Abstract
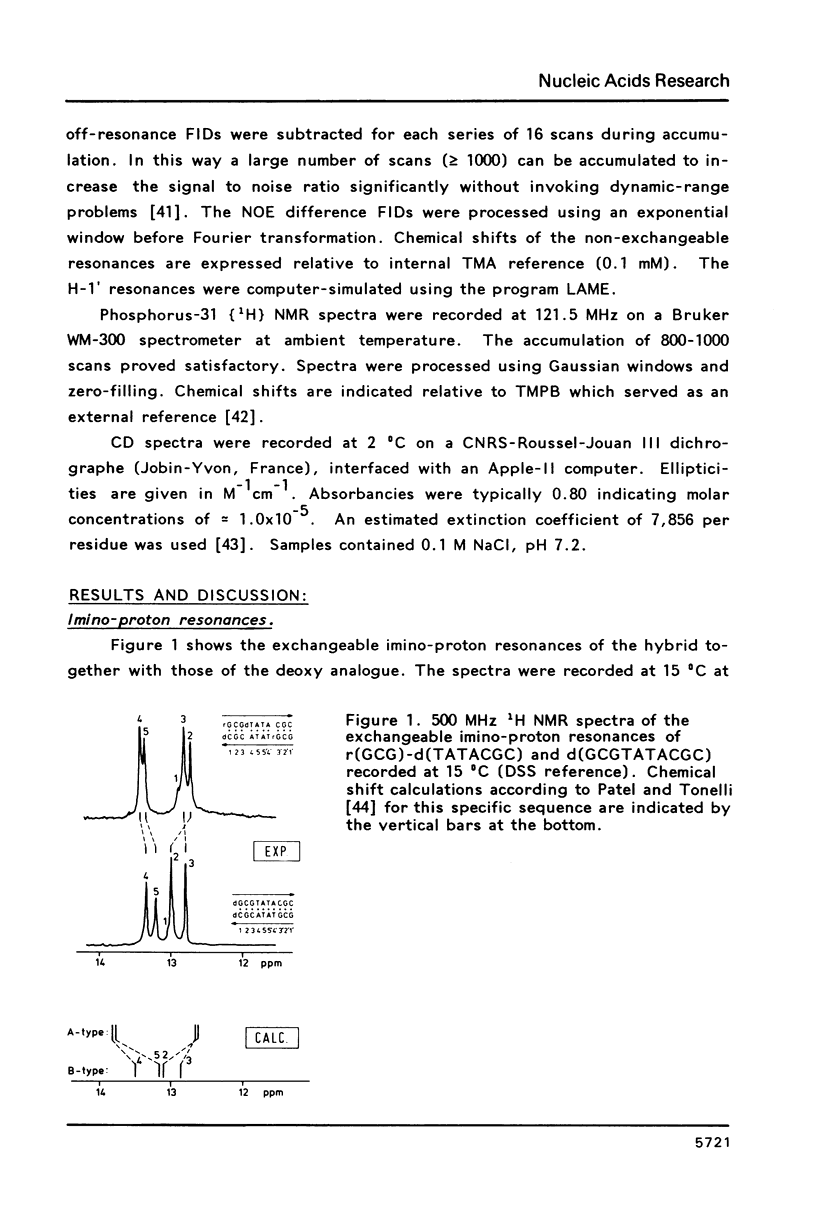
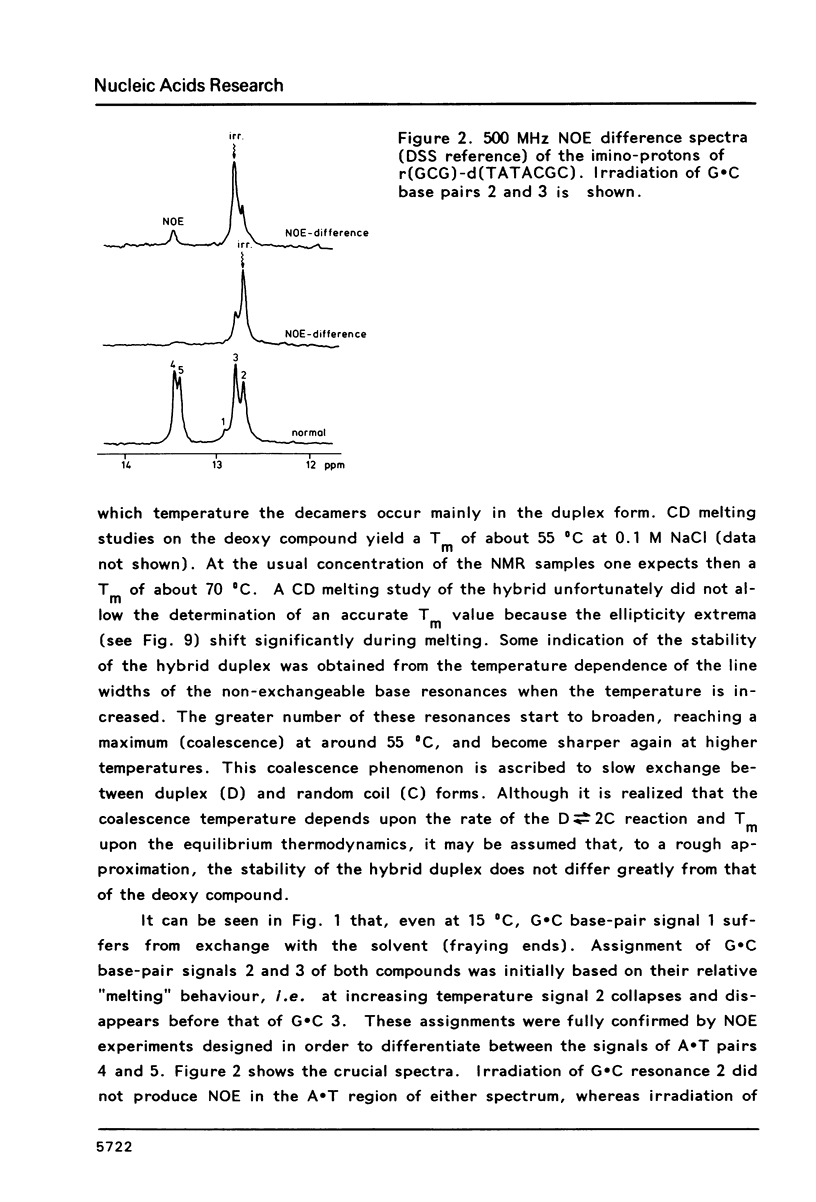
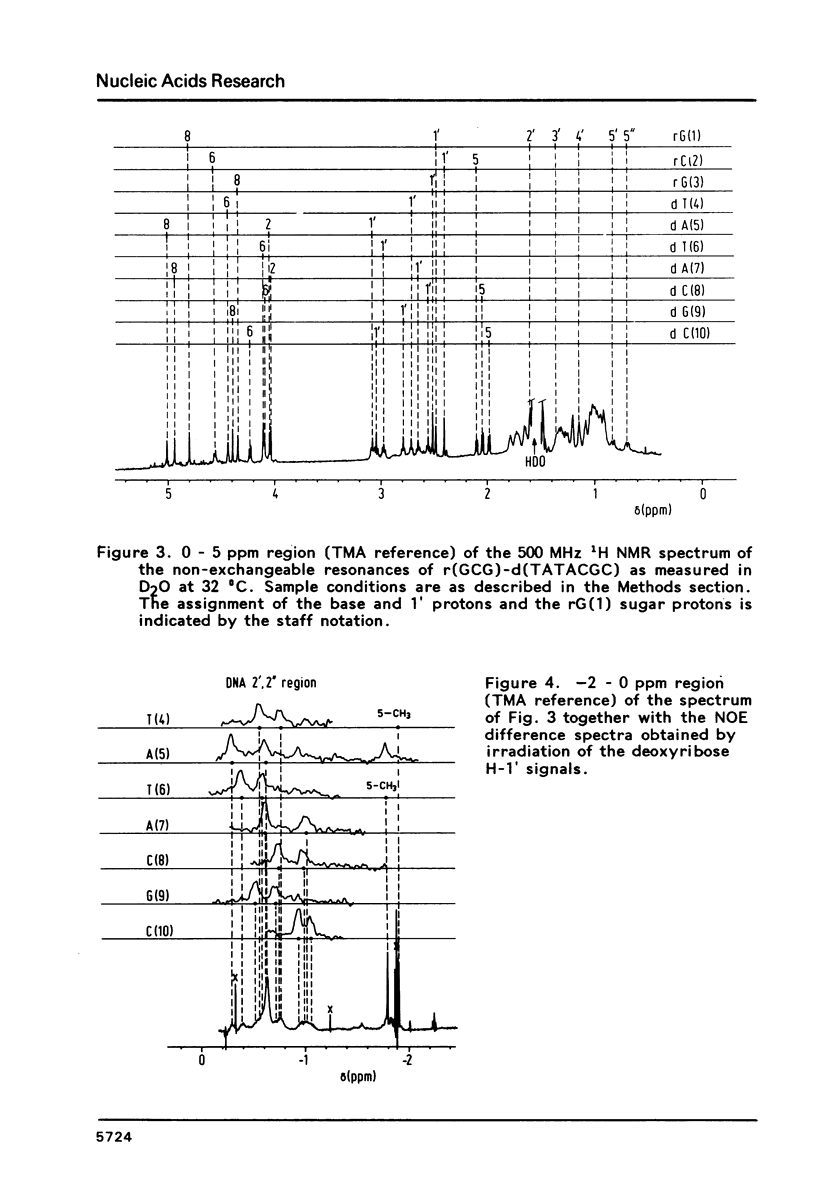
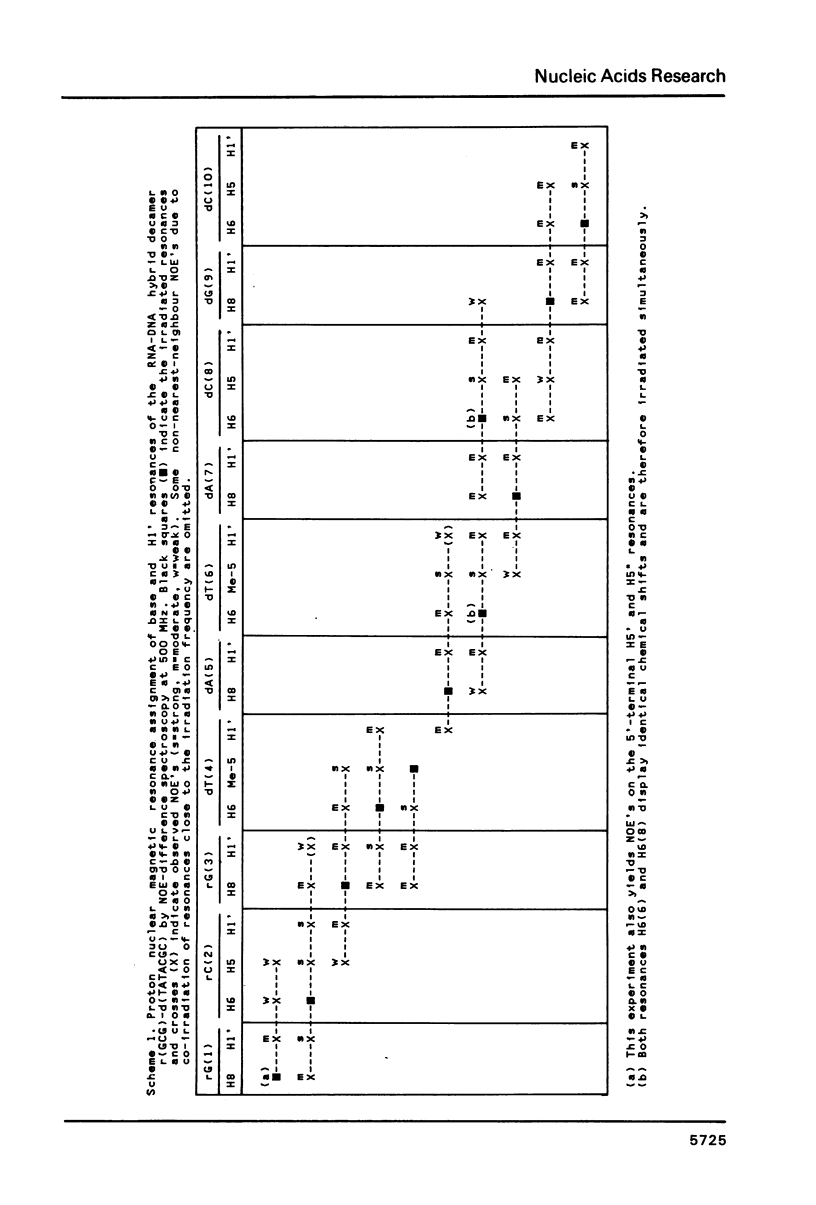
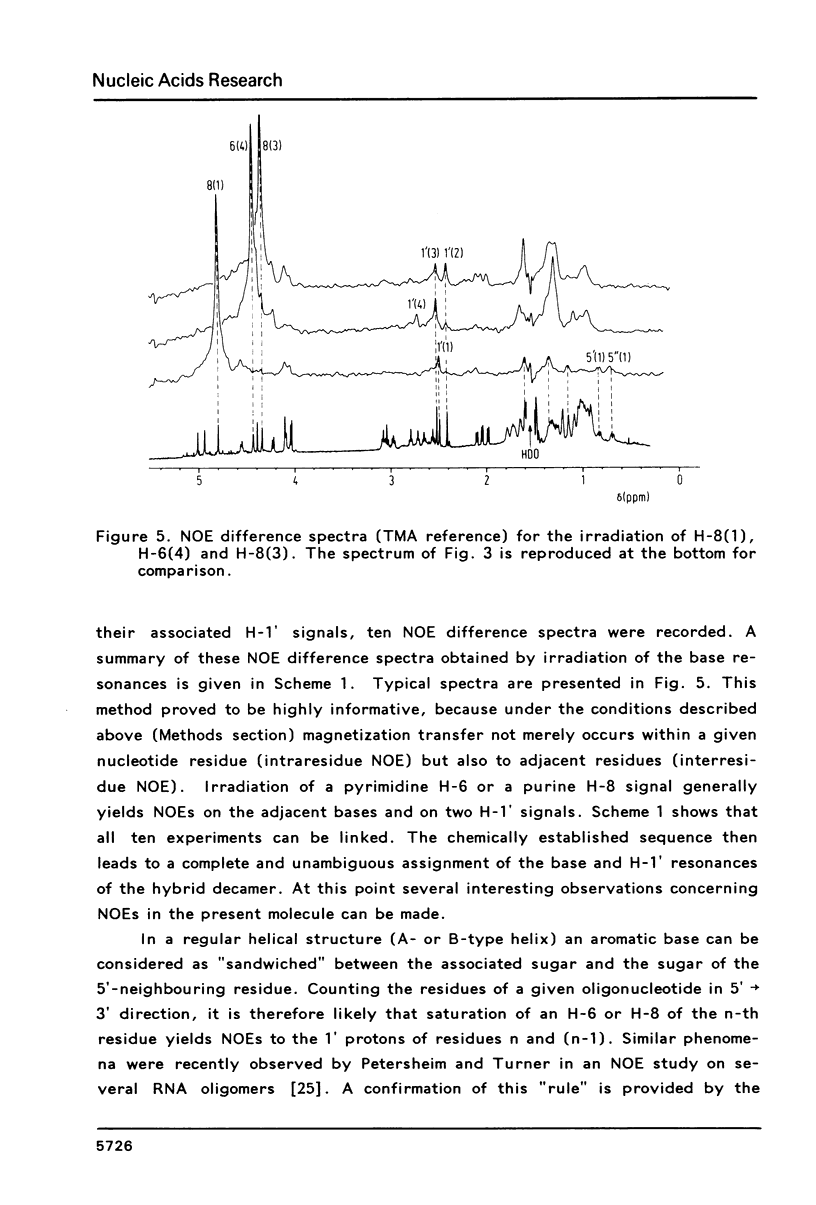
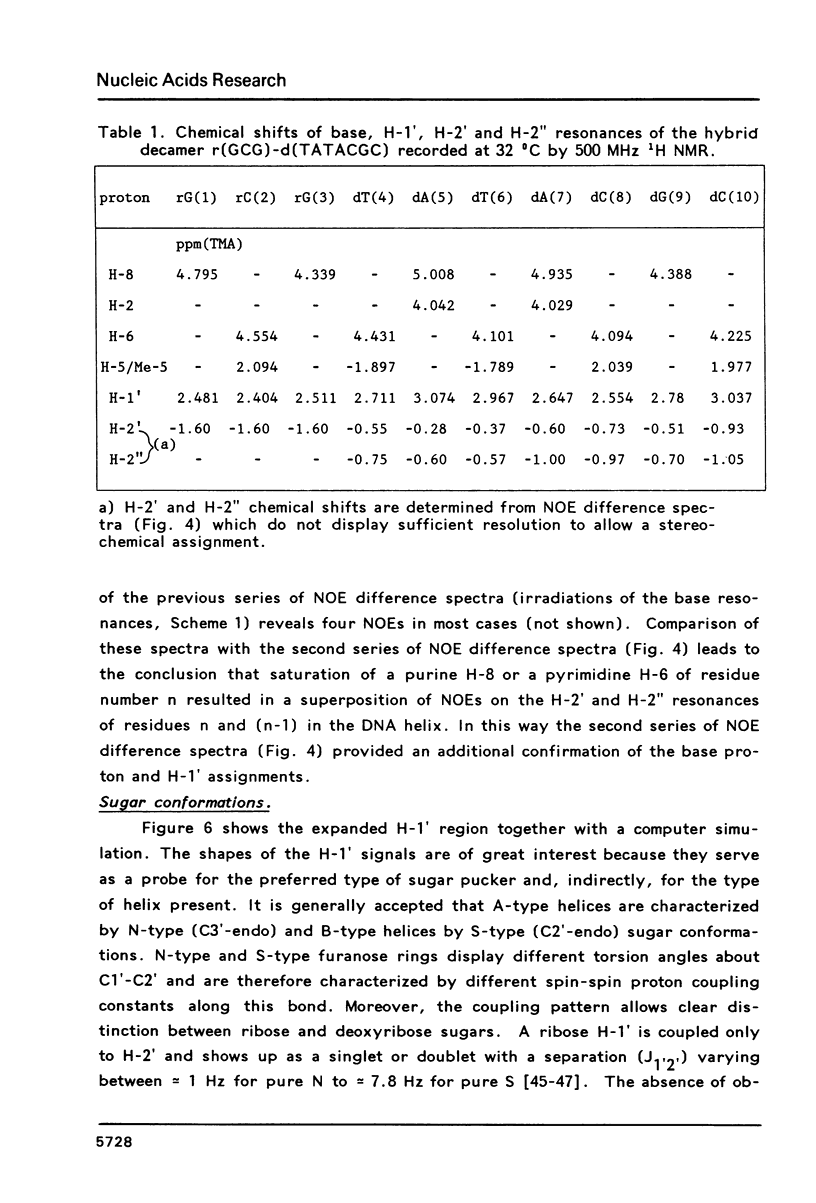
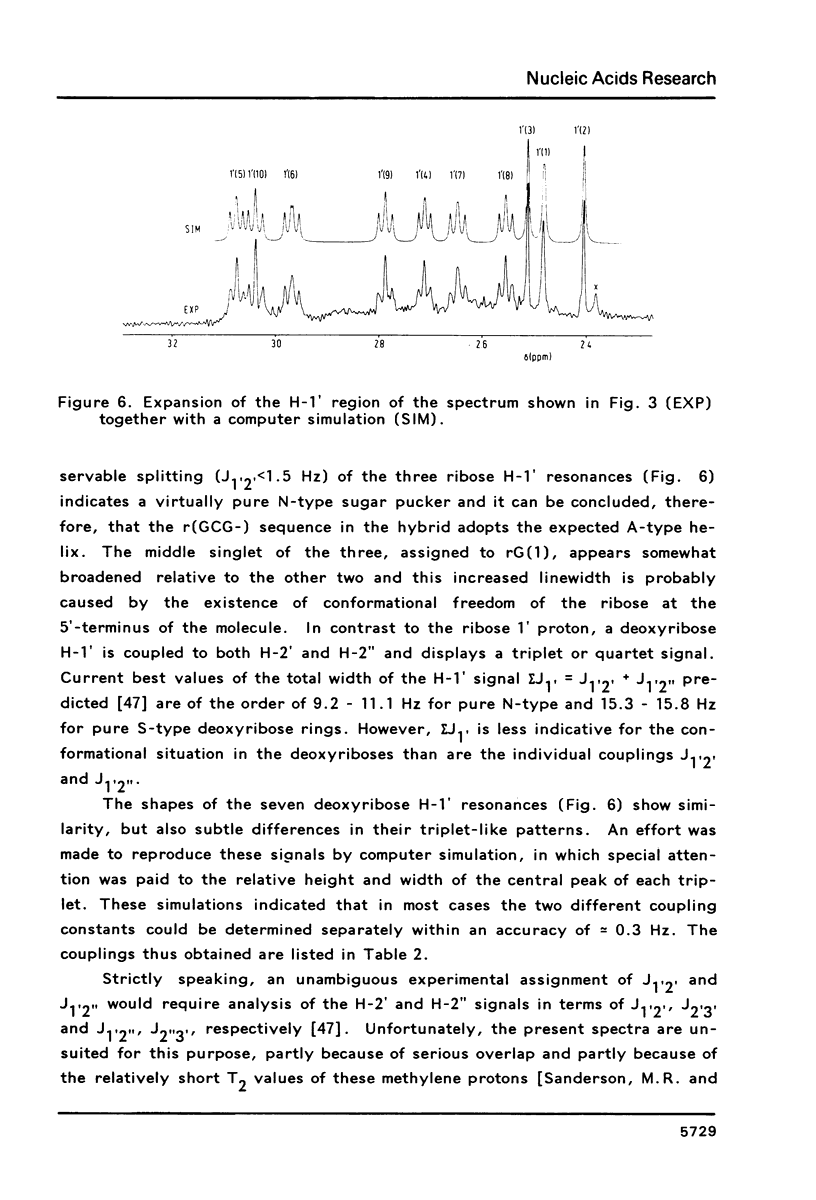
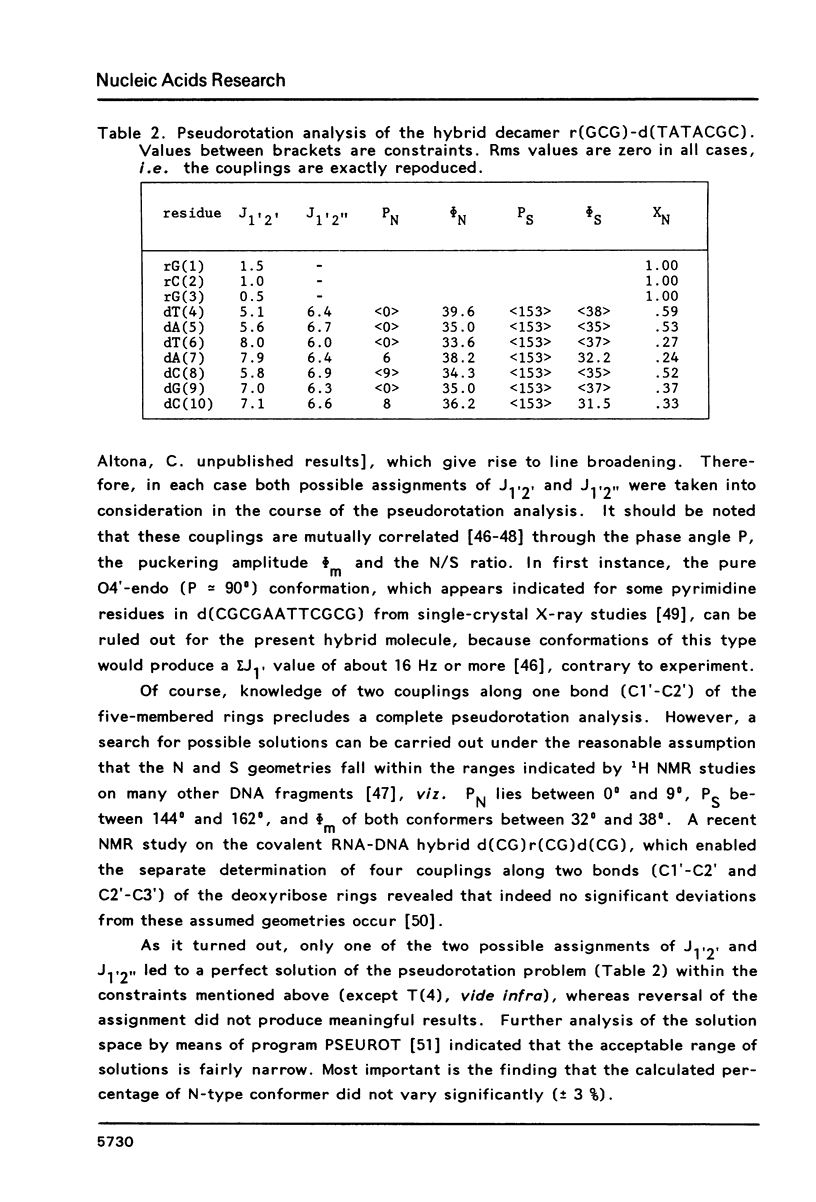
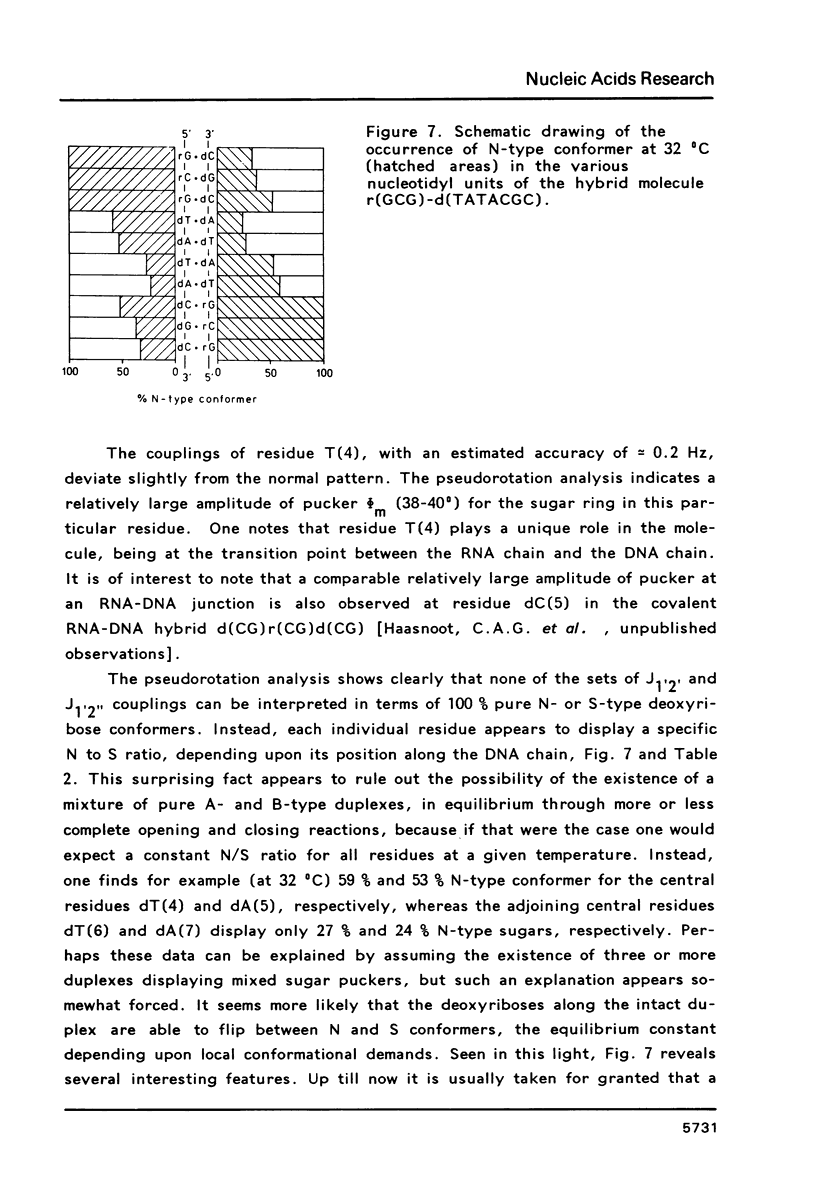
Proton NMR spectra of a covalently linked self-complementary RNA X DNA hybrid, r(GCG)-d(TATACGC), are recorded in H2O and D2O. Imino proton resonances as well as the non-exchangeable base and H-1' resonances are unambiguously assigned by means of nuclear. Overhauser effect measurements. Additional information was obtained by 31P NMR and circular dichroism spectra. The RNA parts in the duplex attain full conformational purity and adopt the usual A-RNA conformation. The DNA residues opposite the RNA tract do not adopt an A-type structure completely. Their respective sugar rings still appear to possess a certain conformational freedom. The same holds true for the central d(-TATA-) sequence which forms a DNA X DNA duplex. There appears to be a structural break in this part: the first two residues, T(4) and A(5), are clearly influenced by the adjacent RNA structure, whereas residues T(6) and A(7) behave quite similar to what usually is found in DNA duplexes in aqueous solution.
