Abstract
Background
Breast cancer 1, early onset (BRCA1) is a vital DNA repair gene, and the single nucleotide polymorphisms (SNPs) of this gene have been studied in diverse cancer types. In this study, we investigated the association between eight common BRCA1 functional SNPs and the risk of differentiated thyroid carcinoma (DTC).
Methods
This cancer center-based case–control study included 303 DTC cases and 511 controls. A polymerase chain reaction-based restriction fragment length polymorphism assay was performed for genotyping. Unconditional logistical regression analysis was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) in single-SNP analysis and haplotype analysis.
Results
A decreased risk of DTC was found for the A1988G heterozygous AG genotype (adjusted OR=0.63, 95% CI: 0.45–0.87, Bonferroni-adjusted p-value=0.036). AATAATA and ATAA haplotypes that carry C33420T variant allele were associated with reduced papillary thyroid cancer risk (adjusted OR=0.52, 95% CI: 0.33–0.84; adjusted OR=0.62, 95% CI: 0.40–0.95, respectively). Also, having a combination of ≥3 favorable genotypes was associated with a DTC risk reduction (adjusted OR=0.69, 95% CI: 0.50–0.95). The A31875G AG/GG genotype was associated with a 69% reduced risk of multifocal primary tumor in DTC patients (adjusted OR=0.31, 95% CI: 0.12–0.81).
Conclusion
BRCA1 genetic polymorphisms may play a role in DTC risk, while the possible associations warrant confirmation in independent studies.
Introduction
Thyroid cancer is the most common endocrine malignancy in the United States; ∼44,670 new thyroid cancer cases were expected in 2010 (1). Thyroid cancer incidence in the United States has approximately doubled in the past decade and has tripled in the United States since the 1970s (2,3). Females are three times as likely as males to have thyroid cancer. In females, in 2010, thyroid cancer accounted for 5% of all new cancer cases and was the fifth most common cancer type (1).
Differentiated thyroid carcinoma (DTC), which includes the pathological subtypes of papillary, follicular, and Hürthle cell carcinoma, is the most common type of thyroid cancer. Various risk factors, including lifestyle, dietary, and hormonal and reproductive factors, have been proposed but remain controversial [reviewed in Ref. (4)]; exposure to ionizing radiation early in life is the only well-established risk factor for DTC (5). However, not everyone exposed to ionizing radiation develops DTC, and most patients with DTC do not report a radiation exposure history, which suggests individual variation in susceptibility to ionizing radiation and DTC.
DNA double-strand breaks (DSBs) are the most biologically significant form of DNA damage induced by ionizing radiation. Unrepaired or misrepaired DSBs due to defective DSB repair mechanisms can cause cell death and enhance genomic instability, which appear to be associated with predisposition to cancer (6). The breast cancer 1, early onset (BRCA1) gene, located on 17q21, encodes a multifunctional tumor-suppressing protein that is involved directly in DSB repair pathway (7). Moreover, through interacting with a multitude of various proteins, BRCA1 protein plays a critical role in maintaining genome stability and promoting cell survival (8). BRCA1 expression has been detected predominantly in the epithelia of various tissues, including the breast, ovary, and thyroid, and correlates closely with proliferation (9,10). It is well recognized that mutations in the BRCA1 gene that truncate or inactivate the protein lead to a cumulative risk of breast and ovarian cancer (8). Besides mutations, many polymorphisms in BRCA1 have been reported (as listed in http://variantgps.nci.nih.gov/cgfseq/pages/snp500.do); several of them have been amply documented with regard to breast and ovarian cancer susceptibility (11–14). While the functional effects of these polymorphisms have not been fully elucidated, it is biologically plausible that some of these polymorphisms may affect DSB repair capacity and thereby modulate individual susceptibility to thyroid cancer. In fact, previous findings from our group suggested less efficiency of DNA damage repair capacity in thyroid cancer patients than in controls (15). However, few studies of the impact of BRCA1 genetic polymorphisms on susceptibility to thyroid cancer have been published.
We sought to test the hypothesis that polymorphic variants in BRCA1 were associated with DTC and papillary thyroid cancer (PTC) risk. We compared the genotype frequency distributions of eight functional BRCA1 single nucleotide polymorphisms (SNPs) in DTC patients and cancer-free controls. As haplotypes of these SNPs may play a more important role than single SNPs, we also analyzed the haplotypes in association with DTC and PTC risk. Clinicopathological characteristics of the disease were examined to explore the clinical relevance of BRCA1 SNPs.
Materials and Methods
Study subjects
Case subjects were identified from patients who presented to The University of Texas M.D. Anderson Cancer Center with a thyroid gland mass from November 1999 to October 2008. Case subjects were recruited prior to definitive surgery or any chemotherapy or radiation therapy, and the final diagnosis was confirmed through histopathological examination. Case subject recruitment was limited to patients who were at least 18 years of age, who had not received a blood transfusion in the past 6 months, who did not have a prior malignancy (except nonmelanoma skin cancer), and who were not taking immunosuppressant medications. Controls were visitors to the same institution with no history of cancer (except nonmelanoma skin cancer) who were recruited from November 1996 to March 2005 to participate as controls in a molecular epidemiologic study of head and neck squamous cell carcinoma. In summary, the genotype analysis included 303 DTC cases and 511 controls. This study was approved by the Institutional Review Board.
After informed consent was obtained, each subject donated 20 mL of blood for laboratory analysis. A self-administered questionnaire was used to collect subjects' demographic data, information about their exposure to carcinogens, and family history of cancer. Ethnicity was self-reported and categorized as non-Hispanic white or other. Subjects who had smoked >100 cigarettes in their lifetimes were classified as smokers, and those who had quit smoking more than one year prior to enrollment in the study were classified as former smokers. Subjects who used alcohol at least once a week for more than one year were classified as drinkers, and those who had quit such alcohol use more than one year prior to enrollment were classified as former drinkers. Radiation exposure was defined as previous whole-body or head-and-neck-specific medical irradiation. Clinicopathological information was obtained from the medial records. T status (T1–T4) and N status (N0, N1a, and N1b) were defined on the basis of the 2002 American Joint Commission on Cancer TNM staging system.
