in all regions. Another issue was that due to the stretching force/movement the coverslips detached from the gel instead of maintaining a “fixed” position. The application of the RFGD treatment to enhance adherence of the gel, in combination with the metal weights successfully resolved this problem. The final presented model proved to provide controlled mechanical conditions and can be used to study the effect of several parameters on PDL cell behavior.
The width of the created 3D space was 150–200 μm wide, which resembles the in vivo human PDL space.15 Further, in vivo PDL cells are embedded in ECM, which consists for the major part of different types of collagen, though predominantly collagen type I. Therefore, collagen type I was also used in this study. Unlike a 2D monolayer culture in which cells typically adopt a spread morphology (in x-y axis), cells residing in the collagen matrix protruded into the z-axis as well, thus showing a true 3D morphology and distribution. This situation is similar to the natural PDL, in which cells align to the collagen fibers that run perpendicular from the alveolar bone to the root cement.15
However, differences remained between cell morphology on the solid surfaces (silicon bottom and coverslip) and in the center. However, these are not surprising, and in agreement with 2D versus 3D culture systems. For example, fibroblasts in 2D cultures exhibit a flat morphology with dorsal-ventral polarity and large lammellipodia, whereas in a 3D matrix, cells show a natural spindle-shaped morphology.16,17 This phenomenon may be related to the ECM environment in which cells that are encapsulated need to overcome surrounding physical impediments to proliferate or migrate.18
Cells in vivo are constantly exposed to mechanical loading resulting from their surrounding ECM. PDL in vivo is functionally subjected to considerable mechanical loading. In addition, in our model, the transfer of load from a deformable substrate, on a gel material with evidently a much lower Young's modulus, was evident, in the relatively thin (200 μm) PDL space. Although there is no covalent link, the affinity of the collagen for the hard surfaces (enhanced by our RFGD treatment) is enough to allow for mechanical interlocking and load transmission. A clear response to the mechanical loading could be demonstrated as cell number increased over time, whereas for the static circumstances this did not occur. The mechanical stresses applied to the cells on this in vitro model are tension forces, which are just a part of the loading stimulations applied to the periodontium. Further enhancements of the model should aim to reproduce more accurately other mechanical events in vivo. Still, in our 3D environment, the PDL cells also oriented perpendicularly to the mechanical force. The higher cell rotation at the edges of the collagen gel may indicate that cells are more challenged to mechanical loading in this specific region. These changes point to the possibility that cells differentiate into different populations similar as in growing PDL, that is, at the edges turn into osteoblast-like cells expressing BSP, but in the center more toward tendon with low Runx-2 expression. This morphological variation may be a sign of different cell phenotypes as a consequence of mechanical stimulation. This phenomenon is in accordance with the in vivo PDL environment, in which cells at the bone and cement borders are prone to differentiate into osteoblasts or cementoblasts, whereas in the center retain a fibroblastic phenotype.
Since the cellular content of PDL is relatively low, our set up likewise started with a low cell number. The mechanical stimulation led to a higher cell proliferation rate compared with the static controls. The increase in proliferation is coherent with other researchers.19 An explanation for this finding other than a direct effect of loading is also the increased influx of oxygen and nutrition supply as a consequence of the stretching movement. In addition, mechanical loading of the cell-gel complex resulted in an increase in ECM-formation-related genes (such as BSP and Collagen I), which agrees with another study, which showed that up regulation of type I collagen mRNA expression occurred in anterior cruciate ligament cells exposed to cyclic stretching.20 This suggests that mechanical signals trigger cell-surface stretch receptors resulting in cascades of genes activation responsible for the synthesis and secretion of key extracellular components.21 The decreased expression of the early osteoblast differentiation gene (Runx-2) and relatively low response in gene expression at later time points can be due to (1) a lower degree of stiffness in the collagen gel compared to 2D stretch substrates, or (2) the observation that the cells in the center differentiate toward PDL-like tissue. Further, it has to be noticed that the gene expression data corroborate with another, more difficult to handle, 3D cell culture model that also showed a markedly low response in gene expression to mechanical forces.8
The main dissimilarity between our model system and natural PDL lies in the comparatively low density of collagen at 0.2%, compared with high cell/collagen density of 42% in the PDL region in vivo.22 The higher collagen content may introduce higher strength, stiffness, and anchoring sites for both mechanical force transfer and cell attachment. It can also be hypothesized that in such an environment the gene expression of cells at the borders is even more sensitive than in the center of the gel, because the mechanical difference is so much more pronounced along the borders.