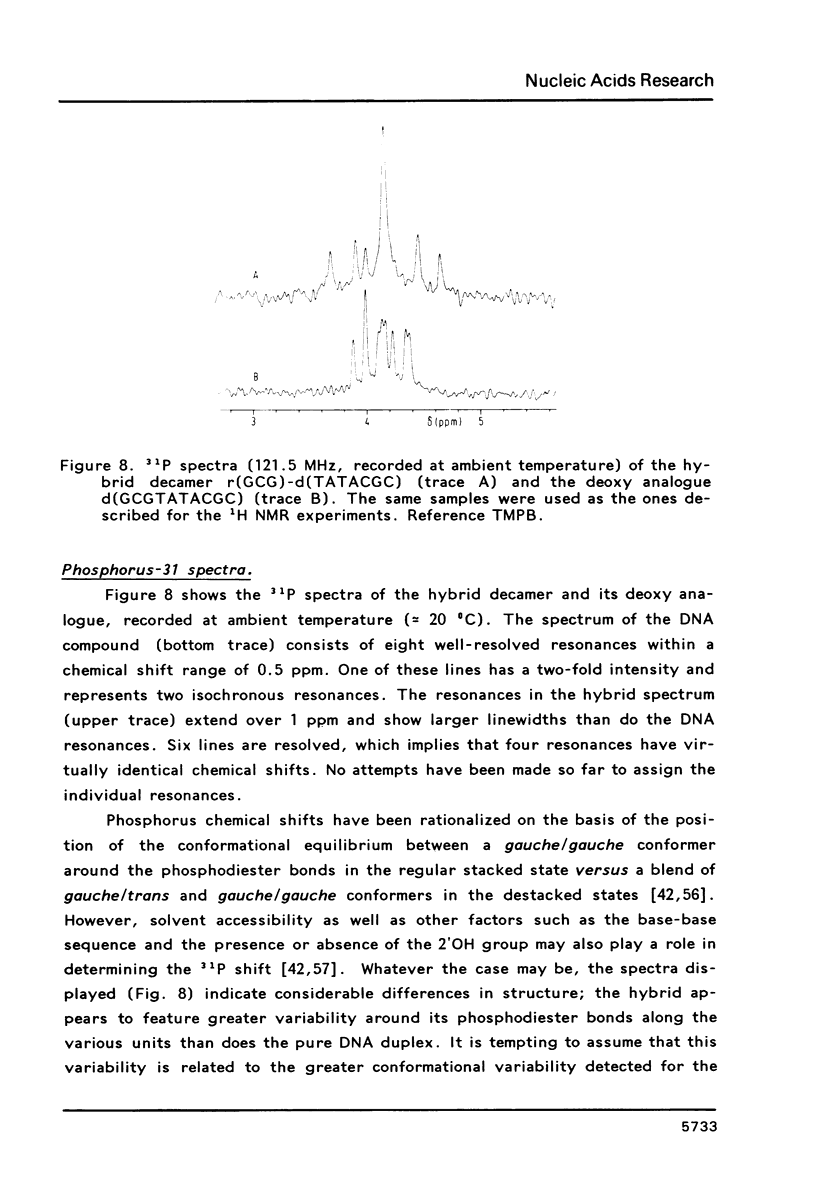
Full text
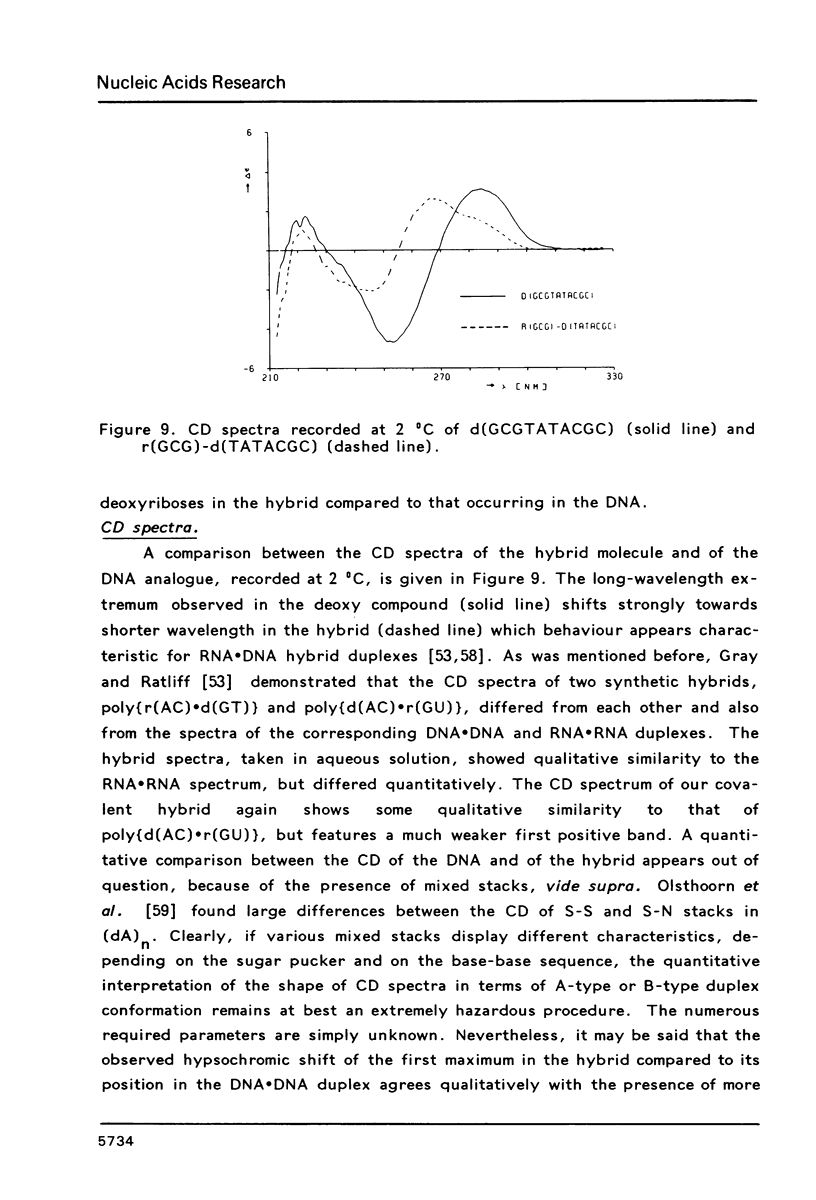
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Bell R. A., Alkema D., Coddington J. M., Hader P. A., Hughes D. W., Neilson T. Prediction of 1H NMR chemical shifts of DNA oligomers. Nucleic Acids Res. 1983 Feb 25;11(4):1143–1149. doi: 10.1093/nar/11.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Schekman R., Kornberg A. A possible role for RNA polymerase in the initiation of M13 DNA synthesis. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2826–2829. doi: 10.1073/pnas.68.11.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D. M., Kan L. S., Leutzinger E. E., Jayaraman K., Miller P. S., Ts'o P. O. Conformational study of two short pentadeoxyribonucleotides, d-CpCpApApG and d-CpTpTpGpG, and their fragments by proton nuclear magnetic resonance. Biochemistry. 1982 Feb 16;21(4):621–630. doi: 10.1021/bi00533a004. [DOI] [PubMed] [Google Scholar]