BRCA1 SNP selection and genotyping
Eight BRCA1 functional SNPs were selected for evaluation as shown in Table 1: six SNPs are located within coding region of BRCA1 that lead to amino-acid changes (nonsynonymous SNPs), and two SNPs are located within the BRCA1 promoter region. All the SNPs selected have a minor allele frequency >5% in the National Cancer Institute SNP500 Cancer project (16). One milliliter of whole blood from each subject was separated into plasma, red cell, and buffy coat components by centrifugation. DNA was extracted from buffy coat using a QIAamp DNA Blood Mini Kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. A standard polymerase chain reaction (PCR)–restriction fragment length polymorphism assay was used to genotype the 8 BRCA1 SNPs of interest. Briefly, 20 ng of genomic DNA was PCR-amplified using the oligonucleotide primers listed in Table 1; the fragments were amplified individually but under the same conditions: an initial denaturing step of 95°C for 5 minutes, followed by 35 cycles at 95°C for 30 seconds, 59°C for 30 seconds, 72°C for 30 seconds, and a final elongation step of 72°C for 5 minutes. PCR products and subsequent digestion products were visualized on a 2% NuSieve agarose gel (Cambrex, Inc., Rockland, ME). Genotypes of each SNP were determined based on the sizes of the restriction digestion products as described in Table 1. Genotyping was performed by laboratory personnel blinded to case–control status, and repeated analysis was performed in a randomly selected 10% of samples.
Table 1.
BRCA1 Single Nucleotide Polymorphisms (GenBank Accession Number AY273801)
| Region | Nucleotide change | DbSNP reference | Amino acid change | Primer sequences (5′→3′) | Restriction enzyme | Restriction fragment length (bp) |
|---|---|---|---|---|---|---|
| Promoter | A1988G | N/A | N/A | S: AAAGACCCAAGGGGTTGGCATC AS: TTAAGATTTGGAAGGTTTTAGATT |
TaqI | AA: 148 AG: 22, 126, 148 GG: 22, 126 |
| Promoter | T2089C | N/A | N/A | S: AAAGACCCAAGGGGTTGGCATC AS: TTAAGATTTGGAAGGTTTTAGATT |
AseI | TT: 148 TC: 24, 124, 148 CC: 24, 124 |
| Exon 11 | A31875G | rs1799950 | Gln315Arg | S: CATGTAATGATAGGCGGACTC AS: GGATTCTCTGAGCATGGCAGGATC |
BamHI | AA: 112 AG: 22, 90, 112 GG: 22, 90 |
| Exon 11 | C33420T | rs799917 | Pro871Leu | S: GTTTCAAAGCGCCAGTCATTGGATC AS: GGACTTTGTTTCTTTAAGGACCCAG |
BamHI | CC: 23, 81 CT: 23, 81, 104 TT: 104 |
| Exon 11 | A33921G | rs16941 | Glu1038Gly | S: GAAAGAGAAATGGGAAATGAGAAC AS: TTCATTAATATTGCTTGAGCGAGCT |
SacI | AA: 108 AG: 23, 85, 108 GG: 23, 85 |
| Exon 11 | A34356G | rs16942 | Lys1183Arg | S: AGAACAGCCTATGGGAAGTA AS: CTCTAATTTCTTGGCCCCTC |
MnII | AA: 25, 206 AG: 25, 63, 143, 206 GG: 25, 63, 143 |
| Exon 13 | T43893C | rs1060915 | Ser1436Ser | S: TTAGAACAGCATGGGAGCCA AS: AAGATATCAGTGTTTGGCGA |
EarI | TT: 60, 77 TC: 60, 77, 137 CC: 137 |
| Exon 16 | A55298G | rs1799966 | Ser1613Gly | S: CCAGAGTCAGCTCGTGTTGGC AS: AATTCTTCTGGGGTCAGGCCAG |
AvaII | AA: 232 AG: 86, 146, 232 GG: 86, 146 |
AS, antisense primer; S, sense primer; SNP, single nucleotide polymorphism.
Statistical analysis
The chi-square test was used to compare frequencies of demographic and environmental exposure parameters between cases and controls. For each SNP, deviation from Hardy–Weinberg equilibrium (HWE) assumption was examined in controls by chi-square test. Among controls, pairwise linkage disequilibrium (LD) measures (D' value) were calculated and an LD plot was generated using Haploview software (17). Differences in frequencies of SNP alleles and genotypes between cases and controls were evaluated using chi-square test or Fisher's exact test as appropriate. Based on an unconditional logistic regression model, odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated in three inheritance models (co-dominant, dominant, and recessive) and were adjusted for potential confounders, including age, sex, ethnicity, smoking status, family history of thyroid cancer, and radiation exposure. The best inheritance models were identified according to the smallest Akaile information criterion (AIC) value. In the codominant model, number of minor alleles was coded as a continuous variable and fitted in the regression model to test for trend.
Haplotype analyses were done using the online SNPStats tool (18). Expectation maximization algorithm was used to estimate haplotype frequencies. Using the most frequent haplotype as the reference group, an additive model was used to introduce haplotype counts, and an unconditional regression model was applied to calculate ORs and 95% CIs with adjustment for potential confounders. In addition, to assess the effect of BRCA1 SNPs in combination and evaluate the dose–response relationship, subjects were trichotomized according to the number of favorable genotypes carried using a dominant model (e.g., favorable genotype was the combined heterozygous and homozygous variant genotypes), which was fitted into a logistic regression model as a continuous variable.
Since sex, age, and ethnic differences could lead to a spurious association between genotypes and disease, we further stratified both genotypes and haplotypes by age (≤50, >50 years), sex (male, female), and ethnicity (non-Hispanic white, other). Subgroup analyses by smoking status (never, ever) and first-degree family history of cancer (yes, no) were also done to examine the possible interaction between these parameters and BRCA1 SNPs. To provide more statistical power for our stratification analysis, we used the dominant or recessive model only according to the AIC value.
