Abstract
Prevalent gene fusions involving regulatory sequences of the androgen receptor (AR) regulated genes (primarily TMPRSS2) and protein coding sequences of nuclear transcription factors of the ETS gene family (predominantly ERG) result in unscheduled androgen dependent ERG expression in prostate cancer (CaP).Cumulative data from a large number of studies in the past six years accentuate ERG alterations in more than half of all CaP patients in Western countries. Studies underscore that ERG functions are involved in the biology of CaP. ERG expression in normal context is selective to endothelial cells, specific hematopoetic cells and pre-cartilage cells. Normal functions of ERG are highlighted in hematopoetic stem cells. Emerging data continues to unravel molecular and cellular mechanisms by which ERG may contribute to CaP. Herein, we focus on biological and clinical aspects of ERG oncogenic alterations, potential of ERG-based stratification of CaP and the possibilities of targeting the ERG network in developing new therapeutic strategies for the disease.
Keywords: ERG, prostate cancer, TMPRSS2-ERG, oncoprotein, androgen receptor, patient stratification
BACKGROUND
Key molecular genetic alterations in prostate cancer
Prostate cancer (CaP) is the most common malignancy that affects men worldwide, with high frequency in the United States, Western Europe[1] and low reported frequency in Asia.[2,3] Risk factors associated with CaP include age, family history and ethnicity.[1,4] Although precise molecular events that contribute to such variation in the CaP incidence are not well established, the differences may be attributed to factors such as genetics, diet, lifestyle, and male hormone levels.[4–6] Despite the recent advances in early detection and continued refinements in treatment strategies, CaP is still the second leading cause of cancer mortality in American men.[1] Discovery of CaP-specific gene expression and/or mutational alterations have contributed to a significant impact on designing molecular markers to distinguish indolent from more aggressive forms of cancers as well as molecular pathways to develop effective novel therapeutic approaches to combat the disease.[7–12]
CaP susceptibility loci with germ-line mutations of RNAseL, ELAC2, MSR1, BRCA 1 and 2, HPCX, KLF6, and HPC20have been reported in primary CaP.[13,14] However, low penetrance and disease heterogeneity have precluded the validation of CaP susceptibility genes. Recent genome wide association studies (GWAS) have identified multiple CaP risk alleles towards defining genetic determinants of CaP risk.[15,16] A “gene less 1.18 Mb region” between FAM84B at centromeric end and C-MYC at telomeric end on chromosome 8q24 has been consistently found to be associated with CaP risk.[17–21] The8q risk allele specific for African ancestry showed an association with higher pathologic stage of CaP in African American men.[22] Functional evaluations of a risk allele on chromosome 10 suggested its impact on regulation of expression of NCOA4 (AR co-activator) and MSMB.[23] Overall, a combinatorial assessment of the risk alleles has shown a significantly increased predictive power of CaP risk.[19,24]
Chromosome loci harboring putative proto-oncogenes or tumor suppressor genes (TSGs) have been extensively evaluated toward identifying specific gene mutations and expression signatures in CaP. Mutations, amplifications or over-expression of the androgen receptor (AR), and mutations in tumor suppressors such as p53 and PTEN, are frequently identified subsets of advanced CaP.[8,9,25–28] Among the recurrent allelic losses of 8p21-22, 6q16, 7q31, 10q23-25 and 16q24 loci detected in primary CaP,[8,29] deleted 8p21-22 locus harbors a widely studied tumor suppressor gene NKX3.1.[30] While early studies showed PTEN mutations in subset of advanced cancers, more recent reports underscore higher frequency of PTEN hemizygous deletions in primary CaP.[31] In addition, frequent gains of chromosome 8q24, as well as over-expression of C-MYC and prostate stem cell antigen (PSCA) within this locus have been reported.[13]
Identification of common CaP specific gene signatures have enriched mechanistic as well as translational research investigations. Expression of genes such as NKX3.1[32] and GSTP1[33] have been studied extensively for their biological roles in onset of CaP. The virtual absence of GSTP1 expression due to promoter methylation has led to blood- and urine-based assays for diagnosis.[34] Overexpression of AMACR and absence of p63 in most prostate tumors have already led to the use of these two proteins in diagnostic pathology.[35] Striking overexpression of a prostate tissue specific gene, DD3/PCA3 in CaP have led to extensive evaluations for its diagnostic utility as a marker in urine based assays.[36] Although CaP specific gene alterations are increasingly studied, the most validated oncogenic alteration to date is ERG. This observation led to multi-faceted investigations towards defining the cancer specific characteristics of ERG, and is discussed in the following sections.
Prevalence of TMPRSS2-ERG fusion in prostate cancer
Identification of ERG proto-oncogene overexpression in CaP transcriptome led to focused evaluations of ERG alterations in CaP.[37–39] Quantitative expression assessment of ERG mRNA in matched benign and malignant prostate cells from a large patient cohort confirmed the tumor cell specific ERG overexpression in 60-70% patients.[39] Over expression of ERG due to fusions between androgen regulated TMPRSS2 gene promoter and the coding regions of ERG has been identified as the most common genomic alteration.[40] These observations also led to the development of a combined CaP gene panel (PCA3, ERG and AMACR) with diagnostic potential in which overexpression of at least one of three genes associated with virtually all of prostate tumor specimens.[39] Discovery of prevalent gene fusions involving promoters of the androgen receptor (AR) regulated genes (TMPRSS2, SLC45A3, NDRG1, Herv-K22q11.23, CANT1 and KLK2) and coding sequences of ETS gene family (ERG, ETV1, ETV4, ETV5)marked a major milestone towards defining molecular mechanisms of prostate carcinogenesis.[11,41] Of the fusions involving TMPRSS2 and ETS factors in CaP, majority (>90%) involve ERG, and ETV1, ETV4 and ETV5 represent very low frequency (1-5%).[11] TMPRSS2 gene is mapped to 21q22.3 between markers ERG and D21S56, and transcribed as 3.8 kb mRNA.TMPRSS2 promoter analysis revealed the presence of a non-canonical ARE as a CIS-regulatory target of AR action.[42] TMPRSS2 is predominantly expressed in prostate tissues with low levels of expression in pancreas, kidney, lung, colon and liver.[43,44] Gene fusions between TMPRSS2 and ERG or ETV1 appears to be CaP specific and are potentially mediated by AR-induced proximity of fusion gene partners in the presence of genotoxic factors[45,46] followed by topoisomerase-2b-mediated recombination event.[47] Comprehensive evaluations of gene fusions involving ETS factors have been covered in excellent reviews.[11,48]
ERG gene structure and transcription
ERG is a member of the ETS gene family[49,50] which is one of the largest families of transcriptional regulators consisting of at least 27 members, subdivided into 5 subfamilies.[51] Conserved PNT/SAM domain and an ETS domain are the common features of members of ETS related proteins. These domains play key roles in regulating downstream target genes that are crucial for several biological processes such as cellular proliferation, differentiation, development, transformation, and apoptosis.[52] ERG consists of 17 exons and is transcribed to generate several alternately spliced forms[53] [Figure 1]. At least five splice variants are translated into proteins: ERG-1 (p41), ERG-2 (p52), ERG-3 (p55), ERG-4 (p49) and ERG-5 (p38)[54] by a combination of alternative mRNA splicing and/or use of alternative polyadenlyation sites.[50,55] Most characteristic of the family is the evolutionarily conserved 85-amino acid ETS domain, which facilitates binding to purine-rich DNA with a GGAA/T core consensus sequence.[51,56]
Figure 1.