- Doornbos J., Barascut J. L., Lazrek H., Imbach J. L., van Westrenen J., Visser G. M., van Boom J. H., Altona C. Conformational analysis of oligoarabinonucleotides. An NMR and CD study. Nucleic Acids Res. 1983 Jul 11;11(13):4583–4600. doi: 10.1093/nar/11.13.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. G., Degnen G. E., Cox E. C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133(3):179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Gorenstein D. G. Nucleotide conformational analysis by 31P nuclear magnetic resonance spectroscopy. Annu Rev Biophys Bioeng. 1981;10:355–386. doi: 10.1146/annurev.bb.10.060181.002035. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Altona C. A conformational study of nucleic acid phosphate ester bonds using phosphorus-31 nuclear magnetic resonance. Nucleic Acids Res. 1979 Mar;6(3):1135–1149. doi: 10.1093/nar/6.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Hartel A. J., Lankhorst P. P., Altona C. Thermodynamics of stacking and of self-association of the dinucleoside monophosphate m2(6)A-U from proton NMR chemical shifts: differential concentration temperature profile method. Eur J Biochem. 1982 Dec 15;129(2):343–357. doi: 10.1111/j.1432-1033.1982.tb07057.x. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Cheng D. M., Jayaraman K., Leutzinger E. E., Miller P. S., Ts'o P. O. Proton nuclear magnetic resonance study of a self-complementary decadeoxyribonucleotide, C-C-A-A-G-C-T-T-G-G. Biochemistry. 1982 Dec 21;21(26):6723–6732. doi: 10.1021/bi00269a017. [DOI] [PubMed] [Google Scholar]
- Luder A., Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendrasingam A., Rhodes N. J., Goodwin D. C., Nave C., Pigram W. J., Fuller W., Brahms J., Vergne J. Conformational transitions in oriented fibres of the synthetic polynucleotide poly[d(AT)].poly[d(AT)] double helix. Nature. 1983 Feb 10;301(5900):535–537. doi: 10.1038/301535a0. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Haasnoot C. A., Van Boom J. H., Altona C. Complete assignment and conformational analysis of a deoxyribotetranucleotide. d(TAAT). A 360 and 500 Mhz NMR study. Biochim Biophys Acta. 1981 Sep 28;655(2):256–264. doi: 10.1016/0005-2787(81)90016-2. [DOI] [PubMed] [Google Scholar]
- Milman G., Langridge R., Chamberlin M. J. The structure of a DNA-RNA hybrid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1804–1810. doi: 10.1073/pnas.57.