We also evaluated the association between BRCA1 SNPs and PTC using the same analyses and covariates as above. All statistical tests were two-sided, and p<0.05 was considered statistically significant. To correct for multiple testing, we estimated the adjusted significance by applying the Bonferroni correction for all the SNPs tested in the analysis. Statistical analysis was performed using SAS software version 9.2 (SAS Institute, Inc., Cary, NC) unless otherwise specified.
Results
Demographic characteristics and environmental exposures of cases and controls are presented in Table 2. There were fewer older adults, men, non-Hispanic whites, and nonsmokers in the case groups (p<0.05). DTC (and PTC) cases reported history of thyroid cancer in first-degree relatives more frequently than did controls (p<0.001). The observed frequency differences in demographic and exposure variables between cases and controls were adjusted for in subsequent logistic regression analysis. Among the 303 DTC patients, 273 (90.1%) were diagnosed with PTC. Among the DTC patients, the majority had T1 (41.4%) and N0 (55.8%) disease when they presented to our institution.
Table 2.
Demographic and Exposure Characteristics of Case and Control Subjects
| |
Controls |
All cases |
|
PTC cases |
|
|||
|---|---|---|---|---|---|---|---|---|
| |
(No.=511) |
(No.=303) |
|
(No.=273) |
|
|||
| Variable | No. | (%) | No. | (%) | pa | No. | (%) | pa |
| Age, years | <0.001 | <0.001 | ||||||
| <30 | 17 | (3.3) | 41 | (13.5) | 39 | (14.3) | ||
| 30–45 | 181 | (35.4) | 121 | (39.9) | 114 | (41.8) | ||
| >45 | 313 | (61.3) | 141 | (46.6) | 120 | (44.0) | ||
| Sex | <0.001 | <0.001 | ||||||
| Male | 245 | (47.9) | 103 | (34.0) | 89 | (32.6) | ||
| Female | 266 | (52.1) | 200 | (66.0) | 184 | (67.4) | ||
| Ethnicity | 0.012 | 0.008 | ||||||
| Non-Hispanic white | 401 | (78.5) | 214 | (70.6) | 191 | (70.0) | ||
| Other | 110 | (21.5) | 89 | (29.4) | 82 | (30.0) | ||
| Family history of cancer | 0.70 | 0.51 | ||||||
| Yes | 257 | (51.4) | 151 | (50.0) | 133 | (48.9) | ||
| No | 243 | (48.6) | 151 | (50.0) | 139 | (51.1) | ||
| 1st-degree relative with DTC | <0.001 | <0.001 | ||||||
| Yes | 6 | (1.2) | 19 | (6.3) | 19 | (7.0) | ||
| No | 494 | (98.8) | 283 | (93.7) | 253 | (93.0) | ||
| Smoking status | 0.001 | 0.003 | ||||||
| Current | 96 | (19.2) | 29 | (9.6) | 28 | (10.3) | ||
| Former | 114 | (22.8) | 69 | (22.8) | 59 | (21.7) | ||
| Never | 290 | (58.0) | 204 | (67.6) | 185 | (68.0) | ||
| Alcohol drinking status | 0.11 | 0.10 | ||||||
| Current | 170 | (34.0) | 95 | (31.4) | 89 | (32.7) | ||
| Former | 74 | (14.8) | 32 | (10.6) | 27 | (9.9) | ||
| Never | 256 | (51.2) | 175 | (58.0) | 156 | (57.4) | ||
| Radiation exposure | 0.11 | 0.13 | ||||||
| No | 504 | (98.6) | 293 | (97.0) | 264 | (97.0) | ||
| Yes | 7 | (1.4) | 9 | (3.0) | 8 | (3.0) | ||
| T status | ||||||||
| T1 | Na | 108 | (41.4) | 105 | (44.1) | |||
| T2 | Na | 45 | (17.2) | 36 | (15.1) | |||
| T3 | Na | 97 | (37.2) | 87 | (36.6) | |||
| T4 | Na | 11 | (4.2) | 10 | (4.2) | |||
| Multifocal primary | ||||||||
| No | Na | 181 | (65.3) | 157 | (62.8) | |||
| Yes | Na | 96 | (34.7) | 93 | (37.2) | |||
| Nodal status | ||||||||
| N0 | Na | 149 | (55.8) | 126 | (51.8) | |||
| N1a | Na | 42 | (15.7) | 42 | (17.3) | |||
| N1b | Na | 76 | (28.5) | 75 | (30.9) | |||
Chi-square test, compared to controls.
DTC, differentiated thyroid carcinoma; na, not applicable; PTC, papillary thyroid carcinoma.
After genotyping 142 randomly selected samples regardless of the disease status, we found that SNP T2089C was in perfect correlation (r2=1) with SNP A1988G; therefore, T2089C was excluded from the subsequent statistical analysis. Table 3 summarizes the allele and genotype frequencies and genotype-specific risks of the 7 SNPs. For all 7 SNPs, the genotype frequencies in controls conformed to HWE (p>0.05). When association analysis was conducted after modeling of the SNPs (codominant, dominant, recessive), for A1988G, A33921G, and T43893C, the best inheritance model was the codominant model; for C33420T, A34356G, and A55298G, the best inheritance model was the dominant model; and for A31875G, the recessive model was taken as the best inheritance model according to the AIC value, but because there was no homozygous variant genotype carrier in the control group, the corresponding OR and 95% CI values were meaningless and are not shown in Table 3.
Table 3.