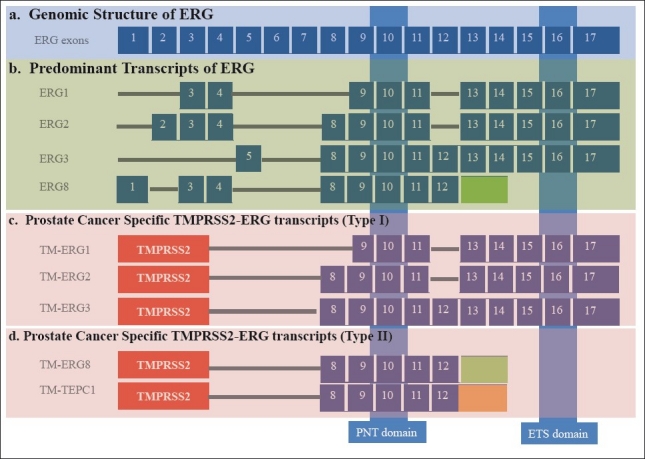
Genomic structure and transcripts of human ERG gene. (a) Genomic structure depicting ERG Exons (blue boxes) numbered from 1-17.[53] (b) Structure of expressed ERG transcripts.[53] (c) Prostate cancer specific TMPRSS2-ERG fusion transcripts containing protein-protein interaction domain (pointed/SAM) and DNA binding (ETS) domain (Type I).[81] (d). TMPRSS2-ERG fusion Type II transcripts containing only pointed/SAM without ETS domain.[81] Note: In prostate cancer, the original ERG exon 8[53] is numbered as 4.[40,78,87]
ERG is among a small number of transcription factors that exhibit an endothelial cell and hematopoietic cell restricted expression pattern in various species. In developing mouse, Erg mRNA is expressed in mesodermal tissues such as endothelial cells, mesenchymal condensations during precartilaginous depositions, and in urogenital regions.[57] Similarly, ERG protein is predominantly detected in endothelial cells, hematopoietic tissues and transiently in pre-cartilage.[58] Erg is expressed transiently during early T-cell development, early pre-B and continue to express in mature B cells.[59,60] Later in development, Erg functions in cell survival maintaining the differentiation of endothelial cells of vascular and lymphatic origins.[61,62] Thus, highly restricted expression of Erg mRNA or ERG protein during early phases of lymphocytic, hematopoietic, chondrocytic and endothelial lineage differentiations appears to be crucial in lineage specification function.[58,63–65] Intriguingly, ERG protein is not detected in any epithelial tissues including prostate epithelium, or in infiltrating lymphocytes that are occasionally seen in the prostate environment.
Normal biological functions of ERG
Biological functions of ERG have been studied in xenopus, zebra fish, mouse and humans.[57,66–71] Angiogenesis is an essential process by which new vessels are developed from preexisting ones, during normal development, as well as in pathologic conditions, including tumor development. Widespread expression of ERG in endothelial cells suggests for its biological roles in these specialized cells. In addition to VE-cadherin, other endothelial specific factors such as, von Willebrand factor, endoglin, and intercellular adhesion molecule-2 are also regulated by ERG supporting its role in endothelial cell differentiation and angiogenesis.[62,72] Endoglin is an accessory receptor for TGF-β and both endoglin and TGF beta receptor type II are positively regulated by Erg.[73,74] Recently, using a functional mutation in mouse models, Erg has been shown to regulate the normal platelet development, stem-cell function, definitive hematopoiesis and the normal megakaryopoiesis.[70] Although, ERG is considered as critical regulator of hematopoiesis, Erg is dispensable during early embryonic hematopoietic development, hematopoietic specification from the mesoderm and is required to sustain definitive hematopoiesis. During this process, ERG acts as a direct regulator of critical transcription factors such as Runx1 and Gata2.[75] During hematopoiesis, adult hematopoetic stem cells require ERG for self-renewal and differentiation.[76] ERG is also documented as a transcription regulator of embryonic stem cell (ES) towards differentiation of early endothelial lineage[77] and exhibits anti-inflammatory responsein endothelial cells by suppressing IL 8.[72]
Prostate cancer associated TMPRSS2- ERG transcripts
Several types of TMPRSS2-ERG fusion transcripts involving various exons of the TMPRSS2 and ERG have been identified in CaP specimens.[66,78–83] These transcripts were identified on the basis of TMPRSS2 fusions with the 5’ end of the ERG and are broadly classified into 8 different groups. In the context of full length transcripts, 2 major forms were identified on the basis of mRNA splicing, cDNA and deduced amino acid sequences.[81] Although, several fusion transcripts are generated from TMPRSS2-ERG fusions, it is not clear whether these transcripts are expressed from a single or multiple foci of CaP. Evaluation of TMPRSS2-ERG transcripts in multi-focal CaP have shown inter-focal heterogeneity with respect to the presence of fusion positive or negative foci in malignant prostate glands.[82,84–86]
Despite the heterogeneity of TMPRSS2-ERG fusions, most common fusion is in between TMPRSS2 exon 1 and ERG exon 4, which results in the deletion of first 32 amino acids from the N-terminus of ERG protein.[87] The expression of TMPRSS2 exon 2 with ERG exon fusion 4 mRNA associated with PSA recurrence and seminal vesicle invasion.[78] The most common full length TMPRSS2-ERG transcripts (Type I) translate into full length proteins (ERG1, ERG2, ERG3) containing protein-protein interacting (pointed/SAM) and DNA-binding (ETS) domains.[81,87] The most predominant of the proteins generated from the fusions is the N-terminal truncated ERG3 protein. Whereas the type II TMPRSS2-ERG transcripts code for ERG8 and a new variant, TEPC, with deletion of 32 amino acids at N-terminus and contain only pointed/SAM domain[81] [Figure 1]. Importantly, higher ratio of type I over type II TMPRSS2-ERG splice forms are shown to correlate well with unfavorable prognostic features of CaP, such as poorly differentiated tumors, higher Gleason sum, positive margin, and biochemical recurrence.[81] Additional studies are needed to assess prognostic association specific TMPRSS2-ERG fusion transcripts with CaP progression. Since ERG is the most common cancer gene activation in CaP, ERG expression and function in normal and other cancer contexts may be illustrative in further understanding the biological roles of ERG in CaP.
Prostate cancer associated functions of ERG
Since the discovery of ERG, several reports have shown that ERG transforms epithelial cells[49,88–91] and functions through mitogenic signals including the MAP kinases.[88] Acute myeloproliferation and megakaryocytic differentiation are the main features of hematologic diseases associated with Down syndrome (trisomy of chromosome 21), in which ERG expression is found to be elevated.[92] Myeloproliferation and acute megakaryocytic leukemia were experimentally demonstrated in a genetically engineered Down syndrome mouse model Ts(17(16))65Dn.[92] Similarly, in cell culture system, over expression of ERG in erythroleukemia cell line, K562 induced erythroid to megakaryoblastic phenotype[91] suggesting a critical role for ERG in malignant hematologic disorders in Down syndrome. In addition, ERG promotes expansion of megakaryocytes from hematopoietic progenitor cells[93] and function as a megakaryocyte oncogene.[94]
In diverse neoplasms, ERG is either over expressed abnormally or fused to other genes due to chromosomal translocations and expressed as a chimeric protein. ERG gene fusions were initially described in Ewing's sarcoma (EWS) and acute myeloid leukemia (AML).[90,95] In a small subset (about 5-10%) of Ewing's sarcoma, EWS-ERG fusions resulted into a chimeric protein containing amino-terminal end of EWS and the carboxy-terminal ERG including the DNA binding ETS domain.[96] Majority (95%) of EWS fusion involve EWS and FLI, the closest homolog of the ERG.[97] Similarly, ERG fuses with TLS/FUS in certain acute myeloid leukemias.[98] These fusions generate chimeric proteins abnormally regulate downstream genes due to altered transactivation and DNA binding activities.