The EM groups showed always a higher proliferation rate, both under stretched and static culture conditions, compared with the BM groups. This finding corroborates with other EMD related studies.23 EMD medium led to a higher BSP gene expression irrespective of stretching. Collagen I expression was lower in the stretched group, but this was compensated by the effect of EMD. The effect of up regulation in the ECM formation related genes in the EMD group agrees with other research.24,25 This indicates that the presence of EMD on PDL cell differentiation is crucial in the initial stages of culture.26 The relatively low response might be associated with the dosage and application method in our 3D mode, as the effect was not as obvious in 2D culture systems. On the other hand, the in vivo situation is also 3D; thus, 2D systems might exaggerate and overstate the actual effect of EMD.
Conclusion
In this study, a 3D model was introduced in the field of PDL-related regenerative research. Besides validating the 3D model to mimic an authentic PDL space, it also provided a useful and well-controlled approach to study cell response to mechanical strain and can serve to investigate cell response to other stimuli. Perhaps, this system can even be used to create a functional PDL that can ultimately be delivered to a defect site.
Supplementary Material
Acknowledgments
This study was financially supported by Dutch Technology Foundation (STW) (grant number NKG.6099) and Royal Netherlands Academy of Arts and Sciences (PSA) (grant number 08-PSA-M-02). Na Yu thanks the China Scholarship Council for their financial support (No.2008627114).
Disclosure Statement
No competing financial interests exist.
References
- 1.Caton J. Nyman S. Zander H. Histometric evaluation of periodontal surgery. II. Connective tissue attachment levels after four regenerative procedures. J Clin Periodontol. 1980;7:224. doi: 10.1111/j.1600-051x.1980.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 2.Fong C.D. Slaby I. Hammarström L. Amelin: an enamel-related protein, transcribed in the cells of epithelial root sheath. J Bone Miner Res. 1996;11:892. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- 3.Hammarström L. Enamel matrix, cementum development and regeneration. J Clin Periodontol. 1997;24:658. doi: 10.1111/j.1600-051x.1997.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 4.Spahr A. Hammarström L. Response of dental follicular cells to the exposure of denuded enamel matrix in rat molars. Eur J Oral Sci. 1999;107:360. doi: 10.1046/j.0909-8836.1999.eos107507.x. [DOI] [PubMed] [Google Scholar]
- 5.Gestrelius S. Andersson C. Lidström D. Hammarström L. Somerman M. In vitro studies on periodontal ligament cells and enamel matrix derivative. J Clin Periodontol. 1997;24:685. doi: 10.1111/j.1600-051x.1997.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 6.Haase H.R. Bartold P.M. Enamel matrix derivative induces matrix synthesis by cultured human periodontal fibroblast cells. J Periodontol. 2001;72:341. doi: 10.1902/jop.2001.72.3.341. [DOI] [PubMed] [Google Scholar]
- 7.Palioto D.B. Coletta R.D. Graner E. Joly J.C. de Lima A.F. The influence of enamel matrix derivative associated with insulin-like growth factor-I on periodontal ligament fibroblasts. J Periodontol. 2004;75:498. doi: 10.1902/jop.2004.75.4.498. [DOI] [PubMed] [Google Scholar]
- 8.Berendsen A.D. Smit T.H. Walboomers X.F. Everts V. Jansen J.A. Bronckers A.L. Three-dimensional loading model for periodontal ligament regeneration in vitro. Tissue Eng Part C Methods. 2009;15:561. doi: 10.1089/ten.TEC.2008.0336. [DOI] [PubMed] [Google Scholar]