BRCA1 Allele and Genotype Frequencies and Risk Estimates of Case and Control Subjects
| |
Controls (No.=511) |
All cases (No.=303) |
|
|
PTC cases (No.=273) |
|
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | No. | (%) | No. | (%) | pa | Adjusted ORb(95% CI) | No. | (%) | pa | Adjusted ORb(95% CI) |
| A1988G | ptrend=0.38 | ptrend=0.47 | ||||||||
| AA | 227 | (44.4) | 159 | (52.5) | 0.024 | 1.00 | 140 | (51.3) | 0.076 | 1.00 |
| AG | 230 | (45.0) | 107 | (35.3) | 0.63 (0.45–0.87) | 100 | (36.6) | 0.66 (0.47–0.92) | ||
| GG | 54 | (10.6) | 37 | (12.2) | 1.14 (0.70–1.87) | 33 | (12.1) | 1.17 (0.70–1.95) | ||
| AG+GG | 284 | (55.6) | 144 | (47.5) | 0.026 | 0.72 (0.53–0.97) | 133 | (48.7) | 0.067 | 0.75 (0.55–1.02) |
| G allele freq. | (33.1) | (29.9) | 0.18 | (30.4) | ||||||
| HWE (P) | 0.76 | |||||||||
| A31875G | ptrend=0.11 | ptrend=0.25 | ||||||||
| AA | 455 | (89.0) | 263 | (86.8) | 0.017 | 1.00 | 239 | (87.5) | 0.041 | 1.00 |
| AG | 56 | (11.0) | 36 | (11.9) | 1.16 (0.72–1.85) | 31 | (11.4) | 1.06 (0.65–1.74) | ||
| GG | 0 | (0.0) | 4 | (1.3) | – | 3 | (1.1) | – | ||
| AG+GG | 56 | (11.0) | 40 | (13.2) | 0.34 | 1.29 (0.82–2.04) | 34 | (12.5) | 0.53 | 1.18 (0.73–1.92) |
| G allele freq. | (5.5) | (7.3) | 0.17 | (6.8) | ||||||
| HWE (P) | 0.39 | |||||||||
| C33420T | ptrend=0.057 | ptrend=0.042 | ||||||||
| CC | 198 | (38.8) | 136 | (44.9) | 0.22 | 1.00 | 122 | (44.7) | 0.27 | 1.00 |
| CT | 226 | (44.2) | 119 | (39.3) | 0.73 (0.52–1.00) | 109 | (39.9) | 0.73 (0.52–1.03) | ||
| TT | 87 | (17.0) | 48 | (15.8) | 0.70 (0.45–1.11) | 42 | (15.4) | 0.66 (0.41–1.06) | ||
| CT+TTc | 313 | (61.2) | 167 | (55.1) | 0.09 | 0.72 (0.53–0.98) | 151 | (55.3) | 0.11 | 0.71 (0.52–0.98) |
| T allele freq. | (39.1) | (35.5) | 0.15 | (35.3) | ||||||
| HWE (P) | 0.11 | |||||||||
| A33921G | ptrend=0.48 | ptrend=0.53 | ||||||||
| AA | 230 | (45.0) | 154 | (50.8) | 0.14 | 1.00 | 136 | (49.8) | 0.27 | 1.00 |
| AG | 227 | (44.4) | 113 | (37.3) | 0.71 (0.52–0.98) | 105 | (38.5) | 0.74 (0.53–1.04) | ||
| GG | 54 | (10.6) | 36 | (11.9) | 1.11 (0.68–1.82) | 32 | (11.7) | 1.08 (0.64–1.80) | ||
| AG+GG | 281 | (55.0) | 149 | (49.2) | 0.11 | 0.78 (0.58–1.05) | 137 | (50.2) | 0.20 | 0.80 (0.59–1.10) |
| G allele freq. | (32.8) | (30.5) | 0.35 | (31.0) | ||||||
| HWE (P) | 0.92 | |||||||||
| A34356G | ptrend=0.15 | ptrend=0.19 | ||||||||
| AA | 222 | (43.4) | 153 | (50.5) | 0.12 | 1.00 | 135 | (49.5) | 0.25 | 1.00 |
| AG | 240 | (47.0) | 121 | (39.9) | 0.68 (0.50–0.94) | 112 | (41.0) | 0.71 (0.51–0.98) | ||
| GG | 49 | (9.6) | 29 | (9.6) | 0.94 (0.55–1.60) | 26 | (9.5) | 0.93 (0.53–1.61) | ||
| AG+GGc | 289 | (56.6) | 150 | (49.5) | 0.051 | 0.72 (0.54–0.98) | 138 | (50.5) | 0.11 | 0.74 (0.54–1.01) |
| G allele freq. | (33.1) | (29.5) | 0.15 | (30.0) | ||||||
| HWE (P) | 0.19 | |||||||||
| T43893C | ptrend=0.85 | ptrend=0.76 | ||||||||
| TT | 167 | (32.7) | 114 | (37.6) | 0.18 | 1.00 | 104 | (38.1) | 0.16 | 1.00 |
| TC | 267 | (52.2) | 138 | (45.6) | 0.74 (0.53–1.03) | 123 | (45.0) | 0.72 (0.51–1.02) | ||
| CC | 77 | (15.1) | 51 | (16.8) | 1.11 (0.71–1.75) | 46 | (16.9) | 1.09 (0.68–1.74) | ||
| TC+CC | 344 | (67.3) | 189 | (62.4) | 0.15 | 0.82 (0.60–1.11) | 169 | (61.9) | 0.13 | 0.80 (0.58–1.10) |
| C allele freq. | (41.2) | (39.6) | 0.53 | (39.4) | ||||||
| HWE (P) | 0.08 | |||||||||
| A55298G | ptrend=0.46 | ptrend=0.49 | ||||||||
| AA | 229 | (44.8) | 151 | (49.8) | 0.32 | 1.00 | 133 | (48.7) | 0.55 | 1.00 |
| AG | 233 | (45.6) | 122 | (40.3) | 0.75 (0.54–1.02) | 114 | (41.8) | 0.79 (0.57–1.09) | ||
| GG | 49 | (9.6) | 30 | (9.9) | 1.09 (0.65–1.85) | 26 | (9.5) | 1.03 (0.60–1.80) | ||
| AG+GGc | 282 | (55.2) | 152 | (50.2) | 0.17 | 0.80 (0.59–1.08) | 140 | (51.3) | 0.30 | 0.83 (0.60–1.13) |
| G allele freq. | (32.4) | (30.0) | 0.35 | (30.4) | ||||||
| HWE (P) | 0.42 | |||||||||
Chi-squared analysis comparing genotype distributions between case and control subjects.