As noted above TMPRSS2-ERG fusions in CaP leading to androgen dependent expression of ERG are exclusive to prostate tumor cells. ERG regulates the expression of C-MYC, a widely studied oncogene, by physically interacting with the ETS binding element within the P2 promoter region.[71] Consistent with the above observations a positive correlation between ERG and C-MYC expression suggests that ERG mediates oncogenic process through C-MYC and may be one of the potential mechanisms in CaP. In addition to the positive regulation of C-MYC, ERG negatively regulates the expression of a number of prostate differentiation genes such as KLK3/ PSA, SLC45A3/Prostein and abrogates the prostate epithelial differentiation program.[71,99] Of note, knock-down of either ERG or C-MYC in TMPRSS2-ERG positive CaP cells showed similar effects on cellular morphology and expression of prostate differentiation related genes.[71]
In the majority of cancers, cell invasion and migration are the key features of aggressive nature of tumors towards metastasis. ERG regulates invasion and migration related genes in CaP such as MMP1, MMP3, MMP9, and ADAM19, the urokinase plasminogen activator (PLAU), and the plasminogen activator inhibitor type1 in CaP.[99–101] ERG enhances cell invasion and metastasis through regulating CXCR4, a chemokine receptor.[27,102] ERG also induces the expression of osteopontin (OPN) through ETS binding sequences within the promoter.[103] OPN, a member of a Small Integrin-Binding Ligand, N-linked Glycoprotein (SIBLING),and a key regulator of metastasis of a wide variety of cancers is up-regulated in several cancers including prostate. Phenotype of human prostate cancer such as metastasis has been correlated with increasing levels of OPN expression.[104]
Accumulating data suggests that ERG mediates epigenetic regulatory function[105] through EZH2, a polycomb group (PcG) protein in CaP.[106] EZH2 promotes cancer formation and progression through activation of oncogenic signaling cascades and inhibition of pro-differentiation pathways.[10] In CaP, NKX3.1 expression is negatively regulated by ERG induced EZH2 interactions.[106] Interestingly, NKX3.1 negatively regulates TMPRSS2 promoter that is frequently fused to ERG.[107] Therefore inhibition of NKX3.1 either by ERG/EZH2 or loss of NKX3.1 due to recurrent 8p21 deletions may fuel TMPRSS2 dependent ERG expression in CaP. Other epigenetic factors include histone acetyl transferases (HATs) and histone deacetylases (HDACs) which are frequently altered in majority of cancers including CaP.[108] ERG binds to and inhibits HAT activity to deregulate protein acetylation and also activates HDAC to deacetylate histone proteins.[109,110] Interestingly, ERG has been shown to play critical role in epithelial-to-mesenchymal transition (EMT) by repressing epithelial specific genes and inducing mesenchymal specific genes through WNT signaling components.[109,111] EMT has received considerable attention as a conceptual paradigm to explain invasive and metastatic behavior during cancer progression. During this transition, the epithelial cells lose their polarity and cohesiveness, acquiring migration and invasive properties.[112] Recent genome wide screening of ERG candidate genes and subsequent validation revealed ERG-enriched targets that include both canonical and non-canonical WNT signaling genes: WNT11, WNT2, WNT9A, CCND1 and FZD7.[113] Both ERG and WNT11 expression were elevated in high-grade prostate tumors.[114,115] FZD4, one of the members of WNT signaling pathway, is often co-expressed with ERG in clinical specimens. Down regulation of ERG or FZD4 releases the transcriptional block on both β1-Integrin and E-cadherin to maintain epithelial phenotype.[109] Interestingly, ERG also up regulates EMT facilitators such as ZEB1 and ZEB2 that negatively control the E-cadherin[111] potentially through SNAIL1 and 2 pathway in CaP.[116] Although EMT is not a prerequisite for invasive cancer development, this process can play an important role in cancer cell dissemination from the tumor due to altered expression of E-cadherins.
ERG has also been shown to interface with genes linked to inflammation and DNA damage repair pathways. ERG activates NF-kB pathway through toll-like receptor 4 suggesting for its role in inflammation related pathways.[117] 15-hydroxy-prostaglandin dehydrogenase (HPGD),a tumor suppressor and prostaglandin catabolizing enzyme, is down regulated in variety of cancers such as lung, colon, breast and bladder cancers. Recent studies have shown a potential link between ERG and prostaglandin signaling and inflammation pathways in which ERG down-regulates the HPGD expression to induce carcinogenesis.[118] Proteomics evaluations of ERG binding proteins show that ERG interacts with Poly (ADP-ribose) polymerase (PARP) and catalytic subunit of DNA protein kinase (DNAPKcs) in a DNA independent manner.[119] This complex formation is required for ETS gene mediated transcription and cell invasion. ERG induced DNA damage in CaP cells can further be potentiated by PARP1 inhibition, an observation similar to effects of these inhibitory compounds in breast cancer with BRCA1/2 mutations. As noted, most of studies addressing biochemical and cell biological functions of ERG in CaP have used VCaP cell line as this is the only well characterised TMPRSS2-ERG positive CaP cell line.[120] Since ERG downstream targets may be cellular context dependent, these data need to be interpreted with caution especially in cases when, findings have not been validated in human CaP specimens or complementary experimental models. Development of additional ERG positive CaP cell lines will also facilitate cell biologic evaluations of ERG.
Although, the presence of elevated expression of ERG in large number of CaP patients have been well characterized by several groups, it is not clear whether ERG is an initiating factor or expressed as a consequence of other aberrant genetic events. Towards this, several groups have developed ERG transgenic mice by prostate targeted expression of ERG driven by rat probasin promoter.[27,87,99,101] Prostatic intraepithelial neoplasia (PIN), a pre-invasive lesions of CaP was reported in the prostates of transgenic mice, which surprisingly did not progress to adenocarcinoma.[99,101] On the contrary, other studies did not observe PIN phenotype, however, developed of adenocarcinoma in combination with either phospho AKT overexpression or with loss of PTEN.[27,87] Similarly, in prostate tissue dissociation/ regeneration system, high levels of ERG expression could induce the initiation of neoplastic transformation of adult prostate epithelial cells and further developed adenocarcinoma in combination with pAKT or AR.[121] Recent evaluations of the association TMPRSS2-ERGfusion with other genomic alterations in human CaP revealed significant associations with deletions of chromosomal regions, 10q23.31 and 17p13.1 harboring PTEN and p53 respectively.[122] Further, ERG fusions showed an intriguing association with CaP specific focal deletion of 3p14.1-p13 harboring several candidate TSGs.[122] While cooperation of ERG with PTEN/p-AKT has been shown in enhancing prostate tumorigenesis, interaction of ERG with other cancer genes needs to be further defined in engineered mouse models. Taken together, the studies focusing on ERG functions provide an emerging picture of the ERG network involved in the regulation of differentiation, cell invasion, epigenetic control, EMT inflammation and DNA damage, all of these support the biological role of ERG in CaP [Figure 2]. Further, interactions/cooperation of ERG with genes (AR, C-MYC, NKX3.1 and PI3K/PTEN axis) functionally significant in CaP, defines potential role of ERG in common CaP pathways. These findings have potential to provide new therapeutic approaches for CaP.
Figure 2.