- 9.Mailhot J.M. Schuster G.S. Garnick J.J. Hanes P.J. Lapp C.A. Lewis J.B. Human periodontal ligament and gingival fibroblast response to TGF-beta 1 stimulation. J Clin Periodontol. 1995;22:679. doi: 10.1111/j.1600-051x.1995.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 10.Verstappen J. Katsaros C. Kuijpers-Jagtman A.M. Torensma R. Von den Hoff J.W. The recruitment of bone marrow-derived cells to skin wounds is independent of wound size. Wound Repair Regen. 2011;19:260. doi: 10.1111/j.1524-475X.2011.00671.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L. Yoshimura Y. Huang Y. Suzuki R. Yokoyama M. Okabe M. Shimamura M. Two independent pathways of maternal cell transmission to offspring: through placenta during pregnancy and by breast-feeding after birth. Immunology. 2000;101:570. doi: 10.1046/j.1365-2567.2000.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter L.C. Walboomers X.F. Bumgardner J.D. Jansen J.A. Intermittent versus continuous stretching effects on osteoblast-like cells in vitro. J Biomed Mater Res A. 2003;67:1269. doi: 10.1002/jbm.a.20028. [DOI] [PubMed] [Google Scholar]
- 13.Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Neidlinger-Wilke C. Wilke H.J. Claes L. Cyclic stretching of human osteoblasts affects proliferation and metabolism: a new experimental method and its application. J Orthop Res. 1994;12:70. doi: 10.1002/jor.1100120109. [DOI] [PubMed] [Google Scholar]
- 15.Melcher A.H. Periodontal ligament. In: Bhaskar S.N., editor. Orban's Oral Histology and Embryology. St. Louis: C.V. Mosby; 1980. pp. 204–239. [Google Scholar]
- 16.Yamada K.M. Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Cukierman E. Pankov R. Stevens D.R. Yamada K.M. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 18.Friedl P. Bröcker E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. 2000;57:41. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G. Crawford R.C. Wang J.H. Proliferation and collagen production of human patellar tendon fibroblasts in response to cyclic uniaxial stretching in serum-free conditions. J Biomech. 2004;37:1543. doi: 10.1016/j.jbiomech.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Kim S.G. Akaike T. Sasagaw T. Atomi Y. Kurosawa H. Gene expression of type I and type III collagen by mechanical stretch in anterior cruciate ligament cells. Cell Struct Funct. 2002;27:139. doi: 10.1247/csf.27.139. [DOI] [PubMed] [Google Scholar]
- 21.Altman G.H. Horan R.L. Martin I. Farhadi J. Stark P.R. Volloch V. Richmond J.C. Vunjak-Novakovic G. Kaplan D.L. Cell differentiation by mechanical stress. FASEB J. 2002;16:270. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- 22.Migkalites C. Orlowski W.A. Study of the noncollagenous components of the periodontium. J Dent Res. 1977;56:1023. doi: 10.1177/00220345770560080501. [DOI] [PubMed] [Google Scholar]
- 23.Lyngstadaas S.P. Lundberg E. Ekdahl H. Andersson C. Gestrelius S. Autocrine growth factors in human periodontal ligament cells cultured on enamel matrix derivative. J Clin Periodontol. 2001;28:181. doi: 10.1034/j.1600-051x.2001.028002181.x. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu E. Nakajima Y. Kato N. Nakayama Y. Saito R. Samoto H. Ogata Y. Regulation of rat bone sialoprotein gene transcription by enamel matrix derivative. J Periodontol. 2004;75:260. doi: 10.1902/jop.2004.75.2.260. [DOI] [PubMed] [Google Scholar]
- 25.He J. Jiang J. Safavi K.E. Spångberg L.S. Zhu Q. Emdogain promotes osteoblast proliferation and differentiation and stimulates osteoprotegerin expression. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;97:239. doi: 10.1016/j.tripleo.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Keila S. Nemcovsky C.E. Moses O. Artzi Z. Weinreb M. In vitro effects of enamel matrix proteins on rat bone marrow cells and gingival fibroblasts. J Dent Res. 2004;83:134. doi: 10.1177/154405910408300210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.