Adjusted for age, sex, ethnicity, first-degree family history of thyroid cancer, smoking status, and radiation exposure.
SNP for which dominant model had the criterion value and was accepted as the best inheritance model.
BRCA1, breast cancer 1, early onset; CI, confidence interval; HWE, Hardy–Weinberg equilibrium; OR, odds ratio.
Overall, 4 SNPs were significantly associated with reduced DTC risk after adjustment for potential confounders. For the BRCA1 promoter SNP A1988G, the heterozygous AG genotype was associated with a 37% reduced risk of DTC (adjusted OR=0.63, 95% CI: 0.45–0.87). For A33921G, the heterozygous AG genotype was associated with a 29% reduced DTC risk (adjusted OR=0.71, 95% CI: 0.52–0.98). For C33420T, the combined CT/TT genotype was associated with a 28% reduced DTC risk (adjusted OR=0.72, 95% CI: 0.53–0.98), and a borderline significant trend was shown for the minor allele to decrease DTC risk (ptrend=0.057). For A34356G, the combined AG/GG genotype was associated with a 28% reduced risk of DTC (adjusted OR=0.72, 95% CI: 0.54–0.98). After correction for multiple testing, only A1988G remained statistically significant (Bonferroni-adjusted p-value=0.036). We also analyzed the association between these SNPs and PTC risk. Similar to the findings for DTC overall, A1988G AG genotype was associated with a 34% reduction in PTC risk (adjusted OR=0.66, 95% CI: 0.47–0.92), and C33420T combined CT/TT genotype was associated with a 29% risk reduction in PTC risk (adjusted OR=0.71, 95% CI: 0.52–0.98). A significant trend was shown for the minor allele of C33420T to decrease PTC risk (ptrend=0.042).
While the statistical power in subgroup analysis is limited, we conducted stratification analysis to explore the possible interaction. Overall, no interaction between the SNPs and selected variables (i.e., age, sex, ethnicity, smoking status, and first-degree family history of cancer) was observed (p>0.05). Stratification analysis by age (≤50, >50 years) revealed that significant association between BRCA1 SNPs and DTC risk was likely confined to young subjects: under the dominant model, the A1988G AG/GG, C33420T CT/TT, and A34356G AG/GG genotypes were associated with reduced risks of DTC in subjects ≤50 years old—34% (adjusted OR=0.66, 95% CI: 0.45–0.96), 34% (adjusted OR=0.66, 95% CI: 0.45–0.98), and 35% (adjusted OR=0.65, 95% CI: 0.45–0.96), respectively—whereas the A31875G AG/GG genotype were associated with increased DTC risk in subjects ≤50 years old (adjusted OR=1.81, 95% CI: 1.04–3.15), but none were statistically significant after correction for multiple testing. Given the low proportion of individuals other than non-Hispanic whites in our study and the complex genetic background of the individuals who were not non-Hispanic whites, the ethnicity-based subgroup analysis was confined to non-Hispanic whites. Two genotypes were associated with reduced DTC risk in non-Hispanic whites: A1988G AG/GG (adjusted OR=0.69, 95% CI: 0.48–0.97) and A34356G AG/GG (adjusted OR=0.66, 95% CI: 0.46–0.93), but these associations were not significant after correction for multiple testing. In subgroup analysis stratified by sex (male, female), smoking status (never, ever), and first-degree family history of cancer (yes, no), no significant association was found in these subgroups (data not shown).
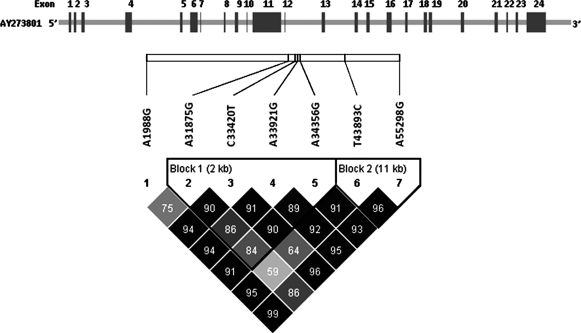
Pairwise LD analysis of the 7 SNPs showed strong LD (D' >0.80) among the 4 SNPs in exon 11 (block 1: A31875G, C33420T, A33921G, A34356G) and between the 2 SNPs in exons 13 and 16 (block 2: T43893C, A55298G) (Fig. 1). At first, we estimated frequencies for the haplotypes encompassing all 7 SNPs. As shown in Table 4, five common haplotypes accounted for >90% of all haplotypes; the remaining haplotypes, which had frequencies of <3%, were classified as others. Compared to haplotype 1 (AACAATA), the most commonly observed haplotype, haplotype 4 (AATAATA), was significantly associated with a 44% reduced risk of DTC (adjusted OR=0.56, 95% CI: 0.35–0.87, p=0.011). Subsequent subgroup analysis found that this inverse association with DTC risk was confined to males (adjusted OR=0.26, 95% CI: 0.08–0.84) and nonsmokers (adjusted OR=0.59, 95% CI: 0.35–0.98). The inverse association remained significant for haplotype 4 and PTC risk (adjusted OR=0.52, 95% CI: 0.33–0.84, p=0.008). In addition, we performed haplotype association analysis within block 1. In agreement with the initial analysis of all 7 SNPs, the ATAA haplotype, which corresponds to haplotype 4, showed a different distribution between cases and controls. The DTC risk estimate for the ATAA haplotype was borderline significant (adjusted OR=0.67, 95% CI: 0.45–1.01, p=0.055). The association was significant for the ATAA haplotype and PTC risk (adjusted OR=0.62, 95% CI: 0.40–0.95, p=0.031). No significant association with DTC (or PTC) risk was found for other haplotypes (data not shown).
FIG. 1.
Linkage disequilibrium analysis of the investigated SNPs in the BRCA1 gene region. The bar and dashed lines above the SNP names indicate the relative location of each SNP along the gene. Below the SNPs is the haplotype block. The number at each intersection is the pairwise D' value of the paired SNPs. SNP, single nucleotide polymorphism.
Table 4.