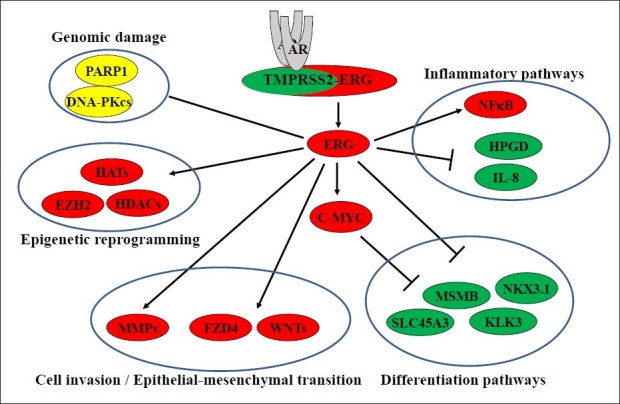
ERG regulated prostate cancer pathways.ERG regulates the expression of target genes associated with cancer initiation and progression pathways such as DNA damage, inflammation,epigenetic control,regulation of differentiation, EMT, cell proliferation and cell invasion. (Red: upregulated;Green:down regulated;Yellow: protein-protein interactions)
ERG as diagnostic/prognostic marker for prostate cancer
Detection of gene fusions has led to a paradigm shift in the diagnosis, classification, and treatment options for hematologic cancers.[123–125] These gene fusions provide CaP specific markers which have promise in improving diagnosis, as well as molecular classification of prostate tumors.[126,127] The feasibility of detecting TMPRSS2-ERG fusion by FISH in prostate biopsies and prostatectomy specimens enhances the detection of CaP in diagnostic and prognostic settings.[128–131] The clinical value of ERG fusion in prostate biopsies needs to be further explored and validated in larger prospective studies.
Interrogation of the presence of TMPRSS2-ERG fusion or ERG mRNA in CaP was initially believed to provide prognostic information. However, in retrospective prostatectomy cohorts conflicting results have been reported regarding associations between ETS fusions and cancer aggressiveness.[11,48] For example, presence of TMPRSS2-ERG fusion predicted cancer recurrence after surgery or lethal outcome in a watchful waiting cohort.[79,132] However, association of the fusion or ERG expression with favorable outcome was also reported.[39,133,134] Since ERG expression in CaP is androgen dependent due to TMPRSS2-ERG fusion, alterations of AR transcription factor activity may result in altered ERG mRNA expression as noted in poorly differentiated tumors.[135] These data also suggest that ERG in combination with a panel of androgen receptor regulated genes (PSA, PMEPA1, NKX3.1, ODC, AMD) may serve as a biomarker panel for Androgen Receptor Function Index (ARFI) in CaP. Thus, ARFI may provide new opportunities in AR function based stratification of CaP, where ERG expression evaluation could play important role in over half of CaP.[135] These findings may provide potential biologic basis for initial observations on association of decreased or no ERG mRNA expression with poor prognosis of CaP.[39] TMPRSS2-ERG fusion isoforms have variable tumor promoting biological activities and certain isoforms are correlated well with more aggressive disease[55] and others with favorable prognosis.[136] Similarly, the ratios of full length splice forms type I and type II also shown to have prognostic association.[81] However, some studies have reported no significant association of TMPRSS2-ERG fusion or ERG expression with disease progression after prostatectomy.[83,137,138] Therefore, larger and better designed studies are needed for further clarification. The observations of combination of TMPRSS2-ERG fusion and PTEN deletions associating with poorer prognosis have been supported with functional studies showing cooperation of these genes in mouse models of CaP.[27,87,121,139] Further assessment of the utility of combinatorial prognostic markers is warranted.
Utility of detection of TMPRSS2-ERG fusion or ERG transcripts in post-digital rectal examination (post-DRE) urine are also being evaluated for improving CaP diagnosis using minimally invasive assays.[140–142] Promising results from evaluations of highly CaP specific non-coding RNA, PCA3, in post-DRE urine specimens, have led the way for evaluation of additional CaP specific expression markers.[143–145] A CaP gene panel (PCA3, ERG and AMACR) with diagnostic potential in which overexpression of at least one of three genes associated with virtually all of the LCM derived prostate tumor specimens suggested for careful evaluation of such panels in post-DRE urine.[39] Evaluation of ERG[141] or TMPRSS2-ERG[140] transcripts in post-DRE urine have provided promising data on diagnostic potential of ERG in this minimally invasive bio-specimen. A recent multi-center study of 1312 men showed promising data with respect to association of TMPRSS2-ERG in post-DRE urine with clinically significant CaP.[142] This study further showed utility of the combination of TMPRSS2-ERG and PCA3 in post-DRE urine in comparison to serum PSA for detecting clinically significant CaP in specimens.[142]
New insights into detection of ERG oncoprotein in prostate cancer
Accurate molecular analysis of ERG oncoprotein in CaP has been a challenge as ETS family of proteins share high homology among the family members. Recent development and evaluation anti-ERG monoclonal antibodies have paved the way for evaluation of ERG protein in routine pathologic specimens. Through exhaustive analysis of 132 whole-mount prostates sections (261 tumor foci and over 200,000 benign glands) for the ERG oncoprotein nuclear expression by an anti-ERG mouse monoclonal antibody (clone 9FY), this study demonstrated 99.9 % specificity for detecting tumor cells in prostate.[138] The ERG oncoprotein expression correlated well with fusion transcript or gene fusion in selected specimens. Strong concordance of ERG positive prostatic intraepithelial neoplasia (PIN) lesions with ERG positive carcinoma (82 out of 85 sections with PIN, 96.5%) affirmed the biological role of ERG in clonal selection of prostate tumors in 65% (86 out of 132) of patients[138] [Figure 3]. These observations lend a support to the functional role of ERG in initiation of preneoplastic lesions.[99,101] Evaluations of anti-ERG rabbit monoclonal antibody (EPR 3864) in CaP tissue microarrays from 207 established correlation between detection of ERG protein expression by IHC and ERG rearrangement by using fluorescence in situ hybridization (FISH). Detection of the ERG protein expression in CaP exhibited 95.7% sensitivity and 96.5% for the presence ERG rearrangement. Further, presence of ERG protein in CaP also correlated with less common ERG rearrangements. Since ERG expression is almost exclusive to prostate tumor cells and IHC is easier to perform in comparison to FISH. It is expected that ERG protein detection in pathologic specimens will greatly facilitate the evaluations of biological and clinical utility of ERG antibodies in CaP. Among the currently known CaP biomarkers, detection ERG oncoprotein offers unprecedented opportunities in the diagnostic setting [Figure 4]. With the availability of highly specific ERG monoclonal antibodies, better and more effective monitoring, treatment, and therapies may also be available in future to patients with CaP.[146,147]
Figure 3.
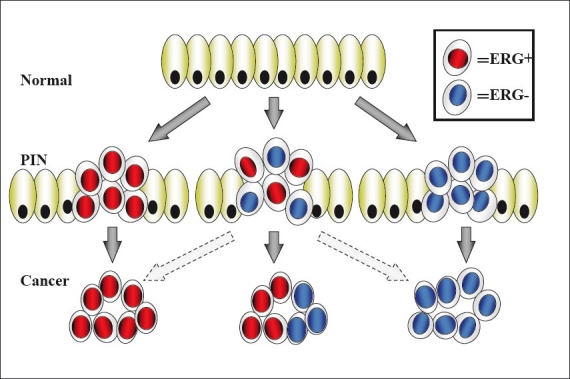
ERG-dependent Clonal Selection of Prostate Tumors. Model describing the ERG-dependent clonal selection of prostate tumors from prostatic intraepithelial neoplasia (PIN) to prostate cancer. Other precursor lesions which may not progress through the PIN morphological stage are not represented by this model. Normal prostate epithelial cells are marked by green color
Figure 4.