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Discontinuous DNA replication. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Bostelaar L. J., De Rooij J. F., Van Boom J. H., Altona C. Circular dichroism study of stacking properties of oligodeoxyadenylates and polydeoxyadenylate. A three-state conformational model. Eur J Biochem. 1981 Apr;115(2):309–321. doi: 10.1111/j.1432-1033.1981.tb05240.x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Bostelaar L. J., Van Boom J. H., Altona C. Conformational characteristics of the trinucleoside diphosphate dApdApdA and its constituents from nuclear magnetic resonance and circular dichroism studies. Extrapolation to the stacked conformers. Eur J Biochem. 1980 Nov;112(1):95–110. doi: 10.1111/j.1432-1033.1980.tb04991.x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Doornbos J., de Leeuw H. P., Altona C. Influence of the 2'-hydroxyl group and of 6-N-methylation on the conformation of adenine dinucleoside monophosphates in solution. A nuclear magnetic resonance and circular dichroism study. Eur J Biochem. 1982 Jul;125(2):367–382. doi: 10.1111/j.1432-1033.1982.tb06693.x. [DOI] [PubMed] [Google Scholar]
- Pardi A., Martin F. H., Tinoco I., Jr Comparative study of ribonucleotide, deoxyribonucleotide, and hybrid oligonucleotide helices by nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3986–3996. doi: 10.1021/bi00517a007. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Tonelli A. E. Assignment of the proton Nmr chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxy oligonucleotides in aqueous solution. Biopolymers. 1974;13(10):1943–1964. doi: 10.1002/bip.1974.360131003. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Nuclear overhauser studies of CCGGAp, ACCGGp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):264–268. doi: 10.1021/bi00271a005. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Proton magnetic resonance melting studies of CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):269–277. doi: 10.1021/bi00271a006. [DOI] [PubMed] [Google Scholar]
- Sanderson M. R., Mellema J. R., van der Marel G. A., Wille G., van Boom J. H., Altona C. Assignment of non-exchangeable base proton and H1' resonances of a deoxyoctanucleoside heptaphosphate d(G-G-C*-C*-G-G-C-C) by using the nuclear Overhauser effect. Nucleic Acids Res. 1983 May 25;11(10):3333–3346. doi: 10.1093/nar/11.10.