Distribution of Haplotypes in the BRCA1 Gene and Risk Estimates of Case and Control Subjects
| |
SNPa |
Frequency |
DTC |
PTC |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Controls (No.=511) | DTC cases (No.=303) | PTC cases (No.=273) | Adjusted ORb(95% CI) | p | Adjusted ORb(95% CI) | p |
| 1 | A | A | C | A | A | T | A | 0.446 | 0.449 | 0.458 | 1.00 | – | 1.00 | – |
| 2 | G | A | T | G | G | C | G | 0.294 | 0.254 | 0.252 | 0.86 (0.65–1.10) | 0.22 | 0.82 (0.62–1.09) | 0.17 |
| 3 | A | A | C | A | A | C | A | 0.076 | 0.088 | 0.084 | 1.32 (0.84–2.01) | 0.20 | 1.22 (0.78–1.92) | 0.37 |
| 4 | A | A | T | A | A | T | A | 0.065 | 0.045 | 0.042 | 0.56 (0.35–0.87) | 0.011 | 0.52 (0.33–0.84) | 0.008 |
| 5 | A | G | C | A | A | T | A | 0.045 | 0.057 | 0.048 | 1.47 (0.87–2.47) | 0.15 | 1.20 (0.68–2.12) | 0.53 |
| Othersc | 0.074 | 0.107 | 0.116 | 1.40 (0.96–2.05) | 0.081 | 1.41 (0.95–2.08) | 0.085 | |||||||
SNPs refer to 1, A1988G; 2, A31875G; 3, C33420T; 4, A33921G; 5, A34356G; 6, T43893C; and 7, A55298G.
Adjusted for age, sex, ethnicity, first-degree family history of thyroid cancer, smoking status, and radiation exposure.
Rare haplotypes with frequencies <0.03.
The effect of number of BRCA1 favorable genotypes on DTC risk is presented in Table 5. We included the four SNPs that were associated with reduced DTC risk and trichotomized subjects by number of BRCA1 favorable genotypes (0, 1–2, or 3–4). Possessing 3–4 favorable genotypes was associated with a 31% reduction in DTC risk (adjusted OR=0.69, 95% CI: 0.50–0.95). This signified a dose–response relationship (p=0.028). When the analysis was restricted to PTC cases and controls, the dose–response relationship remained significant after adjustment (p=0.046).
Table 5.
Combination Effect of BRCA1 Genotypes
| |
Controls (No.=511) |
DTC cases (No.=303) |
|
PTC cases (No.=273) |
|
|||
|---|---|---|---|---|---|---|---|---|
| No. of favorable genotypes | No. | (%) | No. | (%) | Adjusted ORa(95% CI) | No. | (%) | Adjusted ORa(95% CI) |
| 0 | 177 | (34.6) | 126 | (41.6) | 1.00 | 112 | (41.0) | 1.00 |
| 1–2 | 52 | (10.2) | 29 | (9.6) | 0.64 (0.36–1.11) | 25 | (9.2) | 0.62 (0.34–1.12) |
| 3–4 | 282 | (55.2) | 148 | (48.8) | 0.69 (0.50–0.95) | 136 | (49.8) | 0.70 (0.50–0.98) |
| ptrend=0.028 | ptrend=0.046 | |||||||
Adjusted for age, sex, ethnicity, first-degree family history of thyroid cancer, smoking status, and radiation exposure.
There were no significant differences in genotype distributions of the 7 BRCA1 SNPs among DTC patients with different T status and those with different N status (data not shown). However, DTC patients harboring A31875G AA genotype had a significantly higher frequency of multifocal primary tumor than did DTC patients harboring the AG/GG genotype (37.5% vs. 16.2%, p=0.011). The A31875G AG/GG genotype was associated with a 69% reduced risk of multifocal primary tumor in DTC patients (adjusted OR=0.31, 95% CI: 0.12–0.81).
Discussion
Accumulating evidence suggests that DTC susceptibility is likely to be determined by multiple variations in low-penetrance genes that affect a large segment of the general population (19). Our group has been studying genetic polymorphisms in genes involved in multiple DNA repair pathways as markers of host genetic susceptibility to the development of thyroid cancer (20–22). Here, our results suggest that polymorphisms and/or haplotypes of the BRCA1 gene may modulate DTC risk. This study is an extension of our previous work on the radiation-response genes and thyroid cancer risk (22). That previous work revealed a similar inverse association between BRCA1 SNPs and DTC risk, although the association was not statistically significant and the study included only 134 DTC cases and 166 controls and examined only 4 BRCA1 SNPs (22). To the best of our knowledge, the study reported herein is the largest population-based study to evaluate the association between functional BRCA1 genetic polymorphisms and thyroid cancer risk.
In this case–control study, when analyzed individually, BRCA1 promoter SNP A1988G and nonsynonymous coding SNPs C33420T, A33921G, and A34356G exhibited statistically significant protective effects against DTC risk. However, since 7 functional SNPs have been tested for disease association, this may increase the possibility of false-positive results. To address this problem, Bonferroni correction for multiple testing was used and only A1988G was associated with reduced risk of DTC after such adjustment. An alternative approach to the problem of multiple testing is the false-positive report probability (FPRP) introduced by Wacholder et al., which assesses the likelihood that the observed risk association is falsely positive (23). The FPRP is evaluated by incorporating the prior probability and the observed ORs and 95% CIs. Determination of prior probability is based on the functional importance of gene and SNPs, and existing epidemiological data. A high prior probability (0.25) is usually assigned when the biological plausibility is high and existing epidemiological data are fair; a prior probability of 0.1 is assigned when the biological plausibility is high but existing epidemiological data are poor; and a prior probability of 0.01 is assigned when both are poor (24). Based upon the evidence of an association between DNA damage repair capacity and DTC risk (13), BRCA1's function in DNA DSB repair pathway (7), and the selected SNPs that are either nonsynonymous or in promoter region, selection of a relatively high prior probability (0.1) in this study is appropriate. Given an OR of 0.67 (or its reciprocal 1.5) and a prior probability of 0.1, an FPRP below 50% was achieved for these four SNPs. Such FPRP values are considered below the threshold of noteworthiness and suggest that the associations between these SNPs and DTC risk may be important for an index study requiring replication.