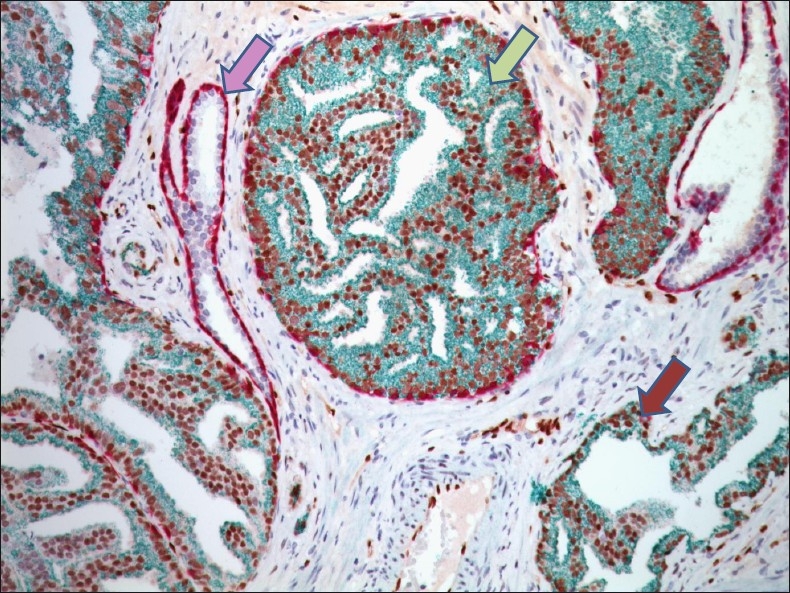
Detection of PIN and prostatic adenocarcinoma by the combination of ERG, AMACR, p63 and CK5 markers in immunohistochemistry.Tumor cells are positive for nuclear ERG (brown) and cytoplasmic AMACR (green), whereas, absence of p63 (purple) and CK5 (purple) indicate the lack of basal cell layer. By contrast, in normal prostatic glands prominent staining with p63 and CK5 distinctively demarcate intact basal cell layer.In PIN disrupted basal cell layer and prominent ERG and AMACR staining is apparent (×400). (Image: Courtesy of Dr. David Tacha, Biocare Medical Inc, Concord, CA, USA)
Since ERG MAb 9FY is highly ERG specific as illustrated by lack of recognition of its closest homolog, FLI,[58,138] the presence of ERG protein in hemangiomas, lymphangiomas, angiosarcomas, epithelioid hemangio-endotheliomas and Kaposi sarcomas[148] serve as an excellent new marker for vascular tumors. Similar studies are also warranted in Acute Myeloid leukemia where ERG has been suggested as prognostic marker based on mRNA based studies.[58,148]
New therapeutic opportunities targeting ERG in prostate cancer
Studies have shown growth inhibitory effects of the ERG si RNA in TMPRSS2-ERG positive VCaP cells and VCaP derived tumors in SCID mice suggesting for therapeutic potential of ERG inhibition in CaP.[66,71] Further, these mechanistic data delineated the effects of ERG siRNA through inhibition of C-MYC and induction of prostate epithelial cell differentiation markers.[71] Recent reports in transgenic mice have shown cooperative effects of ERG overexpression with PTEN/PI3K axis alteartions, leading to progressive features of CaP.[27,87] Thus targeting the inhibition of ERG pathway may provide a promising therapeutic strategy. In addition to siRNA as a potential molecule to interfere with the ERG expression, YK-4-279, a derivative of the lead compound from the small molecule screen, has proven to effectively bind to ERG and subsequently down regulate its transcriptional activity as well as tumor cell invasion in cell culture model.[149,150] Inhibitors of HDACs are currently being considered as one of the potent anti-cancer agents. HDAC inhibitors, such as SAHA, MS-275, TSA and VPA have been evaluated both in vitro and in vivo prostate cancer models[108] and in a number of clinical trials.[151] HDAC inhibitors (VPA, TSA) induce apoptosis of prostate cancer cells (VCaP) through up-regulation of p21/Waf1/CIP1 pathway. These inhibitors alsodown-regulate TMPRSS2-ERG and alter the acetylation status of p53.[110] Targeting nuclear transcription factors is often difficult in designing therapeutic strategies; hence, targeting components of the “ERG Network” may serve as an effective alternative strategy to combat the CaP. Recent findings showed physical interaction of ERG protein with PARP in inducing DNA damage and inhibition of PARP impaired ERG mediated cell invasion and tumorigenesis.[119] These findings suggest a promising therapeutic potential for PARP inhibitors for a large subset of CaP harboring oncogenic activation of the ERG or ETV1. In recent years, PARP inhibitors have been increasingly considered as a viable option in exploiting the DNA-repair defects of BRCA1/2-deficient tumors to induce cell death.[152–154] As CaP is heterogeneous and potentially involves multiple molecular pathways leading to complex phenotypes, development of small molecule inhibitors targeting multiple targets (AR, ERG, PARP, PTEN, PI3K, AKT and mTOR) may incorporate new therapeutic strategies for CaP.[155,156] Importantly, ERG network targeted therapy may be an effective strategy for more than half of CaP in early stages when cancer cells may be more responsive to treatment.
Concluding remarks
Androgen dependent expression of ERG transcription factor as a result of TMPRSS2-ERG fusion is detected in 50-70% of CaP patients in Western countries. Evaluations of ERG fusions represent one of the most studied and validated genomic alterations in CaP. Other gene fusions are low frequency events in CaP and need to be better understood. Since ERG fusions described in CaP are highly specific to this cancer type, numerous studies have evaluated clinical utility of ERG as a diagnostic or prognostic biomarker in CaP. Detection of ERG rearrangement by FISH or immunostaining of ERG protein has been streamlined in pathologic specimens and results from these studies suggest the role of ERG in clonal expansion of ERG positive PIN (pre-invasive lesion) to carcinoma. While ERG alteration is homogenous with in a tumor focus, heterogeneity of ERG alteration is apparent in mutli-focal tumor context by simultaneous presence of ERG positive and negative tumor foci in the malignant prostate of a patient. Detection of ERG alterations in tissue or urine based assays have promise in improving prostate cancer diagnosis and continued investigations are anticipated along these lines. Prognostic value of TMPRSS2-ERG fusion or ERG protein expression is uncertain, however, combination of ERG alteration with other CaP gene alterations such as PTEN may define prognostic marker panels for progressive disease. Additional studies are also warranted to further assess the prognostic properties of specific ERG fusion type or relative abundance of type I and II splice ERG splice variants in CaP. ERG mRNA or ERG protein expression may serve as a surrogate of AR functional status in prostate tumors and therefore evaluation of ERG mRNA or protein expression in prostate tumors has potential in companion diagnostic setting for therapeutics targeting androgen/AR axis.
Functional evaluations of ERG in experimental models suggest causal role of ERG oncogenic activation in prostate tumorigenesis. ERG induces pre-invasive lesions and ERG in combination with PTEN loss, AKT or AR cooperate in neoplastic transformation. ERG knock-down inhibits prostate cancer cell growth. Studies focusing on ERG transcriptional targets in prostate cancer cells suggest role of ERG in regulating genes involved in oncogegnesis, differentiation, cell invasion, DNA damage, epigenetic control, inflammation and epithelial-mesenchyme transition. The emerging “ERG network” defines new facets of ERG functions in CaP and underscores the functional interface of ERG with genes (AR, C-MYC, NKX3.1, and PI3K/PTEN axis) known to have critical functions in CaP. Studies focusing on therapeutic targeting of ERG or its network are promising as shown by therapeutic potential of PARP inhibitors for ERG and ETV1 positive tumors in preclinical models. Taken together, strategies developing ERG based biological classification of prostate tumors and therapeutic targeting of the ERG network in prostate cancer represent new paradigm in prostate cancer stratification and treatment.