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsing E., Wells R. D., Alden C. J., Arnott S. Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J Biol Chem. 1979 Jun 25;254(12):5417–5422. [PubMed] [Google Scholar]
- Selsing E., Wells R. D., Early T. A., Kearns D. R. Two contiguous conformations in a nucleic acid duplex. Nature. 1978 Sep 21;275(5677):249–250. doi: 10.1038/275249a0. [DOI] [PubMed] [Google Scholar]
- Selsing E., Wells R. D. Polynucleotide block polymers consisting of a DNA.RNA hybrid joined to a DNA.DNA duplex. Synthesis and characterization of dGn.rCidCk duplexes. J Biol Chem. 1979 Jun 25;254(12):5410–5416. [PubMed] [Google Scholar]
- Stone M. P., Johnson D. L., Borer P. N. Unusual structures in single-stranded ribonucleic acid: proton nuclear magnetic resonance of AUCCA in deuterium oxide. Biochemistry. 1981 Jun 9;20(12):3604–3610. doi: 10.1021/bi00515a046. [DOI] [PubMed] [Google Scholar]
- Sugino A., Hirose S., Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Dinh S., Neumann J. M., Huynh-Dinh T., Genissel B., Igolen J., Simonnot G. DNA fragment conformations. 1H-NMR studies of helix-coil transition, conformations and dynamic structures of the self-complementary deoxyhexanucleotide d(A-C-A-T-G-T) in aqueous solution. Eur J Biochem. 1982 Jun;124(3):415–425. [PubMed] [Google Scholar]
- Tunis M. J., Hearst J. E. Optical rotatory dispersion of DNA in concentrated salt solutions. Biopolymers. 1968;6(8):1218–1223. doi: 10.1002/bip.1968.360060816. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. A RNA.DNA hybrid that can adopt two conformations: an x-ray diffraction study of poly(rA).poly(dT) in concentrated solution or in fibers. Proc Natl Acad Sci U S A. 1981 Jan;78(1):78–82. doi: 10.1073/pnas.78.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boom J. H., de Rooy J. F. Sequence analysis of synthetic oligonucleotides by high-performance liquid anion-exchange chromatography. J Chromatogr. 1977 Jan 21;131:169–177. doi: 10.1016/s0021-9673(00)80930-9. [DOI] [PubMed] [Google Scholar]
- van Boom J. H., van der Marel G. A., Westerink H., van Boeckel C. A., Mellema J. R., Altona C., Hilbers C. W., Haasnoot C. A., de Bruin S. H., Berendsen R. G. Synthesis and conformational analysis of synthetic DNA fragments. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):403–409. doi: 10.1101/sqb.1983.047.01.047. [DOI] [PubMed] [Google Scholar]