We also identified two haplotypes (AATAATA and ATAA) that were inversely associated with risk; both carry the C33420T variant allele. Given an OR of 0.67 and a prior probability of 0.1, the estimated FPRP value is 30% and 41%, respectively, for the associations between these two haplotypes and PTC risk. Assuming that these associations are true, they may be because either these SNPs are causal loci, or they may be in correlation with the real determinants. Given the location of A1988G in the promoter region, it is possible to influence BRCA1 transcriptional activities. For C33420T, a functional effect is possible since it is located in a putative functional domain of BRCA1, leading to an amino acid change from proline to leucine at position 871. This is a nonconservative change as proline conveys unique structural properties to the polypeptide (25). However, additional studies are needed to determine the functional significance of these genetic variants. Several groups have previously investigated the role of the BRCA1 C33420T SNP in cancer susceptibility. In agreement with our results, a previous case–control study in Chinese women found that the C33420T TT genotype was associated with a significantly decreased risk of cervical cancer (26). However, opposite results and nonsignificant results were observed for breast and ovarian cancer (27–29). While the inconsistency between studies is hard to explain, it is conceivable that BRCA1's inherent tendency to complex with multiple proteins and to mediate various functions might explain variations in the effects of BRCA1 genetic polymorphisms on cancer risk among different cancer sites.
C33420T was in LD with A33921G and A34356G. While we failed to detect an additive effect of these SNPs in haplotype analysis, a dose–response relationship was established, whereby possessing an increasing number of the favorable BRCA1 genotypes (A1988G, C33420T, A33921G, and A34356G) was associated with reduced risk of DTC and PTC. This supports the idea that multiple low-penetrance SNPs might have greater effect on cancer susceptibility when considered in combination than individually.
Examination of BRCA1 SNPs in DTC patients with different clinical characteristics revealed that the A31875G AA genotype carriers had a lower frequency of multifocal primary tumors than the A31875G AG/GG genotype carriers. It is known that multiple cancer loci often arise from independent clonal events, which implicates predisposing influences of multifocality (30). However, it is largely unknown whether genetic susceptibility or environmental exposures are the dominant factors driving formation of multifocal DTC. Our result supports the contribution of genetic susceptibility to multifocal DTC.
This study has several potential limitations. The study sample size was modest, which limits the statistical power in stratification analysis and haplotype analysis, although this is, to the best of our knowledge, the largest study of the association between BRCA1 SNPs and DTC risk. Since we used a hospital-based case–control design, the possibility of selection bias should be considered in interpreting the results. Moreover, both cases and controls were recruited from a single cancer center; thus, study subjects may not have represented a comprehensive genetic profile of DTC in the general population. However, the variant genotype frequencies in the control population did not deviate significantly from the HWE or from reported frequencies in the general population. It is therefore likely that our study population was representative of the general population.
In summary, the data presented provide supporting evidence that germline variants in BRCA1, which belongs to the DNA DSB repair system, may contribute to genetic susceptibility to thyroid cancer. These results suggest that comprehensive genotypic profiling of BRCA1 should be performed in a larger series of DTC patients. Functional studies of BRCA1 genetic variants and their interaction with other genes/proteins could also provide a working knowledge of the effects of DNA repair gene polymorphisms on cancer risk.
Acknowledgments
The authors thank Margaret Lung, Kathryn Patterson, Liliana Mugartegui, and Jenny Vo for their help with subject recruitment; Chong Zhao for DNA extraction and genotyping analysis; and Stephanie Deming for article editing.
This work was supported in part by an American Thyroid Association Thyroid Cancer Grant (to E.M.S.); The University of Texas M.D. Anderson Cancer Center start-up funds (to E.M.S.); National Institutes of Health Grant U01 DE019765-01 (to Dr. Adel K. El-Naggar, E.M.S. is project 2 leader); National Institute of Environmental Health Sciences Grant R01 ES-11740 (to Q.W.); and Cancer Center Support Grant CA016672 to The University of Texas M.D. Anderson Cancer Center (to Dr. John Mendelsohn). L.X. is currently a Cancer Prevention Postdoctoral Fellow at The University of Texas M.D. Anderson Cancer Center supported by Halliburton Employees Fellow in Cancer Prevention funds.
Disclosure Statement
The authors declare no competing financial interests and no other conflicts of interest.