Author's Profile
Dr. Taduru L Sreenath, Center for Prostate Disease Research, Department of Surgery, Uniformed Services University of the Health Sciences 1530 East Jefferson Street Rockville, MD 20852
Dr. Albert Dobi, Center for Prostate Disease Research, Department of Surgery, Uniformed Services University of the Health Sciences 1530 East Jefferson Street Rockville, MD 20852
Dr. Gyorgy Petrovics, Center for Prostate Disease Research, Department of Surgery, Uniformed Services University of the Health Sciences 1530 East Jefferson Street Rockville, MD 20852
Dr. Shiv Srivastava, Center for Prostate Disease Research, Department of Surgery, Uniformed Services University of the Health Sciences 1530 East Jefferson Street Rockville, MD 20852
ACKNOWLEDGMENTS
The authors would like to thank Ms Tia Morris for editing the manuscript. Special thanks to Dr. David Tacha, Biocare Medical Inc, Concord, CA, USA for the multi-color image of ERG-MAb (clone 9FY), AMACR, CK5, p63 immunohistochemistry. The views expressed in this manuscript are those of the authors, and do not reflect the official policy of the Department of the Army, Department of Defense or the US Government.
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Williams H, Powell IJ. Epidemiology, pathology, and genetics of prostate cancer among African Americans compared with other ethnicities. Methods Mol Biol. 2009;472:439–53. doi: 10.1007/978-1-60327-492-0_21. [DOI] [PubMed] [Google Scholar]
- 3.Zeigler-Johnson CM, Rennert H, Mittal RD, Jalloh M, Sachdeva R, Malkowicz SB, et al. Evaluation of prostate cancer characteristics in four populations worldwide. Can J Urol. 2008;15:4056–64. [PMC free article] [PubMed] [Google Scholar]
- 4.Kheirandish P, Chinegwundoh F. Ethnic differences in prostate cancer. Br J Cancer. 2011;105:481–5. doi: 10.1038/bjc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeigler-Johnson CM, Spangler E, Jalloh M, Gueye SM, Rennert H, Rebbeck TR. Genetic susceptibility to prostate cancer in men of African descent: implications for global disparities in incidence and outcomes. Can J Urol. 2008;15:3872–82. [PMC free article] [PubMed] [Google Scholar]
- 6.LangebERG WJ, Isaacs WB, Stanford JL. Genetic etiology of hereditary prostate cancer. Front Biosci. 2007;12:4101–10. doi: 10.2741/2374. [DOI] [PubMed] [Google Scholar]
- 7.Sartor AO, Hricak H, Wheeler TM, Coleman J, Penson DF, Carroll PR, et al. Evaluating localized prostate cancer and identifying candidates for focal therapy. Urology. 2008;72(6 Suppl):S12–24. doi: 10.1016/j.urology.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 8.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Grönberg H, Drake CG, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–69. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter E, Srivastava S, Dobi A. Androgen receptor and prostate cancer. Prostate Cancer Prostatic Dis. 2007;10:114–8. doi: 10.1038/sj.pcan.4500936. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knudsen BS, Vasioukhin V. Mechanisms of prostate cancer initiation and progression. Adv Cancer Res. 2010;109:1–50. doi: 10.1016/B978-0-12-380890-5.00001-6. [DOI] [PubMed] [Google Scholar]
- 13.Salinas CA, Kwon E, Carlson CS, Koopmeiners JS, Feng Z, Karyadi DM, et al. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1203–13. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 14.Kral M, Rosinska V, Student V, Grepl M, Hrabec M, Bouchal J. Genetic determinants of prostate cancer: a review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2011;155:3–9. doi: 10.5507/bp.155.2011.001. [DOI] [PubMed] [Google Scholar]
- 15.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz MM, Freedman ML. Genetics of prostate cancer risk. Mt Sinai J Med. 2010;77:643–54. doi: 10.1002/msj.20222. [DOI] [PubMed] [Google Scholar]
- 17.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 18.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 20.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 21.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple Loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitman EJ, Pomerantz M, Chen Y, Chamberlin MM, Furusato B, Gao C, et al. Prostate cancer risk allele specific for African descent associates with pathologic stage at prostatectomy. Cancer Epidemiol Biomarkers Prev. 2010;19:1–8. doi: 10.1158/1055-9965.EPI-08-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomerantz MM, Shrestha Y, Flavin RJ, Regan MM, Penney KL, Mucci LA, et al. Analysis of the 10q11 cancer risk locus implicates MSMB and NCOA4 in human prostate tumorigenesis. PLoS Genet. 2010;6:e1001204. doi: 10.1371/journal.pgen.1001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 25.Shah S, Small E. Emerging biological observations in prostate cancer. Expert Rev Anticancer Ther. 2010;10:89–101. doi: 10.1586/era.09.161. [DOI] [PubMed] [Google Scholar]
- 26.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong JT. Prevalent mutations in prostate cancer. J Cell Biochem. 2006;97:433–47. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- 30.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 32.Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–27. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa VL, Henrique R, Jerónimo C. Epigenetic markers for molecular detection of prostate cancer. Dis Markers. 2007;23:31–41. doi: 10.1155/2007/356742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins TG, Burns PA, Routledge MN. DNA methylation of GSTP1 as biomarker in diagnosis of prostate cancer. Urology. 2007;69:11–6. doi: 10.1016/j.urology.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Humphrey PA. Diagnosis of adenocarcinoma in prostate needle biopsy tissue. J Clin Pathol. 2007;60:35–42. doi: 10.1136/jcp.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301:1–6. doi: 10.1016/j.canlet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, et al. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: A gene expression analysis on total and microdissected prostate tissue. Am J Pathol. 2002;160:2169–80. doi: 10.1016/S0002-9440(10)61165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–82. [PubMed] [Google Scholar]
- 39.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–52. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 40.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 41.Svensson MA, LaFargue CJ, MacDonald TY, Pflueger D, Kitabayashi N, Santa-Cruz AM, et al. Testing mutual exclusivity of ETS rearranged prostate cancer. Lab Invest. 2011;91:404–12. doi: 10.1038/labinvest.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4. [PubMed] [Google Scholar]
- 44.Vaarala MH, Porvari KS, Kellokumpu S, Kyllönen AP, Vihko PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–40. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 45.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–75. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–39. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 49.Reddy ES, Rao VN, Papas TS. The ERG gene: A 2 human gene related to the ets oncogene. Proc Natl Acad Sci U S A. 1987;84:6131–5. doi: 10.1073/pnas.84.17.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao VN, Papas TS, Reddy ES. erg, a human ets-related gene on chromosome 21: alternative splicing, polyadenylation, and translation. Science. 1987;237:635–9. doi: 10.1126/science.3299708. [DOI] [PubMed] [Google Scholar]
- 51.Watson DK, Turner DP, Scheiber MN, Findlay VJ, Watson PM. ETS transcription factor expression and conversion during prostate and breast cancer progression. Open Cancer J. 2010;3:24–39. [Google Scholar]
- 52.Seth A, Watson DK. Ets transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Owczarek CM, Portbury KJ, Hardy MP, O’Leary DA, Kudoh J, Shibuya K, et al. Detailed mapping of the ERG-ETS2 interval of human chromosome 21 and comparison with the region of conserved synteny on mouse chromosome 16. Gene. 2004;324:65–77. doi: 10.1016/j.gene.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 54.Hewett PW, Nishi K, Daft EL, Clifford Murray J. Selective expression of ERG isoforms in human endothelial cells. Int J Biochem Cell Biol. 2001;33:347–55. doi: 10.1016/s1357-2725(01)00022-x. [DOI] [PubMed] [Google Scholar]
- 55.Duterque-Coquillaud M, Niel C, Plaza S, Stehelin D. New human ERG isoforms generated by alternative splicing are transcriptional activators. Oncogene. 1993;8:1865–73. [PubMed] [Google Scholar]