References
- 1.Jemal A. Siegel R. Xu J. Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Chen AY. Jemal A. Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115:3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 3.Davies L. Welch HG. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA. 2006;295:2164–2167. doi: 10.1001/jama.295.18.2164. [DOI] [PubMed] [Google Scholar]
- 4.Grubbs EG. Rich TA. Li G. Sturgis EM. Younes MN. Myers JN. Edeiken-Monroe B. Fornage BD. Monroe DP. Staerkel GA. Williams MD. Waguespack SG. Hu MI. Cote G. Gagel RF. Cohen J. Weber RS. Anaya DA. Holsinger FC. Perrier ND. Clayman GL. Evans DB. Recent advances in thyroid cancer. Curr Probl Surg. 2008;45:156–250. doi: 10.1067/j.cpsurg.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Ron E. Lubin JH. Shore RE. Mabuchi K. Modan B. Pottern LM. Schneider AB. Tucker MA. Boice JD., Jr. Thyroid cancer after exposure to external radiation: a pooled analysis of seven studies. Radiat Res. 1995;141:259–277. [PubMed] [Google Scholar]
- 6.Jeggo P. The role of the DNA damage response mechanisms after low-dose radiation exposure and a consideration of potentially sensitive individuals. Radiat Res. 2010;174:825–832. doi: 10.1667/RR1844.1. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J. Powell SN. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res. 2005;3:531–539. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 8.O'Donovan PJ. Livingston DM. BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 2010;31:961–967. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]
- 9.Chodosh LA. Expression of BRCA1 and BRCA2 in normal and neoplastic cells. J Mammary Gland Biol Neoplasia. 1998;3:389–402. doi: 10.1023/a:1018784031651. [DOI] [PubMed] [Google Scholar]
- 10.Durocher F. Simard J. Quellette J. Richard V. Fernand L. Pelletier G. Localization of BRCA1 gene expression in adult cynomolgus monkey tissues. J Histochem Cytochem. 1997;45:1173–1188. doi: 10.1177/002215549704500901. [DOI] [PubMed] [Google Scholar]
- 11.Miki Y. Swensen J. Shattuck-Eidens D. Futreal PA. Harshman K. Tavtigian S. Liu Q. Cochran C. Bennett LM. Ding W. Bell R. Rosenthal J. Hussey C. Tran T. McClure M. Frye C. Hattier T. Phelps R. Haugen-Strano A. Katcher H. Yakumo K. Gholami Z. Shaffer D. Stone S. Bayer S. Wray C. Bogden R. Dayananth P. Ward J. Tonin P. Narod S. Bristow PK. Norris FH. Helvering L. Morrison P. Rosteck P. Lai M. Barrett JC. Lewis C. Neuhausen S. Cannon-Albright L. Goldgar D. Wiseman R. Kamb A. Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 12.Seymour IJ. Casadei S. Zampiga V. Rosato S. Danesi R. Falcini F. Strada M. Morini N. Naldoni C. Paradiso A. Tommasi S. Schittulli F. Amadori D. Calistri D. Disease family history and modification of breast cancer risk in common BRCA2 variants. Oncol Rep. 2008;19:783–786. [PubMed] [Google Scholar]
- 13.Janezic SA. Ziogas A. Krumroy LM. Krasner M. Plummer SJ. Cohen P. Gildea M. Barker D. Haile R. Casey G. Anton-Culver H. Germline BRCA1 alterations in a population-based series of ovarian cancer cases. Hum Mol Genet. 1999;8:889–897. doi: 10.1093/hmg/8.5.889. [DOI] [PubMed] [Google Scholar]
- 14.Cox DG. Kraft P. Hankinson SE. Hunter DJ. Haplotype analysis of common variants in the BRCA1 gene and risk of sporadic breast cancer. Breast Cancer Res. 2005;7:R171–R175. doi: 10.1186/bcr973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong P. Zheng R. Wang LE. Bondy ML. Shen H. Borer MM. Wei Q. Sturgis EM. A pilot case-control study of gamma-radiation sensitivity and risk of papillary thyroid cancer. Thyroid. 2005;15:94–99. doi: 10.1089/thy.2005.15.94. [DOI] [PubMed] [Google Scholar]
- 16.Packer BR. Yeager M. Burdett L. Welch R. Beerman M. Qi L. Sicotte H. Staats B. Acharya M. Crenshaw A. Eckert A. Puri V. Gerhard DS. Chanock SJ. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC. Fry B. Maller J. Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Solé X. Guinó E. Valls J. Iniesta R. Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 19.Sturgis EM. Li G. Molecular epidemiology of papillary thyroid cancer: in search of common genetic associations. Thyroid. 2009;19:1031–1034. doi: 10.1089/thy.2009.1597. [DOI] [PubMed] [Google Scholar]
- 20.Ho T. Li G. Lu J. Zhao C. Wei Q. Sturgis EM. Association of XRCC1 polymorphisms and risk of differentiated thyroid carcinoma: a case-control analysis. Thyroid. 2009;19:129–135. doi: 10.1089/thy.2008.0153. [DOI] [PubMed] [Google Scholar]
- 21.Ho T. Li G. Zhao C. Wei Q. Sturgis EM. RET polymorphisms and haplotypes and risk of differentiated thyroid cancer. Laryngoscope. 2005;115:1035–1041. doi: 10.1097/01.MLG.0000162653.22384.10. [DOI] [PubMed] [Google Scholar]
- 22.Sturgis EM. Zhao C. Zheng R. Wei Q. Radiation response genotype and risk of differentiated thyroid cancer: a case-control analysis. Laryngoscope. 2005;115:938–945. doi: 10.1097/01.MLG.0000163765.88158.86. [DOI] [PubMed] [Google Scholar]
- 23.Wacholder S. Chanock S. Garcia-Closas M. El ghormli L. Rithman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matullo G. Guarrera S. Sacerdote C. Polidoro S. Davico L. Gamberini S. Karagas M. Casetta G. Rolle L. Piazza A. Vineis P. Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 25.Chang JS. Yeh RF. Wiencke JK. Wiemels JL. Smirnov I. Pico AR. Tihan T. Patoka J. Miike R. Sison JD. Rice T. Wrensch MR. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X. Han S. Wang S. Chen X. Dong J. Shi X. Xia Y. Wang X. Hu Z. Shen H. Polymorphisms in HPV E6/E7 protein interacted genes and risk of cervical cancer in Chinese women: a case-control analysis. Gynecol Oncol. 2009;114:327–331. doi: 10.1016/j.ygyno.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Nicoloso MS. Sun H. Spizzo R. Kim H. Wickramasinghe P. Shimizu M. Wojcik SE. Ferdin J. Kunej T. Xiao L. Manoukian S. Secreto G. Ravagnani F. Wang X. Radice P. Croce CM. Davuluri RV. Calin GA. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sehl ME. Langer LR. Papp JC. Kwan L. Seldon JL. Arellano G. Reiss J. Reed EF. Dandekar S. Korin Y. Sinsheimer JS. Zhang ZF. Ganz PA. Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clin Cancer Res. 2009;15:2192–2203. doi: 10.1158/1078-0432.CCR-08-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auranen A. Song H. Waterfall C. Dicioccio RA. Kuschel B. Kjaer SK. Hogdall E. Hogdall C. Stratton J. Whittemore AS. Easton DF. Ponder BA. Novik KL. Dunning AM. Gayther S. Pharoah PD. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–618. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 30.Shattuck TM. Westra WH. Ladenson PW. Arnold A. Independent clonal origins of distinct tumor foci in multifocal papillary thyroid carcinoma. N Engl J Med. 2005;352:2406–2412. doi: 10.1056/NEJMoa044190. [DOI] [PubMed] [Google Scholar]