- 56.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. Embo J. 2010;29:2147–60. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M. The Ets family member ERG gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev. 2000;91:331–5. doi: 10.1016/s0925-4773(99)00272-5. [DOI] [PubMed] [Google Scholar]
- 58.Mohamed AA, Tan SH, Mikhalkevich N, Ponniah S, Vasioukhin V, Bieberich CJ, et al. Ets family protein, ERG expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. doi: 10.7150/jca.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 60.Rivera RR, Stuiver MH, Steenbergen R, Murre C. Ets proteins: new factors that regulate immunoglobulin heavy-chain gene expression. Mol Cell Biol. 1993;13:7163–9. doi: 10.1128/mcb.13.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLaughlin F, Ludbrook VJ, Cox J, von Carlowitz I, Brown S, Randi AM. Combined genomic and antisense analysis reveals that the transcription factor ERG is implicated in endothelial cell differentiation. Blood. 2001;98:3332–9. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- 62.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, et al. Transcription factor ERG regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwamoto M, Tamamura Y, Koyama E, Komori T, Takeshita N, Williams JA, et al. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol. 2007;305:40–51. doi: 10.1016/j.ydbio.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson MK, Hernandez-Hoyos G, Diamond RA, RothenbERG EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–48. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 65.Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–42. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–24. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhordain P, Dewitte F, Desbiens X, Stehelin D, Duterque-Coquillaud M. Mesodermal expression of the chicken ERG gene associated with precartilaginous condensation and cartilage differentiation. Mech Dev. 1995;50:17–28. doi: 10.1016/0925-4773(94)00322-e. [DOI] [PubMed] [Google Scholar]
- 68.Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–33. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 69.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–83. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, et al. The transcription factor ERG is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–9. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 71.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–53. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan L, Nikolova-Krstevski V, Zhan Y, Kondo M, Bhasin M, Varghese L, et al. Antiinflammatory effects of the ETS factor ERG in endothelial cells are mediated through transcriptional repression of the interleukin-8 gene. Circ Res. 2009;104:1049–57. doi: 10.1161/CIRCRESAHA.108.190751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pimanda JE, Chan WY, Donaldson IJ, Bowen M, Green AR, Göttgens B. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a -8-kb enhancer. Blood. 2006;107:4737–45. doi: 10.1182/blood-2005-12-4929. [DOI] [PubMed] [Google Scholar]
- 74.Im YH, Kim HT, Lee C, Poulin D, Welford S, Sorensen PH, et al. EWS-FLI1, EWS-ERG, and EWS-ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor beta type II receptor gene. Cancer Res. 2000;60:1536–40. [PubMed] [Google Scholar]
- 75.Taoudi S, Bee T, Hilton A, Knezevic K, Scott J, Willson TA, et al. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 2011;25:251–62. doi: 10.1101/gad.2009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ng AP, Loughran SJ, Metcalf D, Hyland CD, de Graaf CA, Hu Y, et al. ERG is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood. 2011;118:2454–61. doi: 10.1182/blood-2011-03-344739. [DOI] [PubMed] [Google Scholar]
- 77.Nikolova-Krstevski V, Yuan L, Le Bras A, Vijayaraj P, Kondo M, Gebauer I, et al. ERG is required for the differentiation of embryonic stem cells along the endothelial lineage. BMC Dev Biol. 2009;9:72. doi: 10.1186/1471-213X-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–24. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–5. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clark JP, Munson KW, Gu JW, Lamparska-Kupsik K, Chan KG, Yoshida JS, et al. Performance of a single assay for both type III and type VI TMPRSS2: ERG fusions in noninvasive prediction of prostate biopsy outcome. Clin Chem. 2008;54:2007–17. doi: 10.1373/clinchem.2008.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu Y, Dobi A, Sreenath T, Cook C, Tadase AY, Ravindranath L, et al. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin Cancer Res. 2008;14:4719–25. doi: 10.1158/1078-0432.CCR-08-0531. [DOI] [PubMed] [Google Scholar]
- 82.Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 83.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–73. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 85.Furusato B, Shaheduzzaman S, Petrovics G, Dobi A, Seifert M, Ravindranath L, et al. Transcriptome analyses of benign and malignant prostate epithelial cells in formalin-fixed paraffin-embedded whole-mounted radical prostatectomy specimens. Prostate Cancer Prostatic Dis. 2008;11:194–7. doi: 10.1038/sj.pcan.4501007. [DOI] [PubMed] [Google Scholar]
- 86.Andreoiu M, Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Hum Pathol. 2010;41:781–93. doi: 10.1016/j.humpath.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 87.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hart AH, Corrick CM, Tymms MJ, Hertzog PJ, Kola I. Human ERG is a proto-oncogene with mitogenic and transforming activity. Oncogene. 1995;10:1423–30. [PubMed] [Google Scholar]
- 89.Sementchenko VI, Schweinfest CW, Papas TS, Watson DK. ETS2 function is required to maintain the transformed state of human prostate cancer cells. Oncogene. 1998;17:2883–8. doi: 10.1038/sj.onc.1202220. [DOI] [PubMed] [Google Scholar]
- 90.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 91.Rainis L, Toki T, Pimanda JE, Rosenthal E, Machol K, Strehl S, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 2005;65:7596–602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 92.Ng AP, Hyland CD, Metcalf D, Carmichael CL, Loughran SJ, Di Rago L, et al. Trisomy of ERG is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115:3966–9. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- 93.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–47. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salek-Ardakani S, Smooha G, de Boer J, Sebire NJ, Morrow M, Rainis L, et al. ERG is a megakaryocytic oncogene. Cancer Res. 2009;69:4665–73. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 95.Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C, et al. igh expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J Clin Oncol. 2007;25:3337–43. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 96.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–51. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 97.Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, Papas TS, et al. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3:705–13. [PubMed] [Google Scholar]
- 98.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–8. [PubMed] [Google Scholar]
- 99.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Butticè G, Duterque-Coquillaud M, Basuyaux JP, Carrère S, Kurkinen M, Stéhelin D. Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–306. [PubMed] [Google Scholar]
- 101.Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai J, Kandagatla P, Singareddy R, Kropinski A, Sheng S, Cher ML, et al. Androgens Induce Functional CXCR4 through ERG Factor Expression in TMPRSS2-ERG Fusion-Positive Prostate Cancer Cells. Transl Oncol. 2010;3:195–203. doi: 10.1593/tlo.09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flajollet S, Tian TV, Flourens A, Tomavo N, Villers A, Bonnelye E, et al. Abnormal expression of the ERG transcription factor in prostate cancer cells activates osteopontin. Mol Cancer Res. 2011;9:914–24. doi: 10.1158/1541-7786.MCR-10-0537. [DOI] [PubMed] [Google Scholar]
- 104.Thalmann GN, Sikes RA, Devoll RE, Kiefer JA, Markwalder R, Klima I, et al. Osteopontin: possible role in prostate cancer progression. Clin Cancer Res. 1999;5:2271–7. [PubMed] [Google Scholar]
- 105.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150:3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS ONE. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dobi A, Thangapazham R, Srivastava S. San Diego, CA: 2008. Apr 12-16, Conservation of NKX binding sites within the TMPRSS2 promoter and hormone regulation of the ERG gene expression in prostate cancer (Abstract). In: Proceedings of the 99th Annual Meeting of the American Association for Cancer Research. Poster# 60, 2008. [Google Scholar]
- 108.Abbas A, Gupta S. The role of histone deacetylases in prostate cancer. Epigenetics. 2008;3:300–9. doi: 10.4161/epi.3.6.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, et al. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–45. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 110.Fortson WS, Kayarthodi S, Fujimura Y, Xu H, Matthews R, Grizzle WE, et al. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. Int J Oncol. 2011;39:111–9. doi: 10.3892/ijo.2011.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R, et al. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS ONE. 2011;6:e21650. doi: 10.1371/journal.pone.0021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 113.Mochmann LH, Bock J, Ortiz-Tánchez J, Schlee C, Bohne A, Neumann K, et al. Genome-wide screen reveals WNT11, a non-canonical WNT gene, as a direct target of ETS transcription factor ERG. Oncogene. 2011;30:2044–56. doi: 10.1038/onc.2010.582. [DOI] [PubMed] [Google Scholar]
- 114.Zhu H, Mazor M, Kawano Y, Walker MM, Leung HY, Armstrong K, et al. Analysis of Wnt gene expression in prostate cancer: mutual inhibition by WNT11 and the androgen receptor. Cancer Res. 2004;64:7918–26. doi: 10.1158/0008-5472.CAN-04-2704. [DOI] [PubMed] [Google Scholar]
- 115.Uysal-Onganer P, Kawano Y, Caro M, Walker MM, Diez S, Darrington RS, et al. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol Cancer. 2010;9:55. doi: 10.1186/1476-4598-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Drake JM, Strohbehn G, Bair TB, Moreland JG, Henry MD. ZEB1 enhances transendothelial migration and represses the epithelial phenotype of prostate cancer cells. Mol Biol Cell. 2009;20:2207–17. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R, et al. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res. 2011;71:1325–33. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mohamed AA, Tan SH, Sun C, Shaheduzzaman S, Hu Y, Petrovics G, et al. ERG oncogene modulates prostaglandin signaling in prostate cancer cells. Cancer Biol Ther. 2011;11:410–7. doi: 10.4161/cbt.11.4.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–8. [PubMed] [Google Scholar]
- 121.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465–70. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Netto GJ, Epstein JI. Theranostic and prognostic biomarkers: genomic applications in urological malignancies. Pathology. 2010;42:384–94. doi: 10.3109/00313021003779145. [DOI] [PubMed] [Google Scholar]
- 124.Netto GJ. Molecular diagnostics in urologic malignancies: a work in progress. Arch Pathol Lab Med. 2011;135:610–21. doi: 10.5858/2010-0727-RAIR.1. [DOI] [PubMed] [Google Scholar]
- 125.Rowley JD. Chromosomes in leukemia and beyond: from irrelevant to central players. Annu Rev Genomics Hum Genet. 2009;10:1–18. doi: 10.1146/annurev-genom-082908-150144. [DOI] [PubMed] [Google Scholar]
- 126.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–68. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leman ES, GetzenbERG RH. Biomarkers for prostate cancer. J Cell Biochem. 2009;108:3–9. doi: 10.1002/jcb.22227. [DOI] [PubMed] [Google Scholar]
- 128.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 129.Yaskiv O, Zhang X, Simmerman K, Daly T, He H, Falzarano S, et al. The utility of ERG/P63 double immunohistochemical staining in the diagnosis of limited cancer in prostate needle biopsies. Am J Surg Pathol. 2011;35:1062–8. doi: 10.1097/PAS.0b013e318215cc03. [DOI] [PubMed] [Google Scholar]
- 130.Attard G, de Bono JS, Clark J, Cooper CS. Studies of TMPRSS2-ERG gene fusions in diagnostic trans-rectal prostate biopsies. Clin Cancer Res. 2010;16:1340. doi: 10.1158/1078-0432.CCR-09-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–11. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Demichelis F, Fall K, Perner S, Andrén O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 133.Saramäki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG Fusion Identifies a Subgroup of Prostate Cancers with a Favorable Prognosis. Clin Cancer Res. 2008;14:3395–400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 134.Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–6. [PubMed] [Google Scholar]
- 135.Dobi A, Furusato B, Shaheduzzaman S, Chen Y, Vahey M, Nydem T, et al. ERG Expression Levels in Prostate Tumors Reflect Functional Status of the Androgen Receptor (AR) as a Consequence of Fusion of ERG with AR Regulated Gene Promoters. Open Cancer J. 2010;3:101–8. [Google Scholar]
- 136.Boormans JL, Porkka K, Visakorpi T, Trapman J. Confirmation of the association of TMPRSS2(exon 0):ERG expression and a favorable prognosis of primary prostate cancer. Eur Urol. 2011;60:183–4. doi: 10.1016/j.eururo.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 137.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG Status Is Unrelated to PSA Recurrence in Radically Operated Prostate Cancer in the Absence of Antihormonal Therapy. Clin Cancer Res. 2011;17:5878–88. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 138.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–37. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Squire JA. TMPRSS2-ERG and PTEN loss in prostate cancer. Nat Genet. 2009;41:509–10. doi: 10.1038/ng0509-509. [DOI] [PubMed] [Google Scholar]
- 140.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–9. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rice KR, Chen Y, Ali A, Whitman EJ, Blase A, Ibrahim M, et al. Evaluation of the ETS-related gene mRNA in urine for the detection of prostate cancer. Clin Cancer Res. 2010;16:1572–6. doi: 10.1158/1078-0432.CCR-09-2191. [DOI] [PubMed] [Google Scholar]
- 142.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.van Gils MP, Hessels D, Peelen WP, Vergunst H, Mulders PF, Schalken JA. Preliminary evaluation of the effect of dutasteride on PCA3 in post-DRE urine sediments: a randomized, open-label, parallel-group pilot study. Prostate. 2009;69:1624–34. doi: 10.1002/pros.21011. [DOI] [PubMed] [Google Scholar]
- 144.Hessels D, van Gils MP, van Hooij O, Jannink SA, Witjes JA, Verhaegh GW, et al. Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. Prostate. 2010;70:10–6. doi: 10.1002/pros.21032. [DOI] [PubMed] [Google Scholar]
- 145.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol. 2011;8:123–4. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 146.McLeod DG, Dobi A. Words of wisdom.Re: Antibody-Based Detection of ERG Rearrangement-Positive Prostate Cancer. Eur Urol. 2011;60:1123. doi: 10.1016/j.eururo.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 147.Falzarano SM, Zhou M, Carver P, Tsuzuki T, Simmerman K, He H, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;459:441–7. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 148.Miettinen M, Wang ZF, Paetau A, Tan SH, Dobi A, Srivastava S, et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35:432–41. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-RotenbERG JS, Yuan L, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat Med. 2009;15:750–6. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rahim S, Beauchamp EM, Kong Y, Brown ML, Toretsky JA, Üren A. YK-4-279 inhibits ERG and ETV1 mediated prostate cancer cell invasion. PLoS ONE. 2011;6:e19343. doi: 10.1371/journal.pone.0019343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–9. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 152.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leung M, Rosen D, Fields S, Cesano A, Budman DR. Poly(ADP-ribose) polymerase-1 inhibition: preclinical and clinical development of synthetic lethality. Mol Med. 2011;17:854–62. doi: 10.2119/molmed.2010.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sandhu SK, Yap TA, de Bono JS. The Emerging Role of Poly(ADP-Ribose) Polymerase Inhibitors in Cancer Treatment. Curr Drug Targets. 2011 doi: 10.2174/138945011798829438. [In Press] [DOI] [PubMed] [Google Scholar]
- 155.Priolo C, Oh WK, Loda M. Novel therapeutic strategies in prostate cancer: establishing a stratification system for patient selection in targeted trials. IDrugs. 2009;12:165–8. [PubMed] [Google Scholar]
- 156.Mostaghel EA, Montgomery B, Nelson PS. Castration-resistant prostate cancer: targeting androgen metabolic pathways in recurrent disease. Urol Oncol. 2009;27:251–7. doi: 10.1016/j.urolonc